Abstract
To highlight different transcriptional behaviors of the phytoplasma in the plant and animal host, expression of 14 genes of “Candidatus Phytoplasma asteris,” chrysanthemum yellows strain, was investigated at different times following the infection of a plant host (Arabidopsis thaliana) and two insect vector species (Macrosteles quadripunctulatus and Euscelidius variegatus). Target genes were selected among those encoding antigenic membrane proteins, membrane transporters, secreted proteins, and general enzymes. Transcripts were detected for all analyzed genes in the three hosts; in particular, those encoding the antigenic membrane protein Amp, elements of the mechanosensitive channel, and two of the four secreted proteins (SAP54 and TENGU) were highly accumulated, suggesting that they play important roles in phytoplasma physiology during the infection cycle. Most transcripts were present at higher abundance in the plant host than in the insect hosts. Generally, transcript levels of the selected genes decreased significantly during infection of A. thaliana and M. quadripunctulatus but were more constant in E. variegatus. Such decreases may be explained by the fact that only a fraction of the phytoplasma population was transcribing, while the remaining part was aging to a stationary phase. This strategy might improve long-term survival, thereby increasing the likelihood that the pathogen may be acquired by a vector and/or inoculated to a healthy plant.
INTRODUCTION
Phytoplasmas are wall-less plant-pathogenic bacteria, classified as “Candidatus Phytoplasma” spp. (1). They belong to the class Mollicutes and infect a wide variety of plants, causing heavy crop losses in many different countries (2). Phytoplasmas are phloem limited in the infected plant, and they cause severe symptoms (yellowing, dwarfism, and phyllody), often leading to plant death. They are transmitted by phloem-feeding Hemipteran vectors (leafhoppers, planthoppers, and psyllids) in a persistent propagative manner (3).
Phytoplasmas are obligate parasites that depend on host cells for the uptake of essential compounds such as sugars, amino acids, ions, and nucleotide precursors (4). Consistent with this lifestyle, phytoplasmas have very small, A/T-rich genomes, ranging from 530 to 1,350 kb in size (5), that lack essential metabolic pathways, such as ATP synthesis. This genome condensation reflects the phytoplasma adaptation to nutrient-rich environments such as the plant phloem (6) and helps explain why these pathogens are not cultivable under axenic conditions (7).
Although the pathogenicity mechanisms are still largely unclear, phytoplasmas influence plant metabolism both directly, through a set of membrane proteins acting as molecular carriers (6), and indirectly, through secretion of effector proteins (8, 9). In vitro studies have also shown that phytoplasma immunodominant membrane proteins interact with vector proteins (10, 11) and plant proteins (12) and are subjected to strong positive selection (13–15). Moreover, phytoplasmas can modulate their genome expression according to the infection stage and the infected host species, as suggested by microarray analysis (16) and gene expression study of pathogen transcription factors (17) and of genes lying within potential mobile units (18).
Real-time reverse transcription-quantitative PCR (RT-qPCR) is routinely employed for gene expression studies due to its high sensitivity and accuracy (19–22). Strategies employed for bacterial transcript quantification through qPCR are currently based on relative (23–26) or absolute (27–29) quantification approaches. Phytoplasmas live their lives inside very different environments: the plant and the insect vector. The recent availability of phytoplasma genome sequences has provided tools to investigate phytoplasma-host relationships, but little is known about the molecular mechanisms involved in host switching and in the pathogen cycle in the two environments. These points are extremely important, both to provide the first insights into functional genomics of these pathogens and to start devising new tools for fine-tuned control strategies of these important plant pathogens for integration into the current control of vector populations by insecticide treatments. The aim of this work was to identify phytoplasma genes potentially involved in sensing the host environment, thereby discriminating between plant and insect hosts and, in an even more subtle way, between different insect vectors. As phytoplasma colonization of the host is a continuous process from the original low-quantity inoculum to the final high-density population at the end of the infection cycle, a study was designed to measure transcript levels over time in the plant and in two vector insects. qRT-PCR protocols were set up to study the transcription profile of 14 “Ca. Phytoplasma asteris” chrysanthemum yellows phytoplasma (CYP) genes, during infection of Arabidopsis thaliana (L.) Heynh and of the two leafhopper vector species Euscelidius variegatus Kirschbaum and Macrosteles quadripunctulatus Kirschbaum. The two vectors were selected on the basis of their different characteristics with respect to transmitting CYP, as summarized in references 30 and 31: M. quadripunctulatus acquires (100% versus 88%) and transmits (100% versus 82%) CYP with higher efficiency than E. variegatus and supports multiplication of the phytoplasma at a rate higher than that seen with E. variegatus. Consequently, the CYP latent period in the former species is shorter than in the latter one (16 to 18 days versus 30 days). CYP genes were selected for transcript analyses from among predicted secreted proteins, known effectors, and general metabolism enzymes. In the absence of a phytoplasma endogenous control mRNA, the expression level of each pathogen transcript was correlated to the bacterial population measured by qPCR for each experimental date. Absolute quantification of bacterial transcripts was performed (27–29). For each phytoplasma gene, an expression index (EI) was calculated, indicating the transcript copy number per phytoplasma cell at each sampling date and in each infected host, according to the guidelines published for cultivable bacteria (27, 29). Regression analyses were also performed to compare the gene expression trends over time among the three hosts, irrespective of the absolute levels of the individual gene expression.
MATERIALS AND METHODS
Phytoplasma isolate, host plant, and insect vector.
Chrysanthemum yellows phytoplasma (CYP) was originally isolated in Italy from Argyranthemum frutescens (L.) Schultz-Bip and maintained by insect transmission on daisy, Chrysanthemum carinatum Schousboe, the phytoplasma source plant in this work. Arabidopsis thaliana ecotype Col-0 seeds were sown in single pots and kept at 4°C for 3 days. Pots were then placed in a growth chamber at 22 to 24°C with a photoperiod representing a short day (light, 9 h; dark, 15 h [L9:D15]) and were maintained under this condition during the whole experiment. Healthy colonies of Euscelidius variegatus and Macrosteles quadripunctulatus, vectors of CYP (32), were maintained on oat, Avena sativa L., inside plastic and nylon cages in growth chambers at 25°C and a photoperiod of L16:D8. To evaluate phytoplasma gene expression profiles in A. thaliana, experimental plants were inoculated with CYP by the use of M. quadripunctulatus vector. About 100 M. quadripunctulatus nymphs were fed on infected daisies for an acquisition access period (AAP) of 7 days and were then transferred on oat (immune to CYP) for a 25-day latency period (LP). Thirty-six A. thaliana plants were singly exposed to three infective insects for a 72-h inoculation access period (IAP) and were then treated with insecticide. Leaf samples were collected from 10 A. thaliana plants at 10, 14, 21, and 28 days postinoculation (dpi) for nucleic acid extraction. To evaluate the phytoplasma gene expression profile in insect vector, CYP-infected E. variegatus and M. quadripunctulatus were used. About 200 nymphs of each species were collected from healthy colonies, caged together for a 7-day AAP on CYP-source daisies, and then maintained on healthy oat plants. About 15 insects of each species were collected at 7, 14, 21, 28, and 35 days postacquisition (dpa) for nucleic acid extraction.
Extraction of nucleic acids.
Plant samples (about 200 mg of leaves), collected at different dpi, were pooled and divided into 100-mg aliquots stored at −80°C before DNA and RNA extraction.
Total DNA was extracted from 100 mg of plant material using a modified cetyltrimethyl ammonium bromide (CTAB) procedure originally described in reference 33, and the final DNA pellet was dissolved in 50 μl of sterile double-distilled water (ddH2O). Total RNA from plant tissue was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. Insect samples collected at different dpa were stored at −80°C before DNA and RNA extraction. Both total DNA and total RNA were extracted from single insects. A few liquid nitrogen drops were spilled into a 1.5-ml tube containing single leafhopper and the insect was then quickly crushed using a sterile micropestel in 200 μl of TE buffer (10 mM Tris, 1 mM EDTA) prepared with diethyl pyrocarbonate (DEPC) (0.1%) water. The resulting homogenate suspension was rapidly divided for DNA and RNA extraction; 100 μl was added to 400 μl of 3% CTAB buffer and treated as detailed before for DNA extraction, whereas 100 μl was added to 400 μl of TRIzol reagent (Invitrogen, USA) for RNA extraction following the manufacturer's instructions. Total RNA samples, extracted from both plants and insects, were treated with RNase-free DNase I (Life Technologies, Monza, Italy) in the supplied buffer to avoid residual DNA contamination. Following the digestion, the DNase was inactivated by phenol-chloroform extraction according to the manufacturer's instructions. RNA was finally suspended in 30 μl of RNase-free water containing 0.1% DEPC. Nucleic acid extracts were analyzed in a NanoDrop spectrophotometer to evaluate the concentration and purity and stored at −80°C.
Phytoplasma detection and quantification.
The presence of CYP in A. thaliana, E. variegatus, and M. quadripunctulatus samples was verified by qPCR using the protocol described in reference 34. The DNA extracts from 10 infected plants and from 8 infected insects of both species for each sampling date (a total of 40 samples for each of the three species) were used as the template in qPCR to measure the absolute number of phytoplasma genome units (GU) per nanogram of host DNA (35). The quantification of phytoplasma cells was achieved by comparing the quantification cycles (Cqs) of the samples with those of three dilutions of pOP74 plasmid, containing a fragment of the CYP 16S rRNA gene (35). One fg of pOP74 contains 194 molecules of plasmid, with each containing a single copy of the CYP 16S rRNA gene. Because this gene is present in two copies in phytoplasma genomes, 1 fg of pOP74 corresponded to 97 CYP cells. The final concentration of CYP cells was expressed as CYP GU/100 mg of leaf sample or as CYP GU/insect.
cDNA synthesis and mRNA quantification.
For absolute quantification of phytoplasma mRNAs during A. thaliana and insect vector infection, standard curves were produced using serial dilutions of recombinant plasmids carrying a fragment of the corresponding genes (recombinant plasmid DNA [recDNA]). CYP genes were selected based on sequences from a whole-genome shotgun sequencing project that have been deposited at DDBJ/EMBL/GenBank under accession number JSWH00000000. In the absence of a phytoplasma endogenous control mRNA, the expression level of each pathogen transcript was correlated to the bacterial population measured by qPCR for each experimental date. Fragments of 14 selected genes were amplified by conventional PCRs driven by specific primers designed through the use of Primer Express software v3.0.1 (Applied Biosystems, Branchburg, NJ, USA) and of sequences obtained by Illumina sequencing of the CYP genome according to the method described in reference 36 (Table 1). Amplicons were subjected to gel purification using a GeneClean Turbo kit (MP Biomedicals, Solon, OH, USA), cloned into pGEM-T Easy vector (Promega, Madison, WI, USA), and transformed in Escherichia coli DH5α. Plasmids were purified using a Fast Plasmid minikit (Eppendorf AG, Hamburg, Germany) and sequenced with universal primers M13F and M13R. The number of plasmid copies per microliter was derived from the concentration measured at the NanoDrop spectrophotometer, using the following formula: M = C × N/S, where M is the number of molecules per microliter, C the RNA concentration (in ng/μl), S the molecular weight of the fragment, and N a factor derived from the Avogadro constant. recDNA was diluted, distributed in aliquots, and stored at −20°C to be used as a standard for qPCR. Standard curves were constructed by linear regression analysis of the Cq value of each standard dilution replicate over the log of the number of plasmid copies present in each sample. Data acquisition and analysis were handled by the use of CFX Manager software, version 3.0, which automatically calculates the Cq values and the parameters of the standard curves. qPCR efficiency (E) was calculated by the formula E = eln10/−s − 1, where e represents the base of the natural logarithm and a slope (s) value of −3.322 (E = 2) represents 100% efficiency. For each sampling date, cDNA was synthesized from total RNA (500 ng) using a High Capacity cDNA reverse transcription kit (Applied Biosystems, USA) according to the manufacturer's instructions and was stored in sterile microtubes until the qPCRs were performed. SYBR green-based qPCR protocols were optimized for each selected gene by adjusting the concentrations of forward and reverse primers from 100 μM to 300 μM. The final qPCR mix contained 1 μl of cDNA, 1× iQ SYBR green Supermix (Bio-Rad/Life Science Research, Hercules, CA, USA), 100 to 300 nM (each) primers (Table 1), and sterile double-distilled water added to reach a final volume of 25 μl. Reaction conditions were as follows: 5 min at 95°C and 45 cycles of 15 s at 95°C, 30 s at 59°C, and 30 s at 72°C. On each plate, samples were run in duplicate together with four 10-fold serial dilutions of the corresponding standard plasmid. The use of DNA standard curves for transcript quantification allowed comparisons of expression data from different genes (29, 38). Complete qPCR mix with total RNA and sterile distilled water instead of cDNA were used as negative controls in each plate. Reactions were carried out in a CFX Connect real-time PCR detection system (Bio-Rad, USA) supported by CFX Manager software, version 3.0. Melting curves were produced at the end of the PCR to assess the reaction specificity.
TABLE 1.
Chrysanthemum yellows phytoplasma genes selected for gene expression analysis and their function and product localization within the phytoplasma cell compartment, qPCR primer sequences and concentration in the reaction mix, amplicon melting temperature, and qPCR parametersa
| Product category and target gene | Gene product | Product localization | Primer: sequence (reference) | Primer concn (nM) (primer category) | Melting temp (°C) | R2 | E (%) |
|---|---|---|---|---|---|---|---|
| Immunodominant membrane proteins | |||||||
| amp | Antigenic membrane protein | Membrane | Amp64: GCTTTAATGTTTGTTGGCGTTC (37) | 100 | 82.5 | 0.993 | 78.7 |
| CY02-101Rev: AGCTTTTCCAGCATCACCAT | 300 | ||||||
| imp | Immunodominant membrane protein | Membrane | CY06-376Fw: ATTCCAAACTTGCCAGCATT | 300 | 78.5 | 1.000 | 96.0 |
| CY06-376Rev: TCTTGGAGGTTTTTGGCATT | 100 | ||||||
| Generic transporters | |||||||
| mscL | Mechanosensitive channel of large conductance | Membrane | CY05-67Fw: ACTGGGGCTTTAAAAGATTTG | 200 | 77.5 | 0.993 | 75.3 |
| CY05-67Rev: CCTTTTGCAAAGGATTTTTCC | 200 | ||||||
| mdlB | Multidrug transporter | Membrane | CY04-67Fw: CGCAAATGCCTTTTCAAACT | 200 | 81.5 | 0.998 | 83.5 |
| CY04-67Rev: AAGGCGAGGAATAAGCCCTA | 200 | ||||||
| oppC | Oligopeptide transporter | Membrane | CY02-378Fw: CCAAGATGCCATTTCTAGCC | 200 | 80.0 | 0.996 | 84.3 |
| CY02-378Rev: ATTCCGCCTGAAAGTTGATG | 200 | ||||||
| ftsY | SecY translocase component | Membrane | CY03-2292Fw: TGAAGGCATCGAACTTGCTA | 100 | 81.5 | 0.999 | 83.3 |
| CY03-2292Rev: GCTCCTGAAAGTGGAGTTGC | 300 | ||||||
| Specific transporters | |||||||
| artI | Arginine transporter | Membrane | CYartIFw: AATTCCTGTAGCTGCCCAAG | 200 | 80.0 | 1.000 | 88.1 |
| CYartIRev: ATGAGCGGCTTCAATTTGTC | 200 | ||||||
| zntA | Cation transporter | Membrane | CY036-368Fw: GACAAACAACCAGGCGATTT | 100 | 83.5 | 0.997 | 81.5 |
| CY036-368Rev: CTGCAAGACCAAGGACAACA | 300 | ||||||
| Secreted proteins | |||||||
| sap54 | Symptom effector | Secreted | CY01-1720Fw: TCAACAAGTAATGGGGATGAA | 200 | 76.5 | 0.999 | 90.5 |
| CY01-1720Rev: AATTGTTGTATTTCGCTTTCTGT | 200 | ||||||
| sap67 | Unknown | Secreted | CY01-420Fw: TCAAGTGATGGCAATGGGTA | 100 | 78.0 | 0.992 | 78.8 |
| CY01-420Rev1: TTGTATAGAACTTTCTGGTTGTTCAGA | 300 | ||||||
| sap68 | Unknown | Secreted | CY03-1700Fw: ATGGCAATGAATAACGGTCA | 200 | 79.0 | 0.999 | 88.5 |
| CY03-1700Rev: TTGTTGAGCAGCGATACGAC | 200 | ||||||
| tengu | Symptom effector | Secreted | CY04-87Fw1: ATTTGCTGGCTTTTGGGCTA | 200 | |||
| CY04-87Rev1: TTTCAATTAGAGTTATCACGTTTTCAA | 200 | 77.5 | 0.999 | 74.8 | |||
| General metabolism | |||||||
| pgsA | Phosphatidylglycerol synthase | Cytoplasm | CY06-96Fw: TCGTTTGTCTGCTACGCAAG | 200 | 81.0 | 0.996 | 84.6 |
| CY06-96Rev: AAAAGGCAAATGATGGCAAC | 200 | ||||||
| rpsU | rpsU RNA ribosomal subunit | Cytoplasm | CY08-364Fw: GGAGAAACTATCGAAGAAACGCTAC | 200 | 81.5 | 0.995 | 86.0 |
| CY08-364Rev: TTAACGCTTTTTACTACGCATATTTT | 200 |
R2, correlation coefficient; E %, reaction efficiency.
Data analysis.
Absolute quantification of bacterial transcripts was performed. For each phytoplasma gene, an expression index (EI) was calculated, indicating the transcript copy number per phytoplasma cell at each sampling date and in each infected host, according to the guidelines published for cultivable bacteria (26, 28). Regression analyses were also performed to compare the gene expression trends over time among the three hosts, irrespective of the absolute levels of the individual gene expression. To compare the phytoplasma population sizes as well as the EI values of the genes at different times in each host, analysis of variance (ANOVA) was performed on ranks (Kruskal-Wallis test), followed by Tukey or Dunn tests for multiple comparisons. For each gene, in all the three host species, decreased expression from the early to the late sampling dates was observed. To compare the decreases in expression of genes within each functional category in the three host species, linear regression was calculated on the basis of the log-transformed EI of each gene and the sampling times. For A. thaliana samples, the first sampling date (10 dpi) was omitted from regression analysis, due to the high variances among the EI values measured at the initial phase of infection. To compare CYP gene expression levels among the different host species, the EI values from the first two sampling dates (10 and 14 dpi and 7 and 14 dpa for the plant and insect samples, respectively) were pooled, as they did not differ significantly, and were analyzed by ANOVA performed on ranks (Kruskal-Wallis test), followed by the Dunn test for multiple comparisons. For each gene, in all three species, decreased expression was observed from the early to the late sampling dates; to compare the decreases in expression of the genes within each functional category in the three species, linear regression was calculated on the basis of the log-transformed EI of each gene and the sampling times. For A. thaliana samples, the first sampling date (10 dpi) was omitted from regression analysis, due to the high variances among EI values measured at the initial phase of infection. All statistical analyses were performed with SigmaPlot 11.0 (SyStat).
Nucleotide sequence accession numbers.
The strain CYP gene sequences from the whole-genome shotgun sequencing project have been deposited at DDBJ/EMBL/GenBank under accession number JSWH00000000.
RESULTS
Phytoplasma detection and quantification.
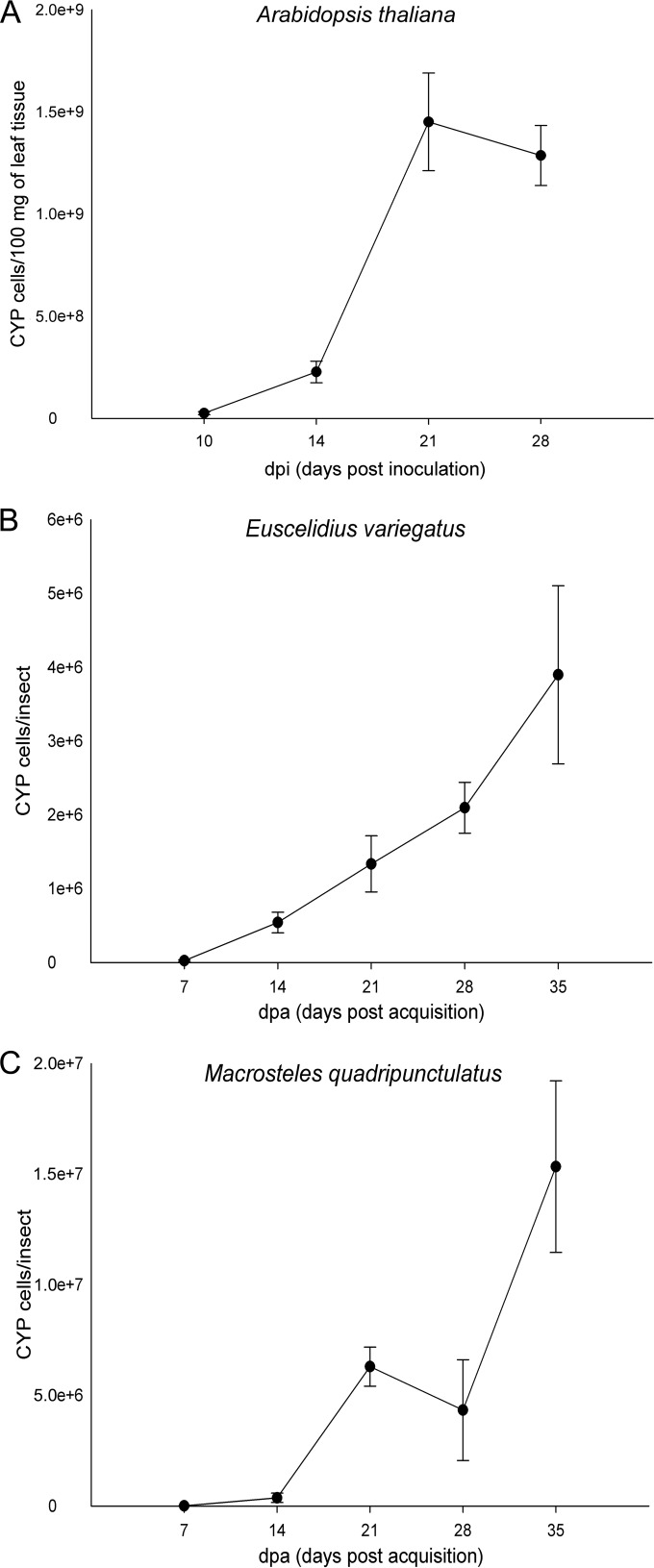
Diagnostic assays confirmed the presence of CYP in all A. thaliana plants at each sampling date. The phytoplasma population, expressed as CYP cells/100 mg of plant tissue, significantly increased from 10 dpi to 21 dpi (Fig. 1), and calculated values ranged from 2.23E+06 at 10 dpi to 2.93E+09 at 21 dpi (see Table S1 in the supplemental material). Diagnostic assays confirmed the presence of CYP in more than 80% of the E. variegatus samples at each sampling date and in all M. quadripunctulatus samples irrespective of the sampling date. The phytoplasma population increased from 7 to 21 dpa in both species (Fig. 1), and calculated values ranged from 1.75E+03 and 3.30E+03 at 7 dpa to 1.17E+07 and 3.57E+07 at 35 dpa for E. variegatus and M. quadripunctulatus, respectively (see Table S1 in the supplemental material).
FIG 1.
CYP population measured in leaf tissues of Arabidopsis thaliana (A) as a function of sampling time (10, 14, 21, and 28 days postinoculation [dpi]) and in individuals of Euscelidius variegatus (B) and Macrosteles quadripunctulatus (C) as a function of sampling time (7, 14, 21, 28, and 35 days postacquisition [dpa]). In all cases, error bars indicate standard errors of the means.
Optimization of qPCR assays.
For each target gene, specific primers were designed using CYP sequences. The primer list and the corresponding amplification conditions (annealing temperature and primer concentration) are reported in Table 1. A specific signal was obtained following melting analysis of the qPCR amplicons, while no amplification was obtained from no-reverse-transcribed-RNA and from no-template controls. Melting peak temperatures ranged from 76.5°C (sap54) to 83.5°C (zntA).
To estimate the expression levels of different CYP genes, the mRNA absolute quantity was divided by the phytoplasma titer measured in the corresponding sample. A plasmid standard curve, ranging from 10E+8 to 10E+4 gene copies, was set up for each target gene. Efficiencies of qPCRs ranged between 74.8% and 96.0% for primers amplifying the tengu and imp genes, respectively, whereas correlation coefficients ranged from 0.992 (sap67) to 1.000 (imp) (Table 1). Unbalanced primer final concentrations were optimized to improve the efficiency of reactions of primers amplifying amp-, imp-, ftsY-, zntA-, and sap67-specific amplicons (Table 1).
Phytoplasma transcript levels in Arabidopsis thaliana.
Mean copy numbers of transcripts per CYP cell in leaf tissues of Arabidopsis thaliana sampled at 10, 14, 21, and 28 dpi are presented in Table S2 in the supplemental material.
Immunodominant membrane proteins.
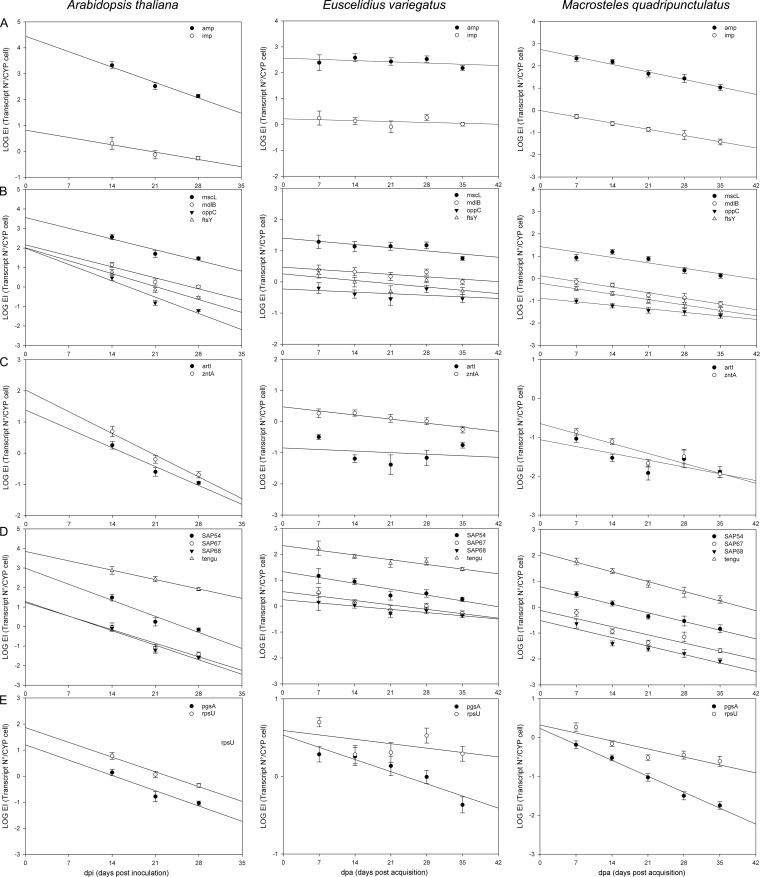
amp showed the highest transcript level among the analyzed CYP genes in A. thaliana challenged over time by phytoplasma infection (see Table S2 in the supplemental material). As with most of other CYP genes, amp transcripts decreased significantly in the late phase of infection (28 dpi) (Fig. 2 and Table 2). The mean expression level of amp was always higher than that of imp. On the other hand, imp transcript levels were constant from 10 to 28 dpi, imp being the most stable CYP gene during phytoplasma infection of A. thaliana as confirmed by the regression analyses (Fig. 2 and Table 2).
FIG 2.
Plot of linear regression analysis of log copy number of transcripts per phytoplasma cell (log expression index [LOG EI]) of chrysanthemum yellows phytoplasma (CYP) genes encoding immunodominant membrane proteins (A), generic transporters (B), specific transporters (C), secreted proteins (D), and proteins involved in general metabolism (E) measured in leaf tissues of Arabidopsis thaliana as a function of sampling time (14, 21, and 28 days postinoculation [dpi]) and in individuals of Euscelidius variegatus and Macrosteles quadripunctulatus as a function of sampling time (7, 14, 21, 28, and 35 days postacquisition [dpa]). In all cases, error bars indicate standard errors of the means.
TABLE 2.
Regression analysis parameters of log expression index values in function of sampling times of chrysanthemum yellows phytoplasma genes, grouped by functional category and measured in Arabidopsis thaliana, Euscelidius variegatus, and Macrosteles quadripunctulatus samplesa
| Functional category | Gene name |
Arabidopsis thaliana |
Euscelidius variegatus |
Macrosteles quadripunctulatus |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Slope | R2 | P | n | Slope | R2 | P | n | Slope | R2 | P | n | ||
| Immunodominant membrane proteins | amp | −0.085 | 0.634 | <0.001 | 30 | −0.007 | 0.023 | 0.357 | 39 | −0.048 | 0.611 | <0.001 | 40 |
| imp | −0.040 | 0.164 | 0.026 | 30 | −0.004 | 0.009 | 0.576 | 38 | −0.040 | 0.586 | <0.001 | 39 | |
| Generic transporters | mscL | −0.079 | 0.497 | <0.001 | 30 | −0.015 | 0.134 | 0.028 | 36 | −0.036 | 0.488 | <0.001 | 39 |
| mdlB | −0.081 | 0.560 | <0.001 | 30 | −0.011 | 0.101 | 0.055 | 37 | −0.036 | 0.489 | <0.001 | 40 | |
| oppC | −0.120 | 0.752 | <0.001 | 30 | −0.006 | 0.023 | 0.364 | 38 | −0.023 | 0.314 | <0.001 | 39 | |
| ftsY | −0.089 | 0.665 | <0.001 | 30 | −0.013 | 0.082 | 0.089 | 36 | −0.035 | 0.477 | <0.001 | 39 | |
| Specific transporters | artI | −0.087 | 0.655 | <0.001 | 29 | −0.006 | 0.012 | 0.604 | 25 | −0.024 | 0.210 | 0.003 | 39 |
| zntA | −0.099 | 0.680 | <0.001 | 29 | −0.020 | 0.281 | <0.001 | 37 | −0.036 | 0.524 | <0.001 | 39 | |
| Secreted proteins | sap54 | −0.118 | 0.613 | <0.001 | 30 | −0.032 | 0.330 | <0.001 | 38 | −0.048 | 0.630 | <0.001 | 40 |
| sap67 | −0.099 | 0.522 | <0.001 | 30 | −0.021 | 0.234 | 0.004 | 34 | −0.043 | 0.555 | <0.001 | 36 | |
| sap68 | −0.107 | 0.680 | <0.001 | 30 | −0.017 | 0.150 | 0.018 | 37 | −0.047 | 0.645 | <0.001 | 38 | |
| tengu | −0.069 | 0.470 | <0.001 | 30 | −0.025 | 0.300 | <0.001 | 36 | −0.053 | 0.661 | <0.001 | 40 | |
| General metabolism | pgsA | −0.084 | 0.555 | <0.001 | 29 | −0.024 | 0.371 | <0.001 | 36 | −0.058 | 0.843 | <0.001 | 38 |
| rpsU | −0.081 | 0.639 | <0.001 | 30 | −0.006 | 0.037 | 0.264 | 36 | −0.030 | 0.519 | <0.001 | 40 | |
Nonsignificant (P > 0.05) regression data are indicated in bold.
Generic transporters.
mscL transcripts were the most abundant within this functional category, showing a mean expression level about 30 times higher than that of mdlB at each sampling date, from 70 to over 100 times higher than that of ftsY, and from 70 to over 500 times higher than that of oppC from 10 to 28 dpi (Fig. 2; see also Table S2 in the supplemental material). In line with this observation, oppC showed a rapid decrease of expression over time, as evidenced by the regression analysis (Fig. 2 and Table 2). Indeed, the oppC slope (absolute value) was the highest among those of the analyzed CYP genes in A. thaliana. Transcript levels of the other generic and specific transporter genes significantly decreased over time but at lower rates (Fig. 2).
Specific transporters.
The transcript levels of zntA were higher than those of artI under all conditions and time points (Fig. 2; see also Table S2 in the supplemental material). Mean expression levels of both artI and zntA decreased significantly in the late phase of infection (28 dpi). Regression analysis (Fig. 2 and Table 2) confirmed a significant decrease of transcript levels for both genes over time, indicating a correlation with sampling time for zntA stronger than that seen with artI and all other generic transporter genes except oppC.
Secreted proteins.
Considering all 14 CYP genes analyzed, tengu and sap54 were the second and the fourth most abundant transcripts under the A. thaliana host conditions, following amp and mscL, respectively. The tengu transcripts were the most abundant in this category (Fig. 2; see also Table S2 in the supplemental material), and tengu showed the second lowest regression slope after that of imp (Fig. 2 and Table 2). In contrast, a drop in the transcript levels of the other secreted proteins was clearly evident in the regression analysis, as slopes for these genes (absolute values) were the highest among the CYP genes analyzed in A. thaliana, just below the slope of oppC. sap54 showed the second highest transcript level and the most evident expression decrease over time (Fig. 2 and Table 2).
General metabolism.
The ribosomal rpsU gene showed a mean transcript level about five times higher than that of pgsA, at each sampling date (Fig. 2; see also Table S2 in the supplemental material). Mean EI values of both rpsU and pgsA decreased significantly in the late infection phases. Regression analysis (Fig. 2 and Table 2) also showed a significant decrease in the transcription of both genes, characterized by very similar slopes.
Phytoplasma gene transcript levels in Euscelidius variegatus.
Mean copy numbers of transcripts per CYP cell in individual E. variegatus sampled at 7, 14, 21, 28, and 35 dpa are presented in Table S3 in the supplemental material.
Immunodominant membrane proteins.
amp showed the highest EI among the analyzed CYP genes in E. variegatus individuals (see Table S3 in the supplemental material). amp and imp EI values did not change significantly during the infection in this vector species as confirmed by the regression analyses (Fig. 2 and Table 2).
Generic transporters.
Among the transporter genes, mscL produced the most abundant transcripts (see Table S3 in the supplemental material). Also, CYP generic transporter genes did not show significant changes of EI during phytoplasma infection of E. variegatus individuals (see Table S3). Regression analysis confirmed this result, as the EI values of all genes but mscL did not vary with time, and the regression of mscL transcript levels over time was barely significant (Fig. 2 and Table 2).
Specific transporters.
The transcript levels of zntA were always higher than those of artI (see Table S3 in the supplemental material). The mean EI of zntA, but not that of artI, decreased significantly in the late phase of infection (35 dpa; see Table S4 in the supplemental material), and its regression showed the highest slope (absolute value) among the other CYP generic and specific transporter genes in the E. variegatus host condition (Fig. 2 and Table 1).
Secreted proteins.
The transcripts of tengu and sap54 were the second and the third most abundant under the E. variegatus host conditions, following those of amp. Indeed, tengu was the most highly transcribed gene within its functional category (see Table S3 in the supplemental material). sap54 was the only gene showing a significant decrease of its transcript levels over time (see Table S3), while regression analyses indicated a significant negative correlation of EI and time for all analyzed secreted protein genes (Fig. 2 and Table 2).
General metabolism.
The ribosomal rpsU and pgsA genes showed analogous transcript levels in the early phase of infection (Fig. 2; see also Table S3 in the supplemental material). While the rpsU EI did not change over time (see Table S3 in the supplemental material), transcript levels of pgsA decreased significantly from the early to late infection phases (Fig. 2 and Table 1).
Phytoplasma gene transcript levels in Macrosteles quadripunctulatus.
Mean copy numbers of transcripts per CYP cell in individual M. quadripunctulatus samples collected at 7, 14, 21, 28, and 35 dpa are presented in Table S4 in the supplemental material.
Immunodominant membrane proteins.
In M. quadripunctulatus, amp showed the highest EI of all the analyzed CYP genes (see Table S4 in the supplemental material). The transcript levels decreased significantly in both late phases of infection (28 and 35 dpa; see Table S4), and regression analyses confirmed this result (Fig. 2 and Table 2). Mean transcript levels of amp were about 400 times higher than those of imp (see Table S4).
Generic transporters.
mscL was the third most highly transcribed CYP gene in infected M. quadripunctulatus, following amp and tengu, and its transcripts were the most abundant within this functional category (see Table S4 in the supplemental material). All generic transporter genes showed a significant decrease of transcript levels over time (see Table S4), with oppC showing the lowest (absolute value) regression slope over time of all CYP genes in the M. quadripunctulatus host (Fig. 2 and Table 1).
Specific transporters.
zntA and artI showed similar transcript levels in infected M. quadripunctulatus samples (see Table S4 in the supplemental material). The mean EI values of both genes decreased significantly in the late infection phase (Fig. 2 and Table 2). A more severe drop of transcript levels was recorded for zntA than for artI.
Secreted proteins.
As seen in A. thaliana, tengu and sap54 were the second and the fourth most highly expressed CYP genes in infected M. quadripunctulatus samples, and the tengu transcripts were the most abundant within this category (see Table S4 in the supplemental material). As seen in A. thaliana, all secreted protein-coding genes showed a significant EI decrease over time (see Table S4), with similar slope values in the regression analysis (Fig. 2 and Table 2).
General metabolism.
As seen in E. variegatus, the ribosomal rpsU and pgsA genes showed similar transcript levels at the earliest sampling date (see Table S4 in the supplemental material). Regression analysis showed a stronger decrease over time for pgsA, with the highest (absolute value) slope of all the analyzed CYP genes in the M. quadripunctulatus host environment (Fig. 2 and Table 1).
Comparison of gene transcript levels among species.
To compare CYP gene transcript profiles among the three hosts (see Table S5 in the supplemental material), the transcript levels of all analyzed CYP genes at the first two sampling dates (10 and 14 dpi and 7 and 14 dpa, for plant and insect samples, respectively) were pooled within each species, as they did not differ significantly (see Tables S2 to S4). Generally, CYP transcript levels were higher in the plant hosts than in the insect hosts and, for the two insect species, were higher in E. variegatus than in M. quadripunctulatus. However, CYP transcript levels did not significantly differ between A. thaliana and E. variegatus samples, except for amp, mscL, artI, and tengu. Transcripts of those four genes, together with mdlB and rpsU, were present at similar levels in the two insect vectors. The EI value for most CYP genes was significantly lower in M. quadripunctulatus than in A. thaliana, with the exception of sap67 and pgsA (see Table S5 in the supplemental material). amp, tengu, and mscL were always the most abundant CYP transcripts in each host species (see Table S5). On the other hand, sap68, pgsA, and sap67 produced the three least abundant CYP transcript levels in A. thaliana, while artI, oppC, and ftsY or zntA showed the lowest transcript levels in E. variegatus and M. quadripunctulatus, respectively. The timing and rate of decrease in CYP gene transcript levels over time differed among the three host species (Table 2). Generally, transcript levels of CYP analyzed genes dropped more severely after infection in the plant (regression slopes ranging from −0.081 to −0.120; Table 2) relative to colonization of the insects (regression slopes ranging from −0.004 to −0.580; Table 2) and more severely in M. quadripunctulatus (regression slopes ranging from −0.024 to −0.058; Table 2) than in E. variegatus (regression slopes ranging from −0.004 to −0.032; Table 2). Indeed, only seven CYP genes showed significantly decreased transcript levels related to increasing time in the latter species. However, for all the phytoplasma genes analyzed in A. thaliana and M. quadripunctulatus, decreased transcript levels were significantly related to increasing time after infection (Table 2). Secreted protein coding genes on average showed the strongest decrease of transcript levels in both vector and plant species, with the exception of tengu in A. thaliana. The transcript levels of oppC and zntA transporters decreased with time in A. thaliana, while the drop in the EI values of generic and specific transporter genes was less evident in the vector species. The pgsA and rpsU EI values displayed very similar decreases over time in A. thaliana samples, whereas, in insects of both species, that pgsA transcript level dropped more severely than that of rpsU (for pgsA and rpsU in E. variegatus and M. quadripunctulatus, regression slopes of −0.024 and −0.006 and regression slopes of −0.058 and −0.030, respectively, were determined; Table 2). Finally, while both the amp and the imp EI values decreased over time in A. thaliana and M. quadripunctulatus, the transcript levels of both genes did not vary in E. variegatus.
DISCUSSION
We report a comparison of transcript levels of selected phytoplasma genes in plant and vector species at different moments of the infection cycle. Target genes were selected according to literature analyses (16) and their potential role in phytoplasma adaptation to different environments: the plant and the insect.
Target gene transcript levels were measured over time by reverse transcription (RT) and qPCR, which allows the quantification of mRNA levels in bacteria (39–42). For data normalization, the transcript levels of bacterial target genes may be related to the number of cells obtained through cell culture (28), to the total RNA mass input in the RT-PCR, or to the internal reference control genes (43). Normalization of bacterial transcripts in relation to genomic DNA (gDNA) from cell culture or recombinant plasmid DNA (recDNA) has also been used (22, 27, 29). For phytoplasmas, methods based on cell culture are not available and total RNA input in the qPCR is always contaminated by large amounts of host RNA. Although the use of internal reference genes has been suggested for phytoplasma transcript normalization (18), this may not be suitable for gene expression analyses over time and in different hosts, as evidenced in other studies of relative quantifications of bacterial gene expression levels (22, 28). In this work, quantification of CYP transcripts was performed through the use of standard recDNA curves (29, 44) and an EI, calculated by dividing the gene transcript copy number and phytoplasma cell number measured at the same sampling date.
The multiplication pattern of CYP in A. thaliana was similar to that in C. carinatum (45), with active phytoplasma multiplication at up to 3 weeks postinoculation, followed by the maintenance of a stationary phytoplasma population size until the end of the experiment (28 dpi).
All selected CYP gene transcripts were present at detectable levels in the three hosts at every sampling date, suggesting their active role in phytoplasma cell cycles. The presence of less than one transcript copy number per CYP cell recorded for all host species, usually at late sampling dates, indicates that not all CYP cells contained each gene transcript at that time of the infection. Generally, transcript levels of most of the 14 selected genes decreased more significantly in the plant and in M. quadripunctulatus from early to late samplings, while the decrease in E. variegatus was less evident. In the former species, phytoplasma multiplication is very active (30) and is associated with some degree of pathogenicity (46). In E. variegatus, transcript levels of most of the selected target genes did not vary significantly during the infection cycle. In this species, phytoplasma multiplication is less efficient (30); in fact, CYP population sizes were always below those measured in M. quadripunctulatus at each sampling date, and no pathogenic effects were recorded (46). In the case of CYP, due to the low phytoplasma population in the infected vectors and plant, the first sampling points were set at 7 and 10 days postinfection of the insect and plant hosts, respectively. Under these conditions, the transcript levels of most analyzed genes decreased with time irrespective of the host species, and our experimental conditions did not allow us to arrive at any conclusion with respect to the very early phases of infection.
Antigenic membrane protein genes.
Phytoplasmas within the “Ca. Phytoplasma asteris” species carry genes that encode two immunodominant membrane proteins, Imp and Amp (14), and both of the genes encoding those proteins are objects of positive selection (13, 47). The amp transcripts were the most abundant of all the analyzed gene transcripts in all three host species, and the amp transcript levels decreased in A. thaliana and M. quadripunctulatus during the CYP infection cycle. Amp interacts specifically with the M. striifrons actin vector (10), possibly to enable phytoplasma motility (11). Indeed, amp transcript levels were stable during infection of E. variegatus, possibly implying a continuous need of the gene product to reach stable colonization of the insect body. Actually, CYP colonization of E. variegatus is slower than that of M. quadripunctulatus (30). Accordingly, amp transcript levels decreased during infection of M. quadripunctulatus. There is no information on the possible role of phytoplasma Amp in plant, but the low EI in M. quadripunctulatus at 28 dpa (when infective insects were caged for inoculation of healthy plants) and the high EI in A. thaliana at the first sampling date (10 dpi) suggest a role for Amp in the host switching for the colonization of the plant. Irrespective of the host species, amp transcripts were always more actively abundant than imp transcripts during the infection cycle, and this was in line with a previous finding determined for a closely related phytoplasma strain (13). Imp transcript levels were constant over time in E. variegatus, but not in M. quadripunctulatus, suggesting different gene regulation patterns in the two vector environments. imp was also the most stable gene with respect to transcripts among the 14 studied in A. thaliana. Nothing is known of the role of “Ca. Phytoplasma asteris” imp in the plant, although this gene is under positive selection in different phytoplasmas (13, 15). Imp of “Ca. Phytoplasma mali” binds to plant actin with a suggested role in phytoplasma motility (12).
Transporter genes.
The “Ca. Phytoplasma asteris” genome includes several transporter genes (48, 49). Transcripts of genes in this category were abundant in the plant host on average, and, in particular, transcripts of mscL and artI were significantly more abundant in A. thaliana than in the two vectors. Also, the mscL gene transcripts were the most abundant transporter gene transcripts during the CYP infection cycle in the three hosts. Microbial cells constitutively express the large mechanosensitive (MS) conductance channel that opens in response to stretch forces in the lipid bilayer, and the gene is upregulated in the presence of osmotic downshocks to protect from cell lysis (50). Phytoplasmas move from the plant to the vector body during feeding and are therefore subjected to severe osmotic stresses, as the osmotic pressure of the plant phloem is, on average, two to five times higher than that of insect body fluids (51). mscL transcription might be induced in the plant, when phytoplasma cells are exposed to high osmolarity, to prepare for the eventuality of hypo-osmotic stress conditions when phytoplasma cells are acquired by the vector. Indeed, de novo gene expression cannot modulate the levels of MS channel proteins on a short time scale (50). Moreover, Oshima et al. (16) showed that phytoplasma growth in planta was partially suppressed by gadolinium chloride, an inhibitor of the MscL osmotic channel, emphasizing its additional role in facilitating phytoplasma growth in plant. Transcripts of other genes in this category (artI and oppC) were among the least abundant during the CYP infection cycle in both vectors. In bacteria, Opp transport systems participate in a wide range of biological events, including biofilm formation (52), antimicrobial-compound production (53), and adaptation to different environments (54–57), through the modulation of the cell membrane lipid profile (58). In bacteria, artI is the periplasmic binding component of the arginine transporter system, specifically binding arginine and ornithine. Arginine is a key amino acid for the bacterial cell, and its metabolism and regulation are linked to the virulence of several pathogenic bacterial species such as Mycobacterium tuberculosis, M. bovis, Listeria monocytogenes, and Legionella pneumophila (59–62). Phloem sap has, in general, a lower concentration of essential amino acids than that found in optimal diets for some phloem feeders (63). We can speculate that the presence of arginine in the insect hosts downregulates the transcription of CYP artI and that the transcription resumes upon inoculation of the phytoplasma in the plant host. Interestingly, oppC, artI, ftsY, and mdlB transcript levels were also stable during the CYP infection cycle in E. variegatus and were among those that decreased least in M. quadripunctulatus. This suggests that basal transcription of these genes occurs in the insect milieu. In contrast, genes in this category were subjected to severe transcript reduction over time in the plant, oppC being the most dramatically affected. Zinc, together with iron, manganese, and copper, is required by all living organisms, and maintaining adequate intracellular levels of transition metals is fundamental to the survival of all organisms. Transcript levels of zntA, predicted to encode the soluble periplasmic metallochaperone that captures zinc and delivers it to the transmembrane component of the transporter, decreased in the three hosts during infection. Pathogens use low-metal conditions as a signal to recognize and respond to the host environment (64), and Salmonella exploits the ZnuABC zinc transporter to maximize zinc availability during growth within the infected animal under conditions where amounts of free metals available for bacterial growth are limited (65). The zntA EI value was lower for M. quadripunctulatus than for E. variegatus, and the transcript levels increased by 10 days after phytoplasma inoculation in the plant host, suggesting a role of zinc in the infection process. There is no information on the possible role of phytoplasma transporter genes in plant host, although the low EI value for M. quadripunctulatus at 28 dpa (when infective insects were caged on healthy plants for IAP) and the high EI value for all of the genes, except ftsY, in A. thaliana at the first sampling date (10 dpi) suggest a role in the colonization of the plant host. Recently, the involvement of transport proteins as additional bacterial cell sensors has been explored (66), as these proteins are well informed about the presence of substrates outside the cell. FtsY protein is involved in cellular secretion, as part of the internal channel of the Sec secretion system that is functional in the “Ca. Phytoplasma asteris” OY strain (67) and in the virulence of Rickettsia spp. (68) and Pseudomonas aeruginosa (69).
Secreted proteins.
Effector proteins are secreted from phytoplasmas via the Sec translocation system and function directly in the host cells (70). About 50 putative secreted proteins are present in phytoplasma genomes, and these effector genes differ from those found in other plant-pathogenic bacteria (71). Some of these have been shown to encode functional effectors, including SAP11, SAP54, SAP67, SAP68, and TENGU (8). SAP11, known to induce a bushy morphology and to enhance vector fitness by blocking jasmonic acid biosynthesis in plants (72), was not found in the draft genome of CYP, and it was not detected by PCR with specific primers designed for the closely related AY-WB phytoplasma (73). Transcripts encoding the four secreted proteins were all present in the three hosts, suggesting a role of these effectors during the CYP life cycle in both plant and insect hosts. Interestingly, sap67 transcripts were present at the same levels in A. thaliana and in the two vector species, while the tengu and sap54 transcripts were among the most abundant transcripts in the two insect vectors. SAP54 and its homolog, PHYL1, of “Ca. Phytoplasma asteris” AY-WB and OY, respectively, induce phyllody-like flower abnormalities (74), probably through the ubiquitin-mediated proteasome-dependent degradation of MADS domain proteins involved in floral development (75, 76). TENGU is a small, secreted peptide encoded by OYP that affects plant morphology with the production of typical phytoplasma witches' broom symptoms, through the inhibition of the auxin-related pathways (77). The high EI values of sap54 and tengu during the infection cycle of the two vectors together with the comparable EI values of sap67 in plant and in the two insects indicate that these genes must have a role in the infection of the animal host.
General metabolism.
Transcripts encoding PgsA, the limiting enzyme in the synthesis of phosphatidylglycerol, the major constituent of bacterial membranes, were present at comparable levels at early phases of infection of the plant and both insect species, supporting its role in the maintenance of essential bacterial phospholipids irrespective of the host species. Transcripts of rpsU, coding 30S ribosomal subunit protein S21, were more abundant in A. thaliana than in M. quadripunctulatus, this being in line with previous results determined for OYP (16). Nevertheless, rpsU transcript levels in A. thaliana and in E. variegatus were similar and were constant in the latter species at all analyzed time points. This gene is known to be stably transcribed at different time points through the entire in vitro life cycle of Bacillus cereus (78), and it can be considered, together with the general and specific transporter genes mdlB, oppC, ftsY, and artI, a candidate for future normalization of CYP gene expression in E. variegatus.
Expression of 14 CYP phytoplasma genes at different times postinfection of A. thaliana and the leafhopper vectors M. quadripunctulatus and E. variegatus was addressed to highlight the different transcription profiles of the bacteria in the plant and animal host. The transcription patterns of genes within the four analyzed categories differed according to the host species, suggesting that the bacteria are able to sense diverse environments and respond accordingly. Moreover, CYP transcript profiles of genes within the same category differed between the two leafhoppers, indicating the ability of CYP to distinguish between leafhopper host environments. Transcripts encoding Amp and the four secreted proteins were present in the three hosts, suggesting the important role of this immunodominant protein and unpredicted functions of these secreted phytoplasma proteins also during leafhopper infection. To explain the observed decrease of phytoplasma transcript levels during the CYP infection cycles in the plant and insect hosts starting from 7 to 10 days postinfection onwards, we might speculate that, during host colonization, new phloem elements and insect cells or organs are progressively invaded, possibly by actively multiplying and transcribing phytoplasma cells, while the transcriptional activity of older cells may slow down. In this hypothesis, phytoplasmas invading the plant phloem would form aggregates (79) that might enter a stationary phase, possibly to improve long-term survival, thereby increasing the likelihood of transmission. In this hypothesis, the number of cells (EI denominator) entering the stationary phase would increase with time after inoculation, while the number of new colony-forming cells would be low on average (as it would represent only a fraction of the measured total phytoplasma DNA). A similar mechanism may happen during colonization of the insect vectors, when phytoplasmas would not be able to escape the vector body and invade new tissues without inoculation to a new plant. In line with this hypothesis, phytoplasmas, being obligate parasites of plant and insects, sense the environment and switch their metabolism accordingly. Despite obvious differences, a very recent pioneer study has proposed that the xylem-limited bacterium Xylella fastidiosa switches its life style from adhesive cells capable of insect transmission to an “exploratory” lifestyle for systemic spread within the plant by production of outer membrane vesicles (80). Interestingly, the release of these vesicles is suppressed by a diffusible signal factor-dependent quorum-sensing system (80).
Supplementary Material
ACKNOWLEDGMENTS
M.R. was supported by a fellowship from the Regione Piemonte P.O.R. 2007/2013 within the project “ELIFITO”; D.P. was supported by a fellowship from the Regione Piemonte within the project “FLADO.”
We thank S. Bertin for insect rearing, P. Caciagli for critical reading of the manuscript, and Dylan Beal for English revision of the text.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03096-14.
REFERENCES
- 1.Dickinson M, Tuffen M, Hodgetts J. 2013. The phytoplasmas: an introduction. Methods Mol Biol 938:1–14. doi: 10.1007/978-1-62703-089-2_1. [DOI] [PubMed] [Google Scholar]
- 2.Foissac X, Wilson MR. 2010. Current and possible future distributions of phytoplasma diseases and their vectors, p 309–324. In Weintraub PG, Jones P (ed), Phytoplasmas: genomes, plant hosts and vectors. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 3.Weintraub PG, Beanland L. 2006. Insect vectors of phytoplasmas. Annu Rev Entomol 51:91–111. doi: 10.1146/annurev.ento.51.110104.151039. [DOI] [PubMed] [Google Scholar]
- 4.Oshima K, Maejima K, Namba S. 14 August 2013, posting date Genomic and evolutionary aspects of phytoplasmas. Front Microbiol doi: 10.3389/fmicb.2013.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcone C, Neimark H, Ragozzino A, Lauer U, Seemuller E. 1999. Chromosome sizes of phytoplasmas composing major phylogenetic groups and subgroups. Phytopathology 89:805–810. doi: 10.1094/PHYTO.1999.89.9.805. [DOI] [PubMed] [Google Scholar]
- 6.Kube M, Mitrovic J, Duduk B, Rabus R, Seemuller E. 2012. Current view on phytoplasma genomes and encoded metabolism. Sci World J 2012:185942. doi: 10.1100/2012/185942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, Davis RE, Wei W, Shao J, Jomantiene R. 2014. Phytoplasma genomes: evolution through mutually complementary mechanisms, gene loss and horizontal acquisition, p 235–271. In Gross D, Lichens-Park A, Kole C (ed), Genomics of plant-associated bacteria. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 8.Sugio A, Hogenhout SA. 2012. The genome biology of phytoplasma: modulators of plants and insects. Curr Opin Microbiol 15:247–254. doi: 10.1016/j.mib.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Sugawara K, Honma Y, Komatsu K, Himeno M, Oshima K, Namba S. 2013. The alteration of plant morphology by small peptides released from the proteolytic processing of the bacterial peptide TENGU. Plant Physiol 162:2005–2014. doi: 10.1104/pp.113.218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki S, Oshima K, Kakizawa S, Arashida R, Jung HY, Yamaji Y, Nishigawa H, Ugaki M, Namba S. 2006. Interaction between the membrane protein of a pathogen and insect microfilament complex determines insect-vector specificity. Proc Natl Acad Sci U S A 103:4252–4257. doi: 10.1073/pnas.0508668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galetto L, Bosco D, Balestrini R, Genre A, Fletcher J, Marzachi C. 2011. The major antigenic membrane protein of “Candidatus Phytoplasma asteris” selectively interacts with ATP synthase and actin of leafhopper vectors. PLoS One 6:e22571. doi: 10.1371/journal.pone.0022571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boonrod K, Munteanu B, Jarausch B, Jarausch W, Krczal G. 2012. An immunodominant membrane protein (Imp) of 'Candidatus Phytoplasma mali' binds to plant actin. Mol Plant Microbe Interact 25:889–895. doi: 10.1094/MPMI-11-11-0303. [DOI] [PubMed] [Google Scholar]
- 13.Kakizawa S, Oshima K, Ishii Y, Hoshi A, Maejima K, Jung HY, Yamaji Y, Namba S. 2009. Cloning of immunodominant membrane protein genes of phytoplasmas and their in planta expression. FEMS Microbiol Lett 293:92–101. doi: 10.1111/j.1574-6968.2009.01509.x. [DOI] [PubMed] [Google Scholar]
- 14.Kakizawa S, Oshima K, Namba S. 2006. Diversity and functional importance of phytoplasma membrane proteins. Trends Microbiol 14:254–256. doi: 10.1016/j.tim.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Siampour M, Izadpanah K, Galetto L, Salehi M, Marzachi C. 2013. Molecular characterization, phylogenetic comparison and serological relationship of the Imp protein of several 'Candidatus Phytoplasma aurantifolia' strains. Plant Pathol 62:452–459. doi: 10.1111/j.1365-3059.2012.02662.x. [DOI] [Google Scholar]
- 16.Oshima K, Ishii Y, Kakizawa S, Sugawara K, Neriya Y, Himeno M, Minato N, Miura C, Shiraishi T, Yamaji Y, Namba S. 2011. Dramatic transcriptional changes in an intracellular parasite enable host switching between plant and insect. PLoS One 6:e23242. doi: 10.1371/journal.pone.0023242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii Y, Kakizawa S, Oshima K. 2013. New ex vivo reporter assay system reveals that sigma factors of an unculturable pathogen control gene regulation involved in the host switching between insects and plants. Microbiologyopen 2:553–565. doi: 10.1002/mbo3.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toruño TY, Music MS, Simi S, Nicolaisen M, Hogenhout SA. 2010. Phytoplasma PMU1 exists as linear chromosomal and circular extrachromosomal elements and has enhanced expression in insect vectors compared with plant hosts. Mol Microbiol 77:1406–1415. doi: 10.1111/j.1365-2958.2010.07296.x. [DOI] [PubMed] [Google Scholar]
- 19.Bustin SA. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 20.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustin SA, Benes V, Nolan T, Pfaffl MW. 2005. Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- 22.Vandecasteele SJ, Peetermans WE, Merckx R, Van Ranst M, Van Eldere J. 2002. Use of gDNA as internal standard for gene expression in staphylococci in vitro and in vivo. Biochem Biophys Res Commun 291:528–534. doi: 10.1006/bbrc.2002.6465. [DOI] [PubMed] [Google Scholar]
- 23.McMillan M, Pereg L. 2014. Evaluation of reference genes for gene expression analysis using quantitative RT-PCR in Azospirillum brasilense. PLoS One 9:e98162–e98162. doi: 10.1371/journal.pone.0098162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badejo AC, Badejo AO, Shin KH, Chai YG. 2013. A gene expression study of the activities of aromatic ring-cleavage dioxygenases in Mycobacterium gilvum PYR-GCK to changes in salinity and pH during pyrene degradation. PLoS One 8:e58066. doi: 10.1371/journal.pone.0058066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cope EK, Goldstein-Daruech N, Kofonow JM, Christensen L, McDermott B, Monroy F, Palmer JN, Chiu AG, Shirtliff ME, Cohen NA, Leid JG. 2011. Regulation of virulence gene expression resulting from Streptococcus pneumoniae and nontypeable Haemophilus influenzae interactions in chronic disease. PLoS One 6:e28523. doi: 10.1371/journal.pone.0028523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stenico V, Baffoni L, Gaggia F, Biavati B. 2014. Validation of candidate reference genes in Bifidobacterium adolescentis for gene expression normalization. Anaerobe 27:34–39. doi: 10.1016/j.anaerobe.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Borges V, Ferreira R, Nunes A, Nogueira P, Borrego MJ, Gomes JP. 2010. Normalization strategies for real-time expression data in Chlamydia trachomatis. J Microbiol Methods 82:256–264. doi: 10.1016/j.mimet.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. 2001. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J Bacteriol 183:7094–7101. doi: 10.1128/JB.183.24.7094-7101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Florindo C, Ferreira R, Borges V, Spellerberg B, Gomes JP, Borrego MJ. 2012. Selection of reference genes for real-time expression studies in Streptococcus agalactiae. J Microbiol Methods 90:220–227. doi: 10.1016/j.mimet.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Bosco D, Galetto L, Leoncini P, Saracco P, Raccah B, Marzachi C. 2007. Interrelationships between “Candidatus Phytoplasma asteris” and its leafhopper vectors (Homoptera: Cicadellidae). J Econ Entomol 100:1504–1511. doi: 10.1603/0022-0493-100.5.1504. [DOI] [PubMed] [Google Scholar]
- 31.Galetto L, Marzachi C, Marques R, Graziano C, Bosco D. 2011. Effects of temperature and CO2 on phytoplasma multiplication pattern in vector and plant. Bull Insectol 64:S151–S152. [Google Scholar]
- 32.Bosco D, Minucci C, Boccardo G, Conti M. 1997. Differential acquisition of chrysanthemum yellows phytoplasma by three leafhopper species. Entomol Exp Appl 83:219–224. doi: 10.1046/j.1570-7458.1997.00175.x. [DOI] [Google Scholar]
- 33.Daire X, Clair D, Reinert W, BoudonPadieu E. 1997. Detection and differentiation of grapevine yellows phytoplasmas belonging to the elm yellows group and to the stolbur subgroup by PCR amplification of non-ribosomal DNA. Eur J Plant Pathol 103:507–514. doi: 10.1023/A:1008641411025. [DOI] [Google Scholar]
- 34.Galetto L, Bosco D, Marzachi C. 2005. Universal and group-specific real-time PCR diagnosis of Flavescence dorée (16Sr-V), bois noir (16Sr-XII) and apple proliferation (16Sr-X) phytoplasmas from field-collected plant hosts and insect vectors. Ann Appl Biol 147:191–201. doi: 10.1111/j.1744-7348.2005.00030.x. [DOI] [Google Scholar]
- 35.Marzachí C, Bosco D. 2005. Relative quantification of chrysanthemum yellows (16SrI) phytoplasma in its plant and insect host using real-time polymerase chain reaction. Mol Biotechnol 30:117–127. doi: 10.1385/MB:30:2:117. [DOI] [PubMed] [Google Scholar]
- 36.Saccardo F, Martini M, Palmano S, Ermacora P, Scortichini M, Loi N, Firrao G. 2012. Genome drafts of four phytoplasma strains of the ribosomal group 16SrIII. Microbiology 158:2805–2814. doi: 10.1099/mic.0.061432-0. [DOI] [PubMed] [Google Scholar]
- 37.Galetto L, Fletcher J, Bosco D, Turina M, Wayadande A, Marzachì C. 2008. Characterization of putative membrane protein genes of the ‘Candidatus Phytoplasma asteris’, chrysanthemum yellows isolate. Can J Microbiol 54:341–351. doi: 10.1139/w08-010. [DOI] [PubMed] [Google Scholar]
- 38.Gomes JP, Hsia RC, Mead S, Borrego MJ, Dean D. 2005. Immunoreactivity and differential developmental expression of known and putative Chlamydia trachomatis membrane proteins for biologically variant serovars representing distinct disease groups. Microb Infect 7:410–420. doi: 10.1016/j.micinf.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Goerke C, Campana S, Bayer MG, Doring G, Botzenhart K, Wolz C. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect Immun 68:1304–1311. doi: 10.1128/IAI.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. 2003. Expression of biofilm-associated genes in Staphylococcus epidermidis during in vitro and in vivo foreign body infections. J Infect Dis 188:730–737. doi: 10.1086/377452. [DOI] [PubMed] [Google Scholar]
- 41.Devers M, Soulas G, Martin-Laurent F. 2004. Real-time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil. J Microbiol Methods 56:3–15. doi: 10.1016/j.mimet.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Corbella ME, Puyet A. 2003. Real-time reverse transcription-PCR analysis of expression of halobenzoate and salicylate catabolism-associated operons in two strains of Pseudomonas aeruginosa. Appl Environ Microbiol 69:2269–2275. doi: 10.1128/AEM.69.4.2269-2275.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takle GW, Toth IK, Brurberg MB. 2007. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol 7:50. doi: 10.1186/1471-2229-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaffl MW. 2004. Quantification strategies in real-time PCR. AZ Quant PCR 1:89–113. [Google Scholar]
- 45.Saracco P, Bosco D, Veratti F, Marzachi C. 2006. Quantification over time of chrysanthemum yellows phytoplasma (16Sr-I) in leaves and roots of the host plant Chrysanthemum carinatum (Schousboe) following inoculation with its insect vector. Physiol Mol Plant Pathol 67:212–219. doi: 10.1016/j.pmpp.2006.02.001. [DOI] [Google Scholar]
- 46.D'Amelio R, Palermo S, Marzachi C, Bosco D. 2008. Influence of Chrysanthemum yellows phytoplasma on the fitness of two of its leafhopper vectors, Macrosteles quadripunctulatus and Euscelidius variegatus. Bull Insectol 61:349–354. [Google Scholar]
- 47.Kakizawa S, Oshima K, Jung HY, Suzuki S, Nishigawa H, Arashida R, Miyata S, Ugaki M, Kishino H, Namba S. 2006. Positive selection acting on a surface membrane protein of the plant-pathogenic phytoplasmas. J Bacteriol 188:3424–3428. doi: 10.1128/JB.188.9.3424-3428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai XD, Zhang JH, Ewing A, Miller SA, Radek AJ, Shevchenko DV, Tsukerman K, Walunas T, Lapidus A, Campbell JW, Hogenhout SA. 2006. Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J Bacteriol 188:3682–3696. doi: 10.1128/JB.188.10.3682-3696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oshima K, Kakizawa S, Nishigawa H, Jung HY, Wei W, Suzuki S, Arashida R, Nakata D, Miyata S, Ugaki M, Namba S. 2004. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat Genet 36:27–29. doi: 10.1038/ng1277. [DOI] [PubMed] [Google Scholar]
- 50.Stokes NR, Murray HD, Subramaniam C, Gourse RL, Louis P, Bartlett W, Miller S, Booth IR. 2003. A role for mechanosensitive channels in survival of stationary phase: regulation of channel expression by RpoS. Proc Natl Acad Sci U S A 100:15959–15964. doi: 10.1073/pnas.2536607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Douglas AE. 2006. Phloem-sap feeding by animals: problems and solutions. J Exp Bot 57:747–754. doi: 10.1093/jxb/erj067. [DOI] [PubMed] [Google Scholar]
- 52.Lee EM, Ahn SH, Park JH, Lee JH, Ahn SC, Kong IS. 2004. Identification of oligopeptide permease (opp) gene cluster in Vibrio fluvialis and characterization of biofilm production by oppA knockout mutation. FEMS Microbiol Lett 240:21–30. doi: 10.1016/j.femsle.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 53.Yazgan A, Ozcengiz G, Marahiel MA. 2001. Tn10 insertional mutations of Bacillus subtilis that block the biosynthesis of bacilysin. Biochim Biophys Acta 1518:87–94. doi: 10.1016/S0167-4781(01)00182-8. [DOI] [PubMed] [Google Scholar]
- 54.Wang XG, Kidder JM, Scagliotti JP, Klempner MS, Noring R, Hu LDT. 2004. Analysis of differences in the functional properties of the substrate binding proteins of the Borrelia burgdorferi oligopeptide permease (opp) operon. J Bacteriol 186:51–60. doi: 10.1128/JB.186.1.51-60.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monnet V. 2003. Bacterial oligopeptide-binding proteins. Cell Mol Life Sci 60:2100–2114. doi: 10.1007/s00018-003-3054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang XG, Lin B, Kidder JM, Telford S, Hu LT. 2002. Effects of environmental changes on expression of the oligopeptide permease (opp) genes of Borrelia burgdorferi. J Bacteriol 184:6198–6206. doi: 10.1128/JB.184.22.6198-6206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borezee E, Pellegrini E, Berche P. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect Immun 68:7069–7077. doi: 10.1128/IAI.68.12.7069-7077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flores-Valdez MA, Morris RP, Laval F, Daffe M, Schoolnik GK. 2009. Mycobacterium tuberculosis modulates its cell surface via an oligopeptide permease (Opp) transport system. FASEB J 23:4091–4104. doi: 10.1096/fj.09-132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Talaue MT, Venketaraman V, Hazbon MH, Peteroy-Kelly M, Seth A, Colangeli R, Alland D, Connell ND. 2006. Arginine homeostasis in J774.1 macrophages in the context of Mycobacterium bovis BCG infection. J Bacteriol 188:4830–4840. doi: 10.1128/JB.01687-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hovel-Miner G, Faucher SP, Charpentier X, Shuman HA. 2010. ArgR-regulated genes are derepressed in the Legionella-containing vacuole. J Bacteriol 192:4504–4516. doi: 10.1128/JB.00465-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 62.Ryan S, Begley M, Gahan CGM, Hill C. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ Microbiol 11:432–445. doi: 10.1111/j.1462-2920.2008.01782.x. [DOI] [PubMed] [Google Scholar]
- 63.Sandstrom J, Moran N. 1999. How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl 91:203–210. doi: 10.1046/j.1570-7458.1999.00485.x. [DOI] [Google Scholar]
- 64.Fones H, Preston GM. 2013. The impact of transition metals on bacterial plant disease. FEMS Microbiol Rev 37:495–519. doi: 10.1111/1574-6976.12004. [DOI] [PubMed] [Google Scholar]
- 65.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, Battistoni A. 2007. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect Immun 75:5867–5876. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tetsch L, Jung K. 2009. The regulatory interplay between membrane-integrated sensors and transport proteins in bacteria. Mol Microbiol 73:982–991. doi: 10.1111/j.1365-2958.2009.06847.x. [DOI] [PubMed] [Google Scholar]
- 67.Kakizawa S, Oshima K, Nighigawa H, Jung HY, Wei W, Suzuki S, Tanaka M, Miyata S, Ugaki M, Namba S. 2004. Secretion of immunodominant membrane protein from onion yellows phytoplasma through the Sec protein-translocation system in Escherichia coli. Microbiology 150:135–142. doi: 10.1099/mic.0.26521-0. [DOI] [PubMed] [Google Scholar]
- 68.Chao C-C, Chelius D, Zhang T, Mutumanje E, Ching W-M. 2007. Insight into the virulence of Rickettsia prowazekii by proteomic analysis and comparison with an avirulent strain. Biochim Biophys Acta 1774:373–381. doi: 10.1016/j.bbapap.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 69.Handfield M, Lehoux DE, Sanschagrin F, Mahan MJ, Woods DE, Levesque RC. 2000. In vivo-induced genes in Pseudomonas aeruginosa. Infect Immun 68:2359–2362. doi: 10.1128/IAI.68.4.2359-2362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hogenhout SA, Loria R. 2008. Virulence mechanisms of Gram-positive plant pathogenic bacteria. Curr Opin Plant Biol 11:449–456. doi: 10.1016/j.pbi.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 71.Sugio A, Maclean AM, Kingdom HN, Grieve VM, Manimekalai R, Hogenhout SA. 2011. Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu Rev Phytopathol 49:175–195. doi: 10.1146/annurev-phyto-072910-095323. [DOI] [PubMed] [Google Scholar]
- 72.Sugio A, Kingdom HN, Maclean AM, Grieve VM, Hogenhout SA. 2011. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc Natl Acad Sci U S A 108:E1254–E1263. doi: 10.1073/pnas.1105664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palmano S, Saccardo F, Martini M, Abbà S, Marzachì C, Ermacora P, Loi N, Firrao G. 2012. Comparative pathogenomics from six new phytoplasma genome drafts, p 48, abstr 35 Abstr 19th Int Organ Mycoplasmol (IOM) Congr, Toulouse, France. [Google Scholar]
- 74.MacLean AM, Sugio A, Makarova OV, Findlay KC, Grieve VM, Toth R, Nicolaisen M, Hogenhout SA. 2011. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol 157:831–841. doi: 10.1104/pp.111.181586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacLean AM, Orlovskis Z, Kowitwanich K, Zdziarska AM, Angenent GC, Immink RGH, Hogenhout SA. 2014. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol 12:e1001835. doi: 10.1371/journal.pbio.1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maejima K, Iwai R, Himeno M, Komatsu K, Kitazawa Y, Fujita N, Ishikawa K, Fukuoka M, Minato N, Yamaji Y, Oshima K, Namba S. 2014. Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J 78:541–554. doi: 10.1111/tpj.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, Komatsu K, Kagiwada S, Yamaji Y, Namba S. 2009. A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc Natl Acad Sci U S A 106:6416–6421. doi: 10.1073/pnas.0813038106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reiter L, Kolsto A-B, Piehler AP. 2011. Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. J Microbiol Methods 86:210–217. doi: 10.1016/j.mimet.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 79.Seemuller E. 1976. Investigation to demonstrate mycoplasmalike organisms in diseased plants by fluorescence microscopy. Acta Hortic 67:109–111. [Google Scholar]
- 80.Ionescu M, Zaini PA, Baccari C, Tran S, da Silva AM, Lindow SE. 2014. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc Natl Acad Sci U S A 111:E3910–E3918. doi: 10.1073/pnas.1414944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.