Abstract
The minichromosome maintenance complex (MCM) represents the replicative DNA helicase both in eukaryotes and archaea. Here, we describe the solution structure of the C-terminal domains of the archaeal MCMs of Sulfolobus solfataricus (Sso) and Methanothermobacter thermautotrophicus (Mth). Those domains consist of a structurally conserved truncated winged helix (WH) domain lacking the two typical ‘wings’ of canonical WH domains. A less conserved N-terminal extension links this WH module to the MCM AAA+ domain forming the ATPase center. In the Sso MCM this linker contains a short α-helical element. Using Sso MCM mutants, including chimeric constructs containing Mth C-terminal domain elements, we show that the ATPase and helicase activity of the Sso MCM is significantly modulated by the short α-helical linker element and by N-terminal residues of the first α-helix of the truncated WH module. Finally, based on our structural and functional data, we present a docking-derived model of the Sso MCM, which implies an allosteric control of the ATPase center by the C-terminal domain.
INTRODUCTION
Maintenance of genetic integrity is an essential process for all living organisms. The minichromosome maintenance complex (MCM) plays a central role in the replication both in eukaryotes and archaea. MCM serves as replicative DNA helicase and forms a hexameric ring-shaped complex on the DNA. The MCM in eukarya consists of six closely related, non-identical proteins whereas the archeal MCM constitutes a homohexameric assembly (1–6). The high sequence homology between MCM proteins of eukarya and archaea suggests that they are derived from a common ancestor. This renders the archaeal MCM a valuable model to study the structure-function relationship of this essential component of the replication machinery.
A canonical MCM protein comprises three domains (Figure 1). The N-terminal part mediates hexamer formation, binds DNA and tightly regulates processivity and polarity of the helicase. The AAA+ domain is responsible for the catalytic activity of the MCM helicase and belongs to the AAA+ superfamily of ATPases. A less well-characterized C-terminal domain is present in all MCM proteins. This domain was shown to reduce the ATPase activity of archeal MCMs (7,8).
Figure 1.
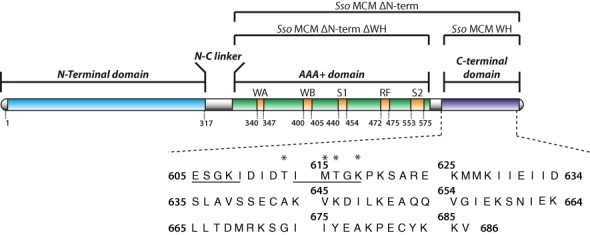
Sso MCM domain organization and amino acid sequence of the C-terminal region. Denomination of deletion mutants used here on top. WA: Walker A motif, WB: Walker B motif, S1: sensor 1 region, RF: arginine finger, S2: sensor 2 region. Asterisks represent point mutations in the Sso MCM C-terminal domain for the analysis of their role in controlling the Sso MCM ATPase and helicase activity. In addition, Sso MCM constructs were generated where the underlined stretches of amino acids were deleted either separately or both in combination, respectively. For a detailed list of constructs used in this study see Supplementary Table S6. Domain organization scheme adapted from (3).
Considerable structural information on the N-terminal and AAA+ domain of archaeal MCMs is available through X-ray and cryo-electronmicroscopy studies (4,9–14). However, no structure of their C-terminal domain, for which a winged helix (WH) topology was predicted based on sequence analysis (5,10), was available. Hence, we addressed the three-dimensional structure of the archaeal MCM C-terminal domain and how this structure might influence the activity of the ATPase center.
Here, we describe the nuclear magnetic resonance (NMR) solution structure of the C-terminal MCM domains of Sulfolobus solfataricus (Sso) and Methanothermobacter thermautotrophicus (Mth). For the Sso MCM C-terminal domain we identify residues involved in the regulation of the ATPase activity. Finally, we present a docking-based model of the Sso MCM, which integrates both published and our data. This model provides a potential mechanistic explanation of how the Sso MCM ATPase activity is allosterically modulated by its C-terminal domain.
MATERIALS AND METHODS
Cloning and mutagenesis
The E. coli expression vectors pET19b and pET28a harboring Sso MCM constructs (full-length Sso MCM (15) and Sso MCMΔN-term, comprising residues V268-V686 (16)) were kindly provided by Francesca Pisani (Institute of Protein Biochemistry, CNR, Naples). Sso MCM (reference sequence UNIPROT Q9UXG1) was mutated at selected positions (Figure 1) by PCR-based site-directed mutagenesis.
The plasmids were amplified by PCR using the following protocol: 20 cycles of denaturing at 95°C for 30 s, annealing at 45°C for 30 s, elongation at 68°C for 7 min, respectively, and a final elongation step at 68°C for 10 min. The resulting PCR products were purified using the MSB Spin PCRapace system (STRATEC Molecular) followed by a DpnI (NEB) digestion for 2 h at 37°C. The digestion product was purified using the MSB Spin PCRapace system and transformed into competent E. coli DH5α cells.
To generate shortened, monomeric Sso MCM constructs (10) amenable to liquid state NMR spectroscopy, the residues 555ILI557 were mutated to 555DSD557 in Sso MCMΔN-term. The subsequent introduction of two stop codons distal to M604 in Sso MCMΔN-term resulted in a construct comprising only the Sso MCM AAA+ domain (Sso MCMΔN-termΔWH). In addition, the thrombin protease cleavage site in pET28a harboring the Sso MCMΔN-term derivatives was replaced by a TEV protease cleavage site. Both Sso MCMΔN-term derivatives produce predominantly monomeric (10) Sso proteins with a N-terminal hexahistidine tag.
A chimeric archaeal MCM was created by swapping the Sso MCM C-terminal domain (E605-V686) with the MCM C-terminal domain (E583-V666) of Mth. The amplified chimeric MCM gene was subcloned into NcoI/XhoI-linearized pET15b. All expression plasmids were sequenced to verify the desired mutations. Oligonucleotide primers were purchased from Eurofins Genomics. The sequences of primers and the position of mutations in the Sso MCM constructs used in this study are listed in Supplementary Tables S1 and S8.
Recombinant protein expression and purification
The Mth and Sso MCM C-terminal domains were expressed and purified as described (17). The full-length Sso MCM and its mutants were expressed in E. coli BL21 CodonPlus(DE3) RIPL® cells (Agilent Technologies). Cells harboring the expression plasmid were grown at 37°C in LB media. Once the cell density reached an OD600 of 0.8/ml of culture, protein expression was induced by adding IPTG (isopropyl-1-thio- D-galactopyranoside) to a final concentration of 1 mM and was continued for 3 h at 37°C. Cells were harvested by centrifugation at 4°C (5.000xg, 30 min) and resuspended in lysis buffer (10 mM Tris-HCl, 150 mM NaCl and 2.5 mM MgCl2, pH 7.2) with a mixture of protease inhibitors, DNAses and RNAses (Roche). Cells were homogenized using a combination of French Press® disintegration and ultrasonification. Cell homogenates were centrifuged (15.400xg, 4°C, 30 min) and the supernatant was subjected to a heat treatment at 80°C for 20 min, and subsequently incubated on ice for 10 min. The heat denatured proteins were removed by centrifugation for 30 min at 37.000xg at 4 °C. The supernatant was separated on a gel filtration column (Superdex 200 26/60, flow rate of 2.5 ml/min, GE Healthcare) equilibrated with lysis buffer. Peak fractions containing the desired protein were collected, analyzed by SDS-PAGE, concentrated by Vivaspin 20 (50 kD molecular weight cutoff, GE Healthcare) and stored at -20°C. The final yield of the recombinant proteins after purification procedure was in the range of 25 mg/l cell culture.
Cells harboring derivatives of the pET28a-Sso MCMΔN-term expression plasmid were grown either in LB media or in M9 media prepared in D2O and supplemented with 1 g/l 15NH4Cl and 2 g/l fully deterated 13C6 glucose for isotopic labeling for NMR experiments. Cells were harvested by centrifugation, resuspended in 50 mM Na-phosphate pH 8, 300 mM NaCl, 5 mM imidazole and a mixture of protease inhibitors, DNAses and RNAses (Roche) and homogenized as described above, but no heat treatment was applied. The supernatant was loaded onto a Ni-NTA agarose (Qiagen) column equilibrated with lysis buffer. After washing with lysis buffer, 10 column volumes of wash buffer (lysis buffer, 20 mM imidazole) were applied to the column. The His-tagged fusion proteins were eluted subsequently (lysis buffer containing 250 mM imidazole). The His-tag was released during dialysis against 10 mM Tris-HCl pH 7.2, 150 mM NaCl, 1 mM DTT and 2.5 mM MgCl2 by TEV protease (1 mg/50 mg protein) at 4°C over night. The resulting cleavage product was separated on a Superdex 200 26/60 gel filtration column (GE Healthcare) equilibrated with 10 mM Tris-HCl pH 7.2, 150 mM NaCl and 2.5 mM MgCl2 at a flow rate of 2.5 ml/min.
Mass spectrometry
The exact molecular weight and the isotope distribution of the Sso MCM C-terminal domain were determined by ESI mass spectrometry using a LTQ Orbitrap XL ETD (Thermo Scientific) coupled to a 2D nanoLC (Eksigent) chromatography system. An amount of 1.5 pmol was loaded on a 75 μm x 10 cm reverse phase column (Nanoseparation) and separated by application of a linear gradient from 20% to 80% buffer B (80% acetonitrile, 0.1% formic acid) against buffer A (5% acetonitrile, 0.1% formic acid).
Radioactive labeling and preparation of DNA substrates
The oligodeoxyribonucleotides used for the preparation of DNA substrates for the ATPase and helicase activity tests are described in Supplementary Table S1. For the helicase assays, oligonucleotides were radioactively labeled prior to annealing. 5′-ends were labeled with 33P-γ-ATP (5000 Ci/mmol; Hartmann Analytic) and T4 PNK for 1 h at 37°C in forward reaction buffer (80 mM Tris-HCl pH 7.4, 10 mM MgCl2 and 5 mM DTT). The reaction was stopped by heating to 72°C for 10 min. Subsequently unincorporated nucleotides were removed by size-exclusion chromatography using Ilustra MicroSpin G-25 columns (GE Healthcare). To prepare Y-shaped DNA substrates, 5′-end-labeled oligonucleotides were mixed with partially complementary unlabeled oligonucleotide at a molar ratio of 1:1.5 in binding buffer (30 mM Tris-HCl, 10 mM MgCl2, pH 7.5), heated to 95°C for 5 min and allowed to slowly cool down to room temperature.
Helicase activity assay
Reaction mixtures (10 μl) were prepared in 30 mM Tris-HCl pH 8.0, 75 mM NaCl, 50 mM potassium acetate;10 mM magnesium-acetate, 1 mM DTT and 5 mM ATP. Each reaction mixture contained either 1 or 2 nM 5′-end labeled DNA substrate and indicated amounts of Sso MCM proteins. The reaction mixtures were incubated for 30 min at 70°C and then rapidly cooled on ice. Then, 10 μl of loading buffer (0.1% bromophenol blue, 0.1% xylene cyanol, 1% SDS; 0.5 mg/ml proteinase K and 40% glycerol) were added. Next, the samples were separated on 8% native polyacrylamide gels in 89 mM Tris base, 89 mM boric acid and 2 mM EDTA (TBE) buffer. After electrophoresis the gels were dried for 1 h at 80°C, exposed to a phosphorimaging screen and visualized with a Phosphor-Imager (Typhoon Trio, GE Healthcare).
ATPase assay
The ATPase activity of purified Sso MCM constructs was assessed by 1D [31P]-NMR spectroscopy. Reactions were performed at 60°C in 550 μl, containing 50 μM protein, 50 mM ATP (JenaBioscience) and 10% D2O in 10 mM Tris-HCl pH 7.2, 150 mM NaCl and 2.5 mM MgCl2. Time series of 1D [31P]-spectra were recorded. The spectra were processed and resulting peak areas were plotted as a function of time. The slope of the linear regression derived from this analysis is a measure for the turnover kinetics of ATP per time. As the MCM concentration was known, the apparent catalytic constant, kcat, app (min-1, turnover number) was calculated by dividing the slope of the linear regression (mM min-1) by the protein concentration (mM).
Biophysical characterization
An initial biophysical analysis of Sso and Mth MCM C-terminal domains is given in the Supplementary Data. Briefly, purified proteins were characterized by circular dichroism (CD) spectroscopy, dynamic and static light scattering. In addition, parameters of protein dynamics (T1, T2 and heteronuclear NOE cross-relaxation, respectively) were assessed by NMR spectroscopy (Supplementary Figures S7 and S8).
Nuclear magnetic resonance spectroscopy
The 1H, 13C and 15N backbone and side chain chemical shift assignments for the MCM C-terminal domain of Sso are available from the BioMagResBank (accession numbers 18986) (17).
NOE-derived distance restraints were derived from three-dimensional [1H, 1H, 13C]-NOESY-HSQC (mixing time: 120 ms) and [1H, 1H, 15N]-NOESY-HSQC (mixing time: 120 ms) experiments at 30°C on Bruker 750 MHz or 600 MHz AvanceIII NMR systems, respectively, equipped with 5 mm triple resonance cryo-probes. 3-(Trimethylsilyl)propane-1-sulfonic acid (DSS) was used for direct 1H chemical shift referencing and 13C as well as 15N chemical shifts were indirectly referenced. Spectra were processed using TOPSPIN V2.1 (Bruker Biospin) and analyzed with CCPNmr Analysis (18).
Direct 31P detection was performed on either Bruker 600 MHz or 500 MHz AvanceIII NMR systems, equipped with 5 mm quadrupole resonance probes (QXI). Spectra were recorded at 600 MHz with a spectral width of 7042 Hz sampled over 1228 complex points in the ω1 (31P) dimension and at 500 MHz with a spectral width of 5868 Hz sampled over 1024 complex points. The acquisition time in the 31P dimension was 0.175 ms and 100 transients were applied. The overall experimental time for a single 1D [31P]-NMR spectrum was 2 min. The 31P radio frequency carrier was set at -10.5 ppm and the recycle time was 1 s. 31P chemical shifts at 60°C were referenced relative to 75% H3PO4 at 0 ppm. Spectra were processed and plotted using TOPSPIN V2.1 (Bruker Biospin).
Structure calculation, coordinates and modeling
Structures were calculated using CYANA v.3.9 (19,20). The 20 of 100 structures with the lowest target function were analyzed and subjected to energy minimization using the program OPAL (21). Upper limit distance constraints were classified according to their intensity in the NOESY spectra corresponding to distance limits of 2.7, 3.3, 4,3, 5.2 and 6.5 Å, respectively. NOE intensities corresponding to fixed Hβ2-Hβ3 and aromatic ring distances were used for calibration. Pseudoatom corrections were added. 3J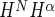 coupling constants were included to calculate torsion angle constraints via local conformational analysis with the FOUND module (22). Furthermore, torsion angle constraints defining the allowed Φ,Ψ-regions in the Ramachandran map were included. Hydrogen bonds that were consistently formed during initial rounds of structure calculation were used as additional constraints in the final structure calculation. The 20 structures with the lowest total energy were deposited in the PDB (entries 2M45 and 2M3A). Figures were produced using UCSF Chimera (23). The statistics of the structure refinement and the quality of the final structures, including PROCHECK classification (24) of Ramachandran statistics, are summarized in Table 1, Supplementary Table S5 and Supplementary Figure S10.
coupling constants were included to calculate torsion angle constraints via local conformational analysis with the FOUND module (22). Furthermore, torsion angle constraints defining the allowed Φ,Ψ-regions in the Ramachandran map were included. Hydrogen bonds that were consistently formed during initial rounds of structure calculation were used as additional constraints in the final structure calculation. The 20 structures with the lowest total energy were deposited in the PDB (entries 2M45 and 2M3A). Figures were produced using UCSF Chimera (23). The statistics of the structure refinement and the quality of the final structures, including PROCHECK classification (24) of Ramachandran statistics, are summarized in Table 1, Supplementary Table S5 and Supplementary Figure S10.
Table 1. NMR and refinement statistics for the Sso MCM C-terminal domain.
| NOE-based distance restraintsa | 1977 |
| Intra-residual (∣i-j∣ = 0) | 296 |
| Sequential (∣i-j∣ = 1) | 541 |
| Medium-range (2 ≤ ∣i-j∣ ≤ 4) | 563 |
| Long-range (∣i-j∣ ≥ 5) | 497 |
| Dihedral angle restraint | 170 |
| Restrained H-bonds | 80 |
| Constraint violations | |
| Distance constraints (Å) | 0.006 ± 0.001 |
| Max. distance constraint violation (Å) | 0.21 ± 0.06 |
| Dihedral angle constraints (°) | 0.08 ± 0.07 |
| Max. dihedral angle violation (°) | 1.28 ± 1.16 |
| CYANA target function (Å2) | 0.42 ± 0.11 |
| AMBER energies (kcal mol−1) | −1370.0 ± 56.9 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.465 |
| mean global r.m.s.d. (Å) | |
| Totalb (backbone) | 4.01 ± 1.58 |
| Totalb (heavy atoms) | 4.13 ± 1.42 |
| Orderedc (backbone) | 0.76 ± 0.19 |
| Orderedc (heavy atoms) | 1.49 ± 0.22 |
| Ramachandran statistics (%)d | |
| Most favored | 88 |
| Additionally allowed | 11 |
| Generously allowed | 1 |
| Disallowed | 0 |
aUpper & lower distance restraints
bSso MCM WH: residues E605-V686
cSso MCM WH: residues K620-V686
dPROCHECK classification
Vector derived residues were not included in the structural statistics.
Docking of the Sso MCM C-terminal domain (PDB: 2M45) with Sso MCM (PDB: 3F9V) was performed with the High Ambiguity Driven Protein-Protein Docking (HADDOCK) (25) installation at the WeNMR grid server (26). The combined 1H and 15N chemical shift perturbation (CSP) of the Sso MCM C-terminal domain was analyzed to achieve constraints for the docking approach. The chemical shifts derived from [1H,15N]-TROSY-HSQC spectra of the isolated Sso MCM C-terminal domain and the same domain covalently linked to the AAA+ domain were compared. Residues for which the CSP was above the 2-fold CSP average were considered further to generate docking constraints. Residues which are in close proximity to the potential domain interface were defined as active interacting residues (605, 606, 615-617, 620, 621, 623, 625, 626). Surrounding residues considered indirectly involved in or affected by the domain interaction were defined as passive interacting residues (673 and 674). Residue G601 of Sso MCM (PDB: 3F9V) was defined as active because it represents the most C-terminal residue of the AAA+ domain preceding the most N-terminal residue (V602) of the C-terminal WH domain. No further restrictions and structural assumptions were applied. The final model (Figure 8) was selected according to its HADDOCK score (Supplementary Table S9).
Figure 8.
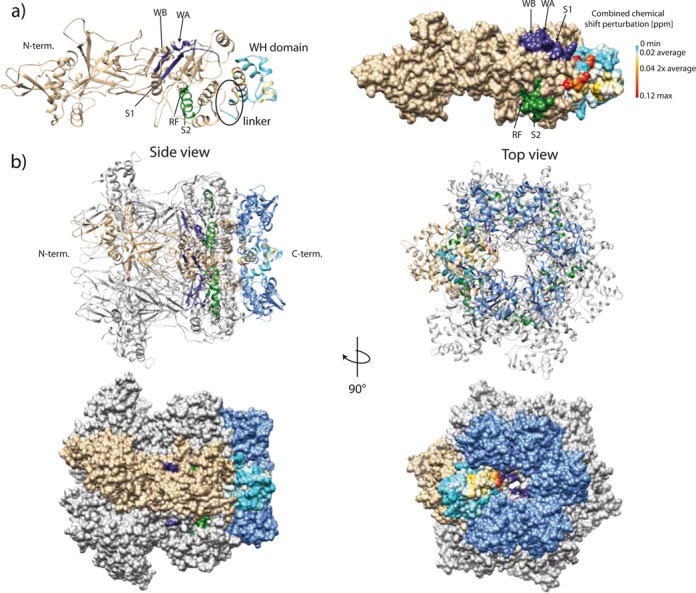
(a) A High Ambiguity Driven Protein-Protein Docking (HADDOCK) derived structural model of Sso MCM including the C-terminal domain. The C-terminal domain is color-coded according to Figure 7. (b) Ribbon and surface diagram the of Sso MCM hexameric model including the C-terminal domain. Based on the hexamer structure of the Sso MCM N-terminal domain (PDB ID: 2VL6; (47)) a hexamer model was generated by applying a 6-fold symmetry to the HADDOCK derived Sso MCM structural model generated from PDB 3F9V(10) and 2M45 (see above). A single MCM subunit is colored in tan. The Sso MCM C-terminal domains are colored in blue. Cis and trans acting elements belonging to one catalytic ATPase center are depicted in purple and green, respectively. WA: Walker A motif, WB: Walker B motif, S1: sensor 1 region, RF: arginine finger, S2: sensor 2 region.
RESULTS AND DISCUSSION
NMR solution structure of the C-terminal domain of the Sso MCM
An initial biophysical characterization of the C-terminal domain of the Sso MCM indicated, that this domain represents a folded and thermostable monomer in solution. Essentially, the same results were obtained for the Mth MCM C-terminal domain (Supplementary Figures S1–S5 and Supplementary Tables S2–S4). In addition, we also found, that the two C-terminal domains did not bind to various DNA substrates differing in sequence and topology (Supplementary Figure S6) in agreement with published data (8,16). The assigned backbone and side-chain chemical shifts were reported previously (BMRB entry: 18986) (17). The structure of the Sso MCM C-terminal domain was calculated on the basis of nuclear Overhauser enhancement (NOE) distance (see also Supplementary Figure S10) and torsion angle constraints. The ensemble of the energy minimized structures with the lowest target function is shown in Figure 2. The structural statistics are given in Table 1 and Supplementary Figure S10.
Figure 2.
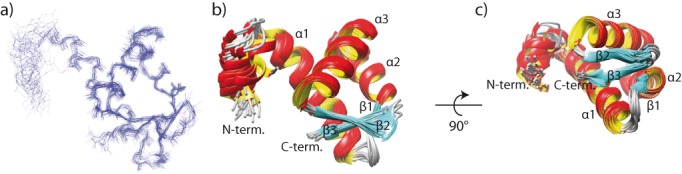
NMR solution structure of the Sso MCM C-terminal WH domain. (a) Superimposed backbone traces for the 20 structures with the lowest energy after OPAL refinement. (b, c) Ribbon representation of the structural ensemble with labeled secondary structure elements. Here, the less ordered residues 600–609 of the Sso MCM C-terminal WH domain are omitted for clarity. The coordinates of the Sso MCM C-terminus have been deposited at the RCSB protein databank (PDB: 2M45).
The NMR-derived solution structure of the Sso MCM C-terminal domain resembles a WH fold with the topology α-β-α-α-β-β. The secondary structural elements are comprised of K620-V638 (α1), A643-K644 (β1), V645-G655 (α2), E657-G673 (α3), and the antiparallel β2 (I675-K679) and β3 (C682-K685) strands (numbering according to full-length sequence).
The very N-terminal residues (E605-P619) of the domain gave rise to only few NOEs and exhibit a reduced degree of structural order and increased dynamics as reflected in the NMR relaxation data (Supplementary Figure S7). Still, this N-terminal extension contains a short α-helical element (I611-T616). This extension links the WH domain to the remainder of the MCM and is called ‘linker’ hereafter. The calculated structural ensemble exhibits a backbone r.m.s.d. of 0.76 Å for the well-structured Sso MCM WH domain (K620-V686). The backbone r.m.s.d. raises to 4.01 Å when including the less ordered N-terminal linker residues (E605-K620). Backbone torsion angles correspond exclusively to the most favored (88%) and additionally allowed (11%) regions of the Ramachandran plot (Table 1). The NMR solution structure of the Mth MCM C-terminal domain adopts the same fold (Supplementary Figure S9) as the Sso MCM WH. Both structures are highly similar in the WH domain and deviate mainly in their loop and N-terminal linker extensions regions from each other (Figure 3). The backbone r.m.s.d. between their secondary structure elements amounts to 0.8 Å, whereas including the loop regions between those increases the backbone r.m.s.d. to 2.8 Å. The N-terminal residues of the Mth MCM WH are highly flexible and reveal no additional α-helical element in this region. The NMR and refinement statistics for the Mth MCM WH are given in Supplementary Figure S10 and Supplementary Table S5. The canonical three stranded WH fold (27,28) contains in addition to the abovementioned secondary structural elements one extended loop region between the β2 and β3 strands and one such loop C-terminal to β3 and is found, e.g. in the WH domain of the eukaryotic MCM6 (29). These two loops are the denominating WH domain wing elements (27–28,30). As compared to this canonical WH fold the MCM C-terminal domains of Sso and of Mth both lack those extended loops (Figure 3) and, hence, form a truncated WH domain. A DALI search (31) revealed a significant number of structurally similar WH domains (Supplementary Tables S6 and S7). However, superposition of the structures with the highest Z-score revealed that except for the Sso and the Mth WH domains only the corresponding domain of the mouse Z-DNA-Binding protein 1 (PDB 2HEO) represents a ‘truncated’ WH domain (Supplementary Figure S11).
Figure 3.
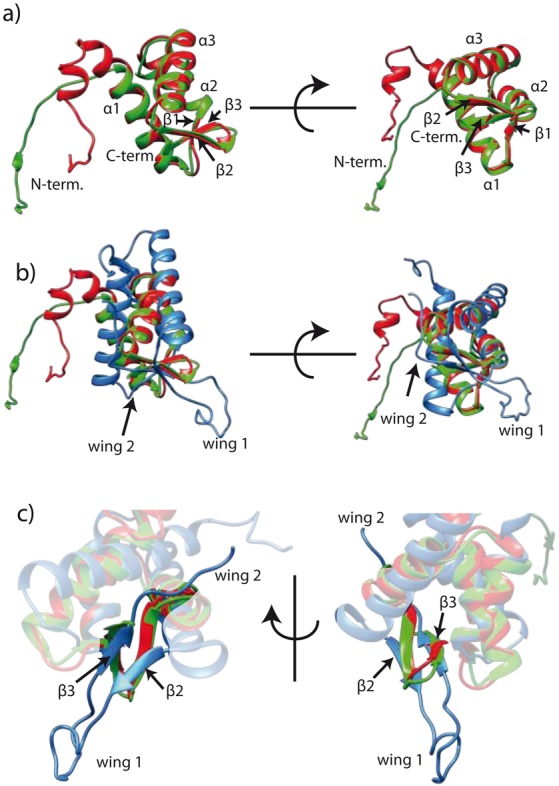
(a) Superimposition of Sso (red) and Mth (green) MCM C-terminal domains. (b) Superimposition of human MCM6 (blue, PDB: 2KLQ) and archaeal MCM (color code as in a)) C-terminal regions. (c) The human MCM6 WH domain shows extended loop regions (wing 1 and 2) which are truncated in the archaeal counterparts. Shown are mean structures derived from NMR structure ensembles. α1-3: α helix 1-3; β1-3: β strand 1-3.
The topology of the Sso MCM WH domain brings the sulfhydryl function of C642 and C682 into close proximity (2 Å, Figure 4), suggesting formation of a disulfide bond with a bond length also found for other proteins (32,33). The significantly downfield shifted resonances of the Cβ-carbons of C642 and C682 (44.1 ppm and 48.0 ppm, respectively; (17)) as expected for oxidized cysteines (34), and our mass spectrometry results (Supplementary Figure S12) are consistent with a disulfide bond. Examination of frequency and position of cysteines in the MCM C-terminal region of archaea (see (35) for selection of species) revealed that only the MCMs of seven closely related archaea including Sso harbor two cysteins in positions compatible with disulfide bond formation in their C-terminal domain (Supplementary Figures S13 and S14). Even though this phylogenetic analysis provides an independent argument in favor of the presence of a disulfide bond in the Sso MCM C-terminal domain, we sought to verify its presence experimentally. However, even in the presence of a 10-fold molar excess of DTT the chemical shift of the cysteine Cβ-carbons did not change as assessed by a [1H,13C]-HSQC (data not shown) implying that this disulfide bond is chemically very stable. Such stability would be required in a reducing intracellular environment. Candidates for intracellular proteins harboring disulfide bonds are more frequently found in thermophilic eubacteria and archaea than in other phylogenetic groups and it has been speculated that intracellular disulfide bond formation might be a feature of proteins to enhance thermal stability (36). Interestingly, the C-terminal domain of Mth does not contain any cysteines and exhibits a reduced thermostability as compared to the Sso C-terminal domain (Supplementary Figure S2 and Supplementary Table S3).
Figure 4.
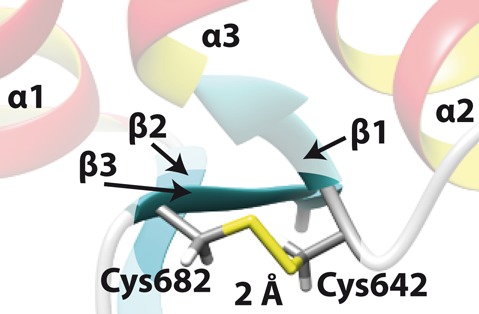
Disulfide bond between C642 and C682 in Sso MCM. The structure derived disulfide bond length is 2 Å. α1-3: α helix 1-3; β1-3: β strand 1-3.
ATPase activity of Sso MCM derivatives
We analyzed the ATPase activity of various Sso MCM mutants (Supplementary Table S8) by applying [31P]-NMR spectroscopy to directly monitor hydrolysis of ATP into ADP and free inorganic phosphate as function of time (Supplementary Figure S15). To quantify Sso MCM's catalytic activity, time series [31P]-spectra were recorded and the well-separated signals of the β-phosphate of ATP and the free inorganic phosphate were used for analysis. As shown in Supplementary Figure S16 this assay was carried out at saturating ATP substrate conditions. Examples for the time-course of the ATP hydrolysis are shown in Figure 5. Time course of ATP hydrolysis and the derived quantitative results as apparent catalytic constant (kcat, app) are summarized in Supplementary Table S8 and Supplementary Figure S17 for all MCM mutants described below. Our Sso MCM preparation exhibited no significant stimulation of its ATPase activity in the presence of DNA (Supplementary Table S8 and Supplementary Figure S17) which is consistent with literature data (37) and in contrast to other archaeal MCMs (38,39). Hence, all further ATPase assays were carried out in absence of DNA and no DNA was detectable in the purified Sso MCM by 31P-NMR spectroscopy (Supplementary Figure S18).
Figure 5.
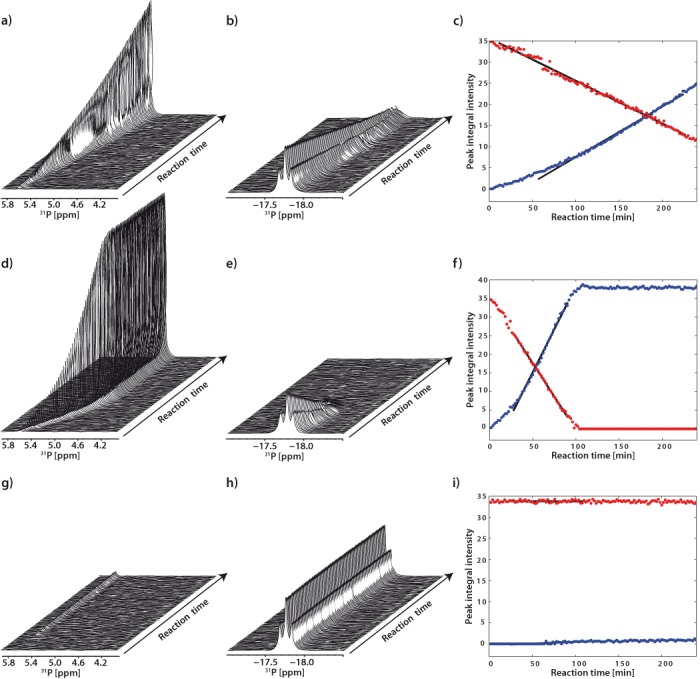
Hydrolysis of ATP. Time series of [31P]-NMR spectra were recorded. For clarity, only the spectral regions containing free inorganic phosphate (a, d, g) and ATP β-phosphate (b, e, h) are shown. Plots covering the full spectral region are given in Supplementary Data (Figure S14). Time -course progress of ATP hydrolysis by Sso MCM (a–c), Sso MCMΔWH (d–f) and as ATPase inactive control, Sso MCM R560A (g–i) is shown. The integrated peak areas of free phosphate (blue dots) and β-phosphate (red dots) of ATP, respectively, are depicted as a function of the reaction time (c, f, i). Black lines depict the result of the regression analysis of the linear part of the kinetics (see also Supplementary Figure S15).
To validate our experimental system we assessed the kcat, app of the full-length Sso MCM which turned out to be 2 min−1 in reasonable agreement with the value of 3.1 min−1 as reported previously (10). The Sso MCM sensor 2 mutant (R560A) and the Walker A mutant (K346A), both lacking ATPase and helicase activity (40), were used as negative control (Figure 5; Supplementary Table S8).
Previous studies addressing the modular organization of Sso MCMs indicated that deletion of their C-terminal domains increases archaeal MCMs’ ATPase activity (7–8,16). Moreover, for the Sso MCM it was shown, that the MCM DNA-binding activity is independent of the presence of its C-terminal domain (8).
Here, we observed a 3- to 5-fold increase in the ATPase activity when the WH domain is deleted alone or with the linker (E605-P619), respectively (Supplementary Figure S17e and f). In addition, deletion of parts (E605-I609, I614-K618) of the linker individually or in combination but leaving the WH domain in place and unmutated also led to a 3- to 4-fold increase of the ATPase activity. However, replacing the linker residues E605-T616 by Ala-Ser-Gly repeats, thereby conserving linker length, or by inserting 5 Ala residues at several positions in the linker, respectively, again results in increase of the ATPase activity. Together, these results suggest that not only the length but also properly placed residue identities within the linker sequence are necessary for the regulatory role of the C-terminal region of the MCM. To further address the influence of this linker (E605-P619) towards the Sso MCM ATPase activity we swapped Sso MCM linker sequences with the respective ones of Mth MCM. Here, we found that swapping the entire C-terminal region has only a slightly higher ATPase stimulating effect than swapping the linker alone. As the conservative substitution in the conserved part of the linker (S606T; Figure 6) produced an ATPase activity which is comparable to wild-type Sso MCM (Supplementary Table S8), we analyzed further the part of the linker with low sequence conservation between Sso (T613-K618) vs. Mth (K591-T596; Figure 6). Incidentally, this part contains the α-helical element (I611-T616) in Sso MCM.
Figure 6.
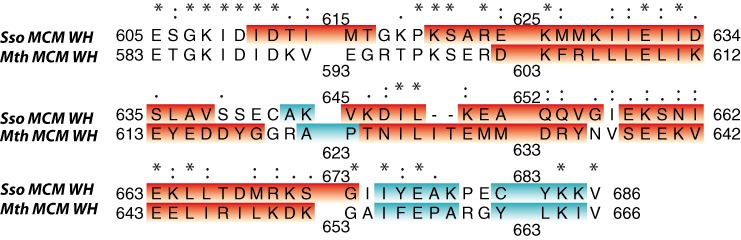
Sequence alignment of the Sso and Mth MCM C-terminal domains. Asterisks represent identical amino acids, colons represent conservative exchanges and a dot indicates conservation of general properties such as charged versus non-charged. Helical (red) and β-strand (cyan) regions present in the NMR-based structure are highlighted.
To analyze the role of this element in Sso MCM, we introduced mutations, which we considered to compromise the local helical structure, for individual residues or stretches of residues (Figure 1). Replacement of M615 by Ala, Gly or Pro, respectively, or of all three of T613,M615,K618 by Ala, Gly or Pro, respectively, increases the ATPase activity by ∼3- to 4-fold. Notably, the amino acids Gly and Pro carry a lower helix propensity than Ala (41) and induce a slightly higher increase of the ATPase activity than Ala, suggesting that the helical nature of this linker element contributes to the ATPase attenuating function of the MCM C-terminal domain.
This mutational analysis of Sso MCM leads to the assumption that the short α-helical element (I611-T616) in the Sso MCM C-terminal WH domain is substantially involved in the modulation the ATPase activity of Sso MCM. As a further control, we ascertained in a helicase assay of selected Sso MCM constructs that a changed ATPase activity is reflected in a correspondingly modulated DNA unwinding activity (Supplementary Figure S19).
In summary, our mutational analysis indicates that integrity and species-specific elements of the entire C-terminal domain (WH domain plus linker) are modulating the ATPase and helicase activity of the Sso MCM. Moreover, it appears that the short α-helical element in the linker plays a complex role in this respect.
Structural model of hexameric Sso MCM including the C-terminal WH domain
As detailed above, the C-terminal domain of archaeal MCMs comprises a well-conserved truncated WH domain and a structurally less conserved linker. Furthermore, we provided evidence, that this WH domain, and in particular the linker connecting it to the AAA+ ATPase domain, control the ATPase activity. Much of this allosteric control could be traced to the short and less conserved α-helical element (I611-T616) of the linker in Sso.
These structural and functional results prompted us to attempt an integration of our and published data on the Sso MCM into a more complete model of the Sso MCM hexameric holoenzyme in order to provide a structural rational for the regulatory activity of the WH domain towards the Sso MCM ATPase center. In order to augment this modeling process, we included NMR data on paramagnetic relaxation enhancement (PRE; (42–44)) and on the combined chemical shift perturbation (CSP; (45,46)) for the Sso MCM C-terminal domain to derive constraints for a HADDOCK-based (25) docking approach. Cosolvens-induced paramagnetic relaxation enhancement (solvent PRE) provides an approach to address macromolecular interactions even if no NOE or CSP data can be obtained (42–44): residues located in the protein interior or at interaction interfaces are shielded from a paramagnetic agent and experience a weak PRE. In contrast, residues located at solvent accessible surfaces (e.g. exposed loop regions) experience a strong PRE. Hence, we used PRE to assess interactions between the WH domain including the linker and the AAA+ domain. The PRE obtained for the isolated Sso MCM C-terminal WH domain is higher for residues connecting secondary structural elements and for the linker region (Supplementary Figure S20), thereby reflecting structural and dynamic properties of the Sso MCM C-terminal WH domain (Figure 2; Supplementary Figure S7). In the presence of a 3-fold molar excess of Sso MCMΔWH the PRE in particular for residues in the linker region and at the N-terminus of helix α1 (K618-R623) is reduced. This suggests that these residues of the WH domain and its linker are protected from solvent by the adjacent AAA+ domain of the MCM molecule. Furthermore, [1H,15N]-TROSY-HSQC spectra (Supplementary Figure S21) of Sso MCM constructs with and without covalently connected C-terminal domain were analyzed to independently identify via CSP residues potentially involved in the interaction between the C-terminal and the AAA+ domain (Figure 7). Interestingly, most residues experiencing a significant CSP reside in the linker region and at the N-terminus of helix α1 of the WH domain. All except two of the residues exhibiting a CSP of above 2-fold average where included as ‘active residues’ into the modeling using HADDOCK. The two C-terminal residues of the WH (G673 and I674) also exhibited a CSP 2-fold above average, but were included as ‘passive’ residues (for details, see Materials and Methods). Finally, the structural model of Sso MCM including the inter-domain interaction between the C-terminal region and the AAA+ domain was deduced by a HADDOCK-based docking approach (Figure 8, Supplementary Table S9). Incidentally, this is the first full-length MCM model including a solved 3D structure of the C-terminal WH domain and structural data related to the MCM of Sso only ((10,47), and present data).
Figure 7.
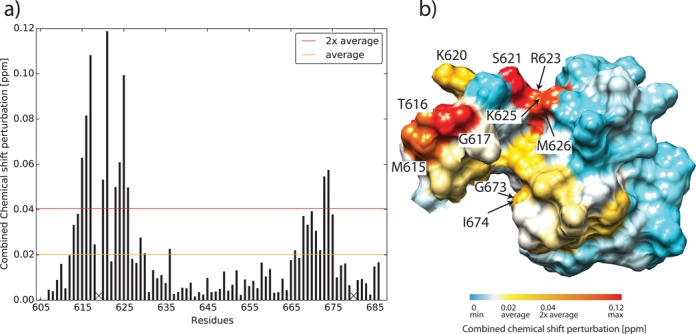
(a) Combined 1H and 15N chemical shift perturbation (CSP) of the Sso MCM C-terminal WH domain. [1H,15N]-TROSY-HSQC spectra of 15N labeled Sso MCM C-terminal WH domain and Sso MCMΔN-term (ILI555DSD) were recorded (Supplementary Figure S19). In order to deduce differences between the isolated Sso MCM C-terminal WH domain and the same domain covalently linked to the AAA+ domain, the combined 1H and 15N chemical shifts of the Sso MCM C-terminal WH domain were compared. 1H/15N scaling factors for glycine and non-glycine residues were 1/5 and 1/8, respectively (45). Proline residues without observable amide resonance are denoted with X. (b) CSP mapping onto the surface representation of Sso MCM C-terminal domain, residues E605-D610 experiencing a CSP<CSPaverage are omitted for clarity. Residues for which CSP>CSPaverage are color-coded ranging from yellow to red. Residues for which CSP>CSP2xaverage are labeled with residue numbers.
In this model, the truncated WH domain caps the AAA+ domain with the linker sandwiched between the two domains (Figure 8a). Of course, this picture only represents a ‘snapshot’ of a probably more dynamic assembly, which was suggested on the basis of unexplained electron density in electronmicroscopy reconstructions (48–51). However, our present data as incorporated in the model favor the ‘end-cap’ arrangement (2,4,51) of the WH domain over a model, which puts the WH domain into the ‘belt region’ of the MCM (48). In addition, CSP analysis of selectively Ile δ-methyl-labeled samples (Supplementary Figure S22) of the WH domain free in solution and attached to the remainder of the MCM together with the CSP and PRE results mentioned above excludes that the WH domain undergoes a significant disorder-to-order transition when linked to the AAA+ domain as proposed earlier (4,51). The model implies a close proximity of the WH domain to the α6-α8 helix bundle (4,10) of the AAA+ domain. This arrangement invites the speculation that the presence of the WH domain and the linker is sensed by α8, transmitted to α6 containing the arginine finger of the ATPase center and the sensor 2 motif, thereby controlling the AAA+ ATPase center allosterically. We concede that this interpretation constitutes a working hypothesis only and needs experimental validation by substantial future structural work. One approach could be the analysis of MCM constructs by solid state NMR using specifically labeled domains in the context of full-length MCM molecules.
CONCLUSION
The C-terminal domain of archaeal MCM complexes represents a truncated WH domain and a less conserved N-terminal extension, linking the truncated WH domain to the adjacent AAA+ domain. The ATPase activity of the AAA+ domain is reduced in presence of this C-terminal domain. Mutational analyses indicate that a short α-helical element of the linker and the N-terminal residues of the first helix of the truncated WH domain apparently exert this allosteric control in the Sso MCM. The inter-domain communication between the MCM AAA+ domain and the C-terminal domain requires idiosyncratic, species-specific determinants, notwithstanding a high structural similarity between archaeal C-terminal truncated WH domain modules as shown here for Sso and Mth. In addition, here the structural description of the Sso MCM monomer is completed by determination of the 3D structure of its C-terminal domain. By integration of the structural and functional data available, a model of the entire Sso MCM could be drafted. This model provides a structural rational for the MCM ATPase allosteric control by its C-terminal domain. Finally, our model offers a basis to further analyze the mechanistic properties of MCM helicases, which are essential and evolutionary conserved components of the replication machinery in eukaryotes and archaea.
ACCESSION NUMBERS
Coordinates of the S. solfataricus and M. thermautotrophicus MCM C-terminal domains have been deposited in the Protein Data Bank under the accession code 2M45 and 2MA3, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to Francesca Pisani (Institute of Protein Biochemistry, Naples) and Silvia Onesti (Elettra Sinchrotone Trieste, Trieste) for providing Sso MCM expression constructs, Silvia Onesti and Helmut Pospiech (FLI) for helpful discussions and Frank Grosse for making available resources for the helicase assays. The WeNMR project (European FP7 e-Infrastructure grant, contract no. 261572, www.wenmr.eu), supported by the European Grid Initiative (EGI) through the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, South Africa, Malaysia, Taiwan, the Latin America GRID infrastructure via the Gisela project and the US Open Science Grid (OSG) are acknowledged for the use of web portals, computing and storage facilities.
FUNDING
Leibniz Graduate School on Ageing and Age-Related Diseases (LGSA) [to C.W.]. Funding for open access charge: Leibniz Institut für Altersforschung, Fritz-Lipmann-Institut (FLI), Jena, Germany. The FLI is a member of the Leibniz Association and is financially supported by the Federal Government of Germany and the State of Thuringia.
Conflict of interest statement. None declared.
REFERENCES
- 1.Forsburg S.L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa A., Onesti S. The MCM complex: (just) a replicative helicase. Biochem. Soc. Trans. 2008;36:136–140. doi: 10.1042/BST0360136. [DOI] [PubMed] [Google Scholar]
- 3.Bochman M.L., Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa A., Onesti S. Structural biology of MCM helicases. Crit. Rev. Biochem. Mol. Biol. 2009;44:326–342. doi: 10.1080/10409230903186012. [DOI] [PubMed] [Google Scholar]
- 5.Slaymaker I.M., Chen X.S. MCM structure and mechanics: what we have learned from archaeal MCM. Subcell. Biochem. 2012;62:89–111. doi: 10.1007/978-94-007-4572-8_6. [DOI] [PubMed] [Google Scholar]
- 6.Vijayraghavan S., Schwacha A. The eukaryotic Mcm2-7 replicative helicase. Subcell. Biochem. 2012;62:113–134. doi: 10.1007/978-94-007-4572-8_7. [DOI] [PubMed] [Google Scholar]
- 7.Jenkinson E.R., Chong J.P. Minichromosome maintenance helicase activity is controlled by N-and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7613–7618. doi: 10.1073/pnas.0509297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barry E.R., McGeoch A.T., Kelman Z., Bell S.D. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res. 2007;35:988–998. doi: 10.1093/nar/gkl1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher R.J., Bishop B.E., Leon R.P., Sclafani R.A., Ogata C.M., Chen X.S. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat. Struct. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 10.Brewster A.S., Wang G., Yu X., Greenleaf W.B., Carazo J.M., Tjajadi M., Klein M.G., Chen X.S. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa A., van Duinen G., Medagli B., Chong J., Sakakibara N., Kelman Z., Nair S.K., Patwardhan A., Onesti S. Cryo-electron microscopy reveals a novel DNA-binding site on the MCM helicase. EMBO J. 2008;27:2250–2258. doi: 10.1038/emboj.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae B., Chen Y.-H., Costa A., Onesti S., Brunzelle J.S., Lin Y., Cann I. K.O., Nair S.K. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure. 2009;17:211–222. doi: 10.1016/j.str.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Slaymaker I.M., Fu Y., Toso D.B., Ranatunga N., Brewster A., Forsburg S.L., Zhou Z.H., Chen X.S. Mini-chromosome maintenance complexes form a filament to remodel DNA structure and topology. Nucleic Acids Res. 2013;41:3446–3456. doi: 10.1093/nar/gkt022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y., Slaymaker I.M., Wang J., Wang G., Chen X.S. The 1.8-Å crystal structure of the N-terminal domain of an archaeal MCM as a right-handed filament. J. Mol. Biol. 2014;426:1512–1523. doi: 10.1016/j.jmb.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 15.Carpentieri F., De Felice M., De Falco M., Rossi M., Pisani F.M. Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J. Biol. Chem. 2002;277:12118–12127. doi: 10.1074/jbc.M200091200. [DOI] [PubMed] [Google Scholar]
- 16.Pucci B., De Felice M., Rocco M., Esposito F., De Falco M., Esposito L., Rossi M., Pisani F.M. Modular organization of the Sulfolobus solfataricus mini-chromosome maintenance protein. J. Biol. Chem. 2007;282:12574–12582. doi: 10.1074/jbc.M610953200. [DOI] [PubMed] [Google Scholar]
- 17.Wiedemann C., Ohlenschläger O., Medagli B., Onesti S., Görlach M. (1)H, (15)N and (13)C chemical shift assignments for the winged helix domains of two archeal MCM C-termini. Biomol. NMR Assign. 2014;8:357–360. doi: 10.1007/s12104-013-9516-0. [DOI] [PubMed] [Google Scholar]
- 18.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 19.Güntert P., Mumenthaler C., Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann T., Güntert P., Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 21.Luginbühl P., Güntert P., Billeter M., Wüthrich K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biomol. NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- 22.Güntert P., Billeter M., Ohlenschläger O., Brown L.R., Wüthrich K. Conformational analysis of protein and nucleic acid fragments with the new grid search algorithm FOUND. J. Biomol. NMR. 1998;12:543–548. doi: 10.1023/A:1008391403193. [DOI] [PubMed] [Google Scholar]
- 23.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski R., Rullmann J.A., MacArthur M., Kaptein R., Thornton J. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 25.de Vries S.J., van Dijk M., Bonvin A.M.J.J. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 26.Wassenaar T.A., van Dijk M., Loureiro-Ferreira N., van der Schot G., de Vries S.J., Schmitz C., van der Zwan J., Boelens R., Giachetti A., Ferella L. WeNMR: structural biology on the grid. J. Grid Comput. 2012;10:743–767. [Google Scholar]
- 27.Gajiwala K.S., Burley S.K. Winged helix proteins. Curr. Opin. Struct. Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 28.Harami G.M., Gyimesi M., Kovács M. From keys to bulldozers: expanding roles for winged helix domains in nucleic-acid-binding proteins. Trends Biochem. Sci. 2013;38:364–371. doi: 10.1016/j.tibs.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Liu C., Wu R., Zhou B., Wang J., Wei Z., Tye B.K., Liang C., Zhu G. Structural insights into the Cdt1-mediated MCM2-7 chromatin loading. Nucleic Acids Res. 2012;40:3208–3217. doi: 10.1093/nar/gkr1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen M.T.N., Jonson P.H., Petersen S.B. Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Eng. 1999;12:535–548. doi: 10.1093/protein/12.7.535. [DOI] [PubMed] [Google Scholar]
- 33.Dombkowski A.A. Disulfide by DesignTM: a computational method for the rational design of disulfide bonds in proteins. Bioinformatics. 2003;19:1852–1853. doi: 10.1093/bioinformatics/btg231. [DOI] [PubMed] [Google Scholar]
- 34.Wishart D.S. Interpreting protein chemical shift data. Prog. Nucl. Magn. Reson. Spectrosc. 2011;58:62–87. doi: 10.1016/j.pnmrs.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Chia N., Cann I., Olsen G.J. Evolution of DNA replication protein complexes in eukaryotes and Archaea. PLoS One. 2010;5:e10866. doi: 10.1371/journal.pone.0010866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallick P., Boutz D.R., Eisenberg D., Yeates T.O. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9679–9684. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGeoch A.T., Trakselis M.A., Laskey R.A., Bell S.D. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 38.Kelman Z., Lee J.K., Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc. Natl. Acad. Sci. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grainge I., Scaife S., Wigley D.B. Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids Res. 2003;31:4888–4898. doi: 10.1093/nar/gkg662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreau M.J., McGeoch A.T., Lowe A.R., Itzhaki L.S., Bell S.D. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 41.Pace C.N., Scholtz J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Respondek M., Madl T., Göbl C., Golser R., Zangger K. Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves. J. Am. Chem. Soc. 2007;129:5228–5234. doi: 10.1021/ja069004f. [DOI] [PubMed] [Google Scholar]
- 43.Madl T., Bermel W., Zangger K. Use of relaxation enhancements in a paramagnetic environment for the structure determination of proteins using NMR spectroscopy. Ang. Chem. Int. Ed. 2009;48:8259–8262. doi: 10.1002/anie.200902561. [DOI] [PubMed] [Google Scholar]
- 44.Madl T., Güttler T., Görlich D., Sattler M. Structural analysis of large protein complexes using solvent paramagnetic relaxation enhancements. Angew. Chem. Int. Ed. 2011;50:3993–3997. doi: 10.1002/anie.201007168. [DOI] [PubMed] [Google Scholar]
- 45.Williamson R.A., Carr M.D., Frenkiel T.A., Feeney J., Freedman R.B. Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry. 1997;36:13882–13889. doi: 10.1021/bi9712091. [DOI] [PubMed] [Google Scholar]
- 46.Görlach M., Wittekind M., Beckman R.A., Mueller L., Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu W., Pucci B., Rossi M., Pisani F.M., Ladenstein R. Structural analysis of the Sulfolobus solfataricus MCM protein N-terminal domain. Nucleic Acids Res. 2008;36:3235–3243. doi: 10.1093/nar/gkn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pape T., Meka H., Chen S., Vicentini G., van Heel M., Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gómez-Llorente Y., Fletcher R.J., Chen X.S., Carazo J.M., San Martín C. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J. Biol. Chem. 2005;280:40909–40915. doi: 10.1074/jbc.M509760200. [DOI] [PubMed] [Google Scholar]
- 50.Costa A., Pape T., van Heel M., Brick P., Patwardhan A., Onesti S. Structural studies of the archaeal MCM complex in different functional states. J. Struct. Biol. 2006;156:210–219. doi: 10.1016/j.jsb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Costa A., Pape T., van Heel M., Brick P., Patwardhan A., Onesti S. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 2006;34:5829–5838. doi: 10.1093/nar/gkl708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


