Abstract
Clostridium thermocellum produces ethanol as one of its major end products from direct fermentation of cellulosic biomass. Therefore, it is viewed as an attractive model for the production of biofuels via consolidated bioprocessing. However, a better understanding of the metabolic pathways, along with their putative regulation, could lead to improved strategies for increasing the production of ethanol. In the absence of an annotated pyruvate kinase in the genome, alternate means of generating pyruvate have been sought. Previous proteomic and transcriptomic work detected high levels of a malate dehydrogenase and malic enzyme, which may be used as part of a malate shunt for the generation of pyruvate from phosphoenolpyruvate. The purification and characterization of the malate dehydrogenase and malic enzyme are described in order to elucidate their putative roles in malate shunt and their potential role in C. thermocellum metabolism. The malate dehydrogenase catalyzed the reduction of oxaloacetate to malate utilizing NADH or NADPH with a kcat of 45.8 s−1 or 14.9 s−1, respectively, resulting in a 12-fold increase in catalytic efficiency when using NADH over NADPH. The malic enzyme displayed reversible malate decarboxylation activity with a kcat of 520.8 s−1. The malic enzyme used NADP+ as a cofactor along with NH4+ and Mn2+ as activators. Pyrophosphate was found to be a potent inhibitor of malic enzyme activity, with a Ki of 0.036 mM. We propose a putative regulatory mechanism of the malate shunt by pyrophosphate and NH4+ based on the characterization of the malate dehydrogenase and malic enzyme.
INTRODUCTION
Clostridium thermocellum is a Gram-positive, anaerobic thermophile capable of reaching one of the highest growth rates on crystalline cellulose (1, 2). Furthermore, given its native ability to produce ethanol and H2, C. thermocellum is seen as an attractive microorganism for the production of biofuels via consolidated bioprocessing of lignocellulosic biomass. Unfortunately, current yields and production rates of ethanol and/or H2 are low due to branched product pathways (3–5) which redirect carbon and electron flux away from a desired biofuel. These unwanted products include lactate, formate, and/or acetate (6–8), as well as secreted amino acids (9, 10). Thus, redirecting carbon and electron flux away from these secreted products toward either ethanol or H2 may improve the economic viability of biofuels production using C. thermocellum.
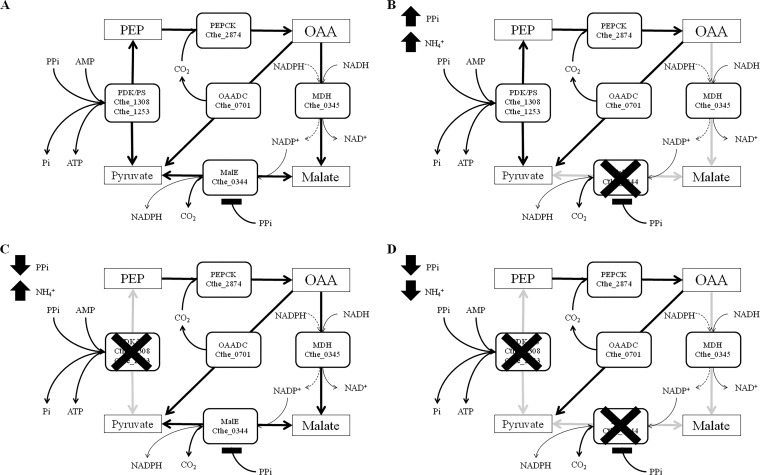
A key pathway node involved in the interconversion of phosphoenolpyruvate (PEP) and pyruvate, which may catalyze either substrate level phosphorylation or transhydrogenation reactions between NADH to NADP+, has been revisited with C. thermocellum (10, 11). The interconversion of PEP and pyruvate may be catalyzed using a number of different putative enzymes. In contrast to most clostridia and other fermentative ethanol and/or H2-producing organisms (7), C. thermocellum ATCC 27405 (GenBank accession number NC_009012.1) does not encode a pyruvate kinase, which catalyzes PEP to pyruvate with the concomitant production of ATP from ADP and Pi. Thus, an alternative means to generate pyruvate during glycolysis must be employed. Instead, C. thermocellum encodes two pyruvate dikinases (Cthe_1308 and Cthe_1253), which can potentially convert PEP to pyruvate and produce ATP from AMP and PPi. However, pyruvate dikinases are believed to play a role in gluconeogenesis, producing PEP, rather than catabolizing it, in many organisms (12, 13). Alternatively, pyruvate production in C. thermocellum could follow an alternative route involving phosphoenolpyruvate carboxykinase (PEPCK) (Cthe_2874), a putative malate dehydrogenase (MDH) (Cthe_0345), and malic enzyme (MalE) (Cthe_0344) (see Fig. 7A). This pathway, referred to as the malate shunt, may be capable of a transhydrogenation reaction, during which electrons are transferred from NADH to NADP+ (11). However, cofactor specificity of both MDH and MalE varies from organism to organism, and thus the specificity of these enzymes must be elucidated to verify the impact they may have on C. thermocellum metabolism.
FIG 7.
Putative pyruvate-producing pathways in C. thermocellum under normal intracellular conditions (A), with high intracellular PPi and NH4+ (B), with low intracellular PPi and high NH4+ (C), and with low PPi and NH4+ (D).
MDHs, which can catalyze the reversible oxidation of malate to oxaloacetate (OAA), play a critical role in many biological processes, such as the tricarboxylic acid cycle, amino acid synthesis, maintenance of reduction and oxidation pools, metabolic stress response, and gluconeogenesis (14–17). They can be divided into two main groups based on cofactor preference: NAD+ dependent (EC 1.1.1.37) and NADP+ dependent (EC 1.1.1.82). Plants have a chloroplastic NADP+-dependent MDH essential for malate shuttling in C3 plants and carbon fixation in C4 plants (18). MDHs have been identified in both dimeric and tetrameric oligomeric states. The tetrameric MDHs, also known as lactate dehydrogenase (LDH)-like MDH, have a quaternary protein structure similar to that of LDH. MDH shares a common ancestry with LDH, rendering the annotation of LDH or MDH based on amino acid sequence unreliable (15). A mutation of a single amino acid in the Escherichia coli MDH can convert its substrate specificity from oxaloacetate to pyruvate, essentially converting the MDH to an LDH (19). This leads to increased uncertainty pertaining to the annotation of LDH or MDH based on amino acid sequence homology and warrants further biochemical characterization for each species of interest.
Like MDH, MalE, which catalyzes the oxidative decarboxylation of malate to pyruvate and CO2, can be classified into three different categories based on the cofactor specificity and the ability to decarboxylate OAA. These include (i) NAD+-dependent, but not OAA-decarboxylating, MalE (EC 1.1.1.39), typically utilized in carbon fixation in plants, (ii) NAD+-dependent OAA-decarboxylating MalE (EC 1.1.1.38), and (iii) NADP+-dependent OAA-decarboxylating MalE (EC 1.1.1.40) (20). While the non-OAA-decarboxylating MalE enzyme is incapable of converting OAA to pyruvate, the two last enzymes can catalyze either malate or OAA to pyruvate, thereby potentially bypassing the malate shunt/transhydrogenase pathway in the case of the latter. Furthermore, cofactor dependence of OAA-decarboxylating malic enzymes can also determine if transhydrogenation from NADH to NADP+ occurs. Thus, characterization of enzyme kinetics and cofactor specificity is critical in validating the proposed transhydrogenation pathway in C. thermocellum.
Recent evidence has indeed supported the idea that PEP is likely converted to pyruvate via the malate shunt in C. thermocellum. Transcriptomic (21) and proteomic (22) studies have shown that Cthe_2874, Cthe_0345, and Cthe_0344 are highly expressed in C. thermocellum, suggesting that they play a key role in pyruvate metabolism. Furthermore, in vitro crude cell extract enzyme activities demonstrated that MDH activity is strictly NADH dependent, whereas MalE activity is strictly NADPH dependent, in C. thermocellum strain DSM 1313 in cell extracts (10). Similar observations were made with C. thermocellum strains AS39 and LQRI, which showed NADH-dependent MDH activity and an ammonium activated NADP+-dependent MalE-like activity in crude cell extracts (11). Subsequent purification of MalE from C. thermocellum AS39 revealed that this NADP+-dependent MalE activity was NH4+ and Mn2+ dependent (23).
To date, however, no one has directly shown which annotated putative genes encoding MDH and MalE are responsible for the assayed enzyme activities in C. thermocellum. While NADH-dependent MDH activity has been demonstrated, current GenBank annotation of the gene Cthe_0345 lists it as an LDH gene belonging to the malate/lactate dehydrogenase Pfam protein domain family database. Furthermore, despite evidence that MalE is NADP+ dependent via enzyme activities in C. thermocellum, the one annotated is indicated as an NAD+-dependent OAA-decarboxylating MalE (EC 1.1.1.38). Furthermore, there have been no studies demonstrating MalE OAA-decarboxylating activity in C. thermocellum. We have therefore cloned, purified, and characterized the putative MDH (Cthe_0345) and MalE (Cthe_0344) to determine cofactor specificity and enzyme kinetics to validate their presence and elucidate their possible roles in the malate shunt pathway and their potential role in C. thermocellum biofuel metabolism.
MATERIALS AND METHODS
Strains and reagents.
Clostridium thermocellum ATCC 27405 genomic DNA was purchased from the American Type Culture Collection. E. coli DH5α was used as the host for plasmid construction and screening. E. coli BL21(DE3) was used as the expression strain for recombinant protein expression. The plasmids used for protein expression were pRSET-A (Life Technologies Corp.) and pET-28a(+) (Millipore Ltd.). All antibiotics used were purchased from Sigma-Aldrich. All PCRs were done using iProof high-fidelity DNA polymerase (Bio-Rad Laboratories Ltd.). PCR purification was done using a QIAquick PCR purification kit (Qiagen Inc.) All restriction enzymes used were purchased from New England BioLabs. Plasmids were extracted using a Qiagen plasmid minikit (Qiagen Inc.). Recombinant proteins were purified using a HiTrap chelating column (GE Healthcare Bio-Sciences Corp.). Malic acid, OAA, NADP+, NADPH, ATP, ADP, AMP, pyrophosphate (PPi), and all buffers used were from Sigma-Aldrich.
Primer and plasmid construction.
Primers for MDH and MalE were designed using the corresponding gene sequence from Integrated Microbial Genomics (24). MDH (Cthe_0345) was amplified by PCR using the sense primer 5′-ATATGGATCCATGGAAATGGTAAAAAGTAGGTC-3′ and the antisense primer 5′-ATATGAATTCTTATAAATTCTTAACTTCGTTCAATAC-3′ (restriction sites are underlined). The PCR product was purified and digested using BamHI and EcoRI restriction enzymes. The digested product was then ligated into pRSET-A by T4 DNA ligase (PROMEGA) to create the recombinant plasmid pAHCT345. MalE (Cthe_0344) was amplified by PCR using the sense primer 5′-CATATGGATTACAGAAAGAATCACTAAG-3′ and the antisense primer 5′-CGGCCGTTATATTCTTGCAACTCCGGTTTTTC-3′. The PCR was purified and digested using NdeI and EagI. The digested product was then ligated into pET-28a(+) to create the recombinant plasmid pKHCT344. Plasmid constructs were verified via sequencing (Macrogen Corp.).
Overexpression and recombinant protein isolation.
E. coli BL21(DE3) containing either the pAHCT345 or pKHCT344 plasmid was cultured overnight in LB medium containing ampicillin (100 μg/ml) for pAHCT345 or kanamycin (30 μg/ml) for pKHCT344 at 37°C. Overnight cultures were reinoculated into fresh LB medium containing the proper antibiotic and grown aerobically at 37°C until an optical density at 600 nm (OD600) of 0.5 to 0.7 was reached. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was then added to a final concentration of 1 mM. The cultures were grown for an additional 12 h at 30°C. Cells were harvested and resuspended in buffer containing 20 mM NaH2PO4 (pH 7.4), 0.5 M NaCl, and 20 mM imidazole. Cells were lysed at 37°C after 15 min of incubation with 1 mg/ml of lysozyme, 1% Triton X-100, 5 μg/ml of DNase, 5 μg/ml of RNase, and 5 mM MgCl2 (25). Cell lysates were centrifuged at 4,500 rpm for 30 min at 4°C. Supernatants were filtered through a 0.22-μm filter prior to being loaded onto an Ni2+ HiTrap metal affinity column and purified by following the manufacturer's instructions (GE Healthcare Bio-Sciences Corp.). The recombinant His6-tagged proteins were eluted using a stepwise imidazole gradient. Imidazole and NaCl were removed from the protein samples using a HiTrap desalting column eluting with 20 mM NaH2PO4 (pH 7.4). Enzyme purity was verified by SDS-PAGE with a 12% resolving gel and a 5% stacking gel. Visualization of the proteins was done by staining the gels with Coomassie brilliant blue R-250. Protein concentration was measured by the Bradford assay using bovine serum albumin as a protein standard (26).
Enzyme assays.
All enzyme activities were measured in a 300-μl well with a total reaction volume of 200 μl up to 50°C. MDH activity was measured in a standard reaction mixture containing 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7.0), 10 mM dithiothreitol (DTT), 2 mM OAA, and 0.2 mM NADH. MalE activity was measured in a standard reaction mixture containing 100 mM MOPS (pH 7.0), 10 mM DTT, 2 mM malate, 0.2 mM NADP+, 5 mM MnCl2, and 20 mM NH4Cl (27). LDH activity was measured in a standard reaction mixture containing 100 mM MOPS (pH 7.0), 10 mM DTT, 10 mM pyruvate, 0.2 mM NADH, and 0.005 mM fructose-1-6-bisphosphate (11). Changes in cofactor (NADH or NADP+) concentration were monitored at 340 nm using a BioTek Synergy 4 plate reader with a molar extinction coefficient of 6,220 M−1 cm−1 (adjusted for light path during calculations).
Kinetic properties.
The kinetic properties for MDH and MalE were determined by varying the substrate or cofactor concentration while keeping the concentration of all other constituents at saturating levels at 25°C and 50°C for the MDH and 50°C for the MalE and at pH 7.0. Inhibition assays were performed using standard reaction conditions with various inhibitor concentrations at 50°C. All kinetic parameters were determined by fitting the data to the Michaelis-Menten equation using Sigma-Plot 11.0 (Systat Software Inc.).
Effects of pH and temperature.
Thermostability was determined by incubating aliquots of recombinant enzyme under the standard reaction conditions, excluding NADH or NADP+. The aliquots were cooled on ice and tested for residual activity at 50°C using the standard conditions. The optimal pH was measured in the following buffers: cacodylic acid (pH 5.6 to 6.5), MOPS (pH 6.5 to 7.5), and Tris-HCl (pH 7.5 to 8.6).
Phylogenetic analysis.
The evolutionary history of both Cthe_0345 (MDH) and Cthe_0344 (MalE) were inferred by using the maximum likelihood method based on the Tamura-Nei model (28). The trees with the highest log likelihood (−29,200.6535 [MDH] and −22,585.0304 [MalE]) are shown. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with superior log likelihood value. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The analyses involved 38 (MDH) and 31 (MalE) nucleotide sequences, respectively. Codon positions included were 1st plus 2nd plus 3rd plus noncoding. There were a total of 796 (MDH) and 849 (MalE) positions in the final data sets. Evolutionary analyses were conducted in MEGA5 (29).
RESULTS
The monomeric molecular masses of the cloned and purified His-tagged MalE and MDH were approximately 48 kDa and 40 kDa, respectively, corresponding to the predicted molecular masses of both proteins containing the 6× His tag. Both enzymes were found to be stable, retaining over 90% activity at 4°C, when stored in desalted elution buffer for a month. However, when the enzymes where stored in the presence of 0.02% or 0.04% sodium azide at 4°C, the activity was reduced by 50% over the same period. A 30% reduction in activity was observed when the enzymes were stored in the presence of 15% glycerol at −20°C for a month.
Effects of pH and temperature on malate dehydrogenase.
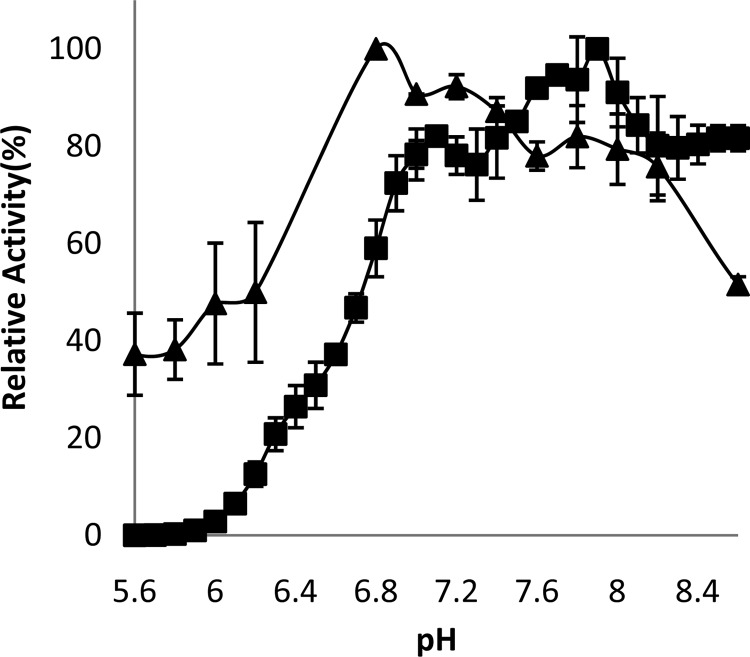
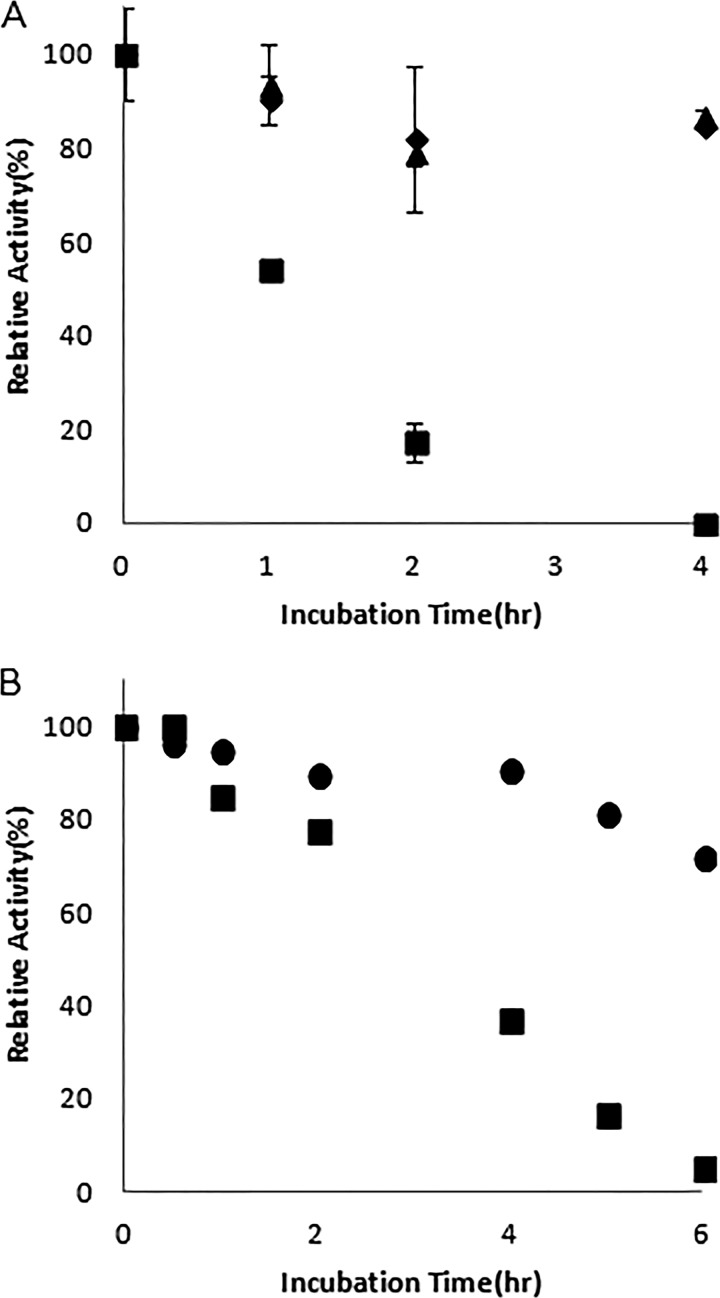
The optimal pH for OAA reduction by MDH was found to be 6.8 when using MOPS as a buffer (Fig. 1). Although MDH-dependent oxidation of malate has been shown in other organisms, typically at a higher pH optimum than that of OAA reduction (30, 31), recombinant C. thermocellum MDH did not oxidize malate in the presence of either NAD+ or NADP+ as a cofactor within the pH range tested (pH 5.6 to 10.5). The MDH was stable at 37°C for 10 h, with a 10% reduction in activity. However, after incubation at 60°C for 1 h, 54% of the initial activity of the purified enzyme was detectable, with none of the initial activity detectable after 4 h (Fig. 2A).
FIG 1.
The relative activity of recombinant Cthe_0344 (MalE) (■) and Cthe_0345 (MDH) (▲) at various pH values with standard assays conditions at 50°C (MalE) and 25°C (MDH).
FIG 2.
Thermostability profile of Cthe_0345(MDH) at 4°C (▲), 25°C (◆), and 60°C (■) (A) and Cthe_0344 (MalE) at 37°C (●) and 60°C (■) (B) under standard assay conditions at pH 7.0 with 20 mM NH4+and 5 mM Mn2+.
Malate dehydrogenase kinetics.
Kinetic parameters for the recombinant MDH are summarized in Table 1. The Km for OAA at 25°C was 0.20 mM using NADH as a cofactor. The kcat for oxaloacetate reduction was 45.8 s−1 with NADH as a cofactor. When NADPH was used as a cofactor, the kcat was reduced to 14.9 s−1 and the Km for oxaloacetate increased to 0.77 mM, resulting in a 12-fold decrease in the catalytic efficiency (kcat/Km). High concentrations of oxaloacetate inhibited the activity of the MDH, with a 30% reduction in activity at 7 mM and a 99% reduction in activity at 25 mM. The addition of ATP, ADP, AMP, or PPi had no observable effect on MDH activity. MDH was also not affected by the addition of fructose 1,6-bisphosphate. No detectable LDH activity was found under any of the conditions tested.
TABLE 1.
Catalytic values for MDHs from Clostridium thermocellum (25°C and 50°C), Vulcanithermus medioatlanticus, Paracoccus denitrificans, Beggiatoa leptomitiformis, Streptomyces coelicolor, Corynebacterium glutamicum, and Streptomyces aureofaciens
| Organism | kcat (s−1) |
Km (mM) |
Catalytic efficiency (mM−1 s−1) | Reference | |
|---|---|---|---|---|---|
| OAA | NADH | ||||
| C. thermocellum (25°C) | 45.82 | 0.20 | NDa | 230.25 | This paper |
| C. thermocellum (50°C) | 389.43 | 0.44 | 0.164 | 879.08 | This paper |
| V. medioatlanticus | 14.72 | 0.048 | 0.0014 | 306.76 | 30 |
| P. denitrificans | 184 | ND | ND | ND | 31 |
| B. leptomitiformis | 60.51 | 0.02 | 0.017 | 3,025.3 | 35 |
| S. coelicolor | 1,870 | 0.19 | 0.083 | 9,894 | 14 |
| C. glutamicum | 510 | 0.057 | 0.041 | 8,947 | 33 |
| S. aureofaciens | ND | 0.1 | 0.085 | ND | 34 |
ND, not determined.
Effects of pH and temperature on malic enzyme activity.
The optimal pH for malate decarboxylation for the recombinant MalE was found to be 7.9, with a plateau of high relative activity between pH 7.1 and 8.5 (Fig. 1). The MalE was stable at 37°C for 4 h, retaining 90.5% of its initial activity. Incubation at 60°C for 4 h led to a 63% reduction in activity (Fig. 2B).
Malic enzyme kinetics.
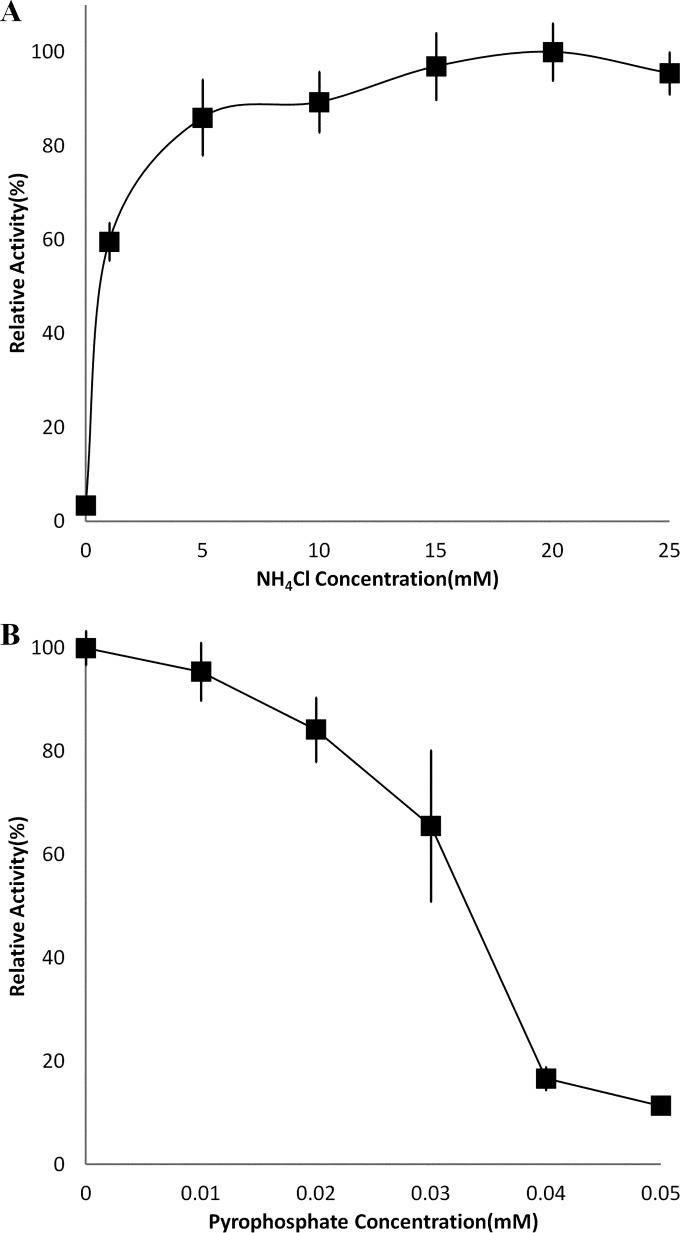
Kinetic parameters for the recombinant MalE are summarized in Table 2. The Km for malate was 0.20 mM using NADP+ as a cofactor, with a kcat of 520.8 s−1 at a temperature of 50°C. No malate decarboxylation activity was detected when NAD+ was used in place of NADP+ as a cofactor. The recombinant MalE did not decarboxylate OAA. Pyruvate carboxylation activity was measurable in the presence of NADPH, with a Km for pyruvate of 0.438 mM and a kcat 71.83 s−1, indicating that this enzyme is reversible. The addition of NH4+ and Mg2+ or Mn2+ increased the activity of the MalE, with Mn2+ being the preferred divalent cation. The activation effect of NH4+ saturated at approximately 15 mM (Fig. 3A). The addition of ATP or AMP resulted in small decreases in activity, 15% and 7%, respectively, at a 1 mM concentration. The addition of PPi, however, led to a sharp decrease in MalE activity, with a calculated Ki of 0.036 mM at 50°C with saturating NH4+ and Mn2+ (Fig. 3B). The addition of PPi had no effect on the Km for malate but decreased the Vmax fairly sharply.
TABLE 2.
Catalytic values for MalEs from Clostridium thermocellum, Streptococcus bovis, Escherichia coli, and Sulfolobus solfataricus
| MalE source | Protein form | kcat (s−1) |
Km (mM) |
Catalytic efficiency (mM−1 s−1) | Reference | |
|---|---|---|---|---|---|---|
| Malate | NADP | |||||
| C. thermocellum | Tetramer | 520.8 | 0.20 | 0.035 | 2,622.36 | This paper |
| S. bovis | Dimer | 90.7 | 0.63 | NDa | 143.97 | 45 |
| E. coli (SfcA) | Tetramer | NAD: 82.7 | 0.66 | 1 | NAD: 125.30 | 20 |
| NADP: 12.4 | NADP: 18.79 | |||||
| E. coli (MaeB) | Octamer | NAD: ND | 3.41 | 0.0415 | NADP: 19.53 | 20 |
| NADP: 66.6 | ||||||
| S. solfataricus | Dimer | 31.5 | 0.018 | 0.003 | 1,750.00 | 44 |
ND, not determined.
FIG 3.
Relative activities of recombinant Cthe_0344 (MalE) at pH 7.0 in the presence of various concentrations of NH4Cl with 5 mM Mn2+ (A) and various concentrations of PPi with 20 mM NH4+ and 5 mM Mn2+ (B).
Phylogenetic analysis.
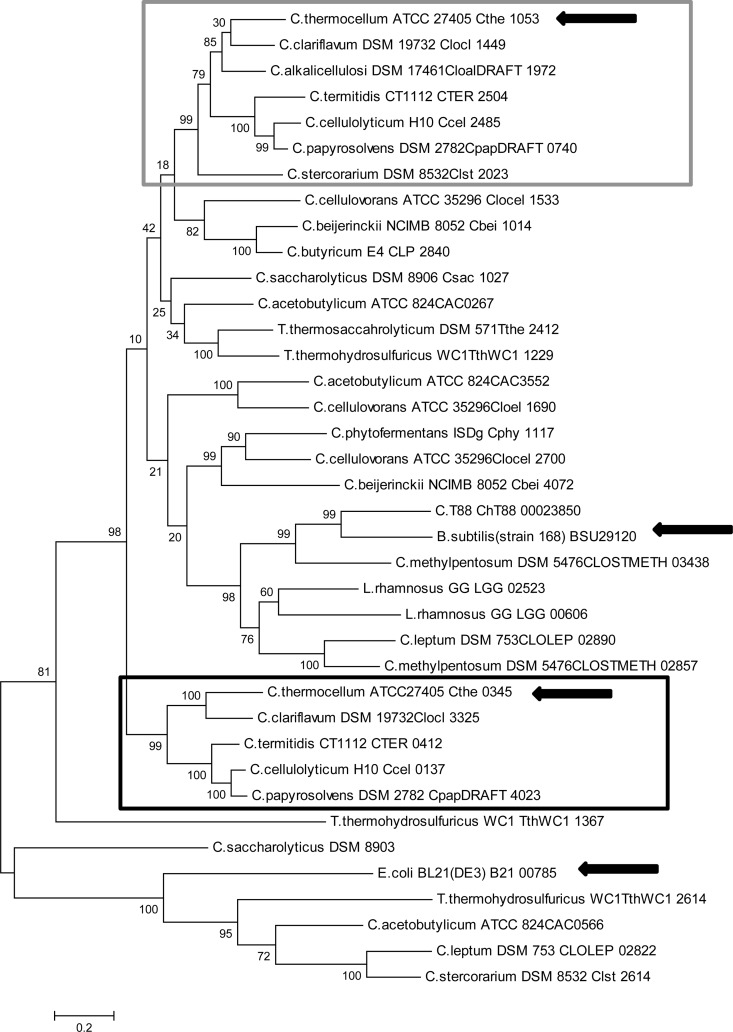
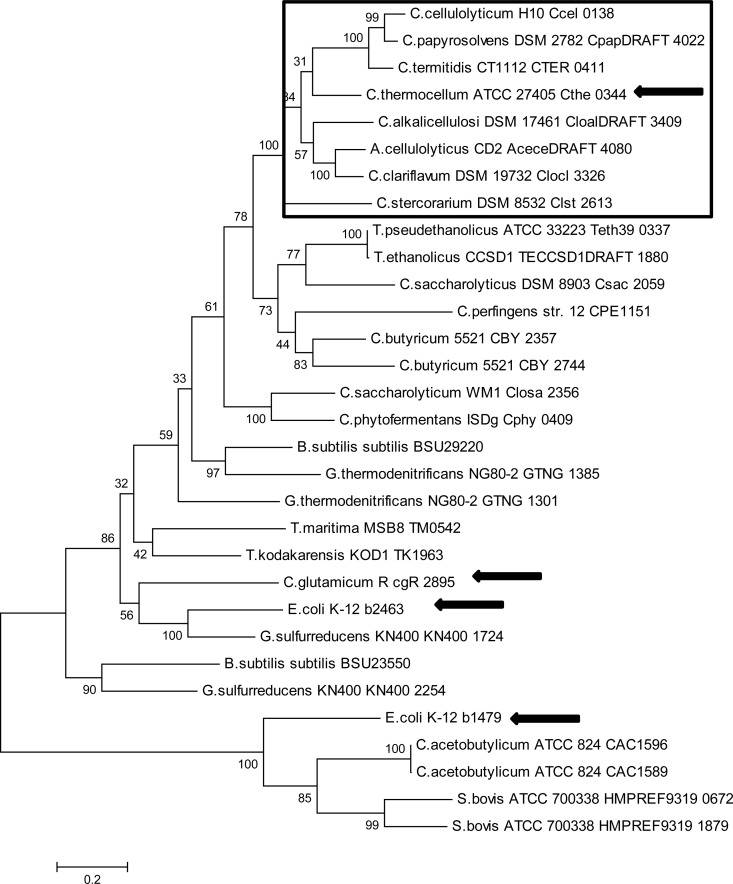
Phylogenetic analysis of the putative MDHs and LDHs from various clostridia showed that while they appear to be derived from a common ancestor, the MDH and LDH from C. thermocellum were separated into different groups (Fig. 4). The grouping around Cthe_0345 (MDH) contains MDHs with the characteristic of having a neighboring malic enzyme gene. The malic enzymes from these organisms have also been found to group with Cthe_0344 (Fig. 5). The alignment of Cthe_0345 (MDH) with the E. coli K-12 MG1655 MDH showed that at position 86, based on Cthe_0345 (MDH) amino acid position, both E. coli and Cthe_0345 (MDH) contained an arginine. However, in positions 18, 90, 212, and 216 the amino acids differed, with Cthe_0345 (MDH) having valine, glutamic acid, aspartic acid, and isoleucine rather than isoleucine, methionine, glycine, and valine in E. coli (Fig. 6).
FIG 4.
Molecular phylogenetic analysis of Cthe_0345 (MDH) by the maximum likelihood method. The gray box indicates a grouping of putative LDH coding genes. The black box indicates a grouping of MDH genes which have a neighboring MalE gene. Arrows indicate sequences for which corresponding enzymes have been purified and characterized. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
FIG 5.
Molecular phylogenetic analysis of Cthe_0344 (MalE) by the maximum likelihood method. The box indicates the group of MalE with a neighboring MDH gene. Arrows indicate sequences for which corresponding enzymes have been purified and characterized. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
FIG 6.
Amino acid alignment of Cthe_0345 (MDH) and Cthe_1053 (LDH) from C. thermocellum and b3236 (MDH) from E. coli K-12 MG1655. Shaded areas indicate positions 18, 86, 90, 212, and 216 based on the amino acid positions of Cthe_0345 (MDH).
DISCUSSION
The conversion of PEP to pyruvate can be viewed as an essential step in C. thermocellum metabolism. However, the lack of pyruvate kinase forces the use of an alternate means of pyruvate production. It has been suggested that pyruvate production is done through a PEPCK, MDH, and MalE or malate shunt pathway, resulting in the transfer of electrons between NADH and NADP+ and the generation of GTP and pyruvate (10, 21, 22). Enzyme assays with cell extracts have showed MDH activity along with NH4+-activated MalE activity (10, 11). However, to date, no genes had been proven to encode MDH and MalE. We have therefore cloned, purified, and characterized the putative MDH (Cthe_0345) and MalE (Cthe_0344) to determine cofactor specificity and enzyme kinetics to validate their presence and elucidate their roles in the malate shunt pathway and their potential role in C. thermocellum biofuel metabolism.
The optimal pH for the MDH-dependent OAA reduction was 6.8, which agrees with the reported values from many thermophilic MDHs (30). MDH-dependent oxidation of malate is generally found to be optimal at a pH higher than that of OAA reduction. However, C. thermocellum MDH did not have any detectable malate oxidation activity regardless of the pH (5.6 to 10.5) or any conditions used during the assays. This seems to indicate that the MDH from C. thermocellum greatly favors oxaloacetate reduction under physiological pH and under all tested conditions. The MDH from C. thermocellum was found to be thermostable, retaining over 50% activity after a 1-h incubation at 60°C. This is higher than what has been observed with the hyperthermophilic Bacillus candolyticus, which lost 50% of its activity after a 1-min incubation at 59°C, but similar to the values for Vulcanithermus medioatlanticus, which retained over 75% activity after 15 min of incubation at 60°C (30, 32). The Km for OAA was significantly higher than for many characterized MDHs, such as the ones from V. medioatlanticus (0.048 mM), Corynebacterium glutamicum (0.057 mM), and Streptomyces aureofaciens (0.1 mM) (30, 33, 34). The kcat for OAA reduction was similar to previously published values for V. medioatlanticus (14.72 s−1) and Beggiatoa leptomitiformis (60.51 s−1) but significantly lower than the published kcat for Phytophthora infestans (3,496.2 s−1) (30, 35–37). The MDH from C. thermocellum preferred the utilization of NADH over NADPH, with a 12-fold reduction in the catalytic efficiency (kcat/Km) with NADPH as a cofactor. This is similar to what was observed in Streptomyces avermitilis, C. glutamicum, and Thermus thermophilus (33, 38, 39).
The monomeric molecular mass of the MalE is much smaller than for most Gram-negative and eukaryotic MalEs but is similar to those for both Gram-positive and archaeal MalEs (20, 40–44). The MalE consists of a 170-kDa tetramer consisting of four 40-kDa monomers (23). The optimal pH was found to be rather high, at 7.9. However, high levels of activity were found between pH 7.1 and 8.5. These values agreed with previously reported values for C. thermocellum (23) and are similar to those for various MalEs from other Gram-positive organisms (40, 45). The Km for malate from the C. thermocellum MalE was found to be lower than for the MalE from Streptococcus bovis (0.63 mM) and E. coli (0.66 mM) but significantly higher than the reported value for the MalE from Sulfolobus solfataricus (0.018 mM) (20, 44, 45). The MalE from C. thermocellum differed from previously reported MalEs in that it was not inhibited to a significant extent by the presence of ATP and AMP, whereas in S. bovis, the addition of 1 mM ATP and 1 mM ADP reduced the MalE activity by 90% and 49%, respectively (45). The recombinant C. thermocellum MalE was inhibited in the presence of relatively low concentrations of PPi. PPi did not seem to affect the binding affinity of malate or NADP+. However, the addition of PPi affected the velocity of the enzyme, leading to a lower Vmax value. Therefore, this indicated that PPi is a noncompetitive inhibitor of MalE. In Trypanosoma cruzi, high levels of PPi induce a shift in phosphoenolpyruvate (PEP) utilization from pyruvate kinase and PEP carboxykinase to pyruvate dikinase (PPDK) (46). In C. thermocellum, the levels of PPDK and MalE were found to be high in both the transcriptome and proteome during growth (21, 22, 47). Therefore, C. thermocellum could utilize intracellular PPi concentration as a signal for directing carbon flow from PEP. Both Burton and Martin (47) and Rydzak et al. (22) have suggested an important role for PPi in exponential phase in this organism. In Caldicellulosiruptor saccharolyticus (48), another cellulolytic thermophilic relative of the clostridia, high levels of PPi have been observed during exponential growth. Increased concentrations of PPi would inhibit the MalE, leading to carbon flowing through the PPDK in order to produce pyruvate. However, deletion of the PPDK gene in C. thermocellum caused no decrease in growth rate or final culture density (49). Together, these findings indicate that C. thermocellum utilizes a robust redundant system of pyruvate generation that can easily adapt to intracellular conditions (Fig. 7). At high concentrations of PPi, the carbon flow would proceed through the PPDK or PEPCK and a membrane-associated OAA decarboxylase complex (OAADC) (Cthe_0699-0701) (Fig. 7B). Once PPi concentrations fall, then the carbon flow would be redirected through the malate shunt and/or the OAADC (Fig. 7C). However, under conditions of both low PPi and low NH4, the carbon would be forced through the OAADC solely for pyruvate generation (Fig. 7D).
The phylogenetic analysis of the Cthe_0345 protein seemed to indicate that some MDHs can be distinguished from LDHs based on their phylogenetic grouping despite the ambiguity in the annotation of MDH and LDH (Fig. 4). Therefore, the presence of a neighboring MalE gene next to a putative MDH or LDH gene would seem to indicate that the MDH or LDH gene is likely an MDH gene. However, the phylogenetic analysis also showed that most MDHs and LDHs are indistinguishable from one another from a phylogenetic point of view. The MalE phylogenetic analysis showed a distinct group of MalEs which have neighboring MDHs, of which the MalE from C. thermocellum is the first representative from this group to be characterized. The substrate specificity of the MDH in E. coli can be modified by a single amino acid change, arginine to glutamine, at position 81 (50). Furthermore, the substrate specificity of the E. coli MDH can be completely shifted to pyruvate with a total of 5 amino acid substitutions (19). Interestingly, when the amino acid sequence of Cthe_0345 (MDH) was compared with that of the E. coli MDH, the amino acids at positions 18, 90, and 216 (based on Cthe_0345 amino acid positions) were found to be valine, glutamic acid, and isoleucine rather than isoleucine, methionine, and valine in E. coli MDH. The amino acids at these positions in Cthe_0345 (MDH) are identical to the amino acid in the same positions for the Cthe_1053 (LDH) and the substitutions made by Yin and Kirsch (19) which shifted substrate specificity from OAA to pyruvate. However, Cthe_0345 (MDH) and Cthe_1053 (LDH) differ at positions 86 and 212. At positions 86, Cthe_0345 (MDH) contains the expected arginine based on the E. coli MDH, whereas Cthe_1053 (LDH) contains a glutamine. Therefore, similar to the case with E. coli, the substrate specificity of the Cthe_0345 (MDH) may be governed by the presence of either an arginine (MDH) or glutamine (LDH) at position 86. However, only 26 of the 38 sequences use in the phylogenetic tree (Fig. 4) had an arginine or glutamine at position 86 when aligned with Cthe_0345 (MDH) (data not shown). While the amino acid at position 86 can hint at the associated activity, it cannot be used reliably to distinguish an MDH from a LDH since not all aligned MDHs and LDHs contain arginine or glutamine at position 86.
Conclusion.
The characterization of the biochemical properties of the MDH and MalE from C. thermocellum has provided insight into their possible catabolic role in this organism and possibly other closely related clostridia. The lack of malate oxidation activity by the MDH under the experimental conditions would indicate that the MDH greatly favors oxaloacetate reduction, therefore primarily production of malate, and has a higher affinity for NADH rather than NADPH. It has been suggested that the activation of MalE by NH4+ is used by C. thermocellum as a signal for the production of cofactors required for biosynthesis (23). Likewise, fluctuating levels of PPi could also possibly be utilized as a signal for C. thermocellum to increase the production of biosynthetic cofactors GTP and NADPH by utilization of the PEPCK and the malate shunt pathway over PPDK. The regulation of the MalE by both NH4+ and PPi indicates that high-energy intermediates and NH4+ may play major role in the regulation of carbon flux through competing pyruvate-producing pathways. Hence, further metabolomic analysis of C. thermocellum would help determine the role of the metabolome as a possible regulator of carbon and electron flux through competing pathways. The combination of biochemical data with metabolomic and proteomic analysis could lead to better understanding of carbon and electron flux regulation in C. thermocellum, leading to the determination of prime candidate genes for genetic engineering.
ACKNOWLEDGMENT
This research was funded through the Genome Canada-funded Microbial Genomics for Biofuels and Co-products Biorefineries project (to R.S. and D.B.L.).
REFERENCES
- 1.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demain AL, Newcomb M, Wu JHD. 2005. Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev 69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rydzak T, Levin DB, Cicek N, Sparling R. 2009. Growth phase-dependent enzyme profile of pyruvate catabolism and end-product formation in Clostridium thermocellum ATCC 27405. J Biotechnol 140:169–175. doi: 10.1016/j.jbiotec.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Rydzak T, Levin DB, Cicek N, Sparling R. 2011. End-product induced metabolic shifts in Clostridium thermocellum ATCC 27405. Appl Microbiol Biotechnol 92:199–209. doi: 10.1007/s00253-011-3511-0. [DOI] [PubMed] [Google Scholar]
- 5.Carere CR, Kalia V, Sparling R, Cicek N, Levin DB. 2008. Pyruvate catabolism and hydrogen synthesis pathway genes of Clostridium thermocellum ATCC 27405. Indian J Microbiol 48:252–266. doi: 10.1007/s12088-008-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Islam R, Cicek N, Sparling R, Levin D. 2006. Effect of substrate loading on hydrogen production during anaerobic fermentation by Clostridium thermocellum 27405. Appl Microbiol Biotechnol 72:576–583. doi: 10.1007/s00253-006-0316-7. [DOI] [PubMed] [Google Scholar]
- 7.Carere CR, Rydzak T, Verbeke TJ, Cicek N, Levin DB, Sparling R. 2012. Linking genome content to biofuel production yields: a meta-analysis of major catabolic pathways among select H2 and ethanol-producing bacteria. BMC Microbiol 12:295. doi: 10.1186/1471-2180-12-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynd L, Grethlein H, Wolkin R. 1989. Fermentation of cellulosic substrates in batch and continuous culture by Clostridium thermocellum. Appl Environ Microbiol 55:3131–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis LD, Holwerda EK, Hogsett D, Rogers S, Shao X, Tschaplinski T, Thorne P, Lynd LR. 2012. Closing the carbon balance for fermentation by Clostridium thermocellum (ATCC 27405). Bioresour Technol 103:293–299. doi: 10.1016/j.biortech.2011.09.128. [DOI] [PubMed] [Google Scholar]
- 10.Deng Y, Olson DG, Zhou J, Herring CD, Joe Shaw A, Lynd LR. 2013. Redirecting carbon flux through exogenous pyruvate kinase to achieve high ethanol yields in Clostridium thermocellum. Metab Eng 15:151–158. doi: 10.1016/j.ymben.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Lamed R, Zeikus JG. 1980. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol 144:569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchins A, Holden J, Adams M. 2001. Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol 183:709–715. doi: 10.1128/JB.183.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao Y, Patnaik R, Roof W. 1993. Control of gluconeogenic growth by pps and pck in Escherichia coli. J Bacteriol 175:6939–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Y-D, Cao Z-Y, Wang Z-D, Chen L-L, Zhu Y-M, Zhu G-P. 2010. Identification and biochemical characterization of a thermostable malate dehydrogenase from the mesophile Streptomyces coelicolor A3(2). Biosci Biotechnol Biochem 74:2194–2201. doi: 10.1271/bbb.100357. [DOI] [PubMed] [Google Scholar]
- 15.Goward C, Nicholls D. 1994. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci 3:1883–1888. doi: 10.1002/pro.5560031027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong Y, Somero GN. 2009. Temperature adaptation of cytosolic malate dehydrogenases of limpets (genus Lottia): differences in stability and function due to minor changes in sequence correlate with biogeographic and vertical distributions. J Exp Biol 212:169–177. doi: 10.1242/jeb.024505. [DOI] [PubMed] [Google Scholar]
- 17.Zaitseva J, Meneely KM, Lamb AL. 2009. Structure of Escherichia coli malate dehydrogenase at 1.45 Å resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun 65:866–869. doi: 10.1107/S1744309109032217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheen J. 1999. C4 gene expression. Annu Rev Plant Physiol Plant Mol Biol 50:187–217. doi: 10.1146/annurev.arplant.50.1.187. [DOI] [PubMed] [Google Scholar]
- 19.Yin Y, Kirsch JF. 2007. Identification of functional paralog shift mutations: conversion of Escherichia coli malate dehydrogenase to a lactate dehydrogenase. Proc Natl Acad Sci U S A 104:17353–17357. doi: 10.1073/pnas.0708265104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bologna FP, Andreo CS, Drincovich MF. 2007. Escherichia coli malic enzymes: two isoforms with substantial differences in kinetic properties, metabolic regulation, and structure. J Bacteriol 189:5937–5946. doi: 10.1128/JB.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raman B, McKeown CK, Rodriguez M, Brown SD, Mielenz JR. 2011. Transcriptomic analysis of Clostridium thermocellum ATCC 27405 cellulose fermentation. BMC Microbiol 11:134. doi: 10.1186/1471-2180-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rydzak T, McQueen PD, Krokhin OV, Spicer V, Ezzati P, Dwivedi RC, Shamshurin D, Levin DB, Wilkins JA, Sparling R. 2012. Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol 12:214. doi: 10.1186/1471-2180-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamed R, Zeikus J. 1981. Thermostable, ammonium-activated malic enzyme of Clostridium thermocellum. Biochim Biophys Acta 660:251–255. doi: 10.1016/0005-2744(81)90167-4. [DOI] [PubMed] [Google Scholar]
- 24.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044AC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniatis T, Sambrook J, Fritsch EF. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 26.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 27.Issakidis E, Miginiac-Maslow M. 1992. Site-directed mutagenesis reveals the involvement of an additional thioredoxin-dependent regulatory site in the activation of recombinant sorghum leaf NADP-malate. J Biol Chem 267:21577–21583. [PubMed] [Google Scholar]
- 28.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the crontrol region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eprintsev AT, Falaleeva MI, Parfyonova NV. 2005. Malate dehydrogenase from the thermophilic bacterium Vulcanithermus medioatlanticus. Biochemistry 70:1027–1030. doi: 10.1007/s10541-005-0220-2. [DOI] [PubMed] [Google Scholar]
- 31.Janiczek O, Kovdf J, Glatz Z. 1993. Purification and properties of malate dehydrogenase from Paracoccus denitrificans. Prep Biochem 23:285–301. doi: 10.1080/10826069308544557. [DOI] [PubMed] [Google Scholar]
- 32.Kristjansson H, Ponnamperuma C. 1980. Purification and properties of malate dehydrogenase from the extreme thermophile Bacillus caldolyticus. Orig Life 10:185–192. doi: 10.1007/BF00928668. [DOI] [PubMed] [Google Scholar]
- 33.Genda T, Nakamatsu T, Ozak H. 2003. Purification and characterization of malate dehydrogenase from Corynebacterium glutamicum. J Biosci Bioeng 95:562–566. doi: 10.1016/S1389-1723(03)80162-7. [DOI] [PubMed] [Google Scholar]
- 34.Mikulásová D, Kollárová M, Miginiac-Maslow M, Decottignies P, Jacquot JP, Kutejová E, Mernik N, Egyudová I, Musrati R, Horecká T. 1998. Purification and characterization of the malate dehydrogenase from Streptomyces aureofaciens. FEMS Microbiol Lett 159:299–305. doi: 10.1016/S0378-1097(97)00567-3. [DOI] [PubMed] [Google Scholar]
- 35.Eprintsev AT, Falaleeva MI, Stepanova IY, Perfenova NV. 2003. Purification and physicochemical properties of malate dehydrogenase from bacteria of the genus Beggiatoa. Biochemistry 68:172–176. doi: 10.1023/A:1022693211134. [DOI] [PubMed] [Google Scholar]
- 36.Maloney AP, Callan SM, Murray PG, Tuohy MG. 2004. Mitochondrial malate dehydrogenase from the thermophilic, filamentous fungus Talaromyces emersonii. Eur J Biochem 271:3115–3126. doi: 10.1111/j.1432-1033.2004.04230.x. [DOI] [PubMed] [Google Scholar]
- 37.López-Calcagno PE, Moreno J, Cedeño L, Labrador L, Concepción JL, Avilán L. 2009. Cloning, expression and biochemical characterization of mitochondrial and cytosolic malate dehydrogenase from Phytophthora infestans. Mycol Res 113:771–781. doi: 10.1016/j.mycres.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z-D, Wang B-J, Ge Y-D, Pan W, Wang J, Xu L, Liu A-M, Zhu G-P. 2011. Expression and identification of a thermostable malate dehydrogenase from multicellular prokaryote Streptomyces avermitilis MA-4680. Mol Biol Rep 38:1629–1636. doi: 10.1007/s11033-010-0273-1. [DOI] [PubMed] [Google Scholar]
- 39.Tomita T, Fushinobu S, Kuzuyama T, Nishiyama M. 2006. Structural basis for the alteration of coenzyme specificity in a malate dehydrogenase mutant. Biochem Biophys Res Commun 347:502–508. doi: 10.1016/j.bbrc.2006.06.131. [DOI] [PubMed] [Google Scholar]
- 40.Gourdon P, Baucher M, Lindley ND, Guyonvarch A. 2000. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl Environ Microbiol 66:2981–2987. doi: 10.1128/AEM.66.7.2981-2987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi K, Doi S, Negoro S, Urabe I, Okada H. 1989. Structure and properties of malic enzyme from Bacillus stearothermophilus. J Biol Chem 264:3200–3205. [PubMed] [Google Scholar]
- 42.Moreadith RW, Lehninger AL. 1984. Purification, kinetic behavior, and regulation of NAD(P)+ malic enzyme of tumor mitochondria. J Biol Chem 259:6222–6227. [PubMed] [Google Scholar]
- 43.Boles E, de Jong-Gubbels P, Pronk JT. 1998. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J Bacteriol 180:2875–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartolucci S, Rella R, Guagliardi A, Raia CA, Gambacorta A, De Rosa M. 1987. Malic enzyme from archaebacterium Sulfolobus solfataricus. Purification, structure, and kinetic properties. J Biol Chem 262:7725–7731. [PubMed] [Google Scholar]
- 45.Kawai S, Suzuki H, Yamamoto K, Inui M, Yukawa H, Kumagai H. 1996. Purification and characterization of a malic enzyme from the ruminal bacterium Streptococcus bovis ATCC 15352 and cloning and sequencing of its gene. Appl Environ Microbiol 62:2692–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acosta H, Dubourdieu M, Quiñones W, Cáceres A, Bringaud F, Concepción JL. 2004. Pyruvate phosphate dikinase and pyrophosphate metabolism in the glycosome of Trypanosoma cruzi epimastigotes. Comp Biochem Physiol B Biochem Mol Biol 138:347–356. doi: 10.1016/j.cbpc.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 47.Burton E, Martin VJJ. 2012. Proteomic analysis of Clostridium thermocellum ATCC 27405 reveals the upregulation of an alternative transhydrogenase-malate pathway and nitrogen assimilation in cells grown on cellulose. Can J Microbiol 58:1378–1388. doi: 10.1139/cjm-2012-0412. [DOI] [PubMed] [Google Scholar]
- 48.Bielen AAM, Willquist K, Engman J, van der Oost J, van Niel EWJ, Kengen SWM. 2010. Pyrophosphate as a central energy carrier in the hydrogen-producing extremely thermophilic Caldicellulosiruptor saccharolyticus. FEMS Microbiol Lett 307:48–54. doi: 10.1111/j.1574-6968.2010.01957.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J, Olson DG, Argyros DA, Deng Y, van Gulik WM, van Dijken JP, Lynd LR. 2013. Atypical glycolysis in Clostridium thermocellum. Appl Environ Microbiol 79:3000–3008. doi: 10.1128/AEM.04037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholls DJ, Miller J, Scawen MD, Clarke AR, Holbrook JJ, Atkinson T, Goward CR. 1992. The importance of arginine 102 for the substrate specificity of Escherichia coli malate dehydrogenase. Biochem Biophys Res Commun 189:1057–1062. doi: 10.1016/0006-291X(92)92311-K. [DOI] [PubMed] [Google Scholar]