Abstract
Strawberries are an important fruit in Belgium in both production and consumption, but little information is available about the presence of Salmonella and Shiga toxin-producing Escherichia coli (STEC) in these berries, the risk factors in agricultural production, and possible specific mitigation options. In 2012, a survey was undertaken of three soil and three soilless cultivation systems in Belgium. No Salmonella spp. were isolated. No STEC was detected in the strawberry samples (0 of 72), but STEC was detected by PCR in 11 of 78 irrigation water and 2 of 24 substrate samples. Culture isolates were obtained for 2 of 11 PCR-positive irrigation water samples and 2 of 2 substrate samples. Multivariable logistic regression analysis revealed elevated generic E. coli numbers (the odds ratio [OR] for a 1 log increase being 4.6) as the most important risk factor for STEC, together with the berry-picking season (elevated risk in summer). The presence of generic E. coli in the irrigation water (≥1 CFU per 100 ml) was mainly influenced by the type of irrigation water (collected rainfall water stored in ponds was more often contaminated than groundwater pumped from boreholes [OR = 5.8]) and the lack of prior treatment (untreated water versus water subjected to sand filtration prior to use [OR = 19.2]). The follow-up study in 2013 at one of the producer locations indicated cattle to be the most likely source of STEC contamination of the irrigation water.
INTRODUCTION
Strawberries are an important product in Belgium with regard to production volumes and sales, but little information is available about the microbiological risks. In 2012, almost 51,000 tons of strawberries were traded in the Belgian auctions, 40,500 tons of which were produced in Belgium. A total of 70% to 80% of the Belgian strawberry production is exported, mainly to other European countries, making strawberries the second most important product, after tomatoes, in sales in the auctions of the Union of Belgian Horticultural Cooperatives (1, 2). Strawberries are a perishable food which can receive no or minimal processing, because of the risk of physical damage to the berries and the subsequent increased risk of spoilage (3). Plants can be grown in soil or soilless cultures in protected environments or in open fields. Berries are harvested throughout the fruiting season and usually manually picked and directly placed in their final packaging for sale to consumers.
Strawberries are generally acknowledged as safe, because of their low pH, ranging from 3.2 to 4.2 (4). However, an outbreak with Escherichia coli O157 in the United States in 2011 with 15 cases, 2 of which were fatal, was caused by strawberries which were contaminated on the field by wildlife contact, namely, deer feces (5). One Salmonella outbreak associated with berries was reported in the European Union in the period 2007 to 2011, but it was linked to fresh raspberry juice and not to strawberries (6). Since there is no routine or regular monitoring of strawberries, very limited information about the prevalence of Salmonella spp. and the levels of generic E. coli on berries is available (3). The few available studies and data suggest a low prevalence of the pathogens Salmonella spp. and E. coli O157 on strawberries (0/173 [7], 0/11 [8], 0/194 [9], 0/31 [10], and 0/36 [11]). The European Food Safety Authority (EFSA) mentioned in an opinion issued in 2014 that, due to this lack of microbial data on berries, it is currently not possible to assess the suitability of a generic E. coli hygiene criterion at the point of primary production for berries (6). It was recommended that each production environment (including open field, enclosed or greenhouse, and wild area environments) should be evaluated for hazards that may compromise hygiene and food safety, in particular, to identify potential sources of fecal contamination.
Shiga toxin-producing E. coli (STEC) is defined as E. coli possessing Shiga toxin gene stx1 or stx2 or both. The majority of STEC strains that are associated with severe human disease are also carriers of the eae (“effacing and attaching”) gene (a gene coding for the protein intimin) or an alternative adherence gene, aggR (which was the case in the notorious outbreak in Germany in 2011 with sprouted seeds [12]). E. coli O157 is the STEC serotype most often implicated in outbreaks, but there are numerous other STEC serotypes that have caused outbreaks with serious human illness, including hemorrhagic colitis (HS) and hemolytic uremic syndrome (HUS) (13, 14). Therefore, the current focus in European Union is on detection of the following top 5 STEC serotypes with the highest pathogenic potential: O157, O26, O103, O111, and O145 (13). Since 2012, there has been a clear molecular approach in screening for pathogenic STEC using (multiplex) PCR, after a prior enrichment step, to look first primarily for the virulence genes, i.e., the presence of the Shiga toxin (stx) genes, usually in combination with the eae gene or aggR gene. If STEC is detected, a serotype PCR analysis for O26, O103, O111, O145, and O157 is recommended to get an indication of the presence of any of these top 5 pathogenic serotypes. It was observed that specific STEC serotypes are associated with specific eae gene variants (eae-β with O26, eae-θ or eae-ε with O103, eae-γ with O157 and O145, and eae-θ, eae-γ, or eae-ε with O111) (14–19). Since PCR detection of virulence genes does not unequivocally demonstrate the presence of all these genes in one E. coli strain, the eae variant(s) may be determined and compared with the detected serotype(s) to gauge the (im)possibility of the stx and eae virulence factors originating from one E. coli strain belonging to the top 5 most pathogenic serotypes. This real-time PCR screening approach with detection of specific eae subtypes associated with the top 5 STEC serotypes has been proposed and applied to cattle feces (20) and in raw-milk cheeses (18) in order to narrow down the number of PCR-positive results. In any case, it is highly recommended that an attempt should be made to confirm positive PCR results by isolating this E. coli strain and confirming its serotype and carriage of virulence genes. Nevertheless, it has been acknowledged that isolation of STEC, which is mostly excreted in small amounts (<100/g) (21), can be a challenging task and may fail in particular in samples with high numbers of competing microbiota (22–24). Moreover, STEC strains are known to easily lose stx genes (already during first subcultivation step), whereby it was noticed that stx genes appeared to be more stable in O157 strains than in non-O157 strains (25). As such, one may choose to react to positive STEC results by PCR without an isolate as a precautionary attitude to ensure that the risk for the consumer is reduced as much as possible, in particular, when the stx genes are detected in the presence of the most pathogenic STEC serotypes and are accompanied by another virulence gene(s) promoting severe human illness, e.g., the matching eae gene variant.
This survey provides more data about the presence and levels of generic E. coli and the prevalence of Salmonella and STEC on strawberries to facilitate future risk assessments and the setting of microbiological criteria. Moreover, the safety and sanitary quality of the production environment for strawberries, in both soil and soilless cultivation types, were investigated in more detail through sampling of the production environment (water, soil, or substrate) and workers' hands to determine the environmental pressure and risk factors for pathogen contamination.
MATERIALS AND METHODS
Main study in 2012.
Six strawberry producers in Flanders, Belgium, were sampled four times during the fruiting and picking season in 2012, i.e., from April to December (Table 1). Seasons were defined as follows: spring includes the months April, May, and June, summer comprises July, August, and September, and fall is October, November, and December. Three farms used cultivation in soil (unprotected in open field and/or protected in plastic tunnels) and three used soilless cultivation (in greenhouses and/or in plastic tunnels in substrate). Climatic data, namely, the average daily temperature (°C) and daily precipitation (>1 mm), were retrieved from http://www.worldweatheronline.com/ for the exact locations of the farms, with the exception of farm 2, for which the nearest available point was at a distance of 7 km.
TABLE 1.
Overview of the six strawberry producers sampled in 2012, their agrotechnical characteristics, and the parameters of the irrigation water
| Producer characteristic | Result for producer: |
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Sampling period | May–November | May–June | May–August | April–December | May–December | May–June |
| No. of visits | 4 | 4 | 4 | 4 | 4 | 4 |
| Cultivation type | Soilless (greenhouse and plastic tunnel) | Soil (plastic tunnel) | Soil (plastic tunnel and open air) | Soilless (greenhouse) | Soilless (greenhouse) | Soil (plastic tunnel and open air) |
| Farm animals present | Cattle and geese | Cattle | None | None | None | Cattle and sheep |
| Irrigation water source | Rain water | Rain water | Groundwater and rain water | Rain water | Rain water | Groundwater |
| Irrigation water storage | Pond with plastic foil | Pond | None (borehole) and pond | Pond | Raised pond with plastic foil | None (borehole) |
| Irrigation water treatment | None | None | None | Sand filtrationa | Sand filtration | None |
| Irrigation water temp in °C: median (minimum–maximum) | 19.6 (13.6–25.4) | 16.5 (14.1–19.4) | 16.2 (15.0–23.1) | 13.8 (12.0–15.0) | 14.8 (6.1–19.3) | 17.2 (15.2–22.0) |
| No. of irrigation water samples with generic E. coli (CFU per 100 ml)/total no. of samples | 16/16 | 8/8 | 1/6 (borehole); 2/2 (pond) | 5/14 | 9/16 | 3/16 |
| No. of irrigation water or culture isolates with STEC (probability of presence per liter)/total no. of samples | ||||||
| stx1/stx2 gene(s) | 5/16 | 5/8 | 0/8 | 1/14 | 0/16 | 0/16 |
| stx1/stx2 and eae genes | 5/16 | 5/8 | 0/8 | 1/14 | 0/16 | 0/16 |
| stx1/stx2 and eae genes, serotype O26, O103, O111, O145, or O157 | 4/16 | 3/8 | 0/8 | 1/14 | 0/16 | 0/16 |
| Culture | 2/16 | 0/8 | 0/8 | 0/14 | 0/16 | 0/16 |
Drain water was reused after reconditioning by sand filtration and UV irradiation.
Follow-up study in 2013.
In the next fruiting season (May 2013 until October 2013), a follow-up study was performed at producer 1 using a more frequent and environmentally focused sampling plan to investigate the source of environmental STEC contamination. In addition to irrigation water and substrate, feces of the beef cattle were sampled, distinguishing between older cattle (>10 months to 3 years of age) and younger cattle (<10 months). Geese with a bathing pond were located next to the collected rainfall water pond used for irrigation. Run-over could occur during rainfall, so the goose feces and goose pond water were also sampled.
Sampling.
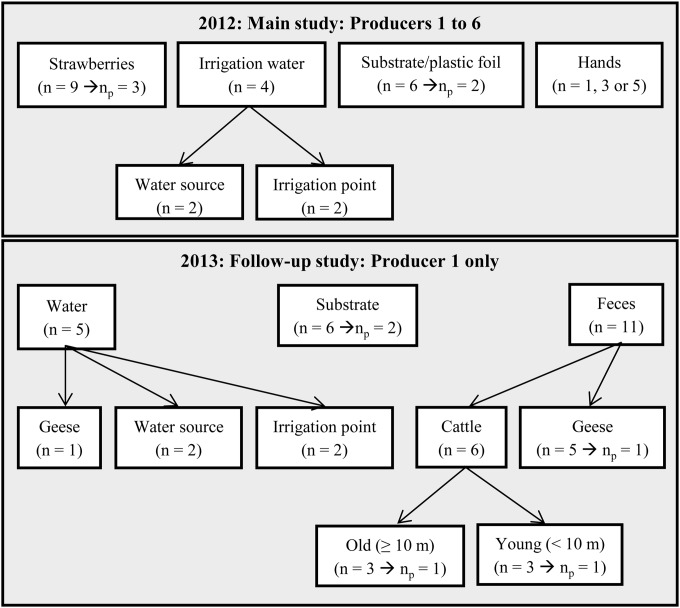
Fig. 1 provides an overview of the numbers and types of samples taken. The numbers of hands sampled, which depended on the number of workers present on the farm, were as follows: <5, 1 sample; 5 to 10, 3 samples; ≥10, 5 samples. The whole surface of hands (approximately 25 cm2) was sampled by rayon swabs (Fortuna Scientific, Singapore) filled with 5 ml physiological peptone salt solution (PPS; Fluka, Switzerland) (8.5 g/liter NaCl, 1 g/liter neutralized bacteriological peptone [Oxoid, United Kingdom]). Strawberry samples consisted of 10 whole berries which were picked with gloves and collected in Twirl'm bags (Labplas, Canada). In the laboratory, strawberry samples were cut into smaller particles with a blender and subsamples of 20 g were pooled into one sample of 60 g per 3 samples. Substrate samples of one handful were collected in Twirl'm bags, and 20-g subsamples from 3 samples were pooled into 1 sample of 60 g in the laboratory. The black plastic foil covering the soil in open fields was sampled by swabbing 1,000 cm2 with a sponge stick with 10 ml buffered peptone water (BPW) (3M, St. Paul, MN, USA). Three sponge swabs were pooled into one sample. Approximately 1.5 liters of water was collected in sterile bottles. Water samples were taken after letting the water run for 1 min, and the temperature of the sampled water was immediately measured. In the laboratory, water samples of 1 liter and 100 ml were filtered on a cellulose nitrate filter (Sartorius Stedim Biotech GmbH, Germany) (pore size, 0.45 μm) for pathogen detection and E. coli enumeration, respectively. If necessary, more filters were used to filter the required water volume when clogging by small particles occurred. The goose pond water was analyzed for pathogens by filtration of 100 ml instead of 1 liter (as higher volumes were not possible due to clogging of the filter). Cattle feces samples (weighing between 14 and 54 g) were collected by research participants walking around in an 8-shaped track in the stable wearing one pair of liquid-absorbing overshoes (Kolmi, Saint Bathélémy d'Anjou, France) as described by Cobbaut et al. (26). Overshoes were collected in filter stomacher bags (FBAG-04; Novolab, Belgium), and samples were pooled from 3 subsamples after enrichment. Five grams of goose droppings was collected in Twirl'em bags, and 5 subsamples were pooled to form 1 sample of 25 g. All samples were transported on ice in a cool box, stored at 4°C, and analyzed within 24 h.
FIG 1.
Overview of the samples taken in 2012, whereby 6 different strawberry producers were visited 4 times, and the follow-up study in 2013 of producer 1, which was visited 12 times. For each visit, the number of samples taken is indicated by n, and if applicable, np indicates the number of pooled samples.
Microbiological analysis.
Samples of strawberries, water, substrate, and plastic foil on the soil were analyzed for the presence or absence of Salmonella spp. and STEC per 60 g, per 3,000 cm2 for the plastic foil, per liter for irrigation water, and per 100 ml for pond water and for the enumeration of E. coli as a hygienic indicator. The farm workers' hands were analyzed for the numbers of E. coli present. Cattle and goose feces were analyzed for the presence or absence of Salmonella spp. and STEC.
E. coli.
A Petrifilm Select E. coli system (LED Techno, Belgium) was used for the enumeration of E. coli in samples of strawberries, substrate, foil, and hands by applying 1 ml of the appropriate 10-fold dilution in BPW followed by incubation at 37°C for 24 h. Filters from water samples were incubated on Rapid'E. coli2 (REC2; Bio-Rad, France) plates at 42°C for 24 h ± 3 h for the enumeration of E. coli.
Salmonella and STEC.
Samples for pathogen detection were enriched in a filter stomacher bag and diluted 5-fold (diluted 10-fold for the goose feces) with BPW at 37°C for 18 to 24 h. In the case of water samples, filters were enriched in 150 ml BPW. Salmonella spp. and Shiga toxin-producing E. coli (STEC) were detected by real-time PCR analysis of the iroB gene for Salmonella spp. and of the stx1, stx2, eae, and aggR genes for STEC with a GeneDisc Salmonella & aggregative E. coli plate as described by Beutin et al. (27). In short, 50 μl of the enriched BPW broth was transferred to a lysis tube (extraction pack Food 1; PALL GeneDisc Technologies), incubated for 10 min at 100°C in a heating block, and centrifuged for 2 min at 10,000 × g (at room temperature). Then, 36 μl of the DNA extract and 36 μl of the master mix (Pall GeneDisc Technologies) were transferred to a GeneDisc plate, which was subsequently loaded in a GeneDisc Cycler. Because false-positive results were obtained during our prior study, Salmonella PCR-positive samples were considered positive only after culture confirmation performed as described in ISO 6579:2002 was applied to validate the selected methods on fresh produce samples (28). In cases in which stx genes were detected, samples were further analyzed for serotypes O26, O103, O111, and O145 and for eae variants β, θ, Υ, and ε by PCR (using a GeneDisc STEC identification plate and a GeneDisc STEC Plus plate [Pall Technologies], respectively). Culture confirmation of PCR STEC-positive samples was performed according to ISO 16654:2001 for E. coli O157 and on CHROMagar STEC (CHROMagar Microbiology, France) and/or Chrom ID STEC (bioMérieux) for non-O157 E. coli followed by serotyping and confirming the presence of the virulence genes in the isolates by PCR in the Laboratory of Hygiene and Technology, Faculty of Veterinary Medicine, Ghent University.
Statistical analysis.
The 95% confidence interval (CI) was calculated for prevalence estimates by the Wilson score method without continuity correction (29). Ordinal classes, starting from the detection limit and comprising 10-fold differences in concentrations, were defined for the enumeration data of generic E. coli (Table 2), because E. coli was undetected in many samples, and different detection limits were obtained, depending on the sample type and method of analysis. Results were processed with SPSS version 21 at a significance level of 95% (P = 0.050). In cases of multiple pairwise comparisons, Bonferroni correction was applied to control the family-wise error rate at 5%. Differences in STEC prevalence were statistically assessed with the chi-square test of independence (likelihood ratio) for categorical variables and with the Mann-Whitney U test for continuous variables (since they were not normally distributed [Kolmogorov-Smirnov test; P < 0.001]). Multiple logistic regression analysis with backward likelihood ratio model selection was performed on all significant factors for significant main effects, and all possible interactions between the obtained effects were checked one by one by addition to the final model. Due to the large number of samples in which generic E. coli was not detected (194/255 = 76%), multiple logistic regression, rather than a generalized linear model with Poisson distribution on the E. coli counts, was also applied for determining the presence of E. coli.
TABLE 2.
Classes in which the generic E. coli enumeration data are grouped for statistical analysis
| E. coli class | Water (log E. coli CFU/100 ml) | Strawberries and substrate (log E. coli CFU/g) | Hands (log E. coli CFU/25 cm2) | Plastic foil on soil (log E. coli CFU/3000 cm2) |
|---|---|---|---|---|
| 1 (= undetected) | <0.0 | <1.0 | <0.7 | <2.6 |
| 2 | ≥0.0 and <1.0 | ≥1.0 | ≥0.7 | ≥2.6 |
| 3 | ≥1.0 and <2.0 | NAa | NA | NA |
| 4 | ≥2.0 and <3.0 | NA | NA | NA |
| 5 | ≥3.0 | NA | NA | NA |
NA, not applicable: no samples (or fewer than five samples, which were then added to the samples in the preceding class).
RESULTS
Main study: sanitary quality of Belgian strawberry production.
No Salmonella spp. were detected on strawberries (0/72; 95% confidence interval [CI], 0.0% to 5.1%), water (0/78; 95% CI, 0.0% to 4.7%), or substrate (0/24; 95% CI, 0.0% to 13.8%) or on the plastic foil covering the soil (0/24; 95% CI, 0.0% to 13.8%).
Shiga toxin-producing E. coli (STEC) was not found on strawberries (0/72; 95% CI, 0.0% to 5.1%) or on the plastic cover (0/24; 95% CI, 0.0% to 13.8%). However, STEC was detected in 14.1% of the irrigation water samples (11/78; 95% CI, 8.1% to 23.5%) and in 8.3% of the substrate samples (2/24; 95% CI, 2.3% to 25.8%). Confirmation with an isolate was achieved for only 4 of 13 PCR-positive samples (Table 3). All four samples were taken on the same day at producer 1 and yielded a STEC O26 strain. The stx toxin genes were always detected simultaneously with the adhesion eae gene, namely, as stx1 and eae (4/11), stx2 and eae (6/11), or stx1 and stx2 and eae (1/11) (Table 3). It should be noted that the eae gene (without the presence of the stx1 or stx2 gene) was detected in 2/72 strawberry samples, 22/78 irrigation water samples, and 5/24 substrate samples and on 6/24 plastic covers, while the aggR gene was never detected (either alone or in combination with stx1 and stx2 genes). A matching combination of an eae variant and a top 5 serotype was found for 10 of the 13 samples positive for STEC (Table 3).
TABLE 3.
Overview of all samples positive for Shiga toxin-producing E. coli, showing the results for all genes detected by PCR, the serotype, the result of culture confirmation, and the enumeration of generic E. coli CFU
| Date | Producer | Sample | GeneDisc (PCR) result |
Culture confirmation | Generic E. coli (CFU/100 ml or g)f | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| stx1 | stx2 | eae | aggR | Serotype(s) among the top 5 | eae variant(s)a | Serotype(s) with corresponding eae varianta | |||||
| 15 May 2012 | 2 | Waterb | − | + | + | − | O145, O26 | θ | − | − | 6.0 × 101 |
| 15 May 2012 | 2 | Waterb | − | + | + | − | O145, O26 | θ | − | − | 5.8 × 101 |
| 31 May 2012 | 2 | Waterb | − | + | + | − | O26, O103, O145 | β, θ | O26, O103 | − | 1.4 × 104 |
| 6 June 2012 | 2 | Waterb | − | + | + | − | O26, O103, O145 | β, θ, ε | O26, O103 | − | 1.1 × 103 |
| 18 June 2012 | 2 | Waterb | − | + | + | − | O26, O103, O145 | β | O26 | − | 1.2 × 103 |
| 4 July 2012 | 1 | Waterb | − | + | + | − | O145, O26 | − | − | − | 2.1 × 101 |
| 14 September 2012 | 1 | Waterb | + | − | + | − | O26, O103, O145 | β, θ, ε, ϒ | O26, O103, O145 | + (O26) | 1.7 × 102 |
| 14 September 2012 | 1 | Waterc | + | − | + | − | O26, O145 | β, ε, ϒ | O26, O145 | + (O26) | 1.6 × 102 |
| 14 September 2012 | 1 | Waterc | + | − | + | − | O26, O103, O145 | β, θ, ϒ | O26, O103, O145 | − | 1.6 × 102 |
| 14 September 2012 | 1 | Waterb | + | − | + | − | O26, O145 | β, ε, ϒ | O26, O145 | − | 1.9 × 102 |
| 8 October 2012 | 4 | Waterc | + | + | + | − | O26, O103, O145 | β, θ, ε | O26, O103 | − | 1.6 × 102 |
| 14 September 2012 | 1 | Substrate | + | − | + | − | O26, O103, O145 | β | O26 | + (O26) | 2.0 × 101 |
| 14 September 2012 | 1 | Substrate | + | + | + | − | O26, O145 | β, ϒ | O26, O145 | + (O26) | 2.0 × 101 |
| 15 May 2013 | 1 | Waterc | + | − | + | − | O26, O145 | β | O26 | − | 1.5 × 102 |
| 15 May 2013 | 1 | Waterb | + | − | + | − | O26, O145 | β, θ | O26 | − | 8.6 × 101 |
| 21 August 2013 | 1 | Waterc | + | − | + | − | − | − | − | − | 1.1 × 101 |
| 18 September 2013 | 1 | Waterb | − | + | + | − | O103, O145 | β, θ | O103 | − | 2.1 × 102 |
| 15 October 2013 | 1 | Waterc | − | + | + | − | O26, O103, O145 | β, θ, ϒ | O26, O103, O145 | − | 1.9 × 103 |
| 15 October 2013 | 1 | Waterb | − | + | + | − | O26, O145 | β, ϒ | O26, O145 | − | 2.3 × 102 |
| 15 October 2013 | 1 | Waterb | − | + | + | − | O26, O145 | β, ϒ | O26, O145 | − | 2.2 × 102 |
| 15 May 2013 | 1 | Substrate | + | − | + | − | O26 | − | − | − | 1.1 × 101 |
| 29 May 2013 | 1 | Geese feces | + | + | + | − | − | − | − | − | NDg |
| 15 May 2013 | 1 | Cattle fecesd | + | + | + | − | O26, O145 | β, θ | O26 | − | ND |
| 15 May 2013 | 1 | Cattle fecese | + | + | + | − | O26, O145 | β, θ | O26 | − | ND |
| 29 May 2013 | 1 | Cattle fecesd | + | + | + | − | O26, O145 | β, θ | O26 | − | ND |
| 29 May 2013 | 1 | Cattle fecese | + | + | + | − | O26, O145, O103, O111 | β, θ, ε | O26, O103, O111 | − | ND |
| 12 June 2013 | 1 | Cattle fecesd | + | + | + | − | O145 | θ, β | − | − | ND |
| 12 June 2013 | 1 | Cattle fecese | + | + | + | − | O26, O145, O103 | β, θ, ϒ | O26, O103, O145 | − | ND |
| 26 June 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | θ, ε | O103 | − | ND |
| 26 June 2013 | 1 | Cattle fecese | + | + | + | − | O26, O145, O103 | β, θ, ε, ϒ | O26, O103, O145 | − | ND |
| 8 July 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | β, θ, ε | O103 | − | ND |
| 8 July 2013 | 1 | Cattle fecese | + | + | + | − | O145, O103 | β, θ, ε, ϒ | O103, O145 | − | ND |
| 24 July 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | β, θ, ε | O103 | − | ND |
| 24 July 2013 | 1 | Cattle fecese | + | + | + | − | O145, O103 | β, θ, ε, ϒ | O103, O145 | − | ND |
| 7 August 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | β, θ, ε | O103 | − | ND |
| 7 August 2013 | 1 | Cattle fecese | + | + | + | − | O145, O103 | β, θ, ε, ϒ | O103, O145 | − | ND |
| 21 August 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | β, θ, ε | O103 | − | ND |
| 21 August 2013 | 1 | Cattle fecese | + | + | + | − | O145, O103 | β, θ, ε, ϒ | O103, O145 | − | ND |
| 4 September 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | β, θ, ε | O103 | − | ND |
| 4 September 2013 | 1 | Cattle fecese | + | + | + | − | O26, O145, O103 | β, θ, ε, ϒ | O26, O103, O145 | − | ND |
| 18 September 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | θ, ϒ | O103, O145 | − | ND |
| 18 September 2013 | 1 | Cattle fecese | + | + | + | − | O145, O103 | β, θ, ε | O103 | − | ND |
| 1 October 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | β, θ, ϒ | O103, O145 | − | ND |
| 1 October 2013 | 1 | Cattle fecese | + | + | + | − | O145, O103 | β, θ, ε | O103 | − | ND |
| 15 October 2013 | 1 | Cattle fecesd | + | + | + | − | O145, O103 | β, ε, ϒ | O103, O145 | − | ND |
| 15 October 2013 | 1 | Cattle fecese | + | + | + | − | O145, O103 | β, θ, ε | O103 | − | ND |
Data are from Bibbal et al. (20).
Irrigation point.
Source.
Feces from cattle >10 months old.
Feces from cattle <10 months old.
Data correspond to the matrix: water (CFU/100 ml) or substrate (CFU/g).
ND, not determined.
Generic E. coli was present on only 2 of 72 strawberry samples at concentrations of 1.0 log CFU/g and 3.0 log CFU/g. E. coli could be enumerated in 7/24 substrate samples (mean concentration of the positive samples, 1.8 ± 1.3 log CFU/g), on 4/24 plastic covers (3.0 ± 0.5 log CFU/3,000 cm2), and on 4/57 of the hands (1.8 ± 1.2 log CFU/25 cm2). E. coli was enumerated most often in irrigation water samples (44/78), with concentrations ranging from 1.0 CFU/100 ml to 4.2 log CFU/100 ml.
Risk factors for the presence of STEC and the fecal indicator E. coli.
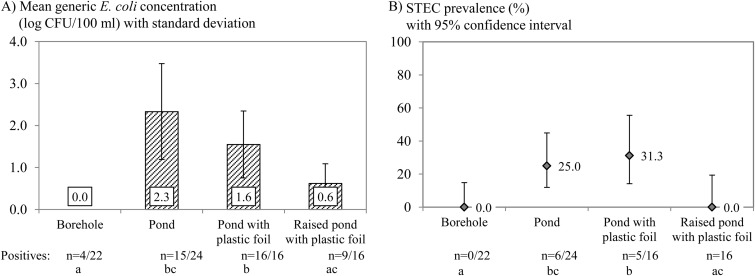
The presence of STEC was significantly correlated with the E. coli class (= the level of E. coli), meaning that samples positive for STEC were associated with a higher average E. coli count. The following factors were significantly correlated with the STEC prevalence and the counts of generic E. coli in univariable statistical analysis: (i) the sample type, (ii) the producer, (iii) the presence of farm animals, (iv) the irrigation water type, (v) treatment of the irrigation water, and (vi) the time of the year (month and season). The differences between the producers suggest that there are specific farm factors influencing the microbiological quality and safety. The presence of farm animals meant that cattle were being reared professionally by the producers and/or that cows or sheep were grazing in a field(s) adjacent to the strawberry production field. The presence of farm animals significantly increased the general prevalence of STEC from 1.0% (1/98; 95% CI, 0.2% to 5.6%) to 12.0% (12/100; 95% CI, 7.0% to 19.8%) and that of generic E. coli from 17.4% (24/138; 95% CI, 12.0% to 24.6%) to 31.6% (37/117; 95% CI, 23.9% to 40.5%). Groundwater samples contained no STEC and exactly 1 CFU of E. coli per 100 ml in only 4 of 22 samples, while STEC was detected in 19.6% (11/56) of the ponds with collected rainfall water samples, and the majority (40/56) were contaminated with, on average, 1.6 log CFU/100 ml E. coli. E. coli counts and the STEC prevalence were lower in a raised pond than in the other ponds (Fig. 2). Treatment of irrigation water by sand filtration was significantly associated with lower numbers of generic E. coli. Untreated water contained, on average, 1.8 ± 1.2 log CFU/100 ml E. coli, while filtered water contained 0.9 ± 0.7 log CFU/100 ml E. coli. Similarly, STEC was also significantly more prevalent in untreated irrigation water, at 20.8% (10/48; 95% CI, 11.7% to 34.3%), than in treated water, at 3.3% (1/30; 95% CI, 0.6% to 16.7%). Seasonal differences were observed: STEC prevalence was highest in September, and generic E. coli counts peaked in September and October.
FIG 2.
Microbiological analysis of different types of irrigation water, namely, groundwater from a borehole and collected rainfall water stored in a pond without protection, in a pond with a plastic foil covering the bottom, and in a raised pond (elevated edges to prevent runoff) with a plastic foil bottom. Different letters indicate statistically significant differences, and n indicates the ratio of the number of positive results to the total number of samples. (A) Mean generic E. coli concentrations (log CFU/100 ml) with error bars indicating the standard deviations of the data from the positive samples. (B) STEC prevalence (%) with error bars indicating the 95% CI for the prevalence estimate.
The following factors did not significantly affect the STEC prevalence or the E. coli counts: (i) the soil cultivation system versus the soilless cultivation system, (ii) the average daily temperature, (iii) the daily rate of precipitation, (iv) the temperature of the irrigation water, (v) the distance between the strawberry fields and the nearest toilet, and (vi) flooding. Minor flooding of the strawberry field had occurred in July 2012 at producer 3, but this event did not have significant effects on the microbiological contamination, presumably because the beds of the strawberry plants were raised and thus there was no contact of water with the plants and strawberries. It may also be interesting that neither the prevalences of STEC (P = 0.118) nor the counts of E. coli (P = 0.080) were significantly different between water samples taken from the source and water samples taken at the actual (starting) point of the drip irrigation.
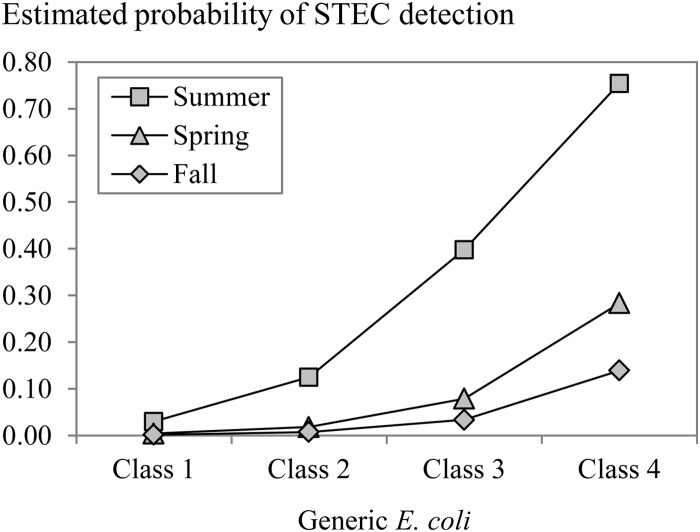
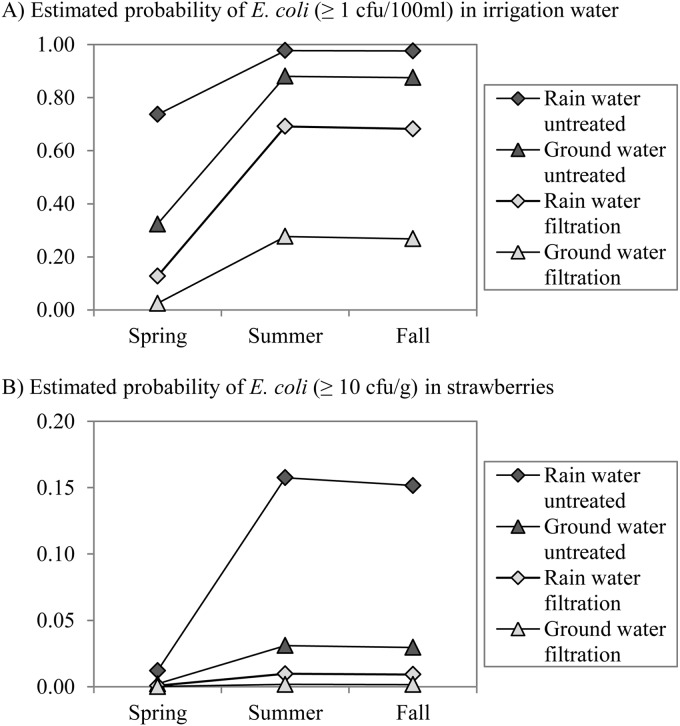
To estimate their quantitative effects and check whether there are interactions, multivariable logistic regression analysis was performed to investigate which significant risk factors from the univariable analysis remained when all factors were considered together. The risk of STEC prevalence was determined according to the generic E. coli count and the season (Fig. 3). The odds ratio (OR) for a 1 log increase of the E. coli level was 4.6. The risk of STEC occurrence was highest in summer and lower in spring (OR = 0.1) and in fall (OR = 0.05). The presence of generic E. coli was determined according to the sample type, the season, the irrigation water type, and the application of water treatment. E. coli was most frequently found in water and then in soil/substrate (OR = 0.1) and was rarest on hands (OR = 0.01) and strawberries (OR = 0.004). Generic E. coli was most prevalent in the summer and fall seasons, and the prevalence was lower in spring (OR = 0.05). E. coli was more frequently present in collected rain water stored in ponds than in groundwater pumped from boreholes (OR = 5.9) and in irrigation water that had not been treated by sand filtration (OR = 19.2) (Fig. 4).
FIG 3.
Estimated probability of STEC presence (per 25 g or 1 liter) determined by multivariable logistic regression in the function of the risk factors, i.e., season and generic E. coli concentration.
FIG 4.
Estimated probability of generic E. coli presence determined by multivariable logistic regression in the function of the risk factors, i.e., season, water type (groundwater or rain water), water treatment (untreated or sand filtration), and sample type, showing the presence in irrigation water (≥1 CFU/100 ml) and the sample type with the highest risk (A) and, in strawberries (≥10 CFU/g), the sample type with the lowest risk (B).
Follow-up study: environmental source of STEC contamination.
Again, as in 2012, no Salmonella was found in the samples taken at producer 1 throughout the 2013 fruiting season (0/48 irrigation water, 0/8 goose pond water, 0/24 substrate, 0/16 goose feces, 0/24 beef cattle feces). Producer 1 operated a mixed farm with production of both strawberries and cattle. STEC was detected in 7/48 irrigation water samples, 0/8 goose pond water samples, 1/24 substrate samples, 1/16 goose feces samples, and 24/24 beef cattle feces samples. Similarly to the previous sampling, stx genes were always accompanied by eae genes. aggR genes were never detected in any sample. eae genes without stx genes were detected in 36/48 irrigation water samples, 16/24 substrate samples, 12/16 goose feces samples, and 7/8 pond water samples. Isolates were obtained from none of the STEC PCR-positive samples (0/33), but the corresponding combination of serotype and eae variant was found in 29 of 33 samples (6/29 irrigation water, 23/29 cattle feces) (Table 3). Similar STEC serotypes were detected by PCR in pens of young (<10 months) and older (≥10 months) cattle. The main serotype which was detected in combination with the corresponding eae variant in cattle feces was O103 (20/24), followed by O145 (10/24), O26 (7/24), and O111 (1/24).
E. coli was enumerated in all water samples (48/48 irrigation water [1.5 ± 0.8 log CFU/100 ml] and 10/10 goose pond water [3.5 ± 0.5 log CFU/100 ml]) and in 14/22 substrate samples (1.7 ± 0.7 log CFU/g). E. coli counts exceeded, respectively, 100 and 1,000 CFU/100 ml in 12/48 and 2/48 samples of the rainfall water collected to be used as irrigation water. The level of generic E. coli was much higher in the bathing pond of geese than in the irrigation water pond, but no STEC were detected.
DISCUSSION
The sanitary quality of Belgian strawberries was good, since no Salmonella or STEC was detected and only two samples contained generic E. coli. The cultivation type (soil versus soilless culture) did not have an impact on the occurrence of STEC or generic E. coli. However, STEC and generic E. coli were regularly present in the production environment, i.e., in the irrigation water and the substrate. The contamination risk of strawberries was strongly reduced because all producers applied drip irrigation to the strawberry plants, so no direct contact between irrigation water and the berries occurred. Contaminated substrate or soil may contaminate the strawberries by splashing of soil particles onto the berries during rainfall. However, most strawberries are grown in protected culture, either in greenhouses or in plastic tunnels in raised beds to facilitate picking. Moreover, even for strawberries grown in soil and in unprotected culture (in the open field without plastic tunnels), the opportunity for soil contact due to rainfall is limited, because the soil in which strawberries are grown is covered by plastic foil. Internalization of pathogens via the irrigation water or the soil through the roots into the strawberries, in particular, with naturally contaminated water with low numbers of pathogens, is highly unlikely (30, 31). As expected from these limited opportunities for contamination, the strawberries in this survey showed satisfactory microbial safety and sanitary quality.
The main risk factors for STEC in the primary production of Belgian strawberries were increased E. coli counts and the summer months. Comparable results were found in Belgian lettuce farms, where elevated E. coli levels increased the probability of the presence of pathogens Campylobacter spp., STEC, and Salmonella spp. (32). Collected rainfall water in ponds which can receive surface runoff water (no elevated borders) and in ponds which are located near cattle grazing or cattle residence areas run the highest risk of contamination with generic E. coli, in particular, in summer and fall. Water treatment prior to irrigation should be regarded as an option, despite the risk-mitigating practices of drip irrigation and protected culture. The risk factors which were identified to indicate a higher likelihood of contamination with pathogens should preferably be addressed by sanitary surveys, training, observational audits, and other methods to verify agricultural and hygiene practices for berries at primary production.
The current European Union legal framework does not include microbiological criteria applicable for fresh strawberries at the primary production stage (hygiene criterion). However, using E. coli as an indicator of recent human or animal fecal contamination is likely to be useful for verification of good agricultural practices (GAP) and good hygienic practices (GHP) applied to berries at individual production sites, for example, to assess the suitability of water used for irrigation (3). Thus, monitoring of E. coli as an indicator organism is appropriate, but it should be done frequently and with consideration of the irrigation water type and the risk factors present on the farm. In the United States, water that may come in contact with the harvestable portion of produce must meet a standard of ≤235 CFU/100 ml generic E. coli throughout the growing season (33). In Europe, microbial criteria have been established in Spain only for the use of treated wastewater for the irrigation of crops that are likely to be eaten uncooked; E. coli should then be ≤100 CFU/100 ml (34). The numbers used for those criteria seem too high to limit the risk of STEC contamination via the irrigation water used by Belgian strawberry producers, because 23% (3/13) of the samples containing between 10 and 100 CFU/100 ml generic E. coli were positive for STEC, while no STEC was found in samples with <10 CFU/100 ml E. coli (0/49). If such criteria are established, they should be set after assessing the local relationship of E. coli with the pathogen(s) of interest.
During the 12 weeks of sampling water and cattle and goose feces in the follow-up study, STEC O145 and STEC O103 were (nearly) always detected, O26 was detected mainly in the first half of the sampling period (May to June) and again at the end (September to October), and O111 was detected only once. No culture isolates were obtained. The STEC PCR results often showed high threshold cycle (CT) values, and it is known to be quite challenging to provide isolates from samples contaminated at low levels, in particular, when high numbers of generic E. coli are present (20, 22). Only half of the STEC O145 samples showed that the serotype was present in combination with its eae variant, while most STEC O26, O103, and O111 samples were presumably eae positive. This combination suggests the potential presence of one E. coli strain harboring virulence genes stx and eae and belonging to the top 5 most pathogenic serotypes which are associated with severe human disease. In our study, the presence of an eae gene or aggR virulence gene for adhesion was also determined, because adhesive STEC strains have enhanced pathogenic potential (13, 35). The aggR gene was never detected throughout this study, in accordance with the current assumption that STEC with aggR, in particular, has a human reservoir with a low likelihood of occurrence in cattle and in the primary production environment (36). In contrast, the eae gene was detected in the absence of stx genes in many samples. The reason may be the presence of adhesive enteropathogenic E. coli (EPEC) or of other bacteria which can also possess the eae gene, such as Citrobacter rodentium (27). STEC was detected in all samples of cattle feces, and in the majority of cases, multiple serotypes were simultaneously detected. Similarly to previous studies, E. coli O157 was not detected in cattle feces samples, supporting the notion that the prevalence of E. coli O157:H7 in cattle is lower in continental Europe than in the United Kingdom and the United States (26, 37). STEC shedding by cattle is very common and highly variable in time, animals' feces samples being positive for STEC in from 10% to 100% of all samples or sampling visits but also variable in concentration levels, which ranged from <2 log CFU/g to >6 log CFU/g (22, 37, 38). The cause of intermitted STEC shedding is currently unknown, but evidence points to cattle being persistently infected with the same STEC strains (22, 26, 38). In contrast to the high prevalence of STEC in cattle feces samples, only one sample of goose feces contained STEC. Moreover, this STEC did not belong to serotype O26, O103, O111, or O145, while the former STEC serotypes were detected in the cattle fecal samples and in the irrigation water. This indicates that the cattle present on the farm, rather than the geese living near the irrigation water source, were the likely reservoir of STEC on the farm and source of contamination the irrigation water. This study confirmed the current opinion that cattle represent the main reservoir for STEC (26, 39). Nevertheless, wild birds, rodents, cats, or dogs may become infected by cattle and possibly be part of the STEC transmission and dissemination (22, 40–42).
ACKNOWLEDGMENTS
The research received funding from the EU FP7 Veg-i-Trade Grant agreement no 244994.
Our special appreciation goes to Elien Verguldt and Astrid De Beleyer, who assisted in the microbiological analysis.
REFERENCES
- 1.VILT. 2013. Internationaal aardbeiencongres zet Vlaamse expertise in de verf. http://www.vilt.be/Aardbeienteelt_Internationaal_aardbeiencongres_zet_Vlaamse_expertise_in_de_verf Accessed 13 May 2014.
- 2.VLAM. 2014. Fruit logistica 2014: Ontmoet 25 Belgische exporteurs op de VLAM-stand. http://pers.vlam.be/de/pers/detail/4687/fruit-logistica-2014-ontmoet-25-belgische-exporteurs-op-de-vlam-stand Accessed 1 July 2014.
- 3.European Food Safety Authority (EFSA). 2014. Scientific opinion on the risk posed by pathogens in food of non-animal origin. Part 2 (Salmonella and Norovirus in berries). EFSA J 12:3706. doi: 10.2903/j.efsa.2014.3706. [DOI] [Google Scholar]
- 4.Knudsen DM, Yamamoto SA, Harris LJ. 2001. Survival of Salmonella spp. and Escherichia coli O157:H7 on fresh and frozen strawberries. J Food Prot 64:1483–1488. [DOI] [PubMed] [Google Scholar]
- 5.Laidler MR, Tourdjman M, Buser GL, Hostetler T, Repp KK, Leman R, Samadpour M, Keene WE. 2013. Escherichia coli O157:H7 infections associated with consumption of locally grown strawberries contaminated by deer. Clin Infect Dis 57:1129–1134. doi: 10.1093/cid/cit468. [DOI] [PubMed] [Google Scholar]
- 6.EFSA. 2013. Scientific opinion on the risk posed by pathogens in food of non-animal origin. Part 1 (outbreak data analysis and risk ranking of food/pathogen combinations). EFSA J 11:3025. doi: 10.2903/j.efsa.2013.3025. [DOI] [Google Scholar]
- 7.Johannessen GS, Loncarevic S, Kruse H. 2002. Bacteriological analysis of fresh produce in Norway. Int J Food Microbiol 77:199–204. doi: 10.1016/S0168-1605(02)00051-X. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee A, Speh D, Dyck EA, Diez-Gonzalez F. 2004. Pre-harvest evaluation of coliforms, Escherichia coli, Salmonella and E. coli O157:H7 in organic and conventional produce grown by Minnesota farmers. J Food Prot 67:894–900. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee A, Speh D, Jones AT, Buesing KM, Diez-Gonzalez F. 2006. Longitudinal microbiological survey of fresh produce grown by farmers in the upper midwest. J Food Prot 69:1928–1936. [DOI] [PubMed] [Google Scholar]
- 10.Bohaychuk VM, Bradbury RW, Dimock R, Fehr M, Gensler GE, King RK, Rieve R, Barrios PR. 2009. A microbiological survey of selected Alberta-grown fresh produce from farmers' markets in Alberta, Canada. J Food Prot 72:415–420. [DOI] [PubMed] [Google Scholar]
- 11.Yoon Y, Kim K, Nam M, Shim WB, Ryu JG, Kim DH, You OJ, Chung DH. 2010. Microbiological assessment in strawberry production and recommendations to establish a Good Agricultural Practice system. Foodborne Pathog Dis 7:1511–1519. doi: 10.1089/fpd.2010.0611. [DOI] [PubMed] [Google Scholar]
- 12.Joris MA, Vanrompay D, Verstraete K, De Reu K, De Zutter L. 2012. Enterohemorrhagic Escherichia coli with particular attention to the German outbreak strain O104:H4. Vlaams Diergeneeskundig Tijdschrift 81:3–10. [Google Scholar]
- 13.EFSA. 2013. Scientific opinion on VTEC-seropathotype and scientific criteria regarding pathogenic assessment. EFSA J 11:3138. doi: 10.2903/j.efsa.2013.3138. [DOI] [Google Scholar]
- 14.Possé B, De Zutter L, Heyndrickx M, Herman L. 2007. Metabolic and genetic profiling of clinical O157 and non-O157 Shiga-toxin-producing Escherichia coli. Res Microbiol 158:591–599. doi: 10.1016/j.resmic.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Beutin L, Krause G, Zimmermann S, Kaulfuss S, Gleier K. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J Clin Microbiol 42:1099–1108. doi: 10.1128/JCM.42.3.1099-1108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanco A, Blanco JE, Mora A, Dahbi G, Alonso AP, Gonzalez EA, Bernardez MI, Blanco J. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-xi). J Clin Microbiol 42:645–651. doi: 10.1128/JCM.42.2.645-651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madic J, de Garam CP, Vingadassalon N, Oswald E, Fach P, Jamet E, Auvray F. 2010. Simplex and multiplex real-time PCR assays for the detection of flagellar (H-antigen) fliC alleles and intimin (eae) variants associated with enterohaemorrhagic Escherichia coli (EHEC) serotypes O26:H11, O103:H2, O111:H8, O145:H28 and O157:H7. J Appl Microbiol 109:1696–1705. doi: 10.1111/j.1365-2672.2010.04798.x. [DOI] [PubMed] [Google Scholar]
- 18.Madic J, Vingadassalon N, de Garam CP, Marault M, Scheutz F, Brugere H, Jamet E, Auvray F. 2011. Detection of Shiga toxin-producing Escherichia coli serotypes O26:H11, O103:H2, O111:H8, O145:H28, and O157:H7 in raw-milk cheeses by using multiplex real-time PCR. Appl Environ Microbiol 77:2035–2041. doi: 10.1128/AEM.02089-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun 68:64–71. doi: 10.1128/IAI.68.1.64-71.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bibbal D, Loukiadis E, Kerouredan M, de Garam CP, Ferre F, Cartier P, Gay E, Oswald E, Auvray F, Brugere H. 2014. Intimin gene (eae) subtype-based real-time PCR strategy for specific detection of Shiga toxin-producing Escherichia coli serotypes O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 in cattle feces. Appl Environ Microbiol 80:1177–1184. doi: 10.1128/AEM.03161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearce MC, Fenlon D, Low JC, Smith AW, Knight HI, Evans J, Foster G, Synge BA, Gunn GJ. 2004. Distribution of Escherichia coli O157 in bovine fecal pats and its impact on estimates of the prevalence of fecal shedding. Appl Environ Microbiol 70:5737–5743. doi: 10.1128/AEM.70.10.5737-5743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joris MA, Verstraete K, De Reu K, De Zutter L. 2013. Longitudinal follow-up of the persistence and dissemination of EHEC on cattle farms in Belgium. Foodborne Pathog Dis 10:295–301. doi: 10.1089/fpd.2012.1277. [DOI] [PubMed] [Google Scholar]
- 23.Sinton LW, Braithwaite RR, Hall CH, Mackenzie ML. 2007. Survival of indicator and pathogenic bacteria in bovine feces on pasture. Appl Environ Microbiol 73:7917–7925. doi: 10.1128/AEM.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Mankin KR, Marchin GL. 2004. Survival of fecal bacteria in dairy cow manure. Trans ASAE 47:1239–1246. doi: 10.13031/2013.16574. [DOI] [Google Scholar]
- 25.Joris MA, Verstraete K, De Reu K, De Zutter L. 2011. Loss of vtx genes after the first subcultivation step of verocytotoxigenic Escherichia coli O157 and non-O157 during isolation from naturally contaminated fecal samples. Toxins 3:672–677. doi: 10.3390/toxins3060672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobbaut K, Houf K, Douidah L, Van Hende J, De Zutter L. 2008. Alternative sampling to establish the Escherichia coli O157 status on beef cattle farms. Vet Microbiol 132:205–210. doi: 10.1016/j.vetmic.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Beutin L, Jahn S, Fach P. 2009. Evaluation of the ‘GeneDisc' real-time PCR system for detection of enterohaemorrhagic Escherichia coli (EHEC) O26, O103, O111, O145 and O157 strains according to their virulence markers and their O- and H-antigen-associated genes. J Appl Microbiol 106:1122–1132. doi: 10.1111/j.1365-2672.2008.04076.x. [DOI] [PubMed] [Google Scholar]
- 28.Delbeke S, Ceuppens S, Holvoet K, Samuels E, Sampers I, Uyttendaele M. 13 October 2014, posting date Multiplex real-time PCR and culture methods for detection of Shiga toxin-producing Escherichia coli and Salmonella Thompson in strawberries, a lettuce mix and basil. Int J Food Microbiol doi: 10.1016/j.ijfoodmicro.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Wilson EB. 1927. Probable inference, the law of succession and statistical inference. J Am Stat Assoc 22:209–212. doi: 10.1080/01621459.1927.10502953. [DOI] [Google Scholar]
- 30.Hirneisen KA, Sharma M, Kniel KE. 2012. Human enteric pathogen internalization by root uptake into food crops. Foodborne Pathog Dis 9:396–405. doi: 10.1089/fpd.2011.1044. [DOI] [PubMed] [Google Scholar]
- 31.Miles JM, Sumner SS, Boyer RR, Williams RC, Latimer JG, McKinney JM. 2009. Internalization of Salmonella enterica serovar Montevideo into greenhouse tomato plants through contaminated irrigation water or seed stock. J Food Prot 72:849–852. [DOI] [PubMed] [Google Scholar]
- 32.Holvoet K, Sampers I, Seynnaeve M, Uyttendaele M. 2014. Relationships among hygiene indicators and enteric pathogens in irrigation water, soil and lettuce and the impact of climatic conditions on contamination in the lettuce primary production. Int J Food Microbiol 171:21–31. doi: 10.1016/j.ijfoodmicro.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 33.FDA Food Safety Modernization Act (FSMA). 2013. Microbial standards for water (agricultural water requirements). http://www.newenglandfarmersunion.org/fsma/agricultural-water-requirements-talking-points/ Accessed 1 July 2014.
- 34.Spanish Official Bulletin (BOE). 2007. Royal Decree 1620/2007 of 7 December, Spanish regulations for water reuse. http://www.boe.es/boe/dias/2007/12/08/pdfs/A50639-50661.pdf Accessed 1 July 2014.
- 35.Tozzi AE, Caprioli A, Minelli F, Gianviti A, De Petris L, Edefonti A, Montini G, Ferretti A, De Palo T, Gaido M, Rizzoni G; Hemolytic Uremic Syndrome Study Group. 2003. Shiga toxin-producing Escherichia coli infections associated with hemolytic uremic syndrome, Italy, 1988–2000. Emerg Infect Dis 9:106–108. doi: 10.3201/eid0901.020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.European Food Safety Authority (EFSA). 2013. Scientific opinion on VTEC-seropathotype and scientific criteria regarding pathogenic assessment. EFSA J 11:3138. doi: 10.2903/j.efsa.2013.3138. [DOI] [Google Scholar]
- 37.Pradel N, Livrelli V, De Champs C, Palcoux JB, Reynaud A, Scheutz F, Sirot J, Joly B, Forestier C. 2000. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J Clin Microbiol 38:1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson SE, Wright EJ, Hart CA, Bennett M, French NP. 2004. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J Appl Microbiol 97:1045–1053. doi: 10.1111/j.1365-2672.2004.02390.x. [DOI] [PubMed] [Google Scholar]
- 39.Chapman PA, Wright DJ, Norman P, Fox J, Crick E. 1993. Cattle as a possible source of verocytotoxin-producing Escherichia coli O157 infection in man. Epidemiol Infect 111:439–447. doi: 10.1017/S0950268800057162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.European Commission (EC). 1997. Report of the Scientific Veterinary Committee on verocytotoxin-producing Escherichia coli (VTEC). http://ec.europa.eu/food/fs/sc/oldcomm4/out15_en.html Accessed 5 April 2014.
- 41.Nielsen EM, Skov MN, Madsen JJ, Lodal J, Jespersen JB, Baggesen DL. 2004. Verocytotoxin-producing Escherichia coli in wild birds and rodents in close proximity to farms. Appl Environ Microbiol 70:6944–6947. doi: 10.1128/AEM.70.11.6944-6947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samadpour N, Stewart J, Steingart K, Addy C, Louderback J, McGinn M, Ellington J, Newman T. 2002. Laboratory investigation of an E-coli O157:H7 outbreak associated with swimming in Battle Ground Lake, Vancouver, Washington. J Environ Health 64:16–20. [PubMed] [Google Scholar]