Abstract
Pluripotency-associated factors and their rivals, lineage specifiers, have long been considered the determining factors for the identity of pluripotent and differentiated cells, respectively. Therefore, factors that are employed for cellular reprogramming in order to induce pluripotency have been identified mainly from embryonic stem cell (ESC)-enriched and pluripotency-associated factors. Recently, lineage specifiers have been identified to play important roles in orchestrating the process of restoring pluripotency. In this review, we summarize the latest discoveries regarding cell fate conversion using pluripotency-associated factors and lineage specifiers. We highlight the value of the “seesaw” model in defining cellular identity, opening up a novel scenario to consider pluripotency and lineage specification.
Keywords: Reprogramming, iPSCs, Pluripotency, Lineage specifier, Seesaw model, Cell fate conversion
Introduction
Understanding how cellular identity is established is a major goal for modern biology. The programming and reprogramming of cellular identity elicit tremendous scientific and public interest. The groundbreaking work of Takahashi and Yamanaka established a precedent with the generation of induced pluripotent stem cells (iPSCs) by the forced expression of only four transcription factors — Oct4, Sox2, Klf4 and c-Myc [1]. Similar to embryonic stem cells (ESCs), iPSCs can proliferate and self-renew indefinitely under appropriate conditions and give rise to all types of cells in the body, which bestows these cells with many potential uses in regenerative medicine. Patients with degenerative diseases such as diabetes and cancer, along with aging individuals could all benefit from iPSC-based therapies [2].
The discovery of iPSCs
Somatic cells can be reprogrammed by nuclear transfer [3] or by fusion with ESCs [4], suggesting that oocytes and ESCs contain factors that can reprogram somatic cells into stem cells. Inspired by this discovery, Yamanaka and his colleagues selected 24 genes that are specifically expressed in ESCs, which also play important roles in the maintenance of ESC identity, as candidate factors to induce pluripotency in mouse somatic cells. By transducing all 24 candidate genes together, G418-resistant colonies were generated by using Fbx15βgeo/βgeo as a selection marker for pluripotency. These cells were further identified to possess ESC properties. To determine which of the 24 candidates were essential, Yamanaka and his colleagues tested the effects of the withdrawal of individual factors from the 24-candidate gene pool on the generation of G418-resistant colonies. Ultimately Oct4, Sox2, Klf4 and c-Myc were identified to be pivotal for the induction of pluripotency in mouse somatic cells [1]. Furthermore, these factors were proven to be able to induce pluripotency in human somatic cells as well [5].
Other laboratories have also been working on inducing pluripotency in somatic cells. By using Lin28 and Nanog together with Oct4 and Sox2, the Thomson laboratory also independently discovered a set of four reprogramming factors that are highly enriched in ESCs [6]. These first proof-of-principle studies opened the realms of iPSC research to a future in regenerative medicine.
Discovering novel pluripotency regulators for the induction of pluripotency
It is coherent to test whether there are novel ESC-associated factors that can regulate iPSC induction. Several reports have suggested factors that are important for the maintenance of ESC identity can also facilitate the induction of pluripotency. For example, Nr5a2, an orphan nuclear receptor that is enriched in ESCs, can replace Oct4 in the induction of pluripotency [7]. Esrrb, another orphan nuclear receptor that plays a pivotal role in the maintenance of pluripotency, can replace Klf4 and c-Myc [8]. PRDM14 and NFRKB were identified as novel determinants of human ESC identity and can substitute for Klf4 in reprogramming [9]. Recently, by a single-cell analysis of the reprogramming process, Lin28, Sall4, Esrrb and Dppa2 were identified as a completely novel set of reprogramming factors [10], which are different from the factors initially identified by Yamanaka et al. These factors are all important for the maintenance of ESC identity [10]. In addition to the highly expressed factors in ESCs, the maternal factor Glis1 in oocytes was reported to be a novel facilitator of reprogramming [11].
Direct reprogramming into iPSCs by lineage specifiers
For years, it was generally believed that ESCs are maintained by a shield of pluripotency factors. These factors function in concert with each other to prevent ESCs from differentiating into any lineage, thus preserving the ESCs at an undifferentiated state [12,13]. A more challenging perspective has been put forward recently. Pluripotency factors might as well function as classical lineage specifiers that direct ESCs to differentiate into a specific lineage and inhibit their commitment to mutually exclusive lineages [14].
Consistent with the notion, in ESCs, Oct4 promotes the differentiation of mesendoderm (ME) and primitive endoderm, while suppressing differentiation of the ectoderm (ECT) [15–17]; Sox2 inhibits ME differentiation but promotes neural ECT differentiation [16,17]. Shu et al. provided the first proof-of-principle report showing that modulating lineage-specifying forces can restore the pluripotency of mouse somatic cells [18].
When screening for factors that may substitute for Oct4 in the induction of pluripotency, Shu et al. found that GATA3, which is known to regulate ME commitment and specification, can substitute for Oct4. Subsequent analysis of other lineage specifiers that mainly function in ME differentiation and early embryonic patterning, which are generally not enriched in ESCs, found that GATA6, SOX7 and PAX1, among others, were also able to substitute for Oct4 to induce pluripotency, whereas ectodermal specifiers could not. All Oct4 substitutes were also able to attenuate the upregulated expression of ECT-associated genes that is triggered by the expression of Sox2, Klf4 and c-Myc (SKM), whereas knockdown of the key ectodermal marker Dlx3 promoted SKM-only reprogramming [18]. These findings suggest that a novel function of Oct4/GATA3 is to suppress ECT differentiation during reprogramming.
Accordingly, the ECT lineage specifiers SOX1, SOX3, RCOR2 and GMNN can replace Sox2 during reprogramming. Similarly, Sox2 and its substitutes attenuate the expression of ME-specific markers induced by expression of Oct4, Klf4 and c-Myc (OKM) [18–20]. Strikingly, co-expression of GATA6 and GMNN can substitute for Oct4 and Sox2 to reprogram mouse fibroblasts into iPSCs in the presence of Klf4 and c-Myc [18].
More recently, Montserrat et al. showed that lineage specifiers can also be used to reprogram human fibroblasts into iPSCs. The authors found that GATA3 can replace OCT4 and the ECT specifier, ZNF521, can replace SOX2. Lastly, they showed that GATA3, together with ZNF521, OTX2 and PAX6, can substitute for both OCT4 and SOX2 for human iPSC induction in the presence of KLF4 and c-MYC [21].
A “seesaw” model for cell fate conversion
A binodal model for cell fate determination, such as GATA1 and PU.1, RUNX2 and PPARγ, has been examined in various instances of pluripotent stem or progenitor cells that assume a binary cell fate decision [22]. Such circuit hints at the concept of a “balanced pluripotent state”. Inspired by these insights, Shu et al. proposed a new model, termed the “seesaw” model, in which the pluripotent state has a precarious balancing equilibrium that results from continuous mutual competition between rival lineage specification forces (Figure 1). This model comprises two coupled modules— the canonical pluripotency module and the lineage-antagonism module. The former module is represented by the mutual activation of Oct4 and Sox2, whereas mutual inhibition of the ME and ECT genes represents the latter module [18].
Figure 1.
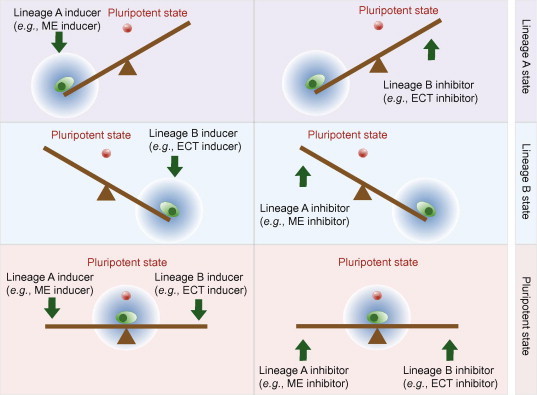
A “seesaw” model for cell fate conversion A modified diagram of the “seesaw” model [18]. Blue clouds indicate the regions that the cell states are likely to sample with noise. The pluripotent state (red ball) is located near the balance region. When the seesaw is balanced between the two differentiation potentials, the cell has a higher probability of entering the pluripotent state. ME stands for mesendoderm and ECT stands for ectoderm.
Both the canonical pluripotency module and the lineage-antagonism module are incorporated leading into the integrated “seesaw” model. The novelty of the “seesaw” model is the proper combination of the two modules. This model led to unexpected insights and scenarios of cell fate conversion. The activation of the cross-activating pluripotency module is important for the reestablishment of the pluripotency network to achieve successful reprogramming. The self-activating pluripotency module gets activated when all of the lineage-specifying forces are counteracted at the dynamic balance point of the “seesaw”. In other words, no particular lineage-specifying activity is dominant in inhibiting the pluripotency module. In this case, the pluripotent state becomes achievable, eliciting the Oct4 and Sox2 self-activating module to coordinate with other pluripotency factors, thus collaboratively restoring the pluripotency network. Once the cross-activating pluripotency module is activated, the ME and ECT lineage fates are blocked by Sox2 and Oct4, respectively. As a result, the pluripotent state is maintained [18].
This innovative model can illustrate the aforementioned points and predicts novel strategies for cell fate conversion, including strategies for the direct conversion of somatic cells into iPSCs by pluripotency factors or lineage specifiers along with the direct conversion of somatic cells into specific lineages by lineage specifiers or pluripotency factors.
Direct reprogramming into other cell types by lineage specifiers or pluripotency factors
The direct reprogramming strategy for cell fate conversion has been widely adapted for some other cell types in addition to iPSCs. The direct conversion of fibroblasts into myoblasts by overexpressing MyoD was reported in 1987 by Davis and colleagues [23]. Recently, increasing numbers of different cell types have been obtained by direct conversion, termed transdifferentiation. For example, a combination of three neuronal lineage-specific transcription factors, Ascl1, Brn2 and Mytl1, is sufficient to induce neurons from fibroblasts [24]. The cardiac-specific transcription factors Gata4, Tbx5 and Mef2c can induce fibroblast transdifferentiation into cardiomyocytes [25]. It is therefore assumed that the more specific and closer the endogenous regulatory network is to the factors, the more efficient the conversion will be [26].
The “seesaw” model also predicts that inhibiting the mutual antagonistic lineage-specifying forces could convert one cell type into another.
Furthermore, consistent with the “seesaw” model, reprogramming factors have been reported to directly produce lineage-committed cells. Two pivotal pluripotency factors, Oct4 and Sox2, were reported to regulate ESC differentiation into different germ layers and to induce direct conversions between different cell types beyond iPSCs.
Previous studies have shown that a twofold increase in Oct4 expression induces ESCs toward mesendodermal specification [15], whereas high levels of Sox2 trigger the neuroectodermal commitment of ESCs. Recently, it was reported that Oct4 and Sox2 also orchestrate a germ-layer fate selection. Oct4 inhibits neuroectodermal differentiation and promotes mesendodermal differentiation, whereas Sox2 promotes neuroectodermal differentiation and inhibits mesendodermal differentiation [16,17].
More recently, overexpression of Oct4 in fibroblasts was shown to lead to transdifferentiation into hematopoietic cells of a mesendodermal lineage [27], whereas overexpression of Sox2 directly converted fibroblasts into neural stem cells. Additionally, decreased expression of Oct4 among the four Yamanaka factors can result in the direct conversion of fibroblasts into neural stem cells [28–30]. These discoveries suggest that pluripotency factors, such as Oct4 and Sox2, can regulate not only pluripotency but also lineage specification.
Conclusion and outlook
After the discovery of the famous Yamanaka factors, a set of transcription factors consisting of Oct4, Sox2, Klf4 and c-Myc, regenerative biology has stepped into a new era. Increasing numbers of pluripotency-related factors have been identified either to replace the Yamanaka factors or to boost the process. Meanwhile, direct transdifferentiation has been successfully demonstrated through a similar strategy, by using lineage-specific transcription factors for specifying each lineage fate. Recently, discoveries and notions related to the “seesaw” model for cell fate conversion have introduced a novel scenario for cell fate conversion causing us to re-evaluate the characteristics of pluripotency factors and lineage specifiers, which are two rivals in the conventional conception of the development of cellular identity. Increasing evidence suggests that pluripotency factors are also lineage specifiers. For example, Oct4 specifies ME differentiation while inhibiting ECT differentiation, and induces hematopoietic transdifferentiation from fibroblasts. Sox2 directs ECT differentiation while prohibiting ME commitment, and induces direct transdifferentiation from fibroblasts into neural stem cells. Overexpression of Nanog, Esrrb, or Tbx3 promotes mesendodermal determination [14]. Lastly, the lineage specifiers depicted as pluripotency rivals, such as GATA3 and PAX6, have been identified to be able to restore pluripotency in somatic cells.
Based on these discoveries, we should reconsider the definitions of pluripotency and lineage specification and present a novel perspective for understanding the determinants of cellular identity, which is one of the most important topics in modern biology.
Competing interests
The author has declared that no competing interests exist.
Acknowledgements
This work was supported by the National Basic Research Program of China (973 Program, Grant No. 2012CB966401), the Key New Drug Creation and Manufacturing Program (Grant No. 2011ZX09102-010-03), the National Science and Technology Major Project (Grant No. 2013ZX10001003), the Ministry of Science and Technology (Grant No. 2011DFA30730 and 2013DFG30680) and 111 Project.
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Robinton D.A., Daley G.Q. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilmut I., Schnieke A.E., McWhir J., Kind A.J., Campbell K.H.S. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 4.Cowan C.A., Atienza J., Melton D.A., Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 7.Heng J.C., Feng B., Han J., Jiang J., Kraus P., Ng J.H. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Feng B., Jiang J., Kraus P., Ng J.H., Heng J.C., Chan Y.S. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 9.Chia N.Y., Chan Y.S., Feng B., Lu X., Orlov Y.L., Moreau D. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 10.Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 12.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loh K.M., Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 16.Thomson M., Liu S.J., Zou L.N., Smith Z., Meissner A., Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Oron E., Nelson B., Razis S., Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Shu J., Wu C., Wu Y., Li Z., Shao S., Zhao W. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang P., Wang Y., Chen J., Li H., Kang L., Zhang Y. RCOR2 is a subunit of the LSD1 complex that regulates ESC property and substitutes for SOX2 in reprogramming somatic cells to pluripotency. Stem Cells. 2011;29:791–801. doi: 10.1002/stem.634. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2007;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 21.Montserrat N., Nivet E., Sancho-Martinez I., Hishida T., Kumar S., Miquel L. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Huang S. Reprogramming cell fates: reconciling rarity with robustness. Bioessays. 2009;31:546–560. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- 23.Davis R.L., Weintraub H., Lassar A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 24.Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vierbuchen T., Wernig M. Direct lineage conversions: unnatural but useful? Nat Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabo E., Rampalli S., Risueño R.M., Schnerch A., Mitchell R., Fiebig-Comyn A. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 28.Han D.W., Tapia N., Hermann A., Hemmer K., Höing S., Araúzo-Bravo M.J. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Ring K.L., Tong L.M., Balestra M.E., Javier R., Andrews-Zwilling Y., Li G. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thier M., Wörsdörfer P., Lakes Y.B., Gorris R., Herms S., Opitz T. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]



