Abstract
Breakthroughs in cell fate conversion have made it possible to generate large quantities of patient-specific cells for regenerative medicine. Due to multiple advantages of peripheral blood cells over fibroblasts from skin biopsy, the use of blood mononuclear cells (MNCs) instead of skin fibroblasts will expedite reprogramming research and broaden the application of reprogramming technology. This review discusses current progress and challenges of generating induced pluripotent stem cells (iPSCs) from peripheral blood MNCs and of in vitro and in vivo conversion of blood cells into cells of therapeutic value, such as mesenchymal stem cells, neural cells and hepatocytes. An optimized design of lentiviral vectors is necessary to achieve high reprogramming efficiency of peripheral blood cells. More recently, non-integrating vectors such as Sendai virus and episomal vectors have been successfully employed in generating integration-free iPSCs and somatic stem cells.
Keywords: Reprogramming, Peripheral blood, Hematopoietic cells, Induced pluripotent stem cells, Cell fate conversion
Introduction
Conventional clinical therapies using small molecules, biologicals and other agents have achieved great success in curing diseases and extending life expectancy. However, many disorders induced by diseased cells, damaged tissues or dysfunctional organs can only be cured by replacing them with functional cells, tissues or organs.
The establishment of the first human embryonic stem cell (hESC) line in 1998 conferred new hopes for patients that require replacement therapy, since ESCs can self-renew in maintenance culture and can also be theoretically differentiated into any cell type in the human body using differentiation induction culture methods [1]. Patient-specific ESC lines would be used to prevent immune rejection by allogeneic transplantation. The somatic cell nuclear transfer (SCNT) technology pioneered by John Gurdon in 1962 and successfully used to clone Dolly the Sheep in 1996 [2] has failed to create patient-specific ESCs until 2013 [3].
Yamanaka and Thomson/Yu took an unconventional approach during the race to create ESC-like cells from somatic cells. They established a small viral vector-based gene expression library by cloning dozens of factors that were highly expressed in ESCs. Twenty-four genes were carefully handpicked in Yamanaka’s library [4], which was more manageable than Thomson/Yu’s library of 50–100 genes [5]. They both screened for a combination of factors that are both necessary and essential for reprogramming fibroblasts into pluripotency, in contrast to the identification of a single master factor in earlier transdifferentiation and reprogramming studies [6]. Yamanaka’s delicate experimental design and serendipity led to the identification of four factors—Oct4 (also known as Pou5f1), Sox2, Klf4 and Myc (also known as c-Myc)—and won him the 2012 Nobel Prize in Physiology or Medicine jointly with John Gurdon for successful cellular reprogramming in 2006 [7–9].
Of interest, two factors in the Thomson/Yu combination are different from the four Yamakana factors: NANOG and LIN28. More studies have used the Yamanaka combination than the Thomson/Yu combination, likely because the Yamanaka factors themselves are more efficient in reprogramming. However, it has been found that the addition of NANOG and LIN28, and especially LIN28, can further increase the efficiency of Yamanaka factor-mediated reprogramming [10]. OCT4 and SOX2 are the two common factors between the combinations of Yamanaka factors and Yu/Thomson factors. Indeed, future study found that OCT4 and SOX2 are the most essential factors for reprogramming, with OCT4 and SOX2 alone being sufficient to reprogram fibroblasts [11] or blood CD34+ cells [12] into iPSCs at high efficiency.
The reprogramming of somatic cells to iPSCs has been considered the most important breakthrough in life sciences since the discovery of DNA structure [7–9]. Cellular reprogramming technology has changed the direction of stem cell research and will undoubtedly continue to impact the field of stem cells and regenerative medicine for decades to come. More recently, the concept of in vitro and in vivo direct reprogramming has been introduced and continues to gain momentum [13]. This technique sidesteps the generation of iPSCs and may be more suitable for some applications in regenerative medicine. Progresses on direct reprogramming will be discussed later in this review.
Blood as a cellular source for reprogramming
Fibroblasts are the cellular source for many reprogramming experiments performed in the last decade, but may not be the best choice for directed reprogramming. Mouse embryonic fibroblasts (MEFs) served as the source cells in Yamanaka’s landmark paper and were used likely because of their common availability in ESC cultures as supporting cells [4]. Consequently, fibroblasts were also used in the majority of following studies on cellular reprogramming. Skin biopsy is currently the best approach to procure human primary fibroblasts. However, skin biopsy is an invasive and non-sterile procedure and requires 2–3 weeks to expand harvested cells before experimentation. Skin cells harbor more mutations due to environmental insults such as UV irradiation than cells from inside the body [14]. In contrast to these shortcomings of dermal fibroblasts, peripheral blood is already widely used in medical diagnostics and is obviously the most accessible resource for cellular reprogramming.
White blood cells are the nucleated cells in peripheral blood (PB) at concentrations of 3.6–11 × 106/ml. Nucleated peripheral blood cells are composed of granulocytes (mostly neutrophils), monocytes, T lymphocytes, B lymphocytes and a few progenitor cells. The major components of PB are red blood cells and platelets, which can be depleted by treatment of red blood cell lysis buffer followed by multiple centrifugations. Alternatively, gradient centrifugation with Ficoll depletes both red blood cells and granulocytes, leading to the enrichment of mononuclear cells (MNCs).
Of interest, Tao Cheng and colleagues reported that terminally-differentiated mouse granulocytes have greater reprogramming efficiency than hematopoietic stem/progenitor cells by SCNT [15]. In contrast to SCNT, reprogramming with exogenously expressed factors is inefficient and requires multiple cell cycles to achieve pluripotency. As such, primary granulocytes, monocytes and B lymphocytes are among the most difficult cells to be reprogrammed due to the lack of reliable protocols to expand these cells. Epstein-Barr virus immortalized lymphoblastoid B cells can be readily expanded in ex vivo culture and thus be reprogrammed to pluripotency [16,17]. Primary progenitor cells and mature T cells in PB can be readily expanded using established methods and are among the most successfully-used sources for reprogramming.
T cells are the most abundant cells after granulocytes in PB (20–30%) and T cells can be readily expanded with IL-2 and anti-CD3/CD28 microbeads [18]. Reprogramming of T cells into pluripotency has been achieved by many labs using different approaches [18–20]. T cell reprogramming has the potential to rejuvenate aged T cells for immunotherapy [21,22]. However, mature T cells harbor a single T cell receptor (TCR) after somatic recombination and have lost the ability to regenerate the T cell repertoire with unlimited possibilities. Thus, most investigators focused on reprogramming of non-lymphoid cells.
In contrast to mature T or B cells, blood progenitors contain an intact genome. In addition, they can be expanded in culture conditions that favor the proliferation of myeloid cells or erythroid cells [12,23]. Blood stem/progenitor cells express surface marker CD34 and reside in the stem cell niche. However, approximately 1% stem/progenitor cells enter circulation each day. Although only 0.01–0.1% cells in PB are CD34+ cells, this population can be enriched by magnetic-activated cell sorting (MACS). Alternatively, culture of MNCs for several days leads to the expansion of CD34+ cells to a 5–20% purity, which can be used for reprogramming without further purification. Interestingly, culturing MNCs in serum-free medium supplemented with cytokines including erythropoietin (EPO) is able to expand erythroid progenitors that express CD36, CD71 and CD235a; these cells have also been successfully reprogrammed to iPSCs by Linzhao Cheng’s group [23,24]. Whether myeloid progenitors or erythroid progenitors are a better source for reprogramming has not been reported.
Critical factors for reprogramming somatic cells to pluripotency
The mechanisms of Yamanaka factor-mediated reprogramming have been intensively studied over the past years. Among the numerous publications, several recent reports deserve special attention. One line of studies suggest that Oct4, Sox2 and Klf4 are pioneer factors that bind at enhancers of genes that promote reprogramming and actively open local chromatin, eventually establishing the transcription networks of pluripotent stem cells (PSCs) [25,26]. A second theory proposes that pluripotency factors function as lineage-specific master transcription factors that prevent commitment of cells to mutually exclusive lineages [27]. In support of this second theory, we found that balanced expression of mesendoderm specifier OCT4 and neuroectoderm specifier SOX2 supports high-efficiency reprogramming of blood CD34+ cells into pluripotency [12]. More intriguingly, Hongkui Deng and associates recently found that OCT4 can be replaced with other mesendoderm specifiers such as GATA3, GATA6 and SOX7, while SOX2 can be replaced by neuroectoderm specifiers like GMNN (also know as Geminin) in reprogramming mouse cells [28]. Moreover, two counteracting lineage specifiers can synergistically induce pluripotency in the absence of both OCT4 and SOX2 [28]. In further support of this finding, Belmonte’s group reported that replacement of OCT4 and SOX2 with mesendoderm-related genes like GATA3 and ectoderm-related genes like ZNF521 can also reprogram human cells [29]. Although OCT4, SOX2 and other factors can be replaced by lineage specifiers, the reprogramming efficiency is generally lower than the commonly used factors that are delineated below.
OCT4 has been found to be the most important factor for cellular reprogramming. Increasing OCT4 in ESCs by ∼50% induces differentiation into both mesoderm and endoderm cells [30]. Recently, we found that a high level of OCT4 alone can directly reprogram CD34+ cells into mesoderm progenitor cells or mesenchymal stem cells (MSCs) [31].
SOX2 is a master regulator of both iPSCs and neural stem cells (NSCs). SOX2-overexpressing ESCs show biased differentiation toward neural lineage [32,33]. It has been reported recently that SOX2 alone can reprogram fibroblasts directly into NSCs [34].
KLF4 is not essential for reprogramming induction in itself, but is critical for generating iPSCs with high quality. Klf4 plays important roles in generating iPSCs that can form high-contribution chimaeras or yield “all-iPSC mice’’ by tetraploid (4n) complementation [35,36]. Mechanistic studies demonstrate super-enhancers in ESCs that determine the cell identity are particularly enriched in Klf4, suggesting that Klf4 enhances the core transcriptional network of iPSCs or ESCs [37]. In addition, a recent study reveals a functional role of Klf4 in mediating higher-order chromatin structure for maintaining and inducing pluripotency [38].
MYC increases reprogramming efficiency by approximately tenfold in many systems [39]. Recent studies indicate that the potency of MYC is accounted for by its role as a nonlinear amplifier of the existing gene expression program [40,41]. However, MYC is a potent oncogene; re-activation of MYC in the progeny of iPSCs induces tumor formation [36,42] and therefore it is preferable to avoid the use of MYC in reprogramming. We have recently found that the use of an improved episomal vector system allows MYC to be completely omitted from the factor combination without significantly decreasing the reprogramming efficiency of PB MNCs [43].
BCL-XL is not commonly used as a factor for facilitating reprogramming. Surprisingly, we found that BCL-XL increases efficiency of Yamanaka factor-mediated reprogramming of adult PB MNCs by tenfold [43]. With the use of the four factors—OCT4, SOX2, KLF4 and BCL-XL, we can generate as many as 20–30 integration-free iPSCs from 1 ml PB [43]. The anti-apoptotic activity of BCL-XL may explain these potent effects. It may also play a role in self-renewal and proliferation of iPSCs, because recurrent amplifications at the BCL-XL-harboring chromosome region 20q11.21 was selected after long-term ESC culture [44].
shTP53 has been used to considerably increase the reprogramming efficiency of blood cells [18,24,45]. Knockdown of p53 unfortunately heightens the risk of generating iPSCs with considerable genetic abnormalities due to the genome guardian nature of p53 [46,47].
Many small molecules have been reported to be able to increase reprogramming efficiency [48]. Among them, sodium butyrate, a histone deacetylase inhibitor, is the most important one for reprogramming of blood cells. Butyrate substantially enhances iPSC derivation from both mouse and human cells by increasing the expression of endogenous pluripotency-associated genes and upregulating the expression of miR-302/367 cluster [49–51].
Other factors such as NANOG [52], LIN28 [53], and miR-302 family [54] also play important roles in boosting reprogramming efficiency. We are currently investigating the ability of these factors to increase PB reprogramming efficiency when introduced to our 4-factor-mediated reprogramming protocol [43].
Approaches for reprogramming blood cells to pluripotency
Highly efficient generation of iPSCs with improved lentiviral vectors
Both gammaretroviral vectors and lentiviral vectors have been used for successful cellular reprogramming. However, VSV-G (the G glycoprotein of the vesicular stomatitis virus) pseudotyped lentiviral vectors are more widely used due to a higher packaging capacity, better titers and the ability to transduce almost any type of cells including non-dividing cells from different species [55,56].
In early studies, investigators used viral vectors for reprogramming of fibroblasts to convert blood CD34+ cells into iPSCs. The vectors that lead to high-level gene expression and thereby efficient reprogramming of fibroblasts do not necessarily induce high-level expression of reprogramming factors in blood CD34+ cells. Several successful endeavors were reported, but the reprogramming efficiency was at 0.01% or lower [57–59]. Our group recently used the long-terminal repeat (LTR) from spleen focus-forming virus (SFFV), a strong promoter in hematopoietic cells, to express reprogramming factors. Subsequently we converted 2% cord blood CD34+ cells into iPSCs with OCT4 and SOX2 alone [12], with efficiency reaching up to 20% when the 4 Yamanaka factors—OCT4, SOX2, MYC and KLF4—are used simultaneously [60].
It is critical that reprogramming factors are expressed at high levels to achieve high-efficiency reprogramming. We found that when the expression levels of reprogramming factors decrease to a “threshold”, successful reprogramming cannot be achieved [12]. This is most likely because more transcription factors in an individual cell can turn on the expression of pluripotency-related genes more efficiently and more rapidly. This dosage-dependent effect of reprogramming factors has been recently confirmed by Hochedlinger’s lab, who reported that elevated expression of Yamanaka factors can successfully reprogram some somatic cells that are refractory to reprogramming mediated by low-levels of Oct4, Sox2 and Klf4 [61].
To achieve high-level cellular reprogramming, the following parameters need to be considered when designing an expression vector.
Promoter
Promoter is one of the most important factors to consider when designing a gene-expression vector. The strength of a promoter is contextual, depending on cell types. There is no single best promoter that is suited for all cell types. Therefore, determination of the relative strength of a particular promoter in cells of interest becomes important. For example, the internal cytomegalovirus (CMV) promoter is a powerful promoter in 293T cells and other cell lines, but showed weak activity in hematopoietic cells [62]. The murine stem cell virus (MSCV) LTR drives high-level expression of transgenes in MSCV-based gammaretroviral vectors, which are widely used in the study of hematopoietic stem cells (HSCs) and leukemia [63–65]. Paradoxically, the MSCV LTR actually underperforms when used within the lentiviral vector backbone. We found that SFFV is much stronger than other promoters in driving transgene expression in human hematopoietic cells in comparison to other promoter candidates, such as CMV immediate-early enhancer/chicken β-actin promoter (CAG), elongation factor 1α (EF1) and phosphoglycerate kinase 1 (PGK) (Figure 1A). Accordingly, the use of the SFFV promoter in the lentiviral construct leads to a more than 100-fold increase in reprogramming efficiency compared with other systems [12,60]. Further identification of other promoters that are stronger than SFFV would have a positive impact on this field.
Figure 1.
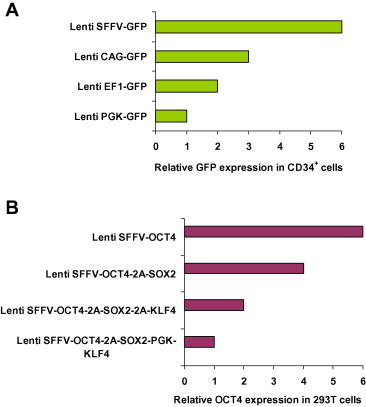
Different lentiviral vector design leads to differential transgene expression levels A. The SFFV promoter is stronger than other commonly-used promoters in human hematopoietic cells. Human cord blood CD34+ cells were transduced with four GFP-expressing lentiviral vectors at the same multiplicity of infection (MOI). Expression of GFP was analyzed by flow cytometry at 3 days after transduction. The backbone of the lentiviral vector was described in detail previously [11]. SFFV, long-terminal repeat from spleen focus-forming virus; CAG, CMV immediate-early enhancer/chicken β-actin promoter; EF1, elongation factor 1α; PGK, phosphoglycerate kinase 1; GFP, green fluorescent protein. B. Linking multiple genes with 2A is a better strategy than dual promoters for the expression of multiple genes. The expression of reprogramming factors OCT4, SOX2 and KLF4 was driven by the SFFV promoter. 293T cells were transduced with four lentiviral vectors at the same MOI. Expression of OCT4 was analyzed by intracellular staining and flow cytometry at 3 days after transduction. The linkage of multiple genes with 2A leads to decreased expression of each one (Lenti SFFV-OCT4 vs. Lenti SFFV-OCT4-2A-SOX2 vs. Lenti SFFV-OCT4-2A-SOX-2A-KLF4). The use of dual promoters considerably decreases the transgene expression compared to the vector design of a single promoter with 2A linkers (Lenti SFFV-OCT4-2A-SOX2-2A-KLF4 vs. Lenti SFFV-OCT4-2A-SOX2-PGK-KLF4). The data shown here are representative of three independent experiments with similar results.
The posttranscriptional regulatory element
The posttranscriptional regulatory element from woodchuck hepatitis virus (WPRE) can stabilize mRNA transcripts, leading to increased expression at the protein level. We and other investigators have found that inclusion of WPRE in the lentiviral constructs increases the transgene expression by approximately fivefold [66,67]. We also found that WPRE can considerably increase gene expression when inserted at the downstream of a transgene in episomal vectors [12].
Synthetic genes
Codon optimization and other strategies may occasionally increase gene expression in mammalian cells. This strategy may not always give rise to desirable results, however. When we used a codon optimization program in an attempt to increase the expression of OCT4 and SOX2 at the protein level, we detected a 20–30% decrease in protein expression compared with wildtype genes, which unsurprisingly leads to a considerable decrease in reprogramming [12]. Conversely, another group reported a 50% increase in gene expression of OCT4, SOX2 and KLF4 at the protein level after codon optimization as detected by Western blotting analysis [68]. However, careful examination of protein expression of these factors by intracellular staining and flow cytometry revealed a 20–30% decrease in protein expression compared to wildtype. As expected, this leads to a considerable decrease in reprogramming efficiency (unpublished observation). Increasing gene expression in mammalian cells by codon optimization remains challenging; currently this is not the best strategy for improving reprogramming efficiency.
Fusion with a transactivation domain
Another strategy to increase the potency of reprogramming factors is by fusing reprogramming factors with a transactivation domain (TAD). It is reported that reprogramming factors that are fused with a TAD from MyoD increases the reprogramming of MEFs by 50-fold [69]. However, we failed to reprogram CD34+ cells using this OCT4 fusion gene. Similarly, TAD of the herpes simplex virus (HSV) type 1 protein VP16 also enhances reprogramming of human and mouse fibroblasts when fused to Oct4, Sox2 and Nanog [70]. It would be interesting to test whether OCT4-VP16 can improve the reprogramming of blood cells.
Stoichiometry of reprogramming factors
The ratios of reprogramming factors are important for successful reprogramming. Studies on the stoichiometry of reprogramming factors found that a combination of equal amounts of Sox2, Klf4, Myc and Oct4 leads to efficient reprogramming of fibroblasts, with an increase of Oct4 by threefold further improving reprogramming efficiency [71]. Another study shows that balanced expression of 4 factors produces iPSCs that efficiently generate ‘‘all-iPSC mice’’ by tetraploid (4n) complementation and maintain normal imprinting at the Dlk1-Dio3 locus, whereas decreased expression of Oct4 and Klf4 and increased expression of Sox2 and Myc lead to the generation of iPSCs with poor quality [36]. Similar to these reports, we found that balanced expression of OCT4 and SOX2 in a single vector substantially increases reprogramming efficiency and minimizes the formation of non-iPSCs [12].
Expression of multiple genes with IRES or dual promoters
One of the commonly used strategies to express multiple genes is by an internal ribosomal entry site (IRES), which mediates translation of the downstream gene via a Cap-independent mechanism [72]. However, Cap-independent translation is considerably less efficient than 5′ Cap-directed translation, and expression levels can vary depending on the specific cell type. In addition to this limitation, composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs, thus in most cases the expression of the gene downstream of IRES ranges from 20% to 50% that of the first gene [73,74]. To our surprise, a recent report showed that expression of Myc and Sox2 downstream of IRES is considerably higher than that of Oct4 and Klf4 upstream of IRES [36], which may be explained by a growth advantage to fibroblasts conferred by high-level Myc expression and thus is selected for. The IRES technique does not work very well in hematopoietic cells, with the observation of Linzhao Cheng and colleagues that transgene expression downstream of IRES is almost undetectable in hematopoietic cells [75]. Instead, the use of another promoter leads to efficient expression of the second transgene [75]. This dual promoter design unfortunately often leads to low-level gene expression at an unpredictable ratio (exemplified in Figure 1B), which is likely due to interference of two promoters with each other in close vicinity.
Expression of multiple genes using 2A sequences
Self-cleaving 2A-like sequences, peptides of 18–22 amino acids, have been used recently to yield equimolar expression of separate proteins on translation from one multicistronic mRNA [76,77]. Cleavage occurs through ribosomal ‘‘skipping’’ during translation, resulting in the release of the upstream protein while translation of the downstream mRNA continues. The protein that is positioned first contains a 2A tag at its C-terminus for all peptide 2A constructs, but these 2A tags do not appear to affect the functionality of expressed reprogramming factors. Commonly used 2A peptides in research are from foot-and-mouth disease virus (F2A), equine Rhinitis A virus (E2A), porcine teschovirus-1 (P2A) and Thosea asigna virus (T2A) [36,78,79]. There is no consensus on the cleavage efficiency of each 2A, though several reports suggest that cleavage of the F2A peptide is the least efficient [36,79,80]. Inclusion of a GSG linker at the N-terminus of the E2A, F2A and T2A peptides has been found to facilitate almost complete cleavage [81–83]. Of interest, expression cassettes containing the T2A sequence were transcribed more efficiently than those containing either F2A or E2A in mouse ESCs [83]. Resultant gene expression has been found similar regardless of the position of a particular gene within a 2A polycistronic construct [83], in contrast to the previous observations of efficient iPSC colony formation when the factors in the expression cassette are ordered as Oct4, Klf4 and Sox2 (OKS), whereas no iPSC colony forms when the factors are ordered as Oct4, Sox2 and Klf4 (OSK) [84]. This discrepancy may be explained by inefficient cleavage of F2A, as our OSK vector linked with E2A can efficiently reprogram blood cells (unpublished observation). Taken together, using 2A peptide sequences in the construction of polycistronic vectors is currently the best design for the expression of several genes at equimolar levels.
Generation of integration-free blood iPSCs using episomal vectors or Sendai virus vectors
iPSCs generated with integrating viral vectors have several safety concerns. (1) The use of the integrating virus is associated with insertional mutagenesis. (2) Reactivation of reprogramming factors may induce tumor formation [36,42]. (3) Residual expression of reprogramming factors interferes with differentiation programs [85,86]. Therefore, it is imperative to use integration-free iPSCs for clinical therapy. Integration-free iPSCs from fibroblasts can be generated by several techniques using transposons, adenovirus vectors, plasmids, minicircle DNAs, artificial chromosome vectors, protein transduction, modified mRNA, Sendai virus vector (SeV) and oriP/EBNA1-based episomal vectors (EV). Of these techniques, blood-derived integration-free iPSCs have only been successfully generated by using the relatively efficient Sendai virus vector and episomal vector approaches.
SeV
SeV is a negative-sense RNA virus where no integration is possible due to the lack of a DNA stage during its life cycle [87]. SeV can infect many types of cells and has been used to generate integration-free iPSCs from fibroblasts, T cells and CD34+ cells [22,39,88,89]. Although SeV does not integrate into the genome and the vector copy gradually decreases, considerable amount of vectors are still present in the cells after 5–10 passages. To solve this problem, a temperature-sensitive SeV has been developed by introducing point mutations in the P and/or L genes of SeV [39]. SeV in the cells decreases to undetectable levels after treating iPSCs at 38 °C for 3–5 days. Recently, a novel replication-defective and persistent SeV (SeVdp) vector has been developed, which is able to accommodate all four Yamanaka factors in one vector for efficient reprogramming of mouse fibroblasts [90]. It is still unknown whether this vector can be used to generate integration-free iPSCs from human blood cells.
EV
The commonly used EV is a plasmid incorporated with 2 elements from Epstein–Barr (EB) virus, i.e., origin of viral replication (oriP) and EB nuclear antigen 1 (EBNA1). OriP allows replication of plasmid in mammalian cells and EBNA1 binds specifically to oriP and tethers the EV plasmid to chromosomal DNA, allowing EBNA1 to mediate replication and partitioning of the episome during division of the host cell. One transfection is sufficient for iPSC reprogramming due to the maintenance of EV plasmids after nucleofection or electroporation, while gradual loss of the vectors during each cell division eventually leads to depletion of residual vectors in the iPSC lines after 2–3 months of culture. EVs were at extremely low efficiency when first used to generate iPSCs from fibroblasts [91]. Further improvements in the vector design and factor combination led to successful generation of integration-free iPSCs from blood cells [18,23]. However, strong oncogenes like SV40 large T antigen (SV40LT) or knockdown of genome guardian p53 in the factor combination were used to boost efficiency in these studies. We managed to generate integration-free iPSCs without using oncogenic factors that may one day compromise patient safety. Our vectors were redesigned with 2 important modifications: (1) the use of the strong promoter SFFV in hematopoietic cells instead of other relatively weak promoters like CAG and (2) inclusion of WPRE at the downstream of the transgene. These changes lead to a more than 10-fold increase in reprogramming efficiency in generating integration-free iPSCs from blood cells [12,43].
Comparison of SeV vs. EV in reprogramming blood cells
Both of these vectors have been successfully used by multiple investigators to generate integration-free iPSCs from blood cells. The major advantage of SeV is that it can efficiently infect blood cells (in particular CD34+ cells), whereas nucleofection is currently the only efficient approach to introduce EV plasmids into blood cells, yet it induces considerable cell death. Overall, EV is more advantageous than SeV for two reasons: (1) the improved EV is approximately tenfold more efficient than SeV in reprogramming blood cells [12,39] and (2) production of EV plasmids is simple and affordable, whereas generation of SeV vectors is relatively more challenging and expensive. However, EVs are still not perfect for reprogramming, and several approaches should be considered to improve cell survival after EV transfection, for example, by dampening the dsDNA transfection-induced innate immune response with small molecules or other agents, or by creating a helper-dependent adenovirus-episomal hybrid vector, which transfects blood CD34+ cells at higher efficiency without attenuating cell survival [92,93].
Two significant papers in reference to the generation of integration-free iPSCs were published during peer review of this manuscript, and their methods may eventually be adapted for generating integration-free iPSCs from blood. Steven Dowdy and colleagues modified a noninfectious, self-replicating Venezuelan equine encephalitis (VEE) virus RNA replicon to express the four Yamanaka factors [94]. In the presence of B18R, a protein from Western vaccinia virus that binds to and neutralizes type I interferons (IFNs) induced VEE virus infection, iPSCs were generated from human newborn or adult human fibroblasts, while discontinuation of B18R leads to depletion of VEE in iPSCs [94]. This technique is similar to SeV, but without the burden of packaging virus. The second recent approach is chemical reprogramming. Hongkui Deng and colleagues reported successful reprogramming of mouse fibroblasts into iPSCs with a combination of seven small-molecule compounds [95]. It takes more than one month for the first mouse iPSC colony to appear using chemical reprogramming. However, these recent developments are important advances in the cellular reprogramming field and should be followed in the near future.
The safety concerns of using iPSCs in clinical therapy
The generation of iPSCs followed by directed differentiation will not only provide large quantities of patient-specific somatic (stem) cells for individualized therapy, but also raises hopes of rejuvenating cells from elderly patients [96,97]. The technological innovations in generating integration-free iPSCs have abrogated concerns associated with the use of integrating viral vectors. However, there are other safety issues that need to be taken into consideration such as epigenetic memory, high mutation rate, immunogenicity and teratoma formation.
Whole genome analysis identified epigenetic memory and aberrant epigenomic reprogramming in initial iPSCs [98,99], which raises a clinical concern on the safety of cells derived from iPSCs. Long-term passage of iPSCs has been found to be able to diminish the epigenetic signature inherited from the parent cells [100]. Furthermore, inclusion of ascorbic acid in the culture medium and the induction of high-level expression of Oct4 and Klf4 are able to prevent aberrant epigenetic variation, allowing for the generation of “all-iPSC” mice [36,101]. The generation of healthy animals from iPSCs demonstrates that the epigenetic variations currently detected may not pose a serious risk [102,103].
Early studies showed that the reprogramming process is mutagenic, with a mutation rate that is 10 times higher than background levels of cells in ex vivo culture [104–106]. However, recent studies indicate that these mutations are largely due to a fixation on rare mutations already present in the parental cells [14,107]. Careful examination of the literature suggests that two potential problems confounded the interpretation of the previously-published data: (1) the use of low-efficiency (0.01% or lower) reprogramming approach, which may lead to a selection for clones harboring rare mutations that enhance reprogramming; (2) the use of source cells that have been cultured for several weeks or longer before reprogramming, which may have introduced rare mutations that are undetectable by current sequencing technology. To control these parameters in our experiments, we used a high-efficiency (2–20%) reprogramming approach and homogenous primary human cord blood (CB) CD34+ cells with only 2 days of culture after cell purification. Our exome sequencing analysis showed only 1.3 coding mutations in each CB iPSC clone, which is considerably lower than previous reports. These results suggest that de novo mutations during CB reprogramming are negligible [60].
It has been surmised that the progeny of patient-specific iPSCs will be well tolerated after transplantation. A single report showed immunogenicity of iPSCs even after syngeneic transplantation [108]. However, further investigation into this issue by two independent labs demonstrated that differentiated cells from iPSCs may actually be negligibly immunogenic [109,110].
The potential for teratoma formation from residential undifferentiated iPSCs is a major hurdle for the clinical application of iPSC-based therapy [111,112]. Several strategies are under development to solve this problem. One study showed that immunodepletion with antibodies against cell surface glycosphingolipid stage-specific embryonic antigen-5 (SSEA-5) and two additional pluripotency surface markers completely prevent teratoma formation from incompletely differentiated ESC cultures [113]. More recently, a chemical approach has been successfully used to prevent tumor formation by selectively eliminating remaining undifferentiated pluripotent cells [114]. The authors found that a single treatment of hESC-derived mixed population with chemical inhibitors of survivin (e.g., quercetin or YM155), a human PSC-specific antiapoptotic factor, induces selective and complete cell death of undifferentiated human PSCs [114]. The use of these selective depletion approaches will considerably decrease the risk of iPSC-based therapy.
Taken together, although early reports cast doubts on the safety of iPSC-based therapy, many recent studies provide hope that these potential problems can be eventually solved.
Applications of integration-free peripheral blood iPSCs
iPSCs are currently applied in disease modeling and toxicology testing [115]. Virus integration and residual expression of reprogramming factors may interfere with the differentiation program [85,86], which may obfuscate the interpretation of the disease modeling results. The ability to efficiently generate integration-free iPSCs from blood cells is predicted to find its application in this category.
For regenerative medicine, iPSCs need to be differentiated into cells of clinical value. Blood iPSCs can be readily differentiated into MSCs, hepatocytes, and cardiomyocytes [43]. Peripheral blood iPSCs show a parental epigenetic memory (as discussed earlier), which may be exploited to preferentially differentiate into hematopoietic cells [116].
HSCs are among the most difficult cells to be differentiated from iPSCs [115], yet recent progress brings us closer to the dream of generating functional HSCs from iPSCs. Mouse repopulating-HSCs can be differentiated from mouse ESCs or iPSCs by overexpressing HOXB4 ectopically [117,118], but long-term multi-lineage HSCs have not been obtained from ESCs or iPSCs without ectopic gene expression. This strategy did not work for human iPSCs, however, due to the differential effects of HOXB4 on cells from different species [64,65]. Transcription factors that specify human HSC identity and/or expand human HSCs are critical for differentiating iPSCs into HSCs with long-term hematopoiesis potential, and the search for new factors deserves special focus. Recently, Dong-Er Zhang and colleagues found that RUNX1a facilitates the differentiation of human iPSCs into HSCs that can engraft immunodeficient mice [119]. Another report from Pereira et al. [120] shows that a combination of 4 factors—Gata2, Gfi1b, c-Fos and Ets6—leads to conversion of mouse fibroblasts to hematopoietic progenitor cells. These studies suggest that a combination of several factors is likely to generate human HSCs from iPSCs. Small molecules may also help in this endeavor: the combination of prostaglandin-E2 (PGE2) and StemRegenin 1 (SR1) substantially increases the efficiency of differentiating iPSCs into cells with long-term repopulating HSC phenotype [121].
Direct reprogramming of fibroblasts and blood cells into somatic (stem) cells
Direct reprogramming of fibroblasts or blood cells into cells of clinical value provides a promising alternative strategy for regenerative therapy and disease modeling. Generation of iPSCs from PB followed by directed differentiation is time-consuming and costly, and may not be the best choice for most patients. Direct reprogramming of fibroblasts into NSCs [34,122], cardiomyocytes [123,124] and hepatocytes [125,126] using retroviruses that express pivotal factors in the target cells has been reported. Another strategy takes advantage of intermediate cells during the process of reprogramming to iPSCs and diverts them to differentiate or transdifferentiate into cells of interest by culturing them in conditions that favor the survival and proliferation of a particular cell type [127,128]. More recently, non-integrating vectors have been used for direct reprogramming. Duanqing Pei and associates combined an episomal system for delivering Yamanaka factors with a chemically-defined culture medium to reprogram epithelial-like cells from human urine into neural progenitor cells [129]. Similarly, Su-Chun Zhang and associates found that transient expression of Yamanaka factors using Sendai virus vectors converts fibroblasts into neural progenitor cells [130]. Similar strategies may also be used to turn blood into NSCs.
We recently found that OCT4 alone is sufficient for reprogramming of blood CD34+ cells into induced MSCs (iMSCs) when culturing cells in MSC-conducive conditions [31]. With the addition of CHIR99021, a GSK3β inhibitor, the reprogramming efficiency can be as high as 16%. Integration-free iMSCs were also generated with an EV vector. Successful programming depends on a vector design that promotes high-level OCT4 expression. The underlying mechanism of high-level OCT4 dependent reprogramming is still under investigation, with it being likely that OCT4 functions as a pioneer transcription factor that opens up chromatin allowing for tissue-specific factors to access [25,26,131]. Building upon this finding, we believe that many cells of therapeutic value, in particular those in the mesoderm lineage such as cardiomyocytes and chondrocytes, can be directly generated from blood by combining OCT4 with transcription factors that specify the particular lineage.
In vivo reprogramming of blood cells for treating systemic disorders
In vitro reprogramming of fibroblasts or blood cells will be able to generate large quantities of cells for replacement therapy. Transplantation of these cells may encounter a major difficulty: how do cells efficiently engraft the host? For example, the majority of MSCs died shortly after systemic injection. Recent progresses on in vivo direct reprogramming will circumvent this major hurdle in regenerative medicine. In vivo reprogramming of pancreatic exocrine cells to beta cells and of cardiac fibroblasts to cardiomyocytes has been successfully achieved, albeit at low efficiency [132–134].
HSCs are the only known cells that constantly migrate out of the marrow and perivascular niches, and travel into circulation to be recruited to damaged tissues. Future investigation into the possibility of in vivo reprogramming of blood stem cells will undoubtedly yield important applications toward systemic diseases. There is still no cure for many genetic disorders such as skeletal dysplasia, Duchene muscular dystrophy and neurodegenerative diseases like amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD). In vivo reprogramming of blood CD34+ cells or T cells into MSCs, muscle stem cells, or NSCs may potentially cure these disorders. This is the “holy grail” of blood reprogramming. Blood cells may be transduced with lentiviral vectors that express reprogramming factors to achieve this goal, or alternatively, transient expression of reprogramming factors with SeV or EV can be used. Targeted integration technologies such as ZFN, TALEN and CRISPR can be employed to increase safety [135]. SeV expresses multiple virus-specific factors, which elicit a strong immune response to transfected cells and may limit the utility of this vector regarding clinical practice. Episomal vectors may be a better choice for in vivo reprogramming, as the only intrinsic factor expressed in EV is EBNA1, which acts to inhibit the CD8-restricted cytotoxic T cell response and evades immune attack [136].
Concluding remarks
The majority of reprogramming studies have been conducted with fibroblasts. Recent success in blood reprogramming encourages more and more investigators to shift their focus from fibroblast reprogramming to in vitro and in vivo cell fate conversion of blood cells. The use of lentiviral vectors for in vivo reprogramming may be used to treat life-threatening diseases that have no safer alternatives, while the use of EV vectors would be preferable for treating less devastating diseases. Currently, the reprogramming efficiency of adult PB with EV plasmids is considerably lower than lentiviral vectors but sufficient for in vitro study. Low efficiency of in vivo reprogramming will make it difficult to deliver significant therapeutic effects. Further investigation in vectorology and resultant technological breakthroughs in non-integrating vectors may eventually develop blood reprogramming technology into paradigm-shifting therapies for the devastating diseases that currently have no cure.
Competing interests
The author has declared that no competing interests exist.
Acknowledgements
The author thanks Amanda Neises, Justin Brier-Jones, Rui-Jun Su and Jason Kiroyan for helpful discussion and critical reading of the manuscript. This work was supported by the Grants for Research and School Partnerships (GRASP) Award from the Loma Linda University and U.S. Army Medical Research Acquisition Activity (USAMRAA) Concept Award (Grant No. W81XWH-11-1-0607).
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Maeshall V.S. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Campbell K.H., McWhir J., Ritchie W.A., Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 3.Tachibana M., Amato P., Sparman M., Gutierrez N.M., Tippner-Hedges R., Ma H. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 6.Graf T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell. 2011;9:504–516. doi: 10.1016/j.stem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 7.Jaenisch R. Nuclear cloning and direct reprogramming: the long and the short path to Stockholm. Cell Stem Cell. 2012;11:744–747. doi: 10.1016/j.stem.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Daley G.Q. Cellular alchemy and the golden age of reprogramming. Cell. 2012;151:1151–1154. doi: 10.1016/j.cell.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Surani M.A. Cellular reprogramming in pursuit of immortality. Cell Stem Cell. 2012;11:748–750. doi: 10.1016/j.stem.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Liao J., Wu Z., Wang Y., Cheng L., Cui C., Gao Y. Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Res. 2008;18:600–603. doi: 10.1038/cr.2008.51. [DOI] [PubMed] [Google Scholar]
- 11.Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 12.Meng X., Neises A., Su R.-J., Payne K.J., Ritter L., Gridley D.S. Efficient reprogramming of human cord blood CD34+ cells into induced pluripotent stem cells with OCT4 and SOX2 alone. Mol Ther. 2012;20:408–416. doi: 10.1038/mt.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladewig J., Koch P., Brustle O. Leveling Waddington: the emergence of direct programming and the loss of cell fate hierarchies. Nat Rev Mol Cell Biol. 2013;14:225–236. doi: 10.1038/nrm3543. [DOI] [PubMed] [Google Scholar]
- 14.Abyzov A., Mariani J., Palejev D., Zhang Y., Haney M.S., Tomasini L. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung L.Y., Gao S., Shen H., Yu H., Song Y., Simith S.L. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat Genet. 2006;38:1323–1328. doi: 10.1038/ng1895. [DOI] [PubMed] [Google Scholar]
- 16.Choi S.M., Liu H., Chaudhari P., Kim Y., Cheng L., Feng J. Reprogramming of EBV-immortalized B-lymphocyte cell lines into induced pluripotent stem cells. Blood. 2011;118:1801–1805. doi: 10.1182/blood-2011-03-340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajesh D., Dickerson S.J., Yu J., Brown M.E., Thomson J.A., Seay N.J. Human lymphoblastoid B-cell lines reprogrammed to EBV-free induced pluripotent stem cells. Blood. 2011;118:1797–1800. doi: 10.1182/blood-2011-01-332064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- 19.Staerk J., Dawlaty M.M., Gao Q., Maetzel D., Hanna J., Sommer C.A. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loh Y.H., Hartung O., Li H., Guo C., Sahalie J.M., Manos P.D. Reprogramming of T cells from human peripheral blood. Cell Stem Cell. 2010;7:15–19. doi: 10.1016/j.stem.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimura T., Kaneko S., Kawana-Tachikawa A., Tajima Y., Goto H., Zhu D. Generation of rejuvenated antigen-specific T cells by reprogramming to pluripotency and redifferentiation. Cell Stem Cell. 2013;12:114–126. doi: 10.1016/j.stem.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Wakao H., Yoshikiyo K., Koshimizu U., Furukawa T., Enomoto K., Matsunaga T. Expansion of functional human mucosal-associated invariant T cells via reprogramming to pluripotency and redifferentiation. Cell Stem Cell. 2013;12:546–558. doi: 10.1016/j.stem.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Chou B.K., Mali P., Huang X., Ye Z., Dowey S.N., Resar L.M. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowey S.N., Huang X., Chou B.K., Ye Z., Cheng L. Generation of integration-free human induced pluripotent stem cells from postnatal blood mononuclear cells by plasmid vector expression. Nat Protoc. 2012;7:2013–2021. doi: 10.1038/nprot.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaret K.S., Carroll J.S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loh K.M., Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Shu J., Wu C., Wu Y., Li Z., Shao S., Zhao W. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montserrat N., Nivet E., Sancho-Martinez I., Hishida T., Kumar S., Miquel L. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 30.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 31.Meng X., Su R.J., Baylink D.J., Neises A., Kiroyan J.B., Lee W.Y. Rapid and efficient reprogramming of human fetal and adult blood CD34+ cells into mesenchymal stem cells with a single factor. Cell Res. 2013;23:658–672. doi: 10.1038/cr.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopp J.L., Ormsbee B.D., Desler M., Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 33.Zhao S., Nichols J., Smith A.G., Li M. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol Cell Neurosci. 2004;27:332–342. doi: 10.1016/j.mcn.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Ring K.L., Tong L.M., Balestra M.E., Javier R., Andrews-Zwilling Y., Li G. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carey B.W., Markoulaki S., Hanna J.H., Faddah D.A., Buganim Y., Kim J. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Z., Gao F., Kim S., Yang H., Lyu J., An W. Klf4 organizes long-range chromosomal interactions with the Oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Ban H., Nishishita N., Fusaki N., Tabata T., Saeki K., Shikamura M. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W. C-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C.Y., Loven J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 43.Su R.-J., Baylink D.J., Neises A., Kiroyan J.B., Meng X., Payne K.J. Efficient generation of integration-free iPS cells from human adult peripheral blood using BCL-XL together with yamanaka factors. PLoS One. 2013;8:e64496. doi: 10.1371/journal.pone.0064496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martins-Taylor K., Xu R.H. Concise review: genomic stability of human induced pluripotent stem cells. Stem Cells. 2012;30:22–27. doi: 10.1002/stem.705. [DOI] [PubMed] [Google Scholar]
- 45.Takenaka C., Nishishita N., Takada N., Jakt L.M., Kawamata S. Effective generation of iPS cells from CD34+ cord blood cells by inhibition of p53. Exp Hematol. 2010;38:154–162. doi: 10.1016/j.exphem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Marion R.M., Strati K., Li H., Murga M., Blanco R., Ortega S. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., Feng H., Gu H., Lewis D.W., Yuan Y., Zhang L. The p53-PUMA axis suppresses iPSC generation. Nat Commun. 2013;4:2174. doi: 10.1038/ncomms3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Li W., Laurent T., Ding S. Small molecules, big roles – the chemical manipulation of stem cell fate and somatic cell reprogramming. J Cell Sci. 2012;125:5609–5620. doi: 10.1242/jcs.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mali P., Chou B.K., Yen J., Ye Z., Zou J., Dowey S. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang G., Taranova O., Xia K., Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285:25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z., Wu W.S. Sodium butyrate promotes generation of human induced pluripotent stem cells through induction of the miR302/367 cluster. Stem Cells Dev. 2013;22:2268–2277. doi: 10.1089/scd.2012.0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saunders A., Faiola F., Wang J. Pursuing self-renewal and pluripotency with the stem cell factor Nanog. Stem Cells. 2013;31:1227–1236. doi: 10.1002/stem.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shyh-Chang N., Daley G.Q. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D.L., Kano Y. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Matrai J., Chuah M.K., VandenDriessche T. Recent advances in lentiviral vector development and applications. Mol Ther. 2010;18:477–490. doi: 10.1038/mt.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar P., Woon-Khiong C. Optimization of lentiviral vectors generation for biomedical and clinical research purposes: contemporary trends in technology development and applications. Curr Gene Ther. 2011;11:144–153. doi: 10.2174/156652311794940782. [DOI] [PubMed] [Google Scholar]
- 57.Loh Y.H., Agarwal S., Park I.H., Urbach A., Huo H., Heffner G.C. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye Z., Zhan H., Mali P., Dowey S., Williams D.M., Jang Y.Y. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunisato A., Wakatsuki M., Kodama Y., Shinba H., Ishida I., Nagao K. Generation of induced pluripotent stem cells by efficient reprogramming of adult bone marrow cells. Stem Cells Dev. 2010;19:229–238. doi: 10.1089/scd.2009.0149. [DOI] [PubMed] [Google Scholar]
- 60.Su R.J., Yang Y., Neises A., Payne K.J., Wang J., Viswanathan K. Few single nucleotide variations in exomes of human cord blood induced pluripotent stem cells. PLoS One. 2013;8:e59908. doi: 10.1371/journal.pone.0059908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi J.K., Hoang N., Vilardi A.M., Conrad P., Emerson S.G., Gewirtz A.M. Hybrid HIV/MSCV LTR enhances transgene expression of lentiviral vectors in human CD34+ hematopoietic cells. Stem Cells. 2001;19:236–246. doi: 10.1634/stemcells.19-3-236. [DOI] [PubMed] [Google Scholar]
- 63.Zhang X.B., Beard B.C., Trobridge G.D., Wood B.L., Sale G.E., Sud R. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J Clin Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X.B., Schwartz J.L., Humphries R.K., Kiem H.P. Effects of HOXB4 overexpression on ex vivo expansion and immortalization of hematopoietic cells from different species. Stem Cells. 2007;25:2074–2081. doi: 10.1634/stemcells.2006-0742. [DOI] [PubMed] [Google Scholar]
- 65.Zhang X.B., Beard B.C., Beebe K., Storer B., Humphries R.K., Kiem H. Differential effects of HOXB4 on nonhuman primate short- and long-term repopulating cells. PLoS Med. 2006;3:e173. doi: 10.1371/journal.pmed.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zufferey R., Donello J.E., Trono D., Hope T.J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramezani A., Hawley T.S., Hawley R.G. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- 68.Warlich E., Kuehle J., Cantz T., Brugman M.H., Maetzig T., Galla M. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19:782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hirai H., Tani T., Katoku-Kikyo N., Kellner S., Karian P., Firpo M. Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells. 2011;29:1349–1361. doi: 10.1002/stem.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Chen J., Hu J.L., Wei X.X., Qin D., Gao J. Reprogramming of mouse and human somatic cells by high-performance engineered factors. EMBO Rep. 2011;12:373–378. doi: 10.1038/embor.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Papapetrou E.P., Tomishima M.J., Chambers S.M., Mica Y., Reed E., Menon J. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci U S A. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martinez-Salas E., Pacheco A., Serrano P., Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J Gen Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 73.Mizuguchi H., Xu Z., Ishii-Watabe A., Uchida E., Hayakawa T. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther. 2000;1:376–382. doi: 10.1006/mthe.2000.0050. [DOI] [PubMed] [Google Scholar]
- 74.Hennecke M., Kwissa M., Metzger K., Oumard A., Kroger A., Schirmbeck R. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29:3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu X., Zhan X., D’Costa J., Tanavde V.M., Ye Z., Peng T. Lentiviral vectors with two independent internal promoters transfer high-level expression of multiple transgenes to human hematopoietic stem-progenitor cells. Mol Ther. 2003;7:827–838. doi: 10.1016/s1525-0016(03)00104-7. [DOI] [PubMed] [Google Scholar]
- 76.Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 77.Ibrahimi A., Vande Velde G., Reumers V., Toelen J., Thiry I., Vandeputte C. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum Gene Ther. 2009;20:845–860. doi: 10.1089/hum.2008.188. [DOI] [PubMed] [Google Scholar]
- 78.Carey B.W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan H.Y., Sivakamasundari V., Xing X., Kraus P., Yap S.P., Ng P. Comparison of IRES and F2A-based locus-specific multicistronic expression in stable mouse lines. PLoS One. 2011;6:e28885. doi: 10.1371/journal.pone.0028885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holst J., Vignali K.M., Burton A.R., Vignali D.A. Rapid analysis of T-cell selection in vivo using T cell-receptor retrogenic mice. Nat Methods. 2006;3:191–197. doi: 10.1038/nmeth858. [DOI] [PubMed] [Google Scholar]
- 83.Gao S.Y., Jack M.M., O’Neill C. Towards optimising the production of and expression from polycistronic vectors in embryonic stem cells. PLoS One. 2012;7:e48668. doi: 10.1371/journal.pone.0048668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 85.Sommer C.A., Sommer A.G., Longmire T.A., Christodoulou C., Thomas D.D., Gostissa M. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seiler K., Soroush Noghabi M., Karjalainen K., Hummel M., Melchers F., Tsuneto M. Induced pluripotent stem cells expressing elevated levels of sox-2, oct-4, and klf-4 are severely reduced in their differentiation from mesodermal to hematopoietic progenitor cells. Stem Cells Dev. 2011;20:1131–1142. doi: 10.1089/scd.2010.0391. [DOI] [PubMed] [Google Scholar]
- 87.Bitzer M., Armeanu S., Lauer U.M., Neubert W.J. Sendai virus vectors as an emerging negative-strand RNA viral vector system. J Gene Med. 2003;5:543–553. doi: 10.1002/jgm.426. [DOI] [PubMed] [Google Scholar]
- 88.Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seki T., Yuasa S., Oda M., Egashira T., Yae K., Kusumoto D. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 90.Nishimura K., Sano M., Ohtaka M., Furuta B., Umemura Y., Nakajima Y. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem. 2011;286:4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dorigo O., Gil J.S., Gallaher S.D., Tan B.T., Castro M.G., Lowenstein P.R. Development of a novel helper-dependent adenovirus-Epstein-Barr virus hybrid system for the stable transformation of mammalian cells. J Virol. 2004;78:6556–6566. doi: 10.1128/JVI.78.12.6556-6566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H., Cao H., Wohlfahrt M., Kiem H.P., Lieber A. Tightly regulated gene expression in human hematopoietic stem cells after transduction with helper-dependent Ad5/35 vectors. Exp Hematol. 2008;36:823–831. doi: 10.1016/j.exphem.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoshioka N., Gros E., Li H.R., Kumar S., Deacon D.C., Maron C. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell. 2013;13:246–254. doi: 10.1016/j.stem.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hou P., Li Y., Zhang X., Liu C., Guan J., Li H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 96.Marion R.M., Blasco M.A. Telomere rejuvenation during nuclear reprogramming. Curr Opin Genet Dev. 2010;20:190–196. doi: 10.1016/j.gde.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 97.Lapasset L., Milhavet O., Prieur A., Besnard E., Babled A., Aït-Hamou N. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nishino K., Toyoda M., Yamazaki-Inoue M., Fukawatase Y., Chikazawa E., Sakaguchi H. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7:e1002085. doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44:398–405. doi: 10.1038/ng.1110. S1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boland M.J., Hazen J.L., Nazor K.L., Rodriguez A.R., Gifford W., Martin G. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 103.Zhao X.Y., Li W., Lv Z., Liu L., Tong M., Hai T. IPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 104.Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hussein S.M., Batada N.N., Vuoristo S., Ching R.W., Autio R., Närvä E. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 106.Ji J., Ng S.H., Sharma V., Neculai D., Hussein S., Sam M. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- 107.Young M.A., Larson D.E., Sun C.W., George D.R., Ding L., Miller C.A. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao T., Zhang Z.N., Rong Z., Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- 109.Araki R., Uda M., Hoki Y., Sunayama M., Nakamura M., Ando S. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- 110.Guha P., Morgan J.W., Mostoslavsky G., Rodrigues N.P., Boyd A.S. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell. 2013;12:407–412. doi: 10.1016/j.stem.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 111.Fong C.Y., Gauthaman K., Bongso A. Teratomas from pluripotent stem cells: A clinical hurdle. J Cell Biochem. 2010;111:769–781. doi: 10.1002/jcb.22775. [DOI] [PubMed] [Google Scholar]
- 112.Kooreman N.G., Wu J.C. Tumorigenicity of pluripotent stem cells: biological insights from molecular imaging. J R Soc Interface. 2010;7:S753–S763. doi: 10.1098/rsif.2010.0353.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tang C., Lee A.S., Volkmer J.P., Sahoo D., Nag D., Mosley A.R. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011;29:829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee M.O., Moon S.H., Jeong H.C., Yi J.Y., Lee T.H., Shim S.H. Inhibition of pluripotent stem cell-derived teratoma formation by small molecules. Proc Natl Acad Sci U S A. 2013;110:14111–14112. doi: 10.1073/pnas.1303669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cherry A.B., Daley G.Q. Reprogrammed cells for disease modeling and regenerative medicine. Annu Rev Med. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim K., Zhao R., Doi A., Ng K., Unternaehrer J., Cahan P. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kyba M., Perlingeiro R.C., Daley G.Q. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 118.Hanna J., Wernig M., Markoulaki S., Sun C.W., Meissner A., Cassady J.P. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 119.Ran D., Shia W.J., Lo M.C., Fan J.B., Knorr D.A., Ferrell P.I. RUNX1a enhances hematopoietic lineage commitment from human embryonic stem cells and inducible pluripotent stem cells. Blood. 2013;121:2882–2890. doi: 10.1182/blood-2012-08-451641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pereira C.F., Chang B., Qiu J., Niu X., Papatsenko D., Hendry C.E. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;3:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gori J.L., Chandrasekaran D., Kowalski J.P., Adair J.E., Beard B.C., D’Souza S.L. Efficient generation, purification, and expansion of CD34+ hematopoietic progenitor cells from nonhuman primate-induced pluripotent stem cells. Blood. 2012;120:e35–e44. doi: 10.1182/blood-2012-05-433797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Han D.W., Tapia N., Hermann A., Hemmer K., Höing S., Araúzo-Bravo M.J. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 123.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang P., He Z., Ji S., Sun H., Xiang D., Liu C. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 126.Sekiya S., Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 127.Kim J., Efe J.A., Zhu S., Talantova M., Yuan X., Wang S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Efe J.A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 129.Wang L., Wang L., Huang W., Su H., Xue Y., Su Z. Generation of integration-free neural progenitor cells from cells in human urine. Nat Methods. 2013;10:84–89. doi: 10.1038/nmeth.2283. [DOI] [PubMed] [Google Scholar]
- 130.Lu J., Liu H., Huang C.T., Chen H., Du Z., Liu Y. Generation of integration-free and region-specific neural progenitors from primate fibroblasts. Cell Rep. 2013;3:1580–1591. doi: 10.1016/j.celrep.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Morris S.A., Daley G.Q. A blueprint for engineering cell fate: current technologies to reprogram cell identity. Cell Res. 2013;23:33–48. doi: 10.1038/cr.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gaj T., Gersbach C.A. Barbas CF 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Levitskaya J., Coram M., Levitsky V., Imreh S., Steigerwald-Mullen P.M., Klein G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]



