Abstract
One strategy for enhancing the establishment of probiotic bacteria in the human intestinal tract is via the parallel administration of a prebiotic, which is referred to as a synbiotic. Here we present a novel method that allows a rational selection of putative probiotic strains to be used in synbiotic applications: in vivo selection (IVS). This method consists of isolating candidate probiotic strains from fecal samples following enrichment with the respective prebiotic. To test the potential of IVS, we isolated bifidobacteria from human subjects who consumed increasing doses of galactooligosaccharides (GOS) for 9 weeks. A retrospective analysis of the fecal microbiota of one subject revealed an 8-fold enrichment in Bifidobacterium adolescentis strain IVS-1 during GOS administration. The functionality of GOS to support the establishment of IVS-1 in the gastrointestinal tract was then evaluated in rats administered the bacterial strain alone, the prebiotic alone, or the synbiotic combination. Strain-specific quantitative real-time PCR showed that the addition of GOS increased B. adolescentis IVS-1 abundance in the distal intestine by nearly 2 logs compared to rats receiving only the probiotic. Illumina 16S rRNA sequencing not only confirmed the increased establishment of IVS-1 in the intestine but also revealed that the strain was able to outcompete the resident Bifidobacterium population when provided with GOS. In conclusion, this study demonstrated that IVS can be used to successfully formulate a synergistic synbiotic that can substantially enhance the establishment and competitiveness of a putative probiotic strain in the gastrointestinal tract.
INTRODUCTION
The mechanistic role of the gastrointestinal (GI) microbiota and its metabolites in maintaining human health has been well demonstrated (1–3). Gut microbes provide several important benefits for their host, including provision of nutrients, development and maturation of the immune system, and protection against pathogens via colonization resistance (4). However, the gut microbiota may also contribute to obesity, inflammatory and autoimmune diseases, and other chronic disease states (5–7). Such diseases are often associated with compositional alterations in the fecal microbiota, a condition referred to as “dysbiosis” (8). Given that the presence of specific types of bacteria and their relative abundance within the gut are considered to affect host health, there is much interest in devising strategies that modulate gut microbiota composition and potentially redress disease-related dysbiotic patterns (9).
Dietary approaches currently available to modulate the gut microbiota include prebiotics (10–12), fermentable fibers (13, 14), probiotics (or live biotherapeutics) (15), and synbiotics, which are a combination of a probiotic and a prebiotic (11, 16). According to Kolida and Gibson (16), synbiotics can be either complementary or synergistic. Complementary synbiotics consist of a probiotic and a prebiotic selected to independently confer benefits to the host. In contrast, synergistic synbiotics are comprised of a prebiotic chosen specifically for the selected probiotic to stimulate its growth, activity, and survival in the gastrointestinal tract (16).
Synergistic synbiotics therefore hold the potential to improve the establishment of a specific bacterial strain when introduced into the gastrointestinal tract. Unfortunately, successful synergistic synbiotic combinations are not well established in the literature despite a large number of studies. To our knowledge, only two reports describe a synbiotic combination in which the prebiotic significantly enhanced the stability, persistence, or metabolic activity of a specific probiotic strain in vivo (17–19). As noted by Kolida and Gibson (16), this low success rate may be explained by the selection of most synbiotic combinations on an arbitrary basis, including shelf life, industrial performance, availability, and cost. Indeed, few synbiotic preparations are formulated based on a rational selection of both the prebiotic and the probiotic (12, 16), such as via in vitro or in vivo screens assessing the ability of the probiotic to utilize the prebiotic (17–21). Even if synbiotic formulations were based on these criteria, synergism between the probiotic strain and the prebiotic was rarely observed in human and animal trials (22–24). These observations suggest that the probiotic strains were unable to utilize the selected prebiotic to expand their populations under the prevailing ecological conditions in the gastrointestinal tract. We therefore propose that synergistic synbiotics are likely to be more successful if selection of the probiotic organism is based on ecological criteria.
In this report, we introduce the concept of in vivo selection (IVS) to identify putative probiotic strains with enhanced ecological performance when used in synbiotic applications. The concept consists of isolating putative probiotic strains from fecal or intestinal samples after enriching for them with dietary administration of the prebiotic. We reasoned that such strains would likely be able to successfully utilize the prebiotic in vivo within the constraints of the competitive gastrointestinal environment. To test IVS, we isolated bifidobacteria from fecal samples of human individuals who had consumed the prebiotic galactooligosaccharide (GOS) during a previous human trial (25). A combination of approaches was used to select a candidate probiotic strain (Bifidobacterium adolescentis strain IVS-1) enriched by GOS in vivo. We then tested the synergistic potential of this strain and GOS when administered as a synbiotic combination in a rat model of high-fat-diet-induced nonalcoholic fatty liver disease (NAFLD). A NAFLD model without severe inflammatory disease was chosen, as inflammation would potentially confound the ecological analysis due to its effects on gut microbiota composition. Although no direct physiological benefits were observed in the rats, the results from the gut microbiota analysis demonstrated that IVS can be used to select a synergistic synbiotic combination that substantially increases the ecological performance of the bacterial strain in vivo.
MATERIALS AND METHODS
Isolation of in vivo-enriched bifidobacteria from humans.
In a previous study (25), fecal samples were collected from subjects who consumed cumulative doses of GOS (0, 2.5, 5, and 10 g per day for 3 weeks each). Throughout the study, fresh fecal samples were collected and immediately plated onto Rogosa LS agar to enumerate bifidobacteria. Bacterial counts were used to identify GOS responders (i.e., individuals who experienced significant increases in numbers of bifidobacteria), and colonies were picked during the period in which 10 g GOS day−1 was consumed. Colonies were purified by successive liquid and plate cultures, and stock cultures were prepared and stored at −80°C. A total of 28 individual colonies (2 to 3 per subject) were propagated. To classify isolates, DNA was extracted by using the phenol-chloroform extraction method (26), and the 16S rRNA gene was amplified by using the 8F and 1391R universal primers. The amplification product was purified (QIAquick PCR purification kit; Qiagen Inc., MD) and sequenced by a commercial provider (Eurofins MWG Operon, Huntsville, AL). Identity was determined by comparing sequences to sequences in the GenBank database; species were assigned based on the best match.
In vitro growth on GOS.
Each isolate was screened for its ability to use GOS as a growth substrate in an MRS broth culture. Growth experiments were performed with basal MRS broth containing 2% (wt/vol) glucose or GOS (Purimune; GTC Nutrition, Golden, CO). The latter contained 92% GOS, with residual carbohydrates being mainly lactose. Control cultures were therefore also grown on basal MRS broth supplemented with the same amount of lactose as that present in the commercial GOS (giving a final concentration of 0.16% lactose). Cultures were incubated anaerobically at 37°C, and growth was determined by optical density measurement at 600 nm. Strains that grew on GOS to cell densities similar to those on glucose were considered GOS fermenters.
Strain-specific primer design and validation.
The genome of B. adolescentis IVS-1 was sequenced to draft status by using a standard shotgun library prep kit on a Roche GS FLX sequencer at the former Core for Applied Genomics and Ecology (CAGE) (University of Nebraska, Lincoln, NE). Sequencing resulted in 65,460 reads that were assembled de novo by using the gsAssembler (Newbler) module of the GS-FLX Off-Instrument software suite. This resulted in draft sequences of 148 contigs with ∼15-fold coverage.
Unique genes in B. adolescentis IVS-1 were identified by comparing the annotated genome with other available B. adolescentis genomes in the JGI database (using the Phylogenetic Profiler for Single Genes tool in IMG). From this analysis, the clustered regularly interspaced short palindromic repeat (CRISPR)-associated helicase Cas3 was selected as the target gene, and a putative primer pair was designed by using Primer 3 software (27). Candidate primers were evaluated for hairpin and dimer formation by using Netprimer (Premier Biosoft International, Palo Alto, CA). The selected forward (F) primer TTGCTTTTGCTCTGGAACATAC and reverse (R) primer GTAATGAGGTAATACTGCGTCC were validated in silico by performing a BLAST search against the NCBI database. These primers were also validated experimentally by quantitative real-time PCR (qRT-PCR) using DNA from 10 different Bifidobacterium strains related to strain IVS-1 (each having >96% identity at the 16S rRNA gene level). These strains included Bifidobacterium adolescentis ATCC 15703, Bifidobacterium adolescentis L2-32, Bifidobacterium longum subsp. longum ATCC 15707, Bifidobacterium longum DJO10A, Bifidobacterium longum ATCC 15697, Bifidobacterium longum subsp. longum F8, Bifidobacterium longum subsp. longum JDM301, Bifidobacterium sp. strain 113, Bifidobacterium sp. strain 12_1_47BFAA, and Bifidobacterium sp. strain HMLN14. Furthermore, to test if primers could select against fecal bacterial communities in both humans and rats, DNA from 23 human fecal samples and 10 Sprague-Dawley rat fecal samples from an independent study were tested. Human fecal materials analyzed included the baseline samples (i.e., before GOS supplementation) from 18 subjects from a previous study by Davis et al. (25) as well as five other human fecal samples from an independent study.
Quantitative real-time PCR.
qRT-PCR was performed by using a Mastercycler Realplex2 instrument (Eppendorf AG, Hamburg, Germany). Each PCR was performed with 25-μl volumes using real-time master mix containing SYBR (5 Prime Inc., Gaithersburg, MD) and either genus-specific primers for Bifidobacterium, F primer TCGCGTC(C/T)GGTGTGAAAG and R primer CACATCCAGC(A/G)TCCAC (25, 26), or the strain-specific primers for B. adolescentis IVS-1 (described above), each at a concentration of 0.8 μM. Annealing temperatures of 58°C and 61°C were used for the genus- and strain-specific PCRs, respectively. Standard curves for absolute quantification of bacterial cell numbers were prepared by using cultures of B. adolescentis IVS-1 grown overnight (14 h), as described previously (25, 26).
Administration of the probiotic, prebiotic, and synbiotic to rats.
A freeze-dried powder of Bifidobacterium adolescentis IVS-1 was produced by a contract manufacturer (Culture Systems, Mishawaka, IN). The powder contained 5 × 1010 CFU g−1 and was stable during the entire course of the study. For delivery to the rats, the powder was suspended in drinking water (double-distilled water) to reach a concentration of 3 × 107 cells ml−1. GOS was diluted in water at a concentration of 0.033 g ml−1, and the synbiotic was prepared by mixing both IVS-1 and GOS in the above-mentioned concentrations. All preparations were prepared fresh daily in drinking water for the duration of the experiment. Cell viability and stability were validated by plating samples on MRS medium at different time points. This analysis revealed that IVS-1 was highly stable in drinking water, with levels dropping <1 log over 24 h. The addition of GOS did not influence the viability of the probiotic in drinking water (data not shown).
Rat study design.
Synergism of the synbiotic preparation was tested in a rat model of NAFLD (28). Four-week-old male Sprague-Dawley rats were obtained from Charles River Laboratories (Wilmington, MA) and acclimated for 5 days prior to study initiation. All animals were housed in pairs in individually vented cages mounted on a rack with positive airflow. The room environment was maintained at 20°C to 21°C with a 12-h light-dark cycle. Prior to the start of the study, all rats received a standard rat chow and autoclaved, double-distilled water ad libitum during the 5-day acclimation period. All animal procedures were approved by University of Nebraska—Lincoln IACUC.
Rats were randomly assigned to one of five treatments, with three to six rats per group. Groups 1 through 4 were fed a high-fat diet (60% kcal from fat) (AIN-58G9 TestDiet) (see Table S1 in the supplemental material), while group 5 received a standard diet (12% fat) (AIN-58G7 TestDiet) for 8 weeks. After 4 weeks of feeding, groups were assigned to one of the following supplement treatments. Rats in groups 1 and 5 received no additional treatment. Group 2 rats received drinking water supplemented with 3.3% GOS to give ∼1 g of GOS day−1 rat−1. Group 3 rats were given drinking water supplemented with ∼1 × 109 CFU of B. adolescentis IVS-1 day−1 rat−1. Group 4 rats received both the GOS and IVS-1 (synbiotic mixture), at the same doses as those given to groups 2 and 3. All treatments were prepared fresh daily and administered for 4 weeks. The daily water intake per rat was significantly different among groups and was used to calculate the absolute doses of probiotic cells per day (P = 0.001) (see Table S2 in the supplemental material). Rats fed the probiotic drank significantly more water (41.9 ± 8.6 ml) than did rats fed the synbiotic (35.4 ± 4.5 ml), resulting in a significantly higher dose of IVS-1 in the probiotic group (1.26 × 109 CFU versus 1.06 × 109 CFU; P = 0.0001). GOS consumption was not significantly different between the prebiotic- and synbiotic-fed groups (P = 0.2063) (see Table S2 in the supplemental material).
Body weights were determined weekly throughout the study. All rats were necropsied after 8 weeks of study. Blood, cecum, colon content, liver, and epididymal fat pads were collected, and the cecum and colon content were immediately frozen in liquid nitrogen and stored at −80°C until further use.
Evaluation of host physiological parameters in rats.
Liver lipid extraction was performed according to methods described previously by Folch and colleagues (29). Aliquots of lipid extract were saponified to quantify triglycerides (TGs) by using the TG diagnostic kit (Thermo dimethyl adipimidate kit; Thermo Electron Clinical Chemistry, Louisville, CO). Data are reported as μg TG mg−1 (wet weight) liver tissue. To evaluate liver damage, plasma alanine aminotransferase (ALT) and alkaline phosphatase (ALP) enzyme levels were measured, which are indicators of hepatocyte damage/leakage and cholangiocyte stress, respectively (30, 31). Blood was collected into heparinized tubes at necropsy, and ALT and ALP levels were quantified by using a Mammalian Liver Profile rotor in a VetScan VS2 analyzer (Abaxis, Union City, CA). Levels of tumor necrosis factor alpha (TNF-α) and monocyte chemoattractant protein 1 (MCP-1) were quantified as measures of systemic inflammation by using a Milliplex rat magnetic bead multiplex assay (Merck Millipore, Billerica, MA) according to the manufacturer's protocol.
Illumina 16S RNA sequencing and sequence analysis.
Colonic and cecal contents were flash-frozen in liquid nitrogen at necropsy, and DNA was extracted as described previously (26), with one modification: the lysis buffer contained 20 mM Tris-HCl (pH 8), 2 mM EDTA, 1.2% Triton X-100 (pH 8.0), and 20 mg ml−1 Lysozyme (MP Biomedicals, Solon, OH). Amplicon sequencing of colonic contents was performed by the University of Minnesota Genomics Center, and all samples were sequenced together in the same run. First, the V5-V6 region of the 16S rRNA gene was amplified with primer pair 784F (5′-RGGATTAGATACCC-3′) and 1064R (5′-CGACRRCCATGCANCACCT-3′) in a 25-μl PCR mixture containing 5 μl of template DNA, 5 μl of 2× HotStarTaq PCR master mix, a final concentration of primers of 500 nM, and 0.025 U μl−1 HotStarTaq polymerase (Qiagen Inc.). Amplification reactions included an initial denaturation step at 95°C for 5 min followed by 20 to 25 cycles of denaturation (50 s at 94°C), annealing (30 s at 40°C), and elongation (30 s at 72°C). Next, samples were diluted 1:100 in water for input into library tailing PCR. The PCR was analogous to the one conducted for initial amplification except for a Taq polymerase concentration of 0.25 U μl−1, and the PCR conditions consisted of an initial denaturation step at 95°C for 5 min followed by 10 to 15 cycles of denaturation (50 s at 94°C), annealing (30 s at 40°C), and elongation (1 min at 72°C).
PCR products were quantified by using the Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Life Technologies). A subset of the amplicon libraries was spot checked on a Bioanalyzer High-Sensitivity DNA chip (Agilent Technologies, Santa Clara, CA) for correct amplicon size. Next, samples were normalized to 2 nM and pooled. The total volume of the libraries was reduced by the use of a SpeedVac, and amplicons were size selected at 420 bp ± 20% by using the Caliper XT system (PerkinElmer, Waltham, MA). Afterwards, library pools were cleaned with 1.8× AMPureXP beads (Beckman Coulter, Brea, CA) and eluted in water. The amount of DNA in the final pool was quantified with PicoGreen and normalized to 2 nM for input into the Illumina MiSeq platform (v3 kit) to produce 300-bp paired-end sequencing products. Clustering was done at 10 pM with a 5% spike of PhiX. The generated sequences were quality filtered with Illumina software at the University of Minnesota Genomics Center. Twenty-two of 24 samples met all quality control criteria and were used for the microbial community analysis.
Microbial community analysis.
Reads were trimmed to 240 bp with the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), and paired-end reads were merged with the merge-illumina-pairs application (https://github.com/meren/illumina-utils/) (P value of 0.03, enforced Q30 check, perfect matching to primers, and no ambiguous nucleotides allowed). Files exceeding 30,000 reads were subsampled to this number in Mothur v.1.31.162 to standardize the sequencing depth across samples. Subsequently, USEARCH v7.0.100163 was used to generate operational taxonomic units (OTUs) with a 98% similarity cutoff. OTU generation included the removal of putative chimeras identified against the Gold reference database, in addition to the chimera removal inherent to the OTU clustering step in UPARSE. After quality control and chimera removal, samples contained an average of 25,718 ± 941 sequences. The resulting sequences were also taxonomically characterized from phylum to genus levels with Ribosomal Database Project (RDP) Classifier with the MultiClassifier v1.1 tool. All phylotypes were computed as percent proportions based on the total number of sequences in each sample.
Statistical analysis.
Results are expressed as means ± standard deviations (SD) unless otherwise stated. To analyze bacterial composition, diversity differences, and host physiological parameters, one-way analyses of variance (ANOVA) with repeated measures in combination with Tukey's post hoc tests were applied. To achieve normality for data that were not normally distributed, values were subjected to log10 transformations. If only two groups were compared, Student's t tests were performed. Spearman's correlations were used to assess correlations between bacterial groups. To account for type I errors, the false discovery rate was used. A P value of <0.05 and correlation coefficient (r) values of >0.60 (in absolute values) were considered significant. Analyses of variance and false discovery rate control were performed by using SAS/STAT (SAS Institute Inc., Cary, NC, USA), while correlations were determined by using GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA, USA).
Nucleotide sequence accession number.
The genome sequence of B. adolescentis IVS-1 has been deposited in the DDBJ/EMBL/GenBank database under accession number JRNZ01000000.
RESULTS
In vivo selection of B. adolescentis IVS-1.
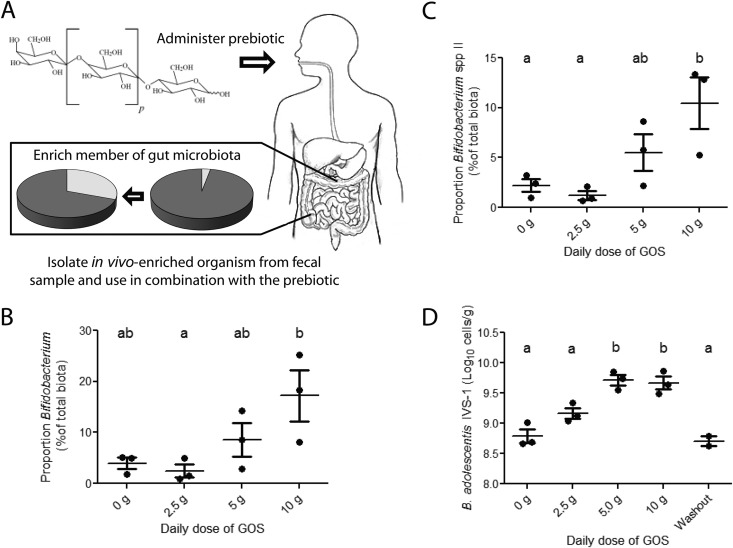
In a previous study (25, 32), we reported a significant and remarkably specific enrichment of Bifidobacterium populations in human subjects during dietary supplementation with GOS (as demonstrated by 454 sequencing, genus-specific qRT-PCR, and quantitative culture), which is in agreement with data from other GOS feeding studies (33–38). Cultural enumeration of fecal samples during the human trial allowed us to identify individuals in which bifidobacteria were enriched by GOS and from whom strains likely to utilize GOS in vivo could be selected. This novel strategy for selection and recovery of autochthonous strains enriched by a prebiotic is referred to as in vivo selection (IVS) (Fig. 1A). Using the IVS approach, a total of 28 presumptive bifidobacterial colonies from 11 subjects were isolated and classified by sequencing of the 16S rRNA genes. Eight isolates were classified as Bifidobacterium adolescentis, eight were classified as Bifidobacterium longum, three were classified as Bifidobacterium pseudocatenulatum, and one was classified as Bifidobacterium bifidum. Of the remaining isolates, four belonged to the Coriobacterium genus, one could be classified only to the family level (Lachnospiraceae), and three could not be sequenced due to insufficient growth. All strains resulting in pure cultures were also screened for their ability to ferment GOS during in vitro growth, and 13 were classified as GOS fermenters, 12 were classified as nonfermenters, and 3 could not be propagated to be tested (data not shown). Out of the 13 strains able to ferment GOS, 5 were classified as B. longum, 5 were classified as B. adolescentis, 1 was classified as B. bifidum, 1 was classified as B. pseudocatenulatum, and another one was classified as Lachnospiraceae. None of the isolated Coriobacterium strains were classified as fermenters.
FIG 1.
In vivo selection to identify putative probiotic strains to be used in synbiotic applications. (A) Concept of in vivo selection. (B) Proportion of fecal bifidobacteria in a human individual consuming GOS (included in chews) in four increasing doses (0, 2.5, 5, and 10 g) during a human feeding trial (25), as determined by 454 pyrosequencing of 16S rRNA tags. (C) Proportion of Bifidobacterium lineage species II in the same individual, as determined by pyrosequencing. (D) Cell numbers of B. adolescentis IVS-1 in the same individual, as quantified by strain-specific qRT-PCR.
Based on the culture data, 454 sequencing (32), and the GOS fermentation tests, we selected one strain and designated it IVS-1. This strain originated from a subject who showed a strong bifidogenic response to GOS (Fig. 1B). Based on 16S rRNA sequencing, IVS-1 had 98.4% identity (100% query coverage and an E value of zero) with the 16S rRNA gene of B. adolescentis ATCC 15703T and was therefore allotted to this species. However, the strain belongs to a distinct phylogenetic cluster (Bifidobacterium species II cluster) detectable by using the V1-V3 region of the 16S rRNA gene (32). This cluster was significantly enriched by GOS in all subjects, including the individual from whom IVS-1 was isolated (Fig. 1C). The ability of B. adolescentis IVS-1 to utilize GOS was demonstrated by growth in MRS broth containing 2% GOS (see Fig. S1 in the supplemental material). The established metabolic benefits of the species B. adolescentis serve as another rationale for the selection of IVS-1 for future applications (39, 40).
To verify that B. adolescentis strain IVS-1 was specifically enriched by GOS in vivo, we devised a strain-specific qRT-PCR approach with primers based on the genome sequence of IVS-1. Primer specificity was validated against 10 closely related Bifidobacterium strains, fecal DNA from all subjects included in the human feeding trial (25) and five additional human individuals, and 10 fecal samples from Sprague-Dawley rats from an independent experiment. A detectable PCR product was obtained only with DNA from B. adolescentis IVS-1 and the fecal sample from which the strain was isolated. This finding indicated that the primers were highly strain specific and that strain IVS-1 was present only in the human subject from whom it was isolated.
The strain-specific qRT-PCR system was then used to quantify the abundance of IVS-1 in fecal samples from this subject during the GOS feeding study. This analysis revealed that IVS-1 levels were increased 8-fold during both the 5-g and 10-g GOS dose periods compared to the 0-g period (P < 0.001) (Fig. 1D), before returning to baseline levels immediately after GOS consumption ended. Collectively, these results demonstrated the utility of IVS to select a bacterial strain enriched in the human gastrointestinal tract through dietary administration of a prebiotic.
Test of the synbiotic combination using rats on a high-fat diet.
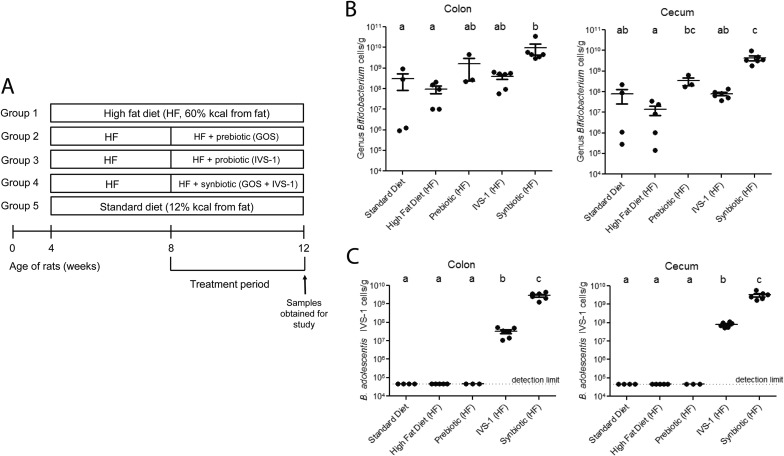
We systematically tested synergism between strain IVS-1 and GOS when used as a synbiotic in rats fed a high-fat diet (Fig. 2A). Decreases in numbers of bifidobacteria are often observed during high-fat-diet feeding (41–43). To determine if our synbiotic strategy could redress this decrease, we employed a rat model of high-fat-diet-induced NAFLD where rats develop steatosis (fatty liver) but do not show an increase in body weight, develop liver inflammation, or progress to nonalcoholic steatohepatitis (NASH) (28). In our study, all high-fat-diet-fed rats developed steatosis (i.e., liver triglyceride levels of >50 μg mg−1 of tissue) and had slightly increased plasma ALP levels compared to rats fed a standard diet (see Table S2 in the supplemental material). Dietary supplements significantly influenced triglyceride liver contents; however, high-fat-diet-fed rats did not develop the histopathological liver inflammation characteristic of NASH (data not shown) and did not have increased plasma ALT levels (see Table S2 in the supplemental material). Plasma tumor necrosis factor alpha (TNF-α) and monocyte chemoattractant protein 1 (MCP-1) levels were not significantly elevated in the high-fat-fed rats compared to the controls (see Table S2 in the supplemental material), indicating a lack of systemic inflammation. Together, these data indicated that all rats receiving a high-fat diet developed NAFLD but not severe inflammatory disease that would confound the evaluation of synbiotic synergy and gut microbial ecology.
FIG 2.
Test of a synbiotic combination of B. adolescentis IVS-1 and GOS in a high-fat-diet rat model. (A) Experimental design of the rat study. Rats were fed either a standard diet or a high-fat diet for 8 weeks, supplemented with or without a probiotic (IVS-1), a prebiotic (GOS), or a synbiotic (IVS-1 plus GOS) for the last 4 weeks. (B) Quantification of absolute cell numbers of bifidobacteria in colonic and cecal contents by genus-specific qRT-PCR. (C) Strain-specific qRT-PCR was used to quantify absolute numbers of B. adolescentis IVS-1 in colonic and cecal contents.
Experiments in rats demonstrate strong synergism between IVS-1 and GOS.
To test the functionality of the prebiotic to support the establishment of B. adolescentis IVS-1 in the rat intestine, rats fed a high-fat diet were administered either IVS-1 alone, GOS alone, or the synbiotic combination; all findings were compared to results for the high-fat controls (Fig. 2A). Consistent with data from previous studies (41, 42), high-fat feeding decreased the abundance of bifidobacteria in both the colon and cecum of the rats, although this reduction did not reach statistical significance (Fig. 2B and Table 1). Genus-specific qRT-PCR analysis revealed that the prebiotic, but not IVS-1, significantly increased the total number of bifidobacteria in the cecum (Fig. 2B). These findings indicate that the introduction of IVS-1 alone did not increase Bifidobacterium abundance above baseline levels (∼108 cells/g), whereas the prebiotic substrate was able to support the resident population. Compared to IVS-1 and GOS alone, the combination of the two dramatically increased the total number of bifidobacteria in the cecum (P < 0.01 between synbiotic and prebiotic treatments; P < 0.001 between synbiotic and probiotic treatments) (Fig. 2B).
TABLE 1.
Proportions of bacterial taxa significantly influenced by dietary treatments
| Taxonomic group | Mean % bacterial abundance ± SDc |
P value determined by ANOVA | ||||
|---|---|---|---|---|---|---|
| Standard diet | Control HF diet | Prebiotic (HF) | Probiotic (HF) | Synbiotic (HF) | ||
| Phyla | ||||||
| Firmicutes | 87.6 ± 5 A | 88.8 ± 9 A | 76.9 ± 8 A | 87.8 ± 5 A | 59.3 ± 7 B | <0.0001 |
| Actinobacteria | 8.9 ± 6 AB | 3.6 ± 2 A | 19.6 ± 8 BC | 7.6 ± 4 A | 39.1 ± 7 C | <0.0001 |
| Families | ||||||
| Clostridiaceae | 3.9 ± 6 | 0.5 ± 1 | 0.8 ± 1 | 5.4 ± 6 A | 0.2 ± 0 B | 0.0061 |
| Incertae sedis XIV | 3.7 ± 6 | 7.5 ± 10 | 11.0 ± 15 | 1.1 ± 2 A | 17.3 ± 12 B | 0.0342 |
| Streptococcaceae | 12.7 ± 5 | 21.3 ± 5 A | 9.3 ± 1 | 8.9 ± 6 B | 6.6 ± 2 B | 0.0045 |
| Erysipelotrichaceae | 16.8 ± 11 | 21.3 ± 17 | 9.2 ± 1 A | 26.5 ± 10 B | 8.3 ± 3 A | 0.0226 |
| Bifidobacteriaceae | 5.9 ± 7 A | 1.3 ± 1 A | 17.0 ± 9 | 4.1 ± 2 A | 37.8 ± 7 B | 0.0017 |
| Coriobacteriaceae | 0.6 ± 0 | 0.3 ± 0 | 1.0 ± 1 | 1.9 ± 3 A | 0.2 ± 0 B | 0.0263 |
| Rikenellaceae | 0.9 ± 1 A | 0.1 ± 0 | 0.3 ± 0 | 0.1 ± 0 | 0.0 ± 0 B | 0.0181 |
| Genera | ||||||
| Clostridium | 3.9 ± 6 | 0.5 ± 1 A | 0.8 ± 1 | 5.3 ± 6 B | 0.2 ± 0 A | 0.0122 |
| Blautia | 3.4 ± 6 | 7.4 ± 10 | 11.0 ± 15 | 0.9 ± 1 A | 17.2 ± 12 B | 0.0431 |
| Holdemania | 0.1 ± 0 | 1.0 ± 2 A | 0.9 ± 0 | 0.0 ± 0 B | 0.0 ± 0 B | 0.0117 |
| Bifidobacterium | 5.9 ± 7 A | 1.3 ± 1 A | 17.0 ± 9 | 4.1 ± 2 A | 37.8 ± 7 B | 0.0017 |
| Lactococcus | 12.4 ± 4 | 21.0 ± 5 A | 9.1 ± 1 | 8.7 ± 6 B | 6.3 ± 2 B | 0.0045 |
| Alistipes | 0.9 ± 1 A | 0.1 ± 0 | 0.3 ± 0 | 0.1 ± 0 | 0.0 ± 0 B | 0.0181 |
| OTUsa | ||||||
| OTU_2 (B. adolescentis, 99%) | 0.0 ± 0 A | 0.0 ± 0 A | 0.0 ± 0 A | 3.4 ± 2 BC | 37.0 ± 7 BD | <0.0001 |
| OTU_1 (L. lactis, 100%) | 12.4 ± 4 | 21.0 ± 5 A | 9.1 ± 1 | 8.6 ± 6 B | 6.3 ± 2 B | 0.0045 |
| OTU_626 (Lachnospiraceaeb) | 0.4 ± 1 A | 0.1 ± 0 A | 0.0 ± 0 B | ND | 0.1 ± 0 | 0.0002 |
| OTU_7 (Turicibacter sanguinis, 97%) | 3.7 ± 3 | 2.3 ± 3 | 2.3 ± 2 | 9.1 ± 7 A | 0.5 ± 1 B | 0.0279 |
| OTU_14 (Blautiab) | 0.0 ± 0 A | 0.1 ± 0 AB | 1.5 ± 1 BC | 0.0 ± 0 AB | 9.7 ± 6 CD | 0.0003 |
| OTU_33 (L. intestinalis, 99%) | ND | 0.0 ± 0 A | ND | 1.1 ± 2 B | 0.0 ± 0 A | 0.0022 |
| OTU_9 (Clostridium sp.b) | 3.8 ± 6 | 0.5 ± 1 A | 0.7 ± 1 | 5.3 ± 6 B | 0.2 ± 0 A | 0.0128 |
| OTU_6 (B. pseudolongum, 97%) | 5.8 ± 7 | 0.9 ± 1 | 16.6 ± 8 A | 0.6 ± 1 | 0.0 ± 0 B | 0.0293 |
| OTU_44 (Clostridium cocleatum, 99%) | ND | 1.0 ± 1 A | ND | 0.0 ± 0 B | ND | 0.0121 |
Percent homologies to the closest type strain in the database are shown in parentheses. If the strain could not be assigned to a type strain (<97% homology), RDP Classifier was used to determine the most likely genus, and the RDP Classifier value is shown (80% cutoff).
OTU without closely related type strain (<97% homology) classified with RDP Classifier.
Values with different uppercase letters are significantly different from each other. HF, high fat; ND, not detected.
Strain-specific qRT-PCR analysis of B. adolescentis IVS-1 clearly demonstrated a synergistic effect of IVS-1 and GOS in the colon and in the cecum. Even though rats receiving IVS-1 alone consumed significantly more IVS-1 on a daily basis than did rats given the synbiotic due to increased drinking water consumption (P = 0.0001) (see Table S2 in the supplemental material), the synbiotic led to an almost 2-log increase in the level of IVS-1 in the colon and cecum (9.47 ± 0.2 log10 cells g−1 and 9.43 ± 0.2 log10 cells g−1, respectively) compared with the probiotic treatment (7.9 ± 0.1 and 7.44 ± 0.3 log10 cells g−1 in the cecum and colon, respectively) (P = 0.0001) (Fig. 2C). No IVS-1 was detected in rats fed the standard diet, the high-fat diet, or the prebiotic alone.
16S rRNA sequencing confirms synergism between IVS-1 and GOS in vivo.
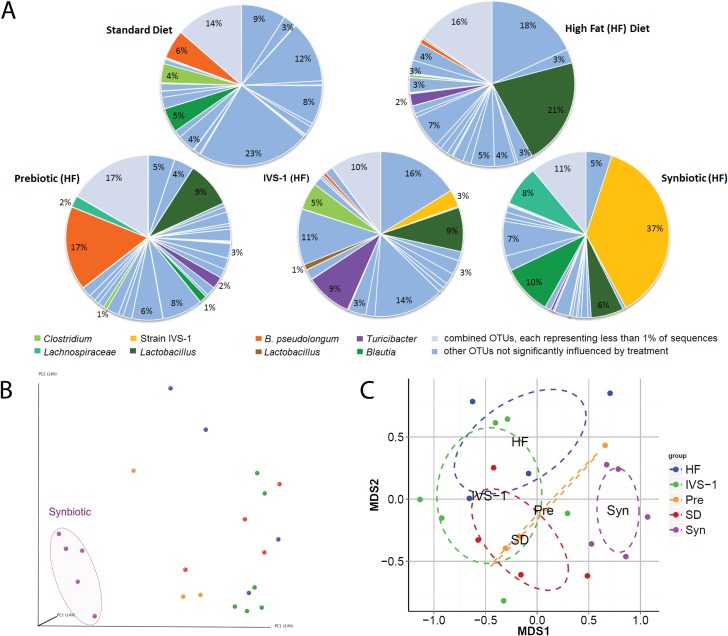
We analyzed the 16S rRNA tags obtained via Illumina sequencing to gain a community-wide perspective on treatment effects on the resident gut microbiota. The ability of pro- and synbiotic treatments to establish IVS-1 in rats was assessed based on the abundance of an operational taxonomic unit (OTU) representing the species B. adolescentis (OTU_2). This species was undetectable in rats that did not receive the probiotic treatment but constituted 3.4% of the microbiota in rats fed IVS-1 (Fig. 3A and Table 1). This finding indicates that the B. adolescentis population observed in rats was due solely to the administration of IVS-1. This finding was expected, as this species is not a member of the normal rat microbiota. Sequences representing B. adolescentis were enriched to 37.0% in rats receiving the synbiotic treatment, indicating a significant enhancement of the probiotic (in terms of abundance) due to the addition of the prebiotic (P = 0.0159). Without GOS, IVS-1 was only the eighth most abundant OTU in the rats' colonic microbiota, while it became the most abundant OTU when given together with GOS, having an abundance almost four times higher than that of the second most abundant OTU (a Blautia species, at 9.7%) (Fig. 3A). This finding demonstrated that IVS-1 could be introduced as the dominant member of the rat gut microbiota when GOS was also provided.
FIG 3.
Characterization of the rat colonic microbiota composition by Illumina sequencing of 16S rRNA tags. (A) Analysis of colonic microbiota at the OTU level. OTUs representing at least 1% of total sequences are shown individually, while OTUs representing <1% are grouped. OTUs in colors other than light blue were significantly influenced by the dietary treatment. (B) Principal coordinate analysis (Bray-Curtis distance) of beta diversity. (C) NMDS plot of beta diversity based on Bray-Curtis distance. SD, standard diet.
Community-wide characterization of effects on gut microbiota.
GOS treatment alone promoted a remarkably specific bifidogenic response, leading to an increase in the abundance of only one OTU related to Bifidobacterium pseudolongum (OTU_6) (Table 1 and Fig. 3A). These findings confirm the highly specific bifidogenic response of GOS, which was previously demonstrated in humans (32).
Although IVS-1 treatment alone did not significantly increase the abundance of the genus Bifidobacterium, it induced a significant increase in the abundance of Bifidobacterium adolescentis at the species level (Table 1). Of note, several unexpected changes were also detected, such as enrichment of the family Clostridiaceae, the genus Clostridium, and an OTU within this genus (OTU_9). Furthermore, the abundance of an OTU related to Lactobacillus intestinalis (OTU_33) increased, while that of an OTU related to Lactococcus lactis (OTU_1) decreased (Table 1).
Synbiotic treatment significantly increased the proportion of Actinobacteria (P < 0.0001), the family Bifidobacteriaceae (P = 0.0017), and the genus Bifidobacterium (P = 0.0017) (Table 1). These shifts were almost completely equivalent to shifts of OTU_2, showing that the above-described alterations at higher taxonomic levels were due to the enrichment of IVS-1. The establishment of IVS-1 was associated with an increase in the abundances of the genus Blautia and one OTU within this genus (OTU_14). In addition, there was a reduction in the abundances of the phylum Firmicutes (P < 0.0001) and families within this phylum, including Clostridiaceae, Streptococcaceae, and Erysipelotrichaceae. The abundances of the genera Clostridium and Lactococcus and OTUs within these genera were also decreased (Table 1).
To assess both the alpha and beta diversities of the community in the colon, different diversity indexes were calculated from the data. Specifically, Shannon's index and the number of observed OTUs were used to determine the alpha diversity, and principal coordinate analysis (PCoA) and nonmetric multidimensional scaling (NMDS) plots based on Bray-Curtis distance were used to visualize the similarity between samples for each treatment.
On average, 135.41 ± 34.4 OTUs per sample were identified. Alpha diversity based on Shannon's index was not significantly influenced by the treatment; however, there was a tendency for reduced diversity in the synbiotic group. This was caused by a slight reduction in community evenness, likely due to the expansion of a single species (B. adolescentis) (data not shown). Two independent approaches were used to analyze the beta diversity of the microbiota communities among treatments. PCoA and NMDS, based on Bray-Curtis distances of beta diversity, revealed that communities from rats fed the synbiotic clustered separately from the microbiomes of rats fed all the other treatments, which clustered together (Fig. 3B and C). This finding demonstrated that only the synbiotic treatment caused a global shift in microbiota structure.
Systematic analyses of associations between members of the gut microbiota.
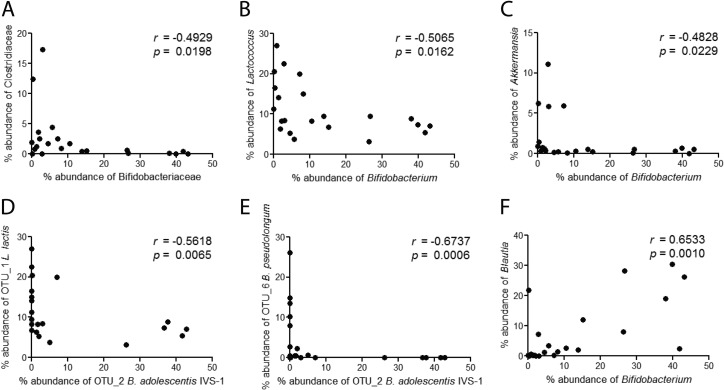
To identify potential interactions between IVS-1 and members of the gut microbiota, and among other bacterial members, we performed correlation analyses on all taxon combinations in the data set. Correlations were performed by using bacterial abundance data from all treatments. Strong negative correlations between the family Bifidobacteriaceae and the family Clostridiaceae (Fig. 4A), the genera Bifidobacterium and Lactococcus (Fig. 4B), and the genera Bifidobacterium and Akkermansia (Fig. 4C) were observed. In addition, strain IVS-1 levels (OTU_2) showed a negative correlation with Lactococcus lactis (Fig. 4D) and a very tight negative association with resident B. pseudolongum (r = −0.64; P = 0.0004) (Fig. 4E). These negative associations suggest direct or indirect competition between these bacterial taxa. Positive associations between both Bifidobacteriaceae and Bifidobacterium and the genus Blautia were detected, suggesting a synergistic relationship, which may be supported by the addition of GOS (Table 1 and Fig. 4F).
FIG 4.
Correlation analysis of colonic taxa present in rats fed a high-fat diet supplemented with or without a probiotic (IVS-1), a prebiotic (GOS), or a synbiotic (IVS-1 plus GOS) or a standard diet. Bacterial quantities are expressed as percent abundances of total bacteria as determined by 16S rRNA sequencing. Spearman's correlations between Bifidobacteriaceae and Clostridiaceae (A), Bifidobacterium and Lactococcus (B), Bifidobacterium and Akkermansia (C), Bifidobacterium adolescentis IVS-1 and Lactococcus lactis (D), Bifidobacterium adolescentis IVS-1 and Bifidobacterium pseudolongum (E), and Bifidobacterium and Blautia (F) were determined.
DISCUSSION
Synergistic synbiotics are a promising concept to modulate the composition of the gut microbiota and promote the establishment of probiotic organisms in the gut (16). Despite this potential, however, there are few in vivo human or animal studies providing evidence that prebiotics can be used to support specific probiotic strains. Unfortunately, most synbiotic studies, including work in rats (44–49), mice (50–52), pigs (53–57), chickens (58, 59), and humans (60), did not employ strain-specific detection methods and therefore did not provide information on the potential synergism between pre- and probiotics. Of the in vivo studies that did discriminate the probiotic strain, most still did not demonstrate that in vivo performance could be enhanced by a prebiotic. This accounts for experiments using synbiotic formulations in humans (61), rats (62, 63), and other animal models (64). These findings suggest that, with few exceptions (17–19), probiotic strains are unable to compete against the resident gut microbiota, which is inherently resistant to outside colonizers (65), even when an exogenous growth substrate in the form of a prebiotic is provided.
Several reasons may explain the low success rates of synergistic synbiotics when evaluated in vivo, even for combinations in which the probiotic strain is able to utilize the prebiotic substrate in vitro. First, to become established in the gut, the probiotic strain must be able to occupy an ecological niche. This means that strains must not only outcompete the resident microbiota for the prebiotic substrate but also secure other nutrients that might be growth limiting (such as amino acids, lipids, vitamins, minerals, and nucleotides, etc.). In addition, probiotic strains must tolerate the prevailing environmental conditions in the digestive tract (including pH, bile acids, IgA, and defensins). Ultimately, in vitro tests are unable to predict the ability of a probiotic to benefit from a prebiotic substrate within the constraints of the competitive gastrointestinal environment. In contrast, the IVS approach described here overcomes many limitations of in vitro tests used to formulate synbiotics because it provides a basis for identifying bacterial strains that are able to utilize the prebiotic substrate under the same ecological conditions in which they are intended to function.
In this study, we employed IVS and selected a synbiotic combination that was tested in a rat model of NAFLD. Although the synbiotic did not influence host phenotypes, it was highly efficient at enhancing population levels of the probiotic strain, making it the most dominant OTU in the gut (Fig. 3A and Table 1). These findings provide a proof of concept for the potential of in vivo selection to identify synbiotic combinations that are, in ecological terms, highly synergistic. In addition to enhancing the abundance of strain IVS-1, the synbiotic used here also redressed the high-fat-diet-induced reduction in the level of bifidobacteria detected in rats that is often reported in the literature (41–43). Therefore, although no metabolic benefits were seen in the rat model used in our study, the synbiotic may be beneficial in other scenarios, as bifidobacteria are considered health-promoting organisms (6, 26, 66–68).
The community-wide analysis provided evidence that synergism between GOS and strain IVS-1 increased the competitive fitness of the strain in the rat intestinal tract. B. pseudolongum, which is a natural member of the rat GI tract (69), was detected in relative abundances of 5.8% and 0.9% in rats fed the standard and high-fat diets, respectively. Although the probiotic treatment did not affect levels of B. pseudolongum, the prebiotic treatment increased the abundance of this species to 16.6%, indicating that B. pseudolongum utilized GOS in vivo. However, the parallel addition of strain IVS-1 with GOS completely excluded B. pseudolongum, and a strong negative correlation between this species and IVS-1 was observed (r = −0.67; P = 0.0006) (Fig. 4E). These findings indicate that IVS-1 not only had a higher affinity for GOS in vivo than the resident Bifidobacterium species but also utilized GOS to increase its competitiveness and effectively outcompete a closely related resident species. This finding is consistent with the niche exclusion model, which states that the organism most efficient at using limited nutrients outcompetes its competitors for the same niche (70). Strong inverse correlations between bifidobacteria and Clostridiaceae, Lactococcus, and Akkermansia (Fig. 4A to C) were also observed. It is likely that these associations are also due to niche competition and are potentially enhanced by GOS administration. Bifidobacteria produce short-chain fatty acids that are inhibitory to other bacteria either by lowering the pH or via direct antimicrobial effects (e.g., acetic acid) (71). In summary, these findings demonstrate that the competitive fitness of strain IVS-1 was increased by GOS, which supports the conclusion that IVS can select synbiotic combinations with extremely high synergism. To what degree the increased competitive interactions between IVS-1 and the resident microbiota impact host health is difficult to predict and likely context dependent, but they clearly should be considered in future studies.
Correlation analyses revealed only one positive association among members of the rat microbiota, between the bifidobacteria (at the family and genus levels) and the genus Blautia. The abundance of OTU_14, an uncultured Blautia strain, was also significantly increased by GOS and in the synbiotic treatment (Table 1). The positive correlation between Bifidobacterium and Blautia (Fig. 4F) indicates a synergistic effect between the two taxa. The significant increase in the abundance of Blautia in the synbiotic treatments further suggests a syntrophic interaction between IVS-1 and Blautia, as GOS is consumed mainly by bifidobacteria (72), and the genus Blautia is not reported to utilize GOS. In contrast, the genus Blautia contains bacteria that are hydrogenotrophic acetogens, which utilize H2 and CO2 as energy sources (73). Although bifidobacteria do not produce these gases, cross-feeding between bifidobacteria and butyrate-producing colon bacteria can result in H2 and CO2 production (74), which might explain the positive correlations between Bifidobacterium and Blautia. However, additional experiments are necessary to establish the mechanism by which GOS can enhance the populations of Blautia in the gut and the positive associations between this genus and IVS-1.
In this study, we have shown how IVS can be used to formulate a highly synergistic synbiotic that can substantially enhance population levels and the competitiveness of a putative probiotic strain in the gastrointestinal tract and establish it as the dominant member of the gut microbiota in a conventional animal model. To our knowledge, this has not yet been reported in the probiotic literature. The process of IVS is broadly applicable and can easily be extended to other host species, body sites, prebiotic substrates (or dietary fibers), or target organisms. For example, it may be possible to use IVS to enhance other putative health-promoting genera such as Akkermansia, which has been shown to respond to prebiotics in vivo (75). While we selected B. adolescentis IVS-1 during a human trial that did not determine the physiological effect of GOS, IVS might be especially powerful when combined with a human clinical trial that determines the beneficial effect of a prebiotic on the host as the primary selection criterion. Therefore, to develop synbiotics for specific health applications, the IVS concept should be extended to select bacterial strains that not only responded to the prebiotic but whose expansion correlated with beneficial physiological effects for the host. Such an approach would have the potential to identify health-promoting strains whose metabolic activity in vivo could be increased through a prebiotic. This might also result in synbiotic applications with greater health effects than those of the prebiotic alone, especially in the subset of humans who do not respond to the prebiotic (14, 32). A human study testing the synbiotic combination identified here (and comparing it with a synbiotic that includes a Bifidobacterium strain that can ferment GOS but was not selected by IVS) is currently in progress. Clearly, the application of IVS is likely to enhance the ecological performance of probiotic strains or live biotherapeutics within the habitats in which they are thought to function, and the technology could be readily applied in the design of microbiota-modulating therapies.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by USDA grant 2012-67017-19344. This work was also supported by the Nebraska Research Initiative's Proof of Concept Program, administered by the University of Nebraska.
We thank the UNL Institutional Animal Care Program staff for excellent animal care and Carol Casey at the Veterans Administration Medical Center for her advice and comments. We also acknowledge Catherine Muller for her contribution to the animal experiments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03903-14.
REFERENCES
- 1.Hildebrand F, Nguyen TLA, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, Liston A, Raes J. 2013. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol 14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee DK, Park SY, An HM, Kim JR, Kim MJ, Lee SW, Cha MK, Kim SA, Chung MJ, Lee KO, Ha NJ. 2011. Antimicrobial activity of Bifidobacterium spp. isolated from healthy adult Koreans against cariogenic microflora. Arch Oral Biol 56:1047–1054. doi: 10.1016/j.archoralbio.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tlaskalová-Hogenová H, Stěpánková R, Kozáková H, Hudcovic T, Vannucci L, Tučková L, Rossmann P, Hrnčíř T, Kverka M, Zákostelská Z, Klimešová K, Přibylová J, Bártová J, Sanchez D, Fundová P, Borovská D, Srùtková D, Zídek Z, Schwarzer M, Drastich P, Funda DP. 2011. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol 8:110–120. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cani PD, Delzenne NM. 2009. Interplay between obesity and associated metabolic disorders: new insights into the gut microbiota. Curr Opin Pharmacol 9:737–743. doi: 10.1016/j.coph.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker AW, Lawley TD. 2013. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res 69:75–86. doi: 10.1016/j.phrs.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Frank DN, Pace NR. 2008. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol 24:4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 10.Al-Sheraji SH, Ismail A, Manap MY, Mustafa S, Yusof RM, Hassan FA. 2013. Prebiotics as functional foods: a review. J Funct Foods 5:1542–1553. doi: 10.1016/j.jff.2013.08.009. [DOI] [Google Scholar]
- 11.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125:1401–1412. [DOI] [PubMed] [Google Scholar]
- 12.Tuohy KM, Probert HM, Smejkal CW, Gibson GR. 2003. Using probiotics and prebiotics to improve gut health. Drug Discov Today 8:692–700. doi: 10.1016/S1359-6446(03)02746-6. [DOI] [PubMed] [Google Scholar]
- 13.Hooda S, Boler BMV, Serao MCR, Brulc JM, Staeger MA, Boileau TW, Dowd SE, Fahey GC Jr, Swanson KS. 2012. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutr 142:1259–1265. doi: 10.3945/jn.112.158766. [DOI] [PubMed] [Google Scholar]
- 14.Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. 2010. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. 2014. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 16.Kolida S, Gibson GR. 2011. Synbiotics in health and disease. Annu Rev Food Sci Technol 2:373–393. doi: 10.1146/annurev-food-022510-133739. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka R, Takayama H, Morotomi M, Kuroshima T, Ueyama S, Matsumoto K, Kuroda A, Masahiko M. 1983. Effects of administration of TOS and Bifidobacterium breve 4006 on the human fecal flora. Bifidobact Microflora 2:17–24. doi: 10.12938/bifidus1982.2.1_17. [DOI] [Google Scholar]
- 18.Ogawa T, Asai Y, Yasuda K, Sakamoto H. 2005. Oral immunoadjuvant activity of a new synbiotic Lactobacillus casei subsp casei in conjunction with dextran in BALB/c mice. Nutr Res 25:295–304. doi: 10.1016/j.nutres.2004.10.012. [DOI] [Google Scholar]
- 19.Ogawa T, Asai Y, Tamai R, Makimura Y, Sakamoto H, Hashikawa S, Yasuda K. 2006. Natural killer cell activities of synbiotic Lactobacillus casei ssp. casei in conjunction with dextran. Clin Exp Immunol 143:103–109. doi: 10.1111/j.1365-2249.2005.02975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabiu BA, Jay AJ, Gibson GR, Rastall RA. 2001. Synthesis and fermentation properties of novel galacto-oligosaccharides by beta-galactosidases from Bifidobacterium species. Appl Environ Microbiol 67:2526–2530. doi: 10.1128/AEM.67.6.2526-2530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modesto M, D'Aimmo MR, Stefanini I, Trevisi P, De Filippi S, Casini L, Mazzoni M, Bosi P, Biavati B. 2009. A novel strategy to select Bifidobacterium strains and prebiotics as natural growth promoters in newly weaned pigs. Livest Sci 122:248–258. doi: 10.1016/j.livsci.2008.08.017. [DOI] [Google Scholar]
- 22.Malinen E, Mättö J, Salmitie M, Alander M, Saarela M, Palva A. 2002. Analysis of Bifidobacterium populations in human faecal samples from a consumption trial with Bifidobacterium lactis Bb-12 and a galacto-oligosaccharide preparation. Syst Appl Microbiol 25:249–258. doi: 10.1078/0723-2020-00117. [DOI] [PubMed] [Google Scholar]
- 23.Alander M, Mättö J, Kneifel W, Johansson M, Koegle B, Crittenden R, Mattila-Sandholm T, Saarela M. 2001. Effect of galacto-oligosaccharide supplementation on human faecal microflora and on survival and persistence of Bifidobacterium lactis Bb-12 in the gastrointestinal tract. Int Dairy J 11:817–825. doi: 10.1016/S0958-6946(01)00100-5. [DOI] [Google Scholar]
- 24.Rattanaprasert M, Roos S, Hutkins RW, Walter J. 2014. Quantitative evaluation of synbiotic strategies to improve persistence and metabolic activity of Lactobacillus reuteri DSM 17938 in the human gastrointestinal tract. J Funct Foods 10:85–94. doi: 10.1016/j.jff.2014.05.017. [DOI] [Google Scholar]
- 25.Davis LMG, Martínez I, Walter J, Hutkins R. 2010. A dose dependent impact of prebiotic galactooligosaccharides on the intestinal microbiota of healthy adults. Int J Food Microbiol 144:285–292. doi: 10.1016/j.ijfoodmicro.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Martínez I, Wallace G, Zhang C, Legge R, Benson AK, Carr TP, Moriyama EN, Walter J. 2009. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol 75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- 28.Ahmed U, Redgrave TG, Oates PS. 2009. Effect of dietary fat to produce non-alcoholic fatty liver in the rat. J Gastroenterol Hepatol 24:1463–1471. doi: 10.1111/j.1440-1746.2009.05870.x. [DOI] [PubMed] [Google Scholar]
- 29.Folch J, Lees M, Sloan Stanely G. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509. [PubMed] [Google Scholar]
- 30.Simental-Mendía LE, Rodríguez-Hernández H, Rodríguez-Morán M, Guerrero-Romero F. 2012. The alanine aminotransferase to triglycerides ratio as a marker to identify nonalcoholic fatty liver disease. Eur J Gastroenterol Hepatol 24:1173–1177. doi: 10.1097/MEG.0b013e3283564ee5. [DOI] [PubMed] [Google Scholar]
- 31.Lee J-T, Pao L-H, Lee M-S, Liao J-W, Shih C-M, Hsiong C-H, Yin F-Y, Shih T-Y, Hu OY-P. 2013. A new approach to facilitate diagnosis of nonalcoholic fatty liver disease through a galactose single point method in rats with fatty liver. Dig Liver Dis 45:134–141. doi: 10.1016/j.dld.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 32.Davis LMG, Martínez I, Walter J, Goin C, Hutkins RW. 2011. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS One 6:e25200. doi: 10.1371/journal.pone.0025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito M, Deguchi Y, Miyamori A, Matsumoto K, Kikuchi H, Kobayashi Y, Yajima T, Kan T. 1990. Effects of administration of galactooligosaccharides on the human faecal microflora, stool weight and abdominal sensation. Microb Ecol Health Dis 3:285–292. doi: 10.3109/08910609009140251. [DOI] [Google Scholar]
- 34.Gibson GR. 1999. Dietary modulation of the human gut microflora using the prebiotics oligofructose and inulin. J Nutr 129:1438–1441. [DOI] [PubMed] [Google Scholar]
- 35.Sako T, Matsumoto K, Tanaka R. 1999. Recent progress on research and applications of non-digestible galacto-oligosaccharides. Int Dairy J 9:69–80. doi: 10.1016/S0958-6946(99)00046-1. [DOI] [Google Scholar]
- 36.Macfarlane S, Macfarlane GT, Cummings JH. 2006. Review article: prebiotics in the gastrointestinal tract. Aliment Pharmacol Ther 24:701–714. doi: 10.1111/j.1365-2036.2006.03042.x. [DOI] [PubMed] [Google Scholar]
- 37.Silk DBA, Davis A, Vulevic J, Tzortzis G, Gibson GR. 2009. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther 29:508–518. doi: 10.1111/j.1365-2036.2008.03911.x. [DOI] [PubMed] [Google Scholar]
- 38.Ben X-M. 2008. Low level of galacto-oligosaccharide in infant formula stimulates growth of intestinal Bifidobacteria and Lactobacilli. World J Gastroenterol 14:6564–6568. doi: 10.3748/wjg.14.6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Wang R, Li X-F, Wang R-L. 2012. Bifidobacterium adolescentis supplementation ameliorates visceral fat accumulation and insulin sensitivity in an experimental model of the metabolic syndrome. Br J Nutr 107:1429–1434. doi: 10.1017/S0007114511004491. [DOI] [PubMed] [Google Scholar]
- 40.Reichold A, Brenner SA, Spruss A, Förster-Fromme K, Bergheim I, Bischoff SC. 2014. Bifidobacterium adolescentis protects from the development of nonalcoholic steatohepatitis in a mouse model. J Nutr Biochem 25:118–125. doi: 10.1016/j.jnutbio.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, Gibson GR, Delzenne NM. 2007. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 42.Everard A, Lazarevic V, Gaïa N, Johansson M, Ståhlman M, Backhed F, Delzenne NM, Schrenzel J, François P, Cani PD. 2014. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J 8:2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cani PD, Delzenne NM, Amar J, Burcelin R. 2008. Role of gut microflora in the development of obesity and insulin resistance following high-fat diet feeding. Pathol Biol 56:305–309. doi: 10.1016/j.patbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Liong MT, Shah NP. 2006. Effects of a Lactobacillus casei synbiotic on serum lipoprotein, intestinal microflora, and organic acids in rats. J Dairy Sci 89:1390–1399. doi: 10.3168/jds.S0022-0302(06)72207-X. [DOI] [PubMed] [Google Scholar]
- 45.Yang S-C, Chen J-Y, Shang H-F, Cheng T-Y, Tsou SC, Chen J-R. 2005. Effect of synbiotics on intestinal microflora and digestive enzyme activities in rats. World J Gastroenterol 11:7413–7417. doi: 10.3748/wjg.v11.i47.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Leu RK, Brown IL, Hu Y, Bird AR, Jackson M, Esterman A, Young GP. 2005. A synbiotic combination of resistant starch and Bifidobacterium lactis facilitates apoptotic deletion of carcinogen-damaged cells in rat colon. J Nutr 135:996–1001. [DOI] [PubMed] [Google Scholar]
- 47.Igarashi M, Liyama Y, Kato R, Tomita M, Asami N. 1994. Effect of Bifidobacterium longum and lactulose on the strength of bone in ovariectomized osteoporosis model rats. Bifidus 7:139–147. [Google Scholar]
- 48.Bielecka M, Biedrzycka E, Majkowska A. 2002. Selection of probiotics and prebiotics for synbiotics and confirmation of their in vivo effectiveness. Food Res Int 35:125–131. doi: 10.1016/S0963-9969(01)00173-9. [DOI] [Google Scholar]
- 49.Bomhof MR, Saha DC, Reid DT, Paul HA, Reimer RA. 2014. Combined effects of oligofructose and Bifidobacterium animalis on gut microbiota and glycemia in obese rats. Obesity 22:763–771. doi: 10.1002/oby.20632. [DOI] [PubMed] [Google Scholar]
- 50.Frece J, Kos B, Svetec IK, Zgaga Z, Beganović J, Lebos A, Susković J. 2009. Synbiotic effect of Lactobacillus helveticus M92 and prebiotics on the intestinal microflora and immune system of mice. J Dairy Res 76:98–104. doi: 10.1017/S0022029908003737. [DOI] [PubMed] [Google Scholar]
- 51.Foye OT, Huang I-F, Chiou CC, Walker WA, Shi HN. 2012. Early administration of probiotic Lactobacillus acidophilus and/or prebiotic inulin attenuates pathogen-mediated intestinal inflammation and Smad 7 cell signaling. FEMS Immunol Med Microbiol 65:467–480. doi: 10.1111/j.1574-695X.2012.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogawa T, Hashikawa S, Asai Y, Sakamoto H, Yasuda K, Makimura Y. 2006. A new synbiotic, Lactobacillus casei subsp. casei together with dextran, reduces murine and human allergic reaction. FEMS Immunol Med Microbiol 46:400–409. doi: 10.1111/j.1574-695X.2006.00046.x. [DOI] [PubMed] [Google Scholar]
- 53.Piva A, Casadei G, Gatta PP, Luchansky JB, Biagi G. 2005. Effect of lactitol, lactic acid bacteria, or their combinations (synbiotic) on intestinal proteolysis in vitro, and on feed efficiency in weaned pigs. Can J Anim Sci 85:345–353. doi: 10.4141/A04-087. [DOI] [Google Scholar]
- 54.Bird AR, Vuaran M, Crittenden R, Hayakawa T, Playne MJ, Brown IL, Topping DL. 2009. Comparative effects of a high-amylose starch and a fructooligosaccharide on fecal bifidobacteria numbers and short-chain fatty acids in pigs fed Bifidobacterium animalis. Dig Dis Sci 54:947–954. doi: 10.1007/s10620-008-0451-3. [DOI] [PubMed] [Google Scholar]
- 55.Böhmer BM, Branner GR, Roth-Maier DA. 2005. Precaecal and faecal digestibility of inulin (DP 10-12) or an inulin/Enterococcus faecium mix and effects on nutrient digestibility and microbial gut flora. J Anim Physiol Anim Nutr (Berl) 89:388–396. doi: 10.1111/j.1439-0396.2005.00530.x. [DOI] [PubMed] [Google Scholar]
- 56.Bomba A, Nemcová R, Gancarcíková S, Herich R, Guba P, Mudronová D. 2002. Improvement of the probiotic effect of micro-organisms by their combination with maltodextrins, fructo-oligosaccharides and polyunsaturated fatty acids. Br J Nutr 88(Suppl 1):S95–S99. doi: 10.1079/BJN2002634. [DOI] [PubMed] [Google Scholar]
- 57.Shim SB, Verstegen MWA, Kim IH, Kwon OS, Verdonk JMAJ. 2005. Effects of feeding antibiotic-free creep feed supplemented with oligofructose, probiotics or synbiotics to suckling piglets increases the preweaning weight gain and composition of intestinal microbiota. Arch Anim Nutr 59:419–427. doi: 10.1080/17450390500353234. [DOI] [PubMed] [Google Scholar]
- 58.Baffoni L, Gaggìa F, Di Gioia D, Santini C, Mogna L, Biavati B. 2012. A Bifidobacterium-based synbiotic product to reduce the transmission of C. jejuni along the poultry food chain. Int J Food Microbiol 157:156–161. doi: 10.1016/j.ijfoodmicro.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 59.Abdel-Raheem S, Abd-Allah S, Hassanein K. 2012. The effects of prebiotic, probiotic and synbiotic supplementation on intestinal microbial ecology and histomorphology of broiler chickens. Int J Agro Vet Med Sci 6:277–289. doi: 10.5455/ijavms.156. [DOI] [Google Scholar]
- 60.Furrie E, Macfarlane S, Kennedy A, Cummings JH, Walsh SV, O'Neil DA, Macfarlane GT. 2005. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut 54:242–249. doi: 10.1136/gut.2004.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gopal PK, Prasad J, Gill HS. 2003. Effects of the consumption of Bifidobacterium lactis HN019 (DR10) and galacto-oligosaccharides on the microflora of the gastrointestinal tract in human subjects. Nutr Res 23:1313–1328. doi: 10.1016/S0271-5317(03)00134-9. [DOI] [Google Scholar]
- 62.Schultz M, Munro K, Tannock GW, Melchner I, Göttl C, Schwietz H, Schölmerich J, Rath HC. 2004. Effects of feeding a probiotic preparation (SIM) containing inulin on the severity of colitis and on the composition of the intestinal microflora in HLA-B27 transgenic rats. Clin Diagn Lab Immunol 11:581–587. doi: 10.1128/CDLI.11.3.581-587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakanishi S, Kataoka K, Kuwahara T, Ohnishi Y. 2003. Effects of high amylose maize starch and Clostridium butyricum on metabolism in colonic microbiota and formation of azoxymethane-induced aberrant crypt foci in the rat colon. Microbiol Immunol 47:951–958. doi: 10.1111/j.1348-0421.2003.tb03469.x. [DOI] [PubMed] [Google Scholar]
- 64.Krause DO, Bhandari SK, House JD, Nyachoti CM. 2010. Response of nursery pigs to a synbiotic preparation of starch and an anti-Escherichia coli K88 probiotic. Appl Environ Microbiol 76:8192–8200. doi: 10.1128/AEM.01427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stecher B, Hardt W-D. 2008. The role of microbiota in infectious disease. Trends Microbiol 16:107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 66.Kalliomäki M, Collado MC, Salminen S, Isolauri E. 2008. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 87:534–538. [DOI] [PubMed] [Google Scholar]
- 67.Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, Looijer-van Langen M, Madsen KL. 2008. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol 295:G1025–G1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 68.Wang Z, Xiao G, Yao Y, Guo S, Lu K, Sheng Z. 2006. The role of bifidobacteria in gut barrier function after thermal injury in rats. J Trauma 61:650–657. doi: 10.1097/01.ta.0000196574.70614.27. [DOI] [PubMed] [Google Scholar]
- 69.Mitsuoka T. 1990. Bifidobacteria and their role in human health. J Ind Microbiol 6:263–267. doi: 10.1007/BF01575871. [DOI] [Google Scholar]
- 70.Petrof EO, Khoruts A. 2014. From stool transplants to next-generation microbiota therapeutics. Gastroenterology 146:1573–1582. doi: 10.1053/j.gastro.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 72.Maathuis AJH, van den Heuvel EG, Schoterman MHC, Venema K. 2012. Galacto-oligosaccharides have prebiotic activity in a dynamic in vitro colon model using a (13)C-labeling technique. J Nutr 142:1205–1212. doi: 10.3945/jn.111.157420. [DOI] [PubMed] [Google Scholar]
- 73.Liu C, Finegold SM, Song Y, Lawson PA. 2008. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydroge. Int J Syst Evol Microbiol 58:1896–1902. doi: 10.1099/ijs.0.65208-0. [DOI] [PubMed] [Google Scholar]
- 74.De Vuyst L, Leroy F. 2011. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol 149:73–80. doi: 10.1016/j.ijfoodmicro.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 75.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.