Abstract
Cardiovascular disease (CVD) is a leading cause of morbidity and mortality. The risk factors for CVD include environmental and genetic components. Human mutations in genes involved in most aspects of cardiovascular function have been identified, many of which are involved in transcriptional regulation. The Mediator complex serves as a pivotal transcriptional regulator that functions to integrate diverse cellular signals by multiple mechanisms including recruiting RNA polymerase II, chromatin modifying proteins and non-coding RNAs to promoters in a context dependent manner. This review discusses components of the Mediator complex and the contribution of the Mediator complex to normal and pathological cardiac development and function. Enhanced understanding of the role of this core transcriptional regulatory complex in the heart will help us gain further insights into CVD.
Keywords: Mediator complex, Cardiac disease, Transcription, Development
Introduction
Cardiovascular disease (CVD) is attributed as the cause of death for approximately 17 million people worldwide in 2008 [1]. Estimates predict that by 2030, over 23 million people will die due to complications involving CVD [1,2]. The risk factors for CVD include genetic abnormalities, metabolic-related diseases and lifestyle choices [3]. Many of the risk factors including physical inactivity, poor diet, obesity and diabetes result in dramatic changes in systemic metabolism [4]. The combinations of CVD and type 2 diabetes risk factors are components of the metabolic syndrome [5]. As the 10 year risk factor for developing CVD doubles in patients with metabolic syndrome, a logical clinical goal is to reduce these risk factors [6].
The intricate patterns of gene expression during normal and disease states are governed by elaborate signaling pathways that converge on the transcriptional machinery. Eukaryotic cells have adapted the transcriptional machinery in order to integrate a plethora of environmental signals, a mechanism vital to maintain homeostasis. The general transcriptional machinery can be recruited to promoters by the Mediator complex [7–9]. The Mediator complex functions as part of the pre-initiation complex that directly binds to and transduces signals from transcription factors [10]. Adaptation to changes in the physiological state of a cell occurs through multiple processes. These include chromatin modification, long non-coding RNAs (ncRNAs), microRNAs (miRNAs) and transcription elongation factors that signal to the transcriptional machinery in part through interaction with the Mediator complex to regulate gene expression (Figure 1) [11,12]. This review focuses on our current understanding of the Mediator complex in cardiovascular development and disease.
Figure 1.
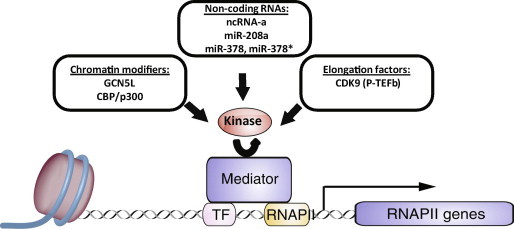
Model representing regulation of Mediator-dependent transcription The Mediator complex coordinates the input from signal-dependent transcription factors (TFs) to recruit RNA polymerase II (RNAPII) and regulate the expression of RNAPII-dependent genes. Chromatin modifiers (GCN5L and CBP/p300), non-coding RNAs including long non-coding RNAs (ncRNA-a) and microRNAs (miR-208a, miR-378 and 378∗), and elongation factors including P-TEFb converge at the Mediator complex to integrate cellular signals resulting in the activation or repression of transcription.
The function of the Mediator complex
The Mediator complex is conserved in eukaryotes and functions to integrate signal-specific events and direct transcription at multiple levels [13,14]. Mediator is a large, multi-subunit complex consisting of more than 26 proteins that forms the pre-initiation complex, recruiting RNA polymerase II (RNAPII) to ensure proper regulation of gene expression [10]. Multiple components of the Mediator complex bind to signal-dependent transcription factors including nuclear hormone receptors, regulating the transcription of target genes [15–17]. This molecular bridge between DNA-bound transcription factors and the general transcription machinery provides an intricate platform for transcriptional control [13,14]. Among the many known functions of the Mediator complex is the recruitment of other transcriptional regulatory complexes including chromatin modifying enzymes such as GCN5L and CBP/p300 [18–21]. Recently, Mediator has been shown to function in conjunction with non-coding RNAs to regulate transcription [22,23]. Thus, Mediator functions to regulate transcription at multiple points in the process of gene expression.
The Mediator complex can be divided into four structural modules. The head, middle and tail modules make up the core Mediator complex [24–28]. The interaction between the core complex and transcription factors such as the thyroid hormone receptor, vitamin D receptor, sterol-response-element-binding protein (SREBP) and nuclear factor-kB (NF-kB) was essential in the early identification of the mammalian Mediator complex [29–32]. The fourth module of the Mediator complex is the kinase submodule which consists of MED13, MED12, cyclin C and cyclin-dependent kinase 8 (CDK8) [33–35]. In mammals, there are homologs of MED13, MED12 and CDK8, which are MED13-like (MED13L), MED12-like (MED12L) and CDK19, respectively [36]. Biochemical analysis of the composition of the Mediator complex demonstrates two major subclasses, the large complex containing all four modules and the smaller complex which lacks the kinase submodule [37]. The reversible interaction and structural studies of the kinase submodule with the core Mediator complex suggest that the interactions between the kinase submodule and RNAPII to the Mediator complex are mutually exclusive [34]. The diverse and still incompletely understood functions of the kinase submodule also include direct interactions with chromatin modifying complexes that further function to repress gene transcription [38–40]. While the majority of studies demonstrate the repressive function of the kinase submodule, recent studies have found that the kinase submodule can also function in some contexts to enhance transcription [41,42].
Mediator in human disease
The number of Mediator complex genetic mutations that are associated with specific human disease is increasing. The list of phenotypes directly associated with the Mediator complex malfunction includes multiple forms of cancer, neurodevelopment and behavioral disorders as well as cardiovascular diseases. A number of excellent reviews describe in detail the conservation, function and mutations associated with the Mediator complex [12,43–48]. Briefly, misregulation of MED1 [49,50], MED12 [51], MED19 [52], MED23 [53], MED28 [54], MED29 [55], CDK8 and CycC [56,57] have all been implicated in various forms of cancer. Multiple human neurodevelopmental diseases are associated with mutations in or altered functions of MED12 [58], MED17 [59], MED23 [60], MED25 [61], CDK19 [62] and CycC [63], while mutations in MED13 [64,65] and MED13L [66,67] have been linked to CVD.
Defects in cardiac development represent approximately 1% of severe birth defects [68,69]. The high prevalence of congenital heart disease raises the question of the underlying cause of the defect. In 2003, mutations in human MED13L, also known as Prosit240 or TRAP240-like, were identified to be correlated with transversion of the great arteries [67]. Recently, two groups have independently reported changes in MED13L expression levels: haplosufficiency or duplication supports the findings that MED13L mutations contribute to congenital heart defects [66,70]. Consistent with these findings, expression studies showed that MED13L is enriched in the heart and further examination revealed relatively high expression of MED13L in the aorta [67]. Together, these studies demonstrate the significant role of MED13L in cardiovascular development. The mechanism for regulation of MED13L expression and function remains unclear and warrants future studies.
Identification of human mutations in the MED13 gene reveals a broader phenotype affecting multiple organs. The clinical report of an 800-kb region of chromosome 17 containing multiple genes including MED13 suggests haplosufficiency in human MED13 may result in developmental abnormalities including small stature, cognitive defects and hearing loss [65]. Further evaluation of MED13 mutations revealed other patients with similar phenotypes [65]. Studies focusing on RNA editing have identified MED13 as an important target for A to I editing in both brain tumor and cyanotic congenital heart disease [65,71]. This information may provide insight into the mechanism for the Mediator complex-dependent regulation of gene expression in normal and diseased states.
Mediator in cardiac disease models
The core Mediator complex is a vital component of eukaryotic transcription [72], potentially explaining why relatively few human mutations have been identified. However, genetic studies of the Mediator complex demonstrate that select components of Mediator are critical for the survival of the multicellular organism but are not necessary for individual cell survival [72–75]. Mutation of peripheral components of the Mediator complex such as MED1 [76,77], MED12 [75], MED13 [78], MED30 [79] and CDK8 [80] in vivo provide tools to study the intricate processes of Mediator-dependent transcriptional regulation.
Despite the link between Mediator and disease described above, few studies have addressed the role of Mediator in CVD. What has been addressed utilizing powerful genetic tools, RNA profiling and metabolic phenotyping has garnered a better understanding of the Mediator complex and its role in CVD.
Mutation of MED30 in the mouse heart results in lethal cardiomyopathy due to altered mitochondria [79]. Homozygous missense mutations in MED30, an isoleucine to phenylalanine substitution at amino acid 44 (I44F) resulting in a hypomorphic allele allowing postnatal survival, leads to cardiomyopathy that can be partially rescued by a ketogenic diet [79]. However, the mutation results in a progressive dilated cardiomyopathy in young adult mice. The cardiomyopathy is manifested by a dramatic decline in mitochondrial oxidative phosphorylation capacity measured by a decrease in respiratory chain function and a decrease in mRNA levels of oxidative phosphorylation genes potentially due to inefficient activation of the transcriptional cofactor PPARGC1A (PGC-1a), a key transcriptional regulator of oxidative phosphorylation gene expression that directly interacts with MED1 [20,81].
Interestingly, a component of the core Mediator complex, MED1, was previously identified in biochemical analysis as a peroxisome proliferator-activated receptor-interacting protein (PRIP) [82,83]. MED1 mutant mice are embryonically lethal at around E11.5 as reported by the Reddy and Roeder groups independently [77,84,85]. Both groups report multiple histological defects in MED1 null mice including thinning of the myocardial compaction layer [50,77]. Further independent studies using a homozygous MED1 hypomorphic allele demonstrate blunted cardiac development in embryos that survive to E13.5 [86]. While these studies clearly demonstrate the requirement for MED1 in development, studies demonstrating the role of MED1 in cardiac specific development and function have not been reported. However, conditional deletion of MED1 in the skeletal muscle and the liver demonstrates a role for MED1 in regulating metabolic homeostasis in obesity studies [87,74].
Mediator regulation of metabolism influences CVD
Recently, multiple organs not traditionally considered to be regulators of metabolism have been implicated in the regulation of whole-body energy expenditure including the heart and the skeletal muscle [78,88,89]. The traditional function of the heart is to supply the rest of the body with nutrients. Maintenance of cardiac contractility requires a high capacity for ATP generation. It is estimated that the heart stores enough energy to sustain function for only a few beats [90]. Thus myocardial metabolism must be highly regulated in response to changes in physiological, pathological and developmental conditions [90]. The heart consumes energy, mainly in the form of fatty acid oxidation, at a high rate and can adapt to changes in the supply of energy by fine-tuning cardiac metabolism and altering gene expression to maximize energy efficiency [90,91]. During cardiovascular diseased states, mitochondrial function is diminished and the heart shifts from β-oxidation toward a glycolytic metabolism [90,92]. Alterations in gene expression occur through signal dependent transcription factors such as SREBP and the cofactors associated with them. SREBP regulates lipid signaling and is thus a key regulator of the enzymes necessary for proper lipid utilization and homeostasis [93].
A recent study has demonstrated that CDK8 regulates SREBP actions by directly phosphorylating the nuclear mSREBP leading to degradation [94]. Further work implicating Mediator in metabolism includes studies in Caenorhabditis elegans demonstrating that MED15 regulates fatty acid metabolism by mediating nuclear hormone receptor-dependent transcription [95]. Additional studies demonstrate that MED23 is involved in the regulation of insulin signaling and cell fate determination in both adipogenesis and smooth muscle cell differentiation [96,97]. MED1, a core protein of the Mediator complex, binds directly to nuclear hormone receptors and is necessary for maintaining energy homeostasis in skeletal muscle [74,82,83]. Taken together, the role of Mediator in regulating metabolism is evident. Further studies are necessary to determine its mechanism and function in cardiovascular diseases.
The Mediator complex functions to regulate RNAPII-dependent gene expression at multiple stages of transcription including elongation by regulating super-elongation complexes through MED23, MED26 and CDK8 [98,99]. Reduction of CDK8 results in a decrease in the phosphorylation of RNAPII C-terminal domain (CTD) and transcriptional elongation [41]. One of the functions of CDK8 is to recruit CDK9, the catalytic subunit of the positive transcription elongation factor (P-TEFb), to the elongation machinery, suggesting a potential role for CDK8 in the regulation of cardiac disease [41]. Interestingly, recent studies demonstrate a direct interaction between MED23 and CDK9 to recruit P-TEFb to MED23-dependent promoters [100]. P-TEFb can phosphorylate RNAPII CTD. Studies of P-TEFb in the heart demonstrate that CDK9 is activated in cardiac hypertrophy [101] and the activation of CDK9 suppresses the expression of PGC1, resulting in mitochondrial dysfunction and thus contributing to apoptotic cardiomyopathy [102].
The function of the Mediator complex in transcriptional regulation in vivo is exceedingly complex and incompletely understood. However, the role of Mediator in the regulation of metabolism is becoming increasingly evident [74,76,78,103]. Our previous work on MED13 in the heart demonstrates the heart’s ability to regulate systemic energy homeostasis [78]. MED13 cardiac transgenic mice demonstrate a resistance to diet-induced obesity due to enhanced energy expenditure, while the cardiac deletion of MED13 results in an enhanced susceptibility to diet-induced obesity. Specifically, MED13 functions to regulate the transcription of a select subset of genes, resulting in altered cardiac gene expression [78]. Although the previous studies of MED13 are of great interest, further studies are needed to define the precise mechanism of gene regulation by the Mediator kinase submodule.
Modulation of the Mediator by ncRNAs
Mediator regulates RNAPII-dependent gene expression which includes both protein-coding RNAs and ncRNAs. Of great interest is a class of ncRNAs known as long ncRNAs (lncRNAs). The vast majority of lncRNAs have unknown functions [104]. However, known functions of lncRNAs vary from modifying gene splicing, to imprinting as well as regulating posttranscriptional events [104]. Recently, Lai et al. reported a subclass of lncRNAs, ncRNA-a, which functions as a transcriptional activator [22]. Interestingly, transcriptional activation by ncRNA-a requires the Mediator complex, specifically MED12. Together, MED12 and ncRNA-a function to regulate chromatin looping. Mutations in MED12 identified in the FG Syndrome reduce the interaction between MED12 and ncRNA-a, suggesting that ncRNA-a may play a significant role in human disease processes [22].
In addition to lncRNAs, a second class of ncRNAs, miRNAs have been shown to be key modifiers of human disease and serve as biomarkers for disease states [3]. miRNAs are small regulatory RNAs about 22 nucleotides in length that function to repress the translation of mRNA, enhance mRNA degradation or in some instances enhance the translation of specific mRNA targets [105]. Multiple studies have shown posttranscriptional modification of Mediator protein expression by miRNAs [78,106–109]. A cardiac specific miRNA, miR-208a regulates stress-dependent cardiomyocyte growth and gene expression partially by the regulation of MED13 [108]. Therapeutic manipulation of miR-208a expression using an antimiR protects against models of CVD and metabolic syndrome [78,110]. Similarly, genetic deletion of miR-378 and 378∗ targets multiple mRNAs including MED13 to regulate systemic energy homeostasis [106]. Together, these studies represent mechanisms to regulate the Mediator complex in response to stress.
Concluding remarks
Genetic evidence demonstrates the importance of the components of the Mediator complex for normal development of multi-tissue organisms. The Mediator complex has been shown to play an important role in numerous aspects of development and diseased states. The recent increase in human genetic disorders revealed to be associated with mutations in various components of the Mediator complex demonstrates the need for the development of animal models to further our understanding of this evolutionarily conserved transcription regulatory complex. Despite the vast amount of studies done deciphering the role of the Mediator complex, few studies have demonstrated tissue specific functions for transcriptional regulation in mammals. Further studies of the many components of Mediator in a context specific manner are warranted. The identification and characterization of novel therapeutic targets regulating components of the Mediator complex such as miR-208a, miR-378 and 378∗, compounds regulating CDK8 activity or chromatin modifying enzymes recruited by Mediator, promise to offer insight into the intricate process of Mediator-dependent transcriptional regulation in normal cardiac development and disease.
Competing interests
The author is a co-inventor of a patent on antimiR-208a for the treatment of metabolic diseases.
Acknowledgements
I would like to thank Duane Hall and Brad Grueter for critical reading of this manuscript. This work is supported by the American Heart Association (Grant No. 13SDG14660064).
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Mendis S., Puska P., Norrving B. World Health Organization; Switzerland: 2011. Global atlas on cardiovascular disease prevention and control. [Google Scholar]
- 2.Go A.S., Mozaffarian D., Roger V.L., Benjamin E.J., Berry J.D., Borden W.B. Heart disease and stroke statistics – 2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quiat D., Olson E.N. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson P.W., Culleton B.F. Epidemiology of cardiovascular disease in the United States. Am J Kidney Dis. 1998;32:S56–S65. doi: 10.1053/ajkd.1998.v32.pm9820465. [DOI] [PubMed] [Google Scholar]
- 5.Grundy S.M., Brewer H.B., Jr, Cleeman J.I., Smith S.C., Jr, Lenfant C., American Heart A. Definition of metabolic syndrome: report of the national heart, lung, and blood institute/American heart association conference on scientific issues related to definition. Circulation. 2004;109:433–438. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 6.Alberti K.G., Eckel R.H., Grundy S.M., Zimmet P.Z., Cleeman J.I., Donato K.A. Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; national heart, lung, and blood institute; American heart association; world heart federation; international atherosclerosis society; and international association for the study of obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 7.Flanagan P.M., Kelleher R.J., 3rd, Sayre M.H., Tschochner H., Kornberg R.D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 8.Maldonado E., Shiekhattar R., Sheldon M., Cho H., Drapkin R., Rickert P. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 9.Thompson C.M., Koleske A.J., Chao D.M., Young R.A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 10.Malik S., Roeder R.G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill J.A., Olson E.N. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 12.Spaeth J.M., Kim N.H., Boyer T.G. Mediator and human disease. Semin Cell Dev Biol. 2011;22:776–787. doi: 10.1016/j.semcdb.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asturias F.J., Jiang Y.W., Myers L.C., Gustafsson C.M., Kornberg R.D. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 14.Myers L.C., Kornberg R.D. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 15.Hu X., Malik S., Negroiu C.C., Hubbard K., Velalar C.N., Hampton B. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103:9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik S., Guermah M., Yuan C.X., Wu W., Yamamura S., Roeder R.G. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol Cell Biol. 2004;24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik S., Roeder R.G. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 18.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S.W., Li G., Lin Y.P., Barrero M.J., Ge K., Roeder R.G. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Wallberg A.E., Yamamura S., Malik S., Spiegelman B.M., Roeder R.G. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 21.Meyer K.D., Donner A.J., Knuesel M.T., York A.G., Espinosa J.M., Taatjes D.J. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 2008;27:1447–1457. doi: 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai F., Orom U.A., Cesaroni M., Beringer M., Taatjes D.J., Blobel G.A. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorsen M., Hansen H., Venturi M., Holmberg S., Thon G. Mediator regulates non-coding RNA transcription at fission yeast centromeres. Epigenetics Chromatin. 2012;5:19. doi: 10.1186/1756-8935-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baek H.J., Malik S., Qin J., Roeder R.G. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flanagan P.M., Kelleher R.J., Sayre M.H., Tschochner H., Kornberg R.D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y.J., Bjorklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 27.Kuras L., Borggrefe T., Kornberg R.D. Association of the mediator complex with enhancers of active genes. Proc Natl Acad Sci U S A. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik S., Roeder R.G. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Boyer T.G., Martin M.E., Lees E., Ricciardi R.P., Berk A.J. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 30.Fondell J.D., Ge H., Roeder R.G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci U S A. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naar A.M., Beaurang P.A., Zhou S., Abraham S., Solomon W., Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 32.Rachez C., Lemon B.D., Suldan Z., Bromleigh V., Gamble M., Naar A.M. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 33.Bourbon H.M., Aguilera A., Ansari A.Z., Asturias F.J., Berk A.J., Bjorklund S. A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 34.Elmlund H., Baraznenok V., Lindahl M., Samuelsen C.O., Koeck P.J., Holmberg S. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akoulitchev S., Chuikov S., Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 36.Sato S., Tomomori-Sato C., Parmely T.J., Florens L., Zybailov B., Swanson S.K. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Taatjes D.J. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ebmeier C.C., Taatjes D.J. Activator–Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci U S A. 2010;107:11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukasawa R., Tsutsui T., Hirose Y., Tanaka A., Ohkuma Y. Mediator CDK subunits are platforms for interactions with various chromatin regulatory complexes. J Biochem. 2012;152:241–249. doi: 10.1093/jb/mvs065. [DOI] [PubMed] [Google Scholar]
- 40.Persaud S.D., Huang W.H., Park S.W., Wei L.N. Gene repressive activity of RIP140 through direct interaction with CDK8. Mol Endocrinol. 2011;25:1689–1698. doi: 10.1210/me.2011-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donner A.J., Ebmeier C.C., Taatjes D.J., Espinosa J.M. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furumoto T., Tanaka A., Ito M., Malik S., Hirose Y., Hanaoka F. A kinase subunit of the human mediator complex, CDK8, positively regulates transcriptional activation. Genes Cells. 2007;12:119–132. doi: 10.1111/j.1365-2443.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- 43.Boube M., Joulia L., Cribbs D.L., Bourbon H.M. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 44.Casamassimi A., Napoli C. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie. 2007;89:1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Conaway R.C., Conaway J.W. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galbraith M.D., Donner A.J., Espinosa J.M. CDK8: a positive regulator of transcription. Transcription. 2010;1:4–12. doi: 10.4161/trns.1.1.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hentges K.E. The Mediator complex: crucial functions in transcription with links to development and disease. Semin Cell Dev Biol. 2011;22:728. doi: 10.1016/j.semcdb.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Kornberg R.D. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Zhu Y., Qi C., Jain S., Le Beau M.M., Espinosa R., 3rd, Atkins G.B. Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc Natl Acad Sci U S A. 1999;96:10848–10853. doi: 10.1073/pnas.96.19.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsumoto K., Yu S., Jia Y., Ahmed M.R., Viswakarma N., Sarkar J. Critical role for transcription coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein/TRAP220 in liver regeneration and PPARalpha ligand-induced liver tumor development. J Biol Chem. 2007;282:17053–17060. doi: 10.1074/jbc.M701956200. [DOI] [PubMed] [Google Scholar]
- 51.Huang S., Holzel M., Knijnenburg T., Schlicker A., Roepman P., McDermott U. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H., Jiang H., Wang W., Gong J., Zhang L., Chen Z. Expression of Med19 in bladder cancer tissues and its role on bladder cancer cell growth. Urol Oncol. 2012;30:920–927. doi: 10.1016/j.urolonc.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Yang X., Zhao M., Xia M., Liu Y.T., Yan J., Ji H.B. Selective requirement for Mediator MED23 in Ras-active lung cancer. Proc Natl Acad Sci U S A. 2012;109:E2813–E2822. doi: 10.1073/pnas.1204311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon N.K., Maresh E.L., Elshimali Y., Li A., Horvath S., Seligson D.B. Elevated MED28 expression predicts poor outcome in women with breast cancer. BMC Cancer. 2010;10:335. doi: 10.1186/1471-2407-10-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuuselo R., Savinainen K., Sandstrom S., Autio R., Kallioniemi A. MED29, a component of the mediator complex, possesses both oncogenic and tumor suppressive characteristics in pancreatic cancer. Int J Cancer. 2011;129:2553–2565. doi: 10.1002/ijc.25924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W., Ji J.Y. Dysregulation of CDK8 and Cyclin C in tumorigenesis. J Genet Genomics. 2011;38:439–452. doi: 10.1016/j.jgg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Firestein R., Bass A.J., Kim S.Y., Dunn I.F., Silver S.J., Guney I. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H., Spaeth J.M., Kim N.H., Xu X., Friez M.J., Schwartz C.E. MED12 mutations link intellectual disability syndromes with dysregulated GLI3-dependent Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2012;109:19763–19768. doi: 10.1073/pnas.1121120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaufmann R., Straussberg R., Mandel H., Fattal-Valevski A., Ben-Zeev B., Naamati A. Infantile cerebral and cerebellar atrophy is associated with a mutation in the MED17 subunit of the transcription preinitiation mediator complex. Am J Hum Genet. 2010;87:667–670. doi: 10.1016/j.ajhg.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hashimoto S., Boissel S., Zarhrate M., Rio M., Munnich A., Egly J.M. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science. 2011;333:1161–1163. doi: 10.1126/science.1206638. [DOI] [PubMed] [Google Scholar]
- 61.Leal A., Huehne K., Bauer F., Sticht H., Berger P., Suter U. Identification of the variant Ala335Val of MED25 as responsible for CMT2B2: molecular data, functional studies of the SH3 recognition motif and correlation between wild-type MED25 and PMP22 RNA levels in CMT1A animal models. Neurogenetics. 2009;10:275–287. doi: 10.1007/s10048-009-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukhopadhyay A., Kramer J.M., Merkx G., Lugtenberg D., Smeets D.F., Oortveld M.A. CDK19 is disrupted in a female patient with bilateral congenital retinal folds, microcephaly and mild mental retardation. Hum Genet. 2010;128:281–291. doi: 10.1007/s00439-010-0848-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueberham U., Hessel A., Arendt T. Cyclin C expression is involved in the pathogenesis of Alzheimer’s disease. Neurobiol Aging. 2003;24:427–435. doi: 10.1016/s0197-4580(02)00132-x. [DOI] [PubMed] [Google Scholar]
- 64.Borik S., Simon A.J., Nevo-Caspi Y., Mishali D., Amariglio N., Rechavi G. Increased RNA editing in children with cyanotic congenital heart disease. Intensive Care Med. 2011;37:1664–1671. doi: 10.1007/s00134-011-2296-z. [DOI] [PubMed] [Google Scholar]
- 65.Boutry-Kryza N., Labalme A., Till M., Schluth-Bolard C., Langue J., Turleau C. An 800 kb deletion at 17q23.2 including the MED13 (THRAP1) gene, revealed by aCGH in a patient with a SMC 17p. Am J Med Genet A. 2012;158A:400–405. doi: 10.1002/ajmg.a.34222. [DOI] [PubMed] [Google Scholar]
- 66.Asadollahi R., Oneda B., Sheth F., Azzarello-Burri S., Baldinger R., Joset P. Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muncke N., Jung C., Rudiger H., Ulmer H., Roeth R., Hubert A. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries) Circulation. 2003;108:2843–2850. doi: 10.1161/01.CIR.0000103684.77636.CD. [DOI] [PubMed] [Google Scholar]
- 68.Bradshaw E.A., Martin G.R. Screening for critical congenital heart disease: advancing detection in the newborn. Curr Opin Pediatr. 2012;24:603–608. doi: 10.1097/MOP.0b013e328357a843. [DOI] [PubMed] [Google Scholar]
- 69.Hoffman J.I., Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 70.Chen C.P., Chen Y.Y., Chern S.R., Wu P.S., Su J.W., Chen Y.T. Prenatal diagnosis and molecular cytogenetic characterization of de novo partial trisomy 12q (12q24.21->qter) and partial monosomy 6q (6q27->qter) associated with coarctation of the aorta, ventriculomegaly and thickened nuchal fold. Gene. 2013;516:138–142. doi: 10.1016/j.gene.2012.12.051. [DOI] [PubMed] [Google Scholar]
- 71.Paz N., Levanon E.Y., Amariglio N., Heimberger A.B., Ram Z., Constantini S. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ito M., Roeder R.G. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- 73.Loncle N., Boube M., Joulia L., Boschiero C., Werner M., Cribbs D.L. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 2007;26:1045–1054. doi: 10.1038/sj.emboj.7601566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen W., Zhang X., Birsoy K., Roeder R.G. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proc Natl Acad Sci U S A. 2010;107:10196–10201. doi: 10.1073/pnas.1005626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rocha P.P., Scholze M., Bleiss W., Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development. 2010;137:2723–2731. doi: 10.1242/dev.053660. [DOI] [PubMed] [Google Scholar]
- 76.Ge K., Guermah M., Yuan C.X., Ito M., Wallberg A.E., Spiegelman B.M. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 77.Ito M., Yuan C.X., Okano H.J., Darnell R.B., Roeder R.G. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 78.Grueter C.E., van Rooij E., Johnson B.A., DeLeon S.M., Sutherland L.B., Qi X. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krebs P., Fan W., Chen Y.H., Tobita K., Downes M.R., Wood M.R. Lethal mitochondrial cardiomyopathy in a hypomorphic Med30 mouse mutant is ameliorated by ketogenic diet. Proc Natl Acad Sci U S A. 2011;108:19678–19682. doi: 10.1073/pnas.1117835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Westerling T., Kuuluvainen E., Makela T.P. Cdk8 is essential for preimplantation mouse development. Mol Cell Biol. 2007;27:6177–6182. doi: 10.1128/MCB.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Riehle C., Abel E.D. PGC-1 proteins and heart failure. Trends Cardiovasc Med. 2012;22:98–105. doi: 10.1016/j.tcm.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan C.X., Ito M., Fondell J.D., Fu Z.Y., Roeder R.G. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Y., Qi C., Jain S., Rao M.S., Reddy J.K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 84.Crawford S.E., Qi C., Misra P., Stellmach V., Rao M.S., Engel J.D. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 85.Zhu Y.J., Crawford S.E., Stellmach V., Dwivedi R.S., Rao M.S., Gonzalez F.J. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J Biol Chem. 2003;278:1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
- 86.Landles C., Chalk S., Steel J.H., Rosewell I., Spencer-Dene B., Lalani el N. The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Mol Endocrinol. 2003;17:2418–2435. doi: 10.1210/me.2003-0097. [DOI] [PubMed] [Google Scholar]
- 87.Bai L., Jia Y., Viswakarma N., Huang J., Vluggens A., Wolins N.E. Transcription coactivator mediator subunit MED1 is required for the development of fatty liver in the mouse. Hepatology. 2011;53:1164–1174. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee N.K., Sowa H., Hinoi E., Ferron M., Ahn J.D., Confavreux C. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bostrom P., Wu J., Jedrychowski M.P., Korde A., Ye L., Lo J.C. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carvajal K., Moreno-Sanchez R. Heart metabolic disturbances in cardiovascular diseases. Arch Med Res. 2003;34:89–99. doi: 10.1016/S0188-4409(03)00004-3. [DOI] [PubMed] [Google Scholar]
- 91.Huss J.M., Kelly D.P. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 92.Rider O.J., Francis J.M., Ali M.K., Holloway C., Pegg T., Robson M.D. Effects of catecholamine stress on diastolic function and myocardial energetics in obesity. Circulation. 2012;125:1511–1519. doi: 10.1161/CIRCULATIONAHA.111.069518. [DOI] [PubMed] [Google Scholar]
- 93.Shao W., Espenshade P.J. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao X., Feng D., Wang Q., Abdulla A., Xie X.J., Zhou J. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J Clin Invest. 2012;122:2417–2427. doi: 10.1172/JCI61462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taubert S., Van Gilst M.R., Hansen M., Yamamoto K.R. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang W., Huang L., Huang Y., Yin J.W., Berk A.J., Friedman J.M. Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev Cell. 2009;16:764–771. doi: 10.1016/j.devcel.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin J.W., Liang Y., Park J.Y., Chen D.R., Yao X., Xiao Q. The mediator MED23 plays opposing roles in directing smooth muscle cell and adipocyte differentiation. Genes Dev. 2012;26:2192–2205. doi: 10.1101/gad.192666.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Conaway R.C., Conaway J.W. The Mediator complex and transcription elongation. Biochim Biophys Acta. 2013;1829:69–75. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takahashi H., Parmely T.J., Sato S., Tomomori-Sato C., Banks C.A., Kong S.E. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang W., Yao X., Huang Y., Hu X.M., Liu R.Z., Hou D.M. Mediator MED23 regulates basal transcription in vivo via an interaction with P-TEFb. Transcription. 2013;4:39–51. doi: 10.4161/trns.22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sano M., Abdellatif M., Oh H., Xie M., Bagella L., Giordano A. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 102.Sano M., Wang S.C., Shirai M., Scaglia F., Xie M., Sakai S. Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J. 2004;23:3559–3569. doi: 10.1038/sj.emboj.7600351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang F., Vought B.W., Satterlee J.S., Walker A.K., Jim Sun Z.Y., Watts J.L. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 104.Lee T.I., Young R.A. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Small E.M., Olson E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carrer M., Liu N., Grueter C.E., Williams A.H., Frisard M.I., Hulver M.W. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378∗. Proc Natl Acad Sci U S A. 2012;109:15330–15335. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mouillet J.F., Chu T., Nelson D.M., Mishima T., Sadovsky Y. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 2010;24:2030–2039. doi: 10.1096/fj.09-149724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 109.Huszar J.M., Payne C.J. MicroRNA 146 (Mir146) modulates spermatogonial differentiation by retinoic acid in mice. Biol Reprod. 2013;88:15. doi: 10.1095/biolreprod.112.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Montgomery R.L., Hullinger T.G., Semus H.M., Dickinson B.A., Seto A.G., Lynch J.M. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]



