Abstract
Genetic manipulation of emerging bacterial pathogens, such as coagulase-negative staphylococci (CoNS), is a major hurdle in clinical and basic microbiological research. Strong genetic barriers, such as restriction modification systems or clustered regularly interspaced short palindromic repeats (CRISPR), usually interfere with available techniques for DNA transformation and therefore complicate manipulation of CoNS or render it impossible. Thus, current knowledge of pathogenicity and virulence determinants of CoNS is very limited. Here, a rapid, efficient, and highly reliable technique is presented to transfer plasmid DNA essential for genetic engineering to important CoNS pathogens from a unique Staphylococcus aureus strain via a specific S. aureus bacteriophage, Φ187. Even strains refractory to electroporation can be transduced by this technique once donor and recipient strains share similar Φ187 receptor properties. As a proof of principle, this technique was used to delete the alternative transcription factor sigma B (SigB) via allelic replacement in nasal and clinical Staphylococcus epidermidis isolates at high efficiencies. The described approach will allow the genetic manipulation of a wide range of CoNS pathogens and might inspire research activities to manipulate other important pathogens in a similar fashion.
INTRODUCTION
Coagulase-negative staphylococci (CoNS), such as human skin-colonizing Staphylococcus epidermidis or Staphylococcus lugdunensis, are frequently isolated from hospital-associated infections and represent emerging bacterial pathogens (1, 2). Many CoNS, in particular hospital-associated S. epidermidis, are resistant to available antibiotics, such as methicillin (≥75% of S. epidermidis isolates), clindamycin, tetracycline, trimethoprim, macrolides, and aminoglycosides (3). Such strains typically cause a variety of complicated infections, particularly infections associated with indwelling medical devices in combination with strong biofilm formation or, even more alarming, life-threatening systemic disease, such as endocarditis or sepsis (2, 4–6).
Genetic engineering of the well-studied pathogen Staphylococcus aureus has revolutionized research activities in the field of staphylococci in the past. However, current knowledge of the physiology and pathogenicity of CoNS is very limited because most available techniques used for genetic manipulation (e.g., electroporation of shuttle plasmids) often fail, most likely because of strong genetic barrier mechanisms, such as restriction-modification (R-M) systems or clustered regularly interspaced short palindromic repeats (CRISPR), previously shown to impede horizontal gene transfer (HGT) events between bacteria (7, 8). However, specifically engineered Escherichia coli strains lacking the dcm gene required for cytosine methylation of DNA have been developed to produce plasmid DNA that is not degraded upon electroporation of S. aureus or of specific S. lugdunensis strains, or even of the clinical S. epidermidis isolate RP62A (9, 10). This approach is promising but requires large amounts of plasmid DNA (typically 5 μg) and seems to be limited to specific CoNS isolates. An alternative to electroporation is protoplast transformation, which is very ineffective in the case of S. epidermidis because of its natural insensitivity to lysostaphin, an enzyme used to degrade staphylococcal cell walls (11). Equally problematic is transduction. Although transducing S. epidermidis phages have been described in the past (12–14), they are suitable solely for DNA transfer among S. epidermidis isolates, but not for DNA introduction from genetically more amenable organisms, such as S. aureus. For such approaches, interspecies transduction events are required, which have been reported, for example, between S. aureus and S. epidermidis isolates via S. aureus phage Φ80 (15). However, because most S. aureus DNA donor strains synthesize ribitol-phosphate wall teichoic acid (WTA) phage receptors distinct from those of most CoNS recipients, phage adsorption of S. aureus transducing phages, such as Φ80 or Φ11, to CoNS is usually blocked, rendering HGT impossible (16–19). Thus, the available techniques are highly challenging and require multiple attempts or, as is more often the case, completely fail with clinical CoNS, which represents a major hurdle for the molecular characterization of CoNS.
Recently, phage-mediated HGT of S. aureus pathogenicity islands (SaPIs) between major bacterial pathogens has been reported (17). Surprisingly, the S. aureus ST395 lineage-specific phage Φ187 has been capable of transferring SaPIs between ST395 isolates and many CoNS because of shared properties in the WTA cell surface receptors (17). Genomic and biochemical analyses of the ST395 WTA biosynthesis pathway have further suggested the occurrence of previous HGT events between ST395 isolates and CoNS, most likely via Φ187-related phages (20). Because most pathogenic CoNS, such as S. epidermidis, have a glycerol-phosphate WTA backbone resembling that of ST395 isolates, Φ187 might be a suitable tool to transfer plasmid DNA to CoNS strains that are otherwise difficult to transform.
Here, a highly efficient method to introduce plasmid DNA into important CoNS pathogens for genetic manipulation is described. The method aims to transfer plasmid DNA via phage Φ187 from a genetically engineered ST395 isolate to major clinical CoNS isolates, and even to strains refractory to electroporation protocols, for their genetic engineering.
MATERIALS AND METHODS
Bacterial strains and growth media.
All the bacterial strains listed in Table 1 were grown at permissive temperatures in basic medium (BM) (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) or in lysogeny broth (Becton Dickinson) supplemented with appropriate antibiotics (10 μg/ml for chloramphenicol, 12.5 μg/ml for tetracycline, or 100 μg/ml for ampicillin). For experiments performed on solid medium, BM agar plates containing 5% sheep blood were used unless otherwise noted.
TABLE 1.
Strains and phages used in this study
| Bacterial strain or phage | Description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α pRB474 | DH5α; bears pRB474 plasmid | Strain collection, Peschel laboratory |
| DB 3.1 pKOR1 | DB3.1 strain; bears pKOR1 plasmid | Olaf Schneewind, Chicago, IL |
| DC10B | E. coli DH10B Δdcm | 9 |
| DC10B pBASE6-sigB-ko2 | DC10B strain; bears pBASE6-sigB-ko2 plasmid | This study |
| S. carnosus TM300 pTX15 | Wild type; bears pTX15 plasmid | Strain collection, Peschel laboratory |
| S. aureus | ||
| RN4220 pRB474 | Wild type; deficient in restriction, capsule, and prophage; bears pRB474 plasmid | This study |
| RN4220 pTX15 | Wild type; deficient in restriction, capsule, and prophage; bears pTX15 plasmid | This study |
| PS187 | Wild type; clinical ST395 isolate | 27 |
| PS187 ΔhsdR ΔsauUSI | PS187 deficient in type IV and type I restriction systems | 17 |
| PS187 ΔhsdR ΔsauUSI pRB474 | PS187 deficient in type IV and type I restriction systems; bears pRB474 plasmid | This study |
| PS187 ΔhsdR ΔsauUSI pTX15 | PS187 deficient in type IV and type I restriction systems; bears pTX15 plasmid | This study |
| PS187 ΔhsdR ΔsauUSI pKOR1 | PS187 deficient in type IV and type I restriction systems; bears pKOR1 plasmid | This study |
| PS187 ΔhsdR ΔsauUSI pBASE-sigB-ko2 | PS187 deficient in type IV and type I restriction systems; bears pBASE-sigB-ko2 plasmid | This study |
| S. epidermidis | ||
| TÜ3298 | Wild type; epidermin producer | 28 |
| ATCC 14990 | Wild type; nasal isolate | ATCC strain collection |
| ATCC 12228 | Wild type | ATCC strain collection |
| RP62A; ATCC 35984 | Wild type; clinical isolate | ATCC strain collection |
| 1457 | Wild type; clinical isolate | 29 |
| O47 | Wild type; clinical isolate | 30 |
| IVK7 | Wild type; nasal isolate | 31 |
| IVK45 | Wild type; nasal isolate | 31 |
| IVK79 | Wild type; nasal isolate | 31 |
| IVK83 | Wild type; nasal isolate | 31 |
| 1 b | Wild type; clinical isolate | 32 |
| 26 b | Wild type; clinical isolate | 32 |
| 33 b | Wild type; clinical isolate | 32 |
| 36 b | Wild type; clinical isolate | 32 |
| 79 b | Wild type; clinical isolate | 32 |
| 84 b | Wild type; clinical isolate | 32 |
| 101 b | Wild type; clinical isolate | 32 |
| 104 b | Wild type; clinical isolate | 32 |
| 1/70102704 | Wild type; clinical isolate | Evgeny Idelevich, Muenster, Germany |
| 5/70107982 | Wild type; clinical isolate | Evgeny Idelevich, Muenster, Germany |
| 144/70215041 | Wild type; clinical isolate | Evgeny Idelevich, Muenster, Germany |
| 181/70247761 | Wild type; clinical isolate | Evgeny Idelevich, Muenster, Germany |
| 204/70131537 | Wild type; clinical isolate | Evgeny Idelevich, Muenster, Germany |
| S. lugdunensis | ||
| HKU09-01 | Wild type; clinical isolate | 33 |
| N920143 | Wild type; clinical isolate | 34 |
| IVK12-3 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| IVK13-1 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| IVK14-2 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| IVK14-6 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| IVK15-2 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| IVK28 | Wild type; nasal isolate | 31 |
| IVK38-2 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| IVK39-2 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| IVK68 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| IVK84 | Wild type; nasal isolate | Bernhard Krismer, Tuebingen, Germany |
| S. caprae | ||
| BK16134/12 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| VA18305/14 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| BK3880/14 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| BK1538/14 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| BK1057/14 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| BK14568/12 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| S. warneri BK15472/12 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| S. haemolyticus 51-14 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| S. saprophyticus BK5803/14 | Wild type; clinical isolate | University Medical Center Hamburg-Eppendorf, Hamburg, Germany |
| S. simulans ATCC27848 | Wild type; human skin isolate | ATCC strain collection |
| S. pseudintermedius ED99 | Wild type; animal isolate | 35 |
| S. epidermidis IVK83 ΔsigB | sigB deletion mutant in IVK83 background | This study |
| S. epidermidis 33 b ΔsigB | sigB deletion mutant in 33 b background | This study |
| S. aureus phage Φ187 | Wild type | 36 |
Molecular genetic methods.
Plasmid transformation was performed according to the method of Winstel et al. (17). Briefly, plasmids were isolated from appropriate E. coli, Staphylococcus carnosus TM300, S. aureus RN4220, or S. aureus PS187 ΔhsdR ΔsauUSI donor strains by using standard techniques, purified, and electroporated into electrocompetent recipient strains using a 1-mm-gap electroporation cuvette. The cells were pulsed at 1,000 V, and subsequently, 950 μl prewarmed BM (37°C) was added. Bacteria were grown for 70 min at 37°C, plated onto selective medium, and grown overnight at 37°C (or 30°C for knockout plasmids). Transformants were counted to calculate the transformation efficiency.
For the construction of ermB-bearing S. epidermidis sigB mutants, an ermB resistance cassette was amplified from plasmid pEC2 (21) with primers lox66erm and lox71erm (Table 2) and ligated with the SmaI-digested pKOR1-like knockout plasmid pBASE6 (22), resulting in plasmid pBASE6-erm/lox2. The flanking regions of sigB were amplified from genomic DNA of S. epidermidis TÜ3298 with primer pairs sigflank1 and sigflank2 (digested with BspEI and Acc65I) (Table 2) and sigflank3 and sigflank4 (digested with NheI and SalI) (Table 2), respectively. The digested flanking regions were consecutively ligated with the identically digested knockout plasmid, resulting in pBASE6-sigB-ko2, which was used to transform E. coli DC10B (9) and S. aureus PS187 ΔhsdR ΔsauUSI cells.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| sigflank 1 | TTTAATATCCGGAGTTGCTTCATTTTGAG |
| sigflank 2 | GCGGTACCGTAAACGAGTTGTTAACAG |
| sigflank 3 | CGTTTGCTAGCTGTTGTTTAATCCATTGG |
| sigflank 4 | AAGCATATGCGTCGACAGGATATAGTTCA |
| lox66erm | TACCGTTCGTATAATGTATGCTATACGAAGTTATGTTAACCCTAAAGTTATGG |
| lox71erm | TACCGTTCGTATAGCATACATTATACGAAGTTATGATCAAATTCCCCGTAGGCGC |
Experiments with phage.
To propagate Φ187, its cognate host strain S. aureus PS187 wild type (w.t.) was grown overnight in BM and diluted in Φ187-containing lysates (1 × 109 PFU/ml; titrated on PS187 wild type as described previously [17]) to a final optical density at 600 nm (OD600) of 0.4. After the addition of CaCl2 (final concentration, 4 mM), the bacteria-phage suspension was incubated without shaking at 37°C for 30 min and subsequently for at least 3 h at 30°C with slight agitation until complete lysis occurred. Cell debris was pelleted via centrifugation (10 min; 5,000 × g), and phage-containing supernatants were filter sterilized (0.02 μm) and stored at 4°C or subsequently used to infect overnight cultures of pKOR1-, pRB474-, or pTX15-bearing S. aureus PS187 ΔhsdR ΔsauUSI grown in BM (obtained via electroporation [see above]). The infection parameters were as described before, except that bacteria were diluted in phage lysates to a final OD600 of 1.0 and incubated at 30°C, which usually required at least 5 h for lysis to occur. Plasmid-bearing phage particle (PBPP) lysates were clarified via centrifugation (10 min; 5,000 × g), filter sterilized, and titrated on PS187 wild type.
For plasmid transfer to CoNS, the bacterial densities of overnight cultures of plasmid recipient strains (e.g., S. epidermidis TÜ3298) were adjusted to an OD600 of 0.5 in BM. Approximately 8.0 × 107 bacteria were centrifuged and resuspended in 200 μl phage buffer (4 mM CaCl2, 1 mM MgSO4, 50 mM Tris-HCl, pH 7.8, 100 mM NaCl, 0.1% gelatin), mixed with 100 μl PBPP lysate (∼1 × 109 PFU/ml), and incubated at 37°C for 15 min with slight agitation, allowing phage particle adsorption to bacterial host cells. Afterward, the bacteria-phage suspension was plated onto selective media containing appropriate antibiotics and incubated for 24 h at 37°C (or 48 h at 30°C for knockout plasmids). The resulting transductants were counted to calculate transduction efficiencies, which are given as transductants per milliliter of phage lysate.
Phage Φ187 adsorption efficiencies were determined as described previously with minor modifications (17). Briefly, adsorption rates were analyzed using a multiplicity of infection (MOI) of 0.1, and the adsorption rate (percent) was calculated by determining the number of unbound PFU in the supernatant and subtracting it from the total number of input PFU as a ratio to the total number of input PFU.
Ultracentrifugation of PBPP lysates.
For enrichment of PBPP, lysates were ultracentrifuged for 2 h at 25,100 rpm (73,000 × g) at 4°C using a Beckman 45Ti ultracentrifuge rotor. Subsequently, the PBPP-containing pellet was resuspended in an appropriate amount of TMN buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgSO4, 500 mM NaCl), and the lysates were titrated on PS187 wild type and used for plasmid transfer experiments, as previously described.
Tests for natural competence and “pseudotransformation.”
To analyze the natural competence of selected S. epidermidis isolates, PBPP lysates (containing pTX15) were treated with 20 U DNase I for 1 h at 37°C to digest extracellular DNA and subsequently used for plasmid transfer to strain TÜ3298, 101 b, or 204/70131537, as previously described. In a second approach, and to inactivate phage particles but not DNA, PBPP lysates (containing pTX15) were heat inactivated for 10 min at 70°C, chilled, and subsequently used for plasmid transfer to selected S. epidermidis strains. Controls were not heat inactivated or lacked DNase I treatment. To determine phage-mediated “pseudotransformation,” selected S. epidermidis strains were incubated with 1.0 μg purified pTX15 plasmid in the presence or absence of Φ187 w.t. particles (∼1 × 109 PFU/ml) for 15 min at 37°C. Φ187 w.t. particles were previously propagated on S. aureus PS187 w.t., which lacks pTX15, pRB474, and pKOR1 plasmids. The bacteria-phage suspension was plated onto selective medium containing the appropriate antibiotic and incubated for 24 h at 37°C. Transductants were counted to calculate the transduction efficiency, as previously described.
Allelic replacement.
Allelic replacement was performed as previously described with minor modifications (23). Briefly, S. aureus PS187 ΔhsdR ΔsauUSI bearing a sigB knockout plasmid was infected with Φ187 to create a PBPP lysate, as previously described, which was subsequently used for plasmid transfer to S. epidermidis. The knockout plasmid was integrated into the genome of the target strain in the presence of chloramphenicol (10 μg/ml) at a temperature of 42°C. Following plasmid integration, two consecutive counterselection steps using anhydrotetracycline at concentrations of 0.2 μg/ml and 1.0 μg/ml were performed. The resulting colonies were transferred to BM agar plates with or without chloramphenicol (10 μg/ml). Chloramphenicol-sensitive colonies were used for PCR analysis to confirm the sigB deletion.
Statistical analysis.
Statistics were performed using GraphPad Prism (version 5.04; GraphPad Software, Inc., La Jolla, CA, USA). Statistically significant differences were calculated using the unpaired two-tailed Student t test.
RESULTS AND DISCUSSION
Inactivation of restriction systems converts S. aureus strain PS187 into a suitable intermediary host for plasmid DNA.
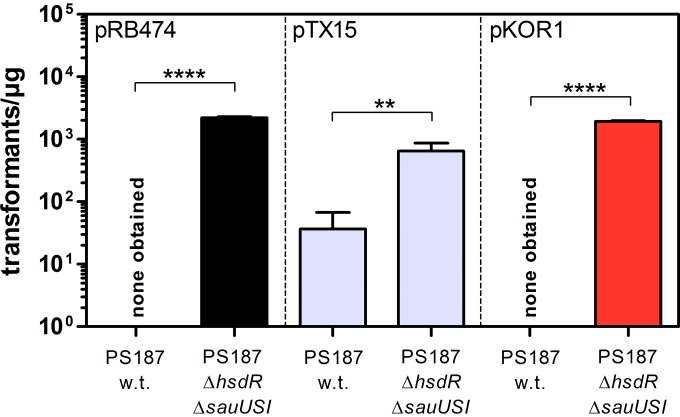
S. aureus ST395 isolate PS187 accepts S. aureus RN4220-derived plasmid DNA at low efficiencies, but inactivation of the restriction systems HsdR and SauUSI has rendered the strain highly susceptible to plasmid transformation (17). Lack of both restriction systems may also enable direct electroporation of strain PS187 with plasmid DNA from sources other than S. aureus. To this end, various plasmids widely used for targeted mutagenesis (pKOR1 [23]), overexpression (pTX15 [24]), or complementation (pRB474 [25]) studies were isolated from either E. coli or S. carnosus using standard techniques and introduced into S. aureus PS187 wild type and its restriction-deficient mutant PS187 ΔhsdR ΔsauUSI via electroporation. PS187 wild type was resistant to electroporation with DNA derived from E. coli, while deletion of restriction systems resulted in robust transformation efficiency (Fig. 1). PS187 wild type could also be transformed at low efficiency with plasmid DNA derived from S. carnosus TM300, although the absence of restriction systems significantly increased the electroporation frequency. Thus, S. aureus PS187 ΔhsdR ΔsauUSI is easily transformable and represents a suitable alternative to the S. aureus strain RN4220 conventionally used as an intermediary host for plasmid DNA.
FIG 1.
Lack of restriction systems converts S. aureus strain PS187 into a suitable intermediary host for plasmid DNA. Shown are the electroporation frequencies of S. aureus PS187 wild type (w.t.) and its restriction system-deficient mutant PS187 ΔhsdR ΔsauUSI. Cells were electroporated with purified E. coli DH5α-derived pRB474, E. coli DB3.1-derived pKOR1, or S. carnosus TM300-derived pTX15 plasmid. The values represent transformants per microgram of DNA and are given as means and standard deviations (SD) (n = 3). Statistically significant differences calculated by the unpaired two-tailed Student t test are indicated: **, P < 0.001 to 0.01; ****, P < 0.0001.
Most CoNS are refractory to common electroporation protocols.
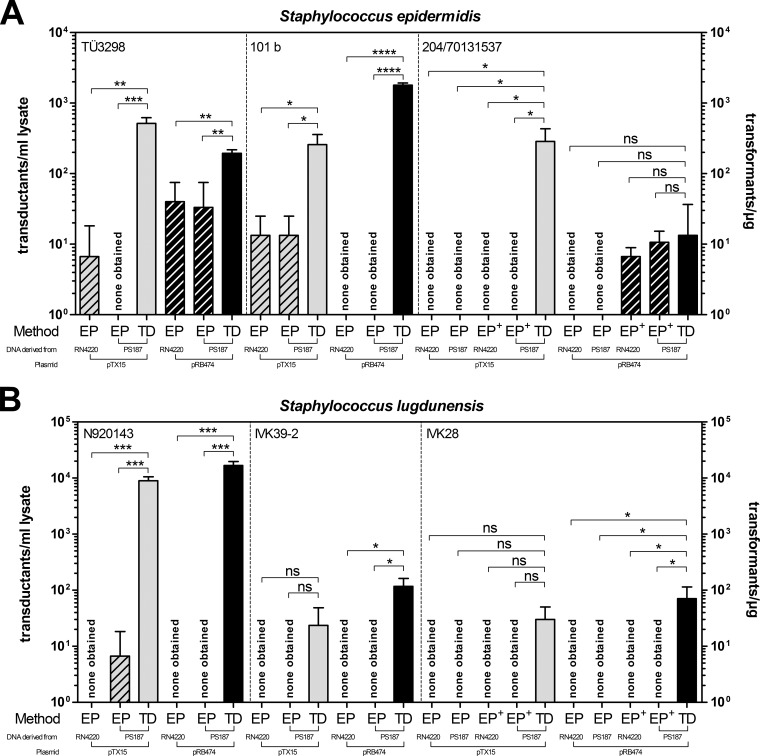
In addition to alterations of WTA structure that diminish phage infection and CRISPR loci that degrade foreign DNA, R-M mechanisms involving distinct DNA methylation patterns represent significant barriers to HGT. The fact that ST395 isolates are capable of exchanging DNA with certain CoNS has led to the assumption that ST395 and CoNS share similar DNA methylation pathways, enabling phage-mediated HGT between donors and recipients with shared WTA structures (17). Therefore, S. aureus PS187 ΔhsdR ΔsauUSI-derived plasmid DNA (or RN4220-derived plasmid DNA, used as a control) might be suitable for electroporation of CoNS. When plasmids pRB474 and pTX15 from one of the two strains were used to electroporate various CoNS isolates, the majority of test strains could hardly be transformed, and some were not even detectably transformable under the electroporation conditions used in this study (Fig. 2A and B). Thus, most CoNS are refractory to electroporation even if plasmid DNA is derived from ST395 clone PS187.
FIG 2.
Benefits of Φ187-mediated plasmid transfer to important CoNS pathogens. Shown is a comparison of plasmid electroporation (EP) (hatched bars) and Φ187-mediated plasmid transfer (transduction [TD]) in various S. epidermidis (A) and S. lugdunensis (B) strains. Purified pTX15 (gray bars) or pRB474 (black bars) plasmids were isolated from S. aureus strain RN4220 or PS187 ΔhsdR ΔsauUSI and used to electroporate electrocompetent CoNS (EP+ indicates application of 2.5 μg DNA). For transduction experiments, PBPP lysates contained ∼1 × 109 PFU/ml. The lysates were titrated on S. aureus PS187 w.t. The values represent transductants per milliliter of phage lysate (for TD experiments; plated on sheep blood-supplemented agar plates) or transformants per microgram of DNA (for EP experiments) and are given as means and SD (n = 3). Statistically significant differences calculated by the unpaired two-tailed Student t test are indicated: ns, not significant (P > 0.05); *, P < 0.01 to < 0.05; **, P < 0.001 to 0.01; ***, P < 0.001; ****, P < 0.0001.
S. aureus phage Φ187 facilitates plasmid transfer to untransformable CoNS via transduction.
As an alternative to electroporation, phage-mediated transduction of DNA can be used to manipulate bacteria. The ability of the ST395-specific S. aureus phage Φ187 to transfer SaPIs from ST395 to CoNS suggested the use of Φ187 as a molecular shuttle for plasmid transfer to CoNS. Therefore, Φ187 was freshly propagated on its cognate host strain S. aureus PS187 wild type and used to infect plasmid-bearing S. aureus PS187 ΔhsdR ΔsauUSI to create PBPP lysates, which were used to infect a variety of CoNS strains. Surprisingly, the novel approach facilitated highly efficient plasmid transfer at frequencies up to ∼104 transductants per ml of PBPP lysate to many CoNS and even to strains usually refractory to electroporation (Fig. 2A and B). Moreover, enrichment of PBPP lysates via ultracentrifugation resulted in both higher phage titers (up to 1 × 1011 PFU/ml) and more efficient plasmid transfer frequencies (see Fig. S1A in the supplemental material). Although plasmid transfer with ultracentrifuged PBPP lysates is recommended, titers of 109 PFU/ml were usually sufficient to accomplish plasmid transfer. Interestingly, the use of BM agar plates containing 5% sheep blood increased the plasmid transfer efficiency (see Fig. S1B in the supplemental material). This effect might be a consequence of supplements other than CaCl2 present in whole blood that facilitate more efficient phage adsorption and infection.
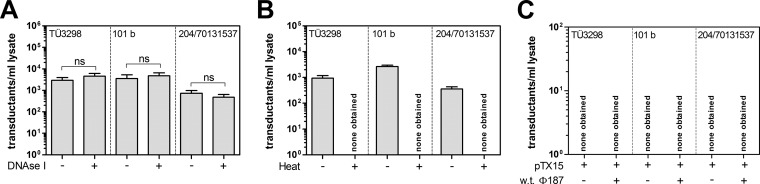
To confirm that plasmid transfer was indeed accomplished via Φ187-mediated transduction and not by a phage-independent mechanism involving, for example, uptake of free DNA (a process that is very rare among staphylococci but has recently been documented in S. aureus under specific conditions [26]), PBPP lysates were treated with DNase I. While DNase I treatment of PBPP lysates did not alter the efficiency of plasmid transfer to different S. epidermidis strains (a test for natural competence), heat inactivation of phage particles in PBPP lysates rendered plasmid transfer undetectable (Fig. 3A and B). Plasmid transfer to CoNS failed even when naked plasmid DNA (pTX15) lacking phage particles or naked DNA was combined with Φ187 particles propagated on PS187 wild type without plasmids (a test for “pseudotransformation” [26]), thereby confirming that plasmid transfer was accomplished via Φ187, which is in line with a transduction process (Fig. 3C). Thus, Φ187 is capable of transducing plasmid DNA from ST395 isolates to CoNS, even those strains that are usually refractory to electroporation.
FIG 3.
Plasmid transfer to CoNS is facilitated by Φ187 via transduction. (A) Analysis of natural competence of selected S. epidermidis isolates. PBPP lysates (containing pTX15) were treated with DNase I (+) or untreated (−) and subsequently used for plasmid transfer to strain TÜ3298, 101 b, or 204/70131537. (B) Effect of heat treatment of PBPP lysates (containing pTX15). PBPP lysates were heat inactivated (+) or not (−), chilled, and subsequently used for plasmid transfer to selected S. epidermidis strains. (C) Analysis of selected S. epidermidis isolates to undergo “pseudotransformation.” Purified pTX15 plasmid was incubated with selected S. epidermidis strains in the presence (+) or in the absence (−) of Φ187 wild-type particles lacking plasmids. The lysates contained ∼1 × 109 PFU/ml. The values represent transductants per milliliter of phage lysate plated on sheep blood-supplemented agar plates and are given as means and SD (n = 3). Statistically significant differences calculated by the unpaired two-tailed Student t test are indicated: ns, not significant.
Clinically relevant CoNS pathogens can be transduced via phage Φ187, facilitating their genetic manipulation.
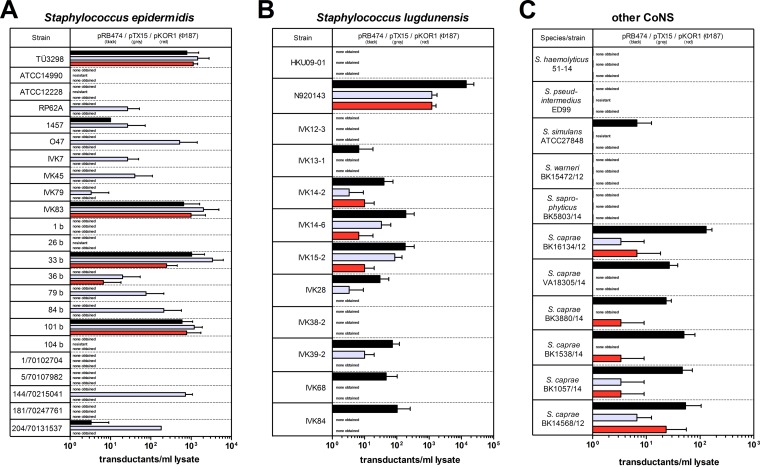
The new approach was tested with a broad panel of CoNS, including S. epidermidis and S. lugdunensis clinical and nasal isolates (details are provided in Table 1). Notably, the majority of S. epidermidis (65.2%) and S. lugdunesis (75.0%) strains could be transduced using various Φ187-based PBPP lysates (Fig. 4A and B). While some S. epidermidis (e.g., TÜ3298) or S. lugdunensis (e.g., N920143) strains underwent highly efficient Φ187-mediated plasmid transfer, some isolates (e.g., S. epidermidis ATCC 14990 or S. lugdunensis HKU09-01) could be transduced only with certain plasmids or were completely resistant, most likely because of genetic barriers inactivating the invading plasmid DNA. However, high-frequency plasmid transfer clearly correlated with strong Φ187 adsorption or, in contrast, failed when no or weak phage adsorption occurred (see Fig. S2A and B in the supplemental material). In agreement with these observations, Φ187 adsorption to clinical Staphylococcus caprae isolates and Staphylococcus simulans correlated with transduceability. In contrast, non-Φ187-binding species, such as Staphylococcus saprophyticus, Staphylococcus warneri, or Staphylococcus haemolyticus, could not be transduced by Φ187, suggesting that similar cell surface receptor properties in the DNA donor and recipient are necessary for Φ187-mediated plasmid transfer to CoNS (Fig. 4C; see Fig. S2C in the supplemental material).
FIG 4.
S. aureus phage Φ187 facilitates plasmid transfer to many untransformable staphylococcal pathogens. Bacteriophage Φ187 propagated on S. aureus donor strain PS187 ΔhsdR ΔsauUSI bearing plasmid pRB474 (black), pTX15 (gray), or pKOR1 (red) was used to transduce a broad panel of S. epidermidis (A) and S. lugdunensis (B) strains and other CoNS (C). The PBPP lysates contained ∼1 × 109 PFU/ml. The values represent transductants per ml of phage lysate plated on sheep blood-supplemented agar plates and are given as means and SD (n = 3). No transductants were observed in controls lacking PBPP lysates, and naturally antibiotic-resistant clones are indicated (resistant).
Finally, a plasmid for targeted mutagenesis was constructed and transduced to clinical and nasal S. epidermidis isolates using this approach. The gene encoding the alternative transcription factor sigma B (sigB) was used as a test gene. Notably, rapid gene deletion was achieved at high efficiency (Table 3). Thus, Φ187 is a suitable tool for transferring plasmids to important clinical CoNS pathogens and represents a novel tool for their genetic manipulation.
TABLE 3.
Genetic engineering of S. epidermidis using phage Φ187-mediated plasmid transfer
| Bacterial strain | Counterselection efficiency (%)a | sigB mutagenesis efficiency (%)b |
|---|---|---|
| S. epidermidis 33b | 100 | 85.7 |
| S. epidermidis IVK83 | 96.4 | 42.8 |
Counterselection efficiency was calculated after anhydrotetracycline selection by dividing the number of chloramphenicol-sensitive clones by the total number of tested isolates.
Mutagenesis efficiency was calculated after PCR analysis by dividing the number of sigB deletion mutants by the total number of tested isolates.
In summary, the new approach represents a major technical advance for plasmid transfer to important CoNS pathogens, allowing their genetic manipulation in the future. Although prior work from 1965 reported interspecies plasmid transfer events between S. aureus and S. epidermidis (15), the phenomenon seems to be limited to a small minority of S. epidermidis isolates and has hardly been used since. In contrast, the majority of important clinical CoNS isolates, including strong biofilm-producing S. epidermidis strains 1457 and RP62A and, equally important, genome-sequenced strains, can be transduced by the approach presented here in a highly efficient and reliable fashion. In addition, the transfer of knockout plasmids via Φ187 will strongly facilitate the genetic manipulation of CoNS in the future. Additionally, Φ187 might be capable of transducing chromosomal markers from ST395 to CoNS in conserved genomic regions shared by ST395 and CoNS to facilitate rapid gene disruption in CoNS. Although some CoNS isolates and species could not be transduced, most likely because of distinct phage receptor properties, lack of plasmid replication in the recipient strains, or other genetic barriers limiting HGT, the technique described here is applicable to most important and clinically relevant CoNS, including S. epidermidis and S. lugdunensis. This approach may prompt research activities to manipulate other important bacterial pathogens in a similar fashion.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karsten Becker and Evgeny Idelevich for clinical S. epidermidis strains.
This work was supported by German Research Council grants TRR34 to A.P. and Ro2413/4-1 to H.R. and by German Center for Infection Research (DZIF) grants to H.R. and A.P.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04190-14.
REFERENCES
- 1.Otto M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol 7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Eiff C, Peters G, Heilmann C. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis 2:677–685. doi: 10.1016/S1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- 3.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M, SENTRY Participants Group . 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis 32(Suppl 2):S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 4.Otto M. 2013. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med 64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 5.Fey PD. 2010. Modality of bacterial growth presents unique targets: how do we treat biofilm-mediated infections? Curr Opin Microbiol 13:610–615. doi: 10.1016/j.mib.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoenfelder SM, Lange C, Eckart M, Hennig S, Kozytska S, Ziebuhr W. 2010. Success through diversity—how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int J Med Microbiol 300:380–386. doi: 10.1016/j.ijmm.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 9.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heilbronner S, Hanses F, Monk IR, Speziale P, Foster TJ. 2013. Sortase A promotes virulence in experimental Staphylococcus lugdunensis endocarditis. Microbiology 159:2141–2152. doi: 10.1099/mic.0.070292-0. [DOI] [PubMed] [Google Scholar]
- 11.Augustin J, Gotz F. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett 54:203–207. [DOI] [PubMed] [Google Scholar]
- 12.Minshew BH, Rosenblum ED. 1972. Transduction of tetracycline resistance in Staphylococcus epidermidis. Antimicrob Agents Chemother 1:508–511. doi: 10.1128/AAC.1.6.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nedelmann M, Sabottke A, Laufs R, Mack D. 1998. Generalized transduction for genetic linkage analysis and transfer of transposon insertions in different Staphylococcus epidermidis strains. Zentralbl Bakteriol 287:85–92. doi: 10.1016/S0934-8840(98)80151-5. [DOI] [PubMed] [Google Scholar]
- 14.Rosendorf LL, Kayser FH. 1974. Transduction and plasmid deoxyribonucleic acid analysis in a multiply antibiotic-resistant strain of Staphylococcus epidermidis. J Bacteriol 120:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto S, Niwa C, Kuwahara S. 1965. Transduction of drug-resistances in Staphylococcus. II. Transduction of chloramphenicol-resistance in both Staphylococcus aureus and Staphylococcus epidermidis by typing phage 80. Jpn J Microbiol 9:15–19. [PubMed] [Google Scholar]
- 16.Endl J, Seidl PH, Fiedler F, Schleifer KH. 1984. Determination of cell wall teichoic acid structure of staphylococci by rapid chemical and serological screening methods. Arch Microbiol 137:272–280. doi: 10.1007/BF00414557. [DOI] [PubMed] [Google Scholar]
- 17.Winstel V, Liang C, Sanchez-Carballo P, Steglich M, Munar M, Broker BM, Penades JR, Nubel U, Holst O, Dandekar T, Peschel A, Xia G. 2013. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun 4:2345. doi: 10.1038/ncomms3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winstel V, Xia G, Peschel A. 2014. Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int J Med Microbiol 304:215–221. doi: 10.1016/j.ijmm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Xia G, Corrigan RM, Winstel V, Goerke C, Grundling A, Peschel A. 2011. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J Bacteriol 193:4006–4009. doi: 10.1128/JB.01412-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winstel V, Sanchez-Carballo P, Holst O, Xia G, Peschel A. 2014. Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. mBio 5:e00869-14. doi: 10.1128/mBio.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruckner R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett 151:1–8. doi: 10.1016/S0378-1097(97)00116-X. [DOI] [PubMed] [Google Scholar]
- 22.Geiger T, Francois P, Liebeke M, Fraunholz M, Goerke C, Krismer B, Schrenzel J, Lalk M, Wolz C. 2012. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog 8:e1003016. doi: 10.1371/journal.ppat.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Peschel A, Ottenwalder B, Gotz F. 1996. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol Lett 137:279–284. doi: 10.1111/j.1574-6968.1996.tb08119.x. [DOI] [PubMed] [Google Scholar]
- 25.Bruckner R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187–192. doi: 10.1016/0378-1119(92)90048-T. [DOI] [PubMed] [Google Scholar]
- 26.Morikawa K, Takemura AJ, Inose Y, Tsai M, Nguyen T, Le T, Ohta T, Msadek T. 2012. Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog 8:e1003003. doi: 10.1371/journal.ppat.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asheshov EA, Jevons MP. 1963. The effect of heat on the ability of a host strain to support the growth of a Staphylococcus phage. J Gen Microbiol 31:97–107. doi: 10.1099/00221287-31-1-97. [DOI] [PubMed] [Google Scholar]
- 28.Allgaier H, Jung G, Werner RG, Schneider U, Zahner H. 1986. Epidermin: sequencing of a heterodetic tetracyclic 21-peptide amide antibiotic. Eur J Biochem 160:9–22. doi: 10.1111/j.1432-1033.1986.tb09933.x. [DOI] [PubMed] [Google Scholar]
- 29.Mack D, Siemssen N, Laufs R. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun 60:2048–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heilmann C, Gerke C, Perdreau-Remington F, Gotz F. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun 64:277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krismer B, Liebeke M, Janek D, Nega M, Rautenberg M, Hornig G, Unger C, Weidenmaier C, Lalk M, Peschel A. 2014. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog 10:e1003862. doi: 10.1371/journal.ppat.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, Wurster S, Scherpe S, Davies AP, Harris LG, Horstkotte MA, Knobloch JK, Ragunath C, Kaplan JB, Mack D. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 33.Tse H, Tsoi HW, Leung SP, Lau SK, Woo PC, Yuen KY. 2010. Complete genome sequence of Staphylococcus lugdunensis strain HKU09-01. J Bacteriol 192:1471–1472. doi: 10.1128/JB.01627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heilbronner S, Holden MT, van Tonder A, Geoghegan JA, Foster TJ, Parkhill J, Bentley SD. 2011. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol Lett 322:60–67. doi: 10.1111/j.1574-6968.2011.02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devriese LA, Vancanneyt M, Baele M, Vaneechoutte M, De Graef E, Snauwaert C, Cleenwerck I, Dawyndt P, Swings J, Decostere A, Haesebrouck F. 2005. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int J Syst Evol Microbiol 55:1569–1573. doi: 10.1099/ijs.0.63413-0. [DOI] [PubMed] [Google Scholar]
- 36.Pantucek R, Doskar J, Ruzickova V, Kasparek P, Oracova E, Kvardova V, Rosypal S. 2004. Identification of bacteriophage types and their carriage in Staphylococcus aureus. Arch Virol 149:1689–1703. doi: 10.1007/s00705-004-0335-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.