Abstract
The bacterial second messengers (p)ppGpp and bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) regulate important functions, such as transcription, virulence, biofilm formation, and quorum sensing. In mycobacteria, they regulate long-term survival during starvation, pathogenicity, and dormancy. Recently, a Pseudomonas aeruginosa strain lacking (p)ppGpp was shown to be sensitive to multiple classes of antibiotics and defective in biofilm formation. We were interested to find out whether Mycobacterium smegmatis strains lacking the gene for either (p)ppGpp synthesis (ΔrelMsm) or c-di-GMP synthesis (ΔdcpA) would display similar phenotypes. We used phenotype microarray technology to compare the growth of the wild-type and the knockout strains in the presence of several antibiotics. Surprisingly, the ΔrelMsm and ΔdcpA strains showed enhanced survival in the presence of many antibiotics, but they were defective in biofilm formation. These strains also displayed altered surface properties, like impaired sliding motility, rough colony morphology, and increased aggregation in liquid cultures. Biofilm formation and surface properties are associated with the presence of glycopeptidolipids (GPLs) in the cell walls of M. smegmatis. Thin-layer chromatography analysis of various cell wall fractions revealed that the levels of GPLs and polar lipids were reduced in the knockout strains. As a result, the cell walls of the knockout strains were significantly more hydrophobic than those of the wild type and the complemented strains. We hypothesize that reduced levels of GPLs and polar lipids may contribute to the antibiotic resistance shown by the knockout strains. Altogether, our data suggest that (p)ppGpp and c-di-GMP may be involved in the metabolism of glycopeptidolipids and polar lipids in M. smegmatis.
INTRODUCTION
Nucleotide-based second messengers regulate various biological processes in all domains of life. Pentaphosphate guanosine (pppGpp) or tetraphosphate guanosine (ppGpp), collectively referred to as (p)ppGpp, and bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) are two such bacterial second messengers (1, 2). The alarmone (p)ppGpp is synthesized when bacteria are stressed or starved and regulates important processes, like stringent response, quorum sensing, virulence, and biofilm formation (2–5). In mycobacteria, (p)ppGpp is synthesized and broken down by the dual-function enzyme Rel, encoded by the rel gene (6). The Mycobacterium smegmatis rel gene knockout (ΔrelMsm) strain is compromised for long-term survival during nutrient starvation and progressive hypoxia (7). Similarly, Mycobacterium tuberculosis lacking the RelMtb protein has reduced long-term survival in the lungs of mice and in a mouse hypoxic granuloma model and fails to form tubercle lesions in a guinea pig model of infection (8–11). Both the M. tuberculosis and M. smegmatis genomes encode a second (p)ppGpp synthetase called small alarmone synthetase (12, 13). However, its role in mycobacterial physiology has not been completely elucidated.
The signaling nucleotide c-di-GMP is synthesized by diguanylate cyclases containing GGDEF domains and is broken down by phosphodiesterases containing either EAL or HD-GYP domains (2, 14–16). It regulates processes such as biofilm formation, virulence, and cell division (17, 18). In M. smegmatis, c-di-GMP is synthesized and broken down by a bifunctional protein, DcpA (formerly called the MSDGC1 protein), encoded by the dcpA gene, which harbors both GGDEF and EAL domains (19, 20). When dcpA is removed, the resulting ΔdcpA strain exhibits reduced long-term survival during nutrient starvation (19). C-di-GMP binds the transcription factor LtmA and regulates the expression of lipid transport and metabolism genes in M. smegmatis (21). It also regulates pathogenicity and dormancy in M. tuberculosis (22).
Apart from long-term survival, pathogenicity, and virulence, other phenotypes or processes regulated by (p)ppGpp and c-di-GMP in mycobacteria so far remain unexplored. Moreover, we also wanted to investigate whether the two GTP-derived nucleotide second messengers regulate similar or different processes in mycobacteria. Recently, it was shown that a strain of Pseudomonas aeruginosa lacking (p)ppGpp is sensitive to multiple classes of antibiotics and is defective in biofilm formation (23). Hence, we investigated the question of how a strain of M. smegmatis impaired in (p)ppGpp or c-di-GMP signaling would behave in the presence of antibiotics. To address this question, we used phenotype microarray (PM) technology, which allows one to study hundreds of biochemical phenotypes of bacteria simultaneously (24, 25). It has been successfully used to study the metabolic profiles of Escherichia coli (25), Bacillus subtilis (26), Staphylococcus aureus (27), M. tuberculosis and Mycobacterium bovis (28). It consists of a specialized reader-incubator and a set of 20 96-well plates numbered PM1 to PM20, which are coated with various sources of carbon, nitrogen, sulfur, phosphorus, osmolytes, and antimicrobials (24, 25). For our studies, we used 10 PM plates, PM11 to PM20, that were coated with various antimicrobials. Hence, the present work was designed to investigate the roles of (p)ppGpp and c-di-GMP in mycobacterial survival and physiology using PM technology.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A list of all the strains used in this study is provided in Table 1. M. smegmatis mc2155 (the wild type [WT]) (29) and its isogenic variants ΔrelMsm, the rel complemented strain (relComp) (30), ΔdcpA, and the dcpA complemented strain (dcpAComp) were grown in MB7H9 (Difco) broth with 2% (wt/vol) glucose as a carbon source and 0.05% (vol/vol) Tween 80 or on MB7H9 medium containing 1.5% (wt/vol) agar. The antibiotics kanamycin and hygromycin were used at a concentration of 40 μg/ml when required.
TABLE 1.
Strains used in the study
| Strain | Description | Source or reference |
|---|---|---|
| M. smegmatis mc2155 | Wild-type strain | 29 |
| ΔrelMsm | M. smegmatis strain in which the rel gene has been replaced with a Hygr cassette | Laboratory strain (7) |
| ΔdcpA | M. smegmatis strain in which the dcpA gene has been replaced with a Kanr cassette | Laboratory strain (19) |
| relComp | ΔrelMsm strain of M. smegmatis complemented with the rel gene; Kanr | Gift from Christina Stallings (30) |
| dcpAComp | ΔdcpA strain complemented with the dcpA gene; Kanr Hygr | Laboratory strain (19) |
PM procedure.
All the M. smegmatis strains (WT, ΔrelMsm, relComp, ΔdcpA, and dcpAComp) were initially grown on MB7H9 agar containing 2% glucose for 48 h. Cell suspensions with a transmittance of 81% were made in MB7H9 broth from these plates using a sterile swab dipped in 0.1% Tween 80. To these suspensions, the redox indicator tetrazolium violet was added to a final concentration of 0.01%. One hundred microliters of the suspension was inoculated into the wells of the PM plates. The plates were subsequently incubated at 37°C for 48 h in a Biolog OmniLog incubator, which measures the growth of bacteria every 15 min. As bacteria respire, tetrazolium violet is reduced to a purple color, which is directly proportional to the growth of the bacteria. The intensity of the purple color is recorded as a dye reduction value, which is then plotted as the area under the curve (AUC) by the Biolog parametric software. The AUC for the test strain (e.g., a mutant) is then overlaid on the AUC of the reference strain (e.g., the WT) by parametric software. This results in a green-yellow-red display, where the overlap between the two AUCs is shown in yellow. If the AUC of the test strain is more than that of the reference strain, then it is said to have “gained the phenotype” for that particular compound present in the PM plate, and this is depicted in green in the overlaid AUC plot. In other words, the test strain has enhanced survival or growth compared with the reference strain. On the other hand, if the test strain has an AUC value less than that of the reference strain, then it is said to have “lost the phenotype,” and this is shown as red in the overlaid AUC plots. In other words, the test strain has poor growth or survival compared with the reference strain. The gain or loss of phenotype is always measured with respect to the reference strain. Dye reduction curve values were plotted using ggplot2 in the R statistical software (http://www.r-project.org). Each antimicrobial was present in four different concentrations in the PM plates, and hence, a total of 240 different compounds were present in the 10 PM plates used in this study. A list of the compounds is presented in Data Set S1 in the supplemental material. PM analysis of the WT, the ΔrelMsm, and the ΔdcpA strains was carried out at least four times and that of the complemented strains twice.
Determination of MICs.
MIC values were determined using the resazurin microtiter plate assay (REMA), which is a very widely used method to determine MICs for various mycobacteria (31). Briefly, the transmittance of the culture was adjusted to a McFarland turbidity standard of 1 and then diluted 1:10. One hundred microliters of the diluted culture (approximately 3 × 106 CFU) was inoculated into 96-well microtiter plates containing 2-fold dilution series of the antibiotics ampicillin, amoxicillin, streptomycin, rifampin, erythromycin, tetracycline, and norfloxacin. The plates were sealed with Parafilm M (Bemis Flexible Packaging, Neenah, WI) and incubated in a humidified incubator at 37°C for 36 h. Postincubation, 15 μl of 0.01% resazurin was added to the wells of the microtiter plates, and the plates were further incubated for 4 h. If live bacteria are present in any of the wells, the dye turns pink; otherwise, it stays blue. The MIC was defined as the minimum antibiotic concentration at which the color of the resazurin did not change. The experiment was performed twice. We also plated an aliquot from such wells onto MB7H9 agar to determine the number of surviving CFU (CFU/ml).
Biofilm formation and quantification assay.
Biofilms were grown in Sauton's fluid base medium supplemented with 2% glucose as a carbon source, as previously described by Mathew et al. (32). Twenty-five milliliters of the medium was poured into sterile petri dishes. Two hundred microliters of the saturated culture was inoculated into petri dishes containing medium. The plates were then incubated at 37°C in a humidified incubator for 6 days and imaged with a 500-ms exposure of white light in a Sygene G-box gel documentation system.
Quantification of biofilms was done as described previously by O'Toole et al. (33) and as adapted successfully by Bharati et al. (19) in the case of M. smegmatis. Briefly, the stationary-phase cultures were washed with Sauton's medium, diluted to a final optical density at 600 nm (OD600) of 0.1 in Sauton's medium containing 2% glucose, and inoculated into the wells of 96-well polystyrene plates. Each well received an inoculum of 200 μl, and 8 wells were assayed for each strain. The plates were sealed with Parafilm M and incubated at 37°C for 3 to 4 days. After the incubation, biofilms were removed from the wells, and the wells were washed twice with water. The adherent biofilm was quantified by staining with 1% crystal violet for 45 min at room temperature. The residual dye was washed with water, and the plate was allowed to dry. The bound dye was solubilized in 300 μl 80% (vol/vol) ethanol, and the A570 was measured using a microtiter plate reader (SpectraMax 340PC384; Molecular Devices).
Primary adherence assay.
The protocol for the primary adherence assay was adapted from that of Mohamed et al. (34). Briefly, the stationary-phase cultures of all the strains were diluted to an OD600 of 0.1. Two milliliters of the diluted cultures was poured into the wells of 12-well polystyrene plates and incubated for 4 h. The wells were then washed with phosphate-buffered saline (PBS) twice, fixed with Bouin's solution (acetic acid, 5%; formaldehyde, 9%; picric acid, 0.9%) for 15 min, and finally washed with 1× PBS. Adherent bacteria were then visualized under a 40× objective in an Olympus IX81 microscope. Five different fields were chosen randomly and observed. The experiment was performed twice.
Colony morphology and sliding motility.
To determine colony morphology, cells were plated on MB7H9 agar plates supplemented with 2% glucose and incubated at 37°C for 6 to 7 days. Postincubation, images of the individual colonies were taken. To score for sliding motility, we followed the protocol described by Martinez et al. (35). Briefly, cells were grown until stationary phase in MB7H9 medium supplemented with 2% glucose and 0.05% Tween 80. Three microliters of the culture was spotted in the middle of MB7H9 plates solidified with 0.3% agarose without any carbon source. The plates were incubated at 37°C for 4 days in a humidified incubator as described previously (35). After the incubation, the plates were imaged with a 500-ms exposure of white light in a Sygene G-box gel documentation system.
Aggregation assay.
The protocol for the aggregation assay was adapted from that of Deshayes et al. (36). Bacteria were grown until stationary phase in MB7H9 medium supplemented with 2% glucose and 0.05% Tween 80. Individual cells were separated from the aggregates by centrifuging the cells at 10 × g for 1 min. The OD600 values of the supernatants were compared to those of cultures in which the aggregates were broken by vortexing with glass beads. The aggregation index is defined as the ratio between the OD600 of the supernatant and the OD600 of the vortexed culture.
GPL isolation.
Glycopeptidolipids (GPLs) from M. smegmatis cultures were purified using the protocol described by Khoo et al. (37). Stationary-phase cultures were centrifuged at 5,000 × g for 10 min, and the cell pellet was lyophilized. One hundred milligrams of lyophilized cells was suspended in 2:1 (vol/vol) chloroform-methanol and kept at 37°C for 8 h without agitation, followed by 12 h with agitation. The suspension was subsequently centrifuged at 10,000 × g for 10 min. The lower organic phase was collected in a round-bottom flask and dried to completion using a RotaVac vacuum evaporator. The GPLs were then extracted in a minimum volume of 2:1 (vol/vol) chloroform-methanol and deacetylated by adding methanolic NaOH to a final concentration of 0.1 N and incubating at room temperature for 30 min. The alkali-stable GPLs were then concentrated to complete dryness on a RotaVac vacuum evaporator, resuspended in equal volumes of 4:2:1 chloroform-methanol-water, and stored at room temperature for 1 h. The suspension was subjected to centrifugation for 30 min at 12,000 × g at 15°C. The lower organic layer was collected, dried by vacuum evaporation, and resuspended in a minimum volume of 2:1 chloroform-methanol and 5 ml of saturated brine. After storing the suspension for 10 min at room temperature, it was centrifuged at 12,000 × g for 30 min at 15°C. The lower organic phase containing the purified GPLs was collected and dried by vacuum evaporation, resuspended in 2:1 chloroform-methanol, and stored at 4°C until further use. The thin-layer chromatograms of GPLs were developed in 9:1 chloloform-methanol and visualized by charring at 120°C after spraying the chromatogram with 20% ethanolic sulfuric acid.
Purification of polar and apolar lipids.
Polar and apolar lipids were isolated following the protocol of Parish and Stoker (38). Briefly, 50 mg of lyophilized cells was suspended in 2 ml of methanol-0.3% NaCl (100:10) with 1 ml of petroleum ether and rotated for 15 min. After removing the upper petroleum ether layer carefully, 1 ml of petroleum ether was again added to the residue and further rotated for 15 min. Both petroleum ether extracts were then combined and evaporated under liquid nitrogen to yield apolar lipids, which were resuspended in dichloromethane prior to thin-layer chromatography (TLC) analysis. To extract polar lipids, the methanolic saline extract was heated in a boiling-water bath for 5 min, allowed to cool to room temperature, mixed with 2.3 ml of chloroform-methanol-0.3% NaCl (9:10:3), and rotated for 60 min. The biomass was separated from the extract by centrifugation and retained and was further extracted with 0.75 ml of chloroform-methanol-0.3% NaCl (5:10:4) for 30 min. The two solvent extracts were combined, mixed with 1.3 ml of chloroform and 1.3 ml of 0.3% NaCl, rotated for 30 min, and subsequently centrifuged at 3,500 × g to separate the lower organic and the upper aqueous phases. The aqueous phase was discarded, and the organic phase was dried to yield polar lipids. The dried lipids were then dissolved in 300 μl of chloroform-methanol (2:1) prior to TLC analysis. The polar lipids were separated by developing the thin-layer chromatogram in chloroform-methanol-water (60:30:6) and visualized by charring the TLC sheet at 110°C after spraying with 5% ethanolic phosphomolybdic acid.
Hydrophobicity assay.
The protocol for the hydrophobicity assay was adapted from that of Mohamed et al. (39). Briefly, stationary-phase cultures were diluted to an OD600 of 0.6, and 1.8 ml of the diluted culture was mixed with 0.2 ml of p-xylene. The mixture was then vortexed for 90 s and allowed to settle at room temperature for 20 min. The aqueous phase was then carefully removed, and its OD600 was measured. The percentage of cell hydrophobicity was calculated as follows: percent cell hydrophobicity = [1 − (ODfinal/ODinitial)] × 100.
RESULTS
The ΔrelMsm and the ΔdcpA strains show enhanced survival after exposure to several antibiotics present in the PM plates.
Postincubation, we compared the AUCs of the knockout strains with those of the WT or the respective complemented strains, and also those of the complemented strains with that of the WT, using Biolog's parametric software. The differences between the AUCs of the strains are listed in Data Set S1 in the supplemental material. Since the (p)ppGpp0 ΔrelA spoT strain of P. aeruginosa was sensitive to multiple antibiotics, we anticipated that both the ΔrelMsm and the ΔdcpA strains of M. smegmatis would be sensitive to the antibiotics present in the PM plates. Surprisingly, the comparisons of AUCs revealed that both the ΔrelMsm and the ΔdcpA strains displayed enhanced survival in the presence of many antibiotics compared with the WT or the respective complemented strains (Table 2; see Fig. S1 to S4 in the supplemental material). We predicted that the growth profiles of the complemented strains would look similar to that of the WT. This was the case when the AUC of the dcpAComp strain was compared with that of the WT (see Fig. S4 in the supplemental material). However, the growth of the relComp strain differed from that of the WT. Since the complemented rel gene was not under its native promoter, the amount of Rel protein expressed may not be equal to that in the WT, thus explaining the anomaly (see Fig. S5 in the supplemental material). The numbers of antimicrobials for which the test strain had either lost (poor growth) or gained (better growth) phenotypes compared with the reference strain are listed in Table 2.
TABLE 2.
Numbers of phenotypes lost (poor growth) or gained (better growth) when the AUCs of the strains under study were compared with each other
| Strains compared (test strain vs. reference strain) | No. of phenotypes |
|
|---|---|---|
| Gained | Lost | |
| ΔrelMsm vs. WT | 199 | 39 |
| ΔdcpA vs. WT | 213 | 23 |
| ΔrelMsm vs. relComp | 221 | 16 |
| ΔdcpA vs. dcpAComp | 195 | 43 |
| relComp vs. WT | 89 | 147 |
| dcpAComp vs. WT | 142 | 91 |
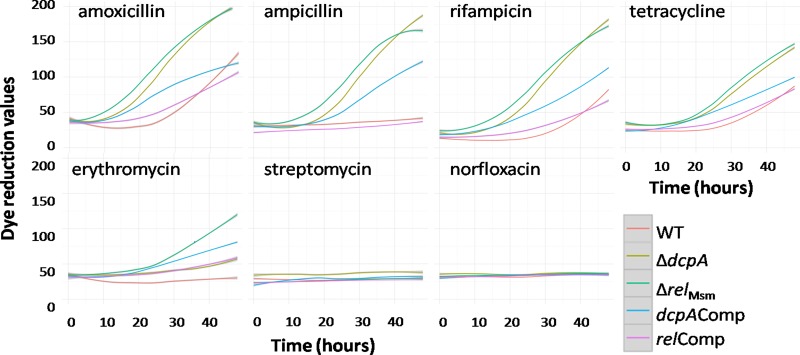
To depict the trends in the PM data, we plotted the dye reduction values for seven antibiotics belonging to different antibiotic classes using ggplot2 in the R statistical software (Fig. 1) and determined their MICs using the REMA method to validate the PM data (Table 3). Both the ΔrelMsm and the ΔdcpA strains exhibited enhanced growth compared with the WT or the respective complemented strains in the presence of the β-lactam antibiotics amoxicillin and ampicillin in the PM plates (Fig. 1). As mycobacteria are inherently resistant to β-lactam antibiotics, the MICs for amoxicillin and ampicillin were indeterminable (Table 3). This was corroborated by the CFU on the MB7H9 agar plates when an aliquot was plated from the wells containing amoxicillin and ampicillin at concentrations as high as 1,024 μg/ml (see Table S1 in the supplemental material). In the wells containing the RNA polymerase inhibitor rifampin, both the ΔrelMsm and the ΔdcpA strains displayed enhanced survival compared with the WT or the respective complemented strains in the PM plates (Fig. 1). The 2-fold-higher MICs of the knockout strains substantiate this observation (Table 3). However, the complemented strains had MICs equal to those of the knockout strains, and this could be due to the fact that the complemented genes were not under their native promoters (Table 3). In the wells containing the protein synthesis inhibitor tetracycline, the knockout strains had 2-fold-higher MICs than the WT or the respective complemented strains, thus reflecting the trend in the PM data (Fig. 1 and Table 3).
FIG 1.
Growth profiles of strains in the presence of seven antibiotics on PM plates. The y axis represents the dye reduction values generated due to bacterial respiration, and x axis represents the incubation times of the PM plates. The graphs were plotted using the ggplots function of the R statistical package.
TABLE 3.
MICs of representative antibiotics for the WT and its isogenic variants determined using the REMA method
| Antibiotic | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| WT | ΔrelMsm | relComp | ΔdcpA | dcpAComp | |
| Rifampin | 8 | 16 | 16 | 16 | 16 |
| Streptomycin | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Erythromycin | 16 | 64 | 16 | 64 | 32 |
| Tetracycline | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 |
| Norfloxacin | 4 | 4 | 4 | 4 | 4 |
| Ampicillin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
| Amoxicillin | >1,024 | >1,024 | >1,024 | >1,024 | >1,024 |
For the macrolide antibiotic erythromycin, both knockout strains showed better survival than the WT, but the dcpAComp strain displayed better growth than the ΔdcpA knockout strain (Fig. 1). The MIC of the ΔrelMsm strain was 4-fold higher than those of the WT and the relComp strains (Table 3). The MIC of the ΔdcpA strain was 4-fold higher than that of the WT and 2-fold higher than that of the dcpAComp strain (Table 3). None of the strains showed growth in the wells containing the aminoglycoside antibiotic streptomycin and the DNA topoisomerase inhibitor norfloxacin in the PM plates (Fig. 1), and this is seen as no change in the MICs of any of the strains for these antibiotics (Table 3). Other than the β-lactam antibiotics, CFU were not detected for any antibiotics (see Table S1 in the supplemental material). Thus, the MICs determined using the REMA method (Table 3) and CFU (see Table S1 in the supplemental material) more or less reflect the trends seen in the PM data. Taken together, the PM data and the MICs suggest that the ΔrelMsm and ΔdcpA strains are more resistant to antibiotics than the WT or the respective complemented strains.
The knockout strains are defective in biofilm formation and display altered cell surface phenotypes.
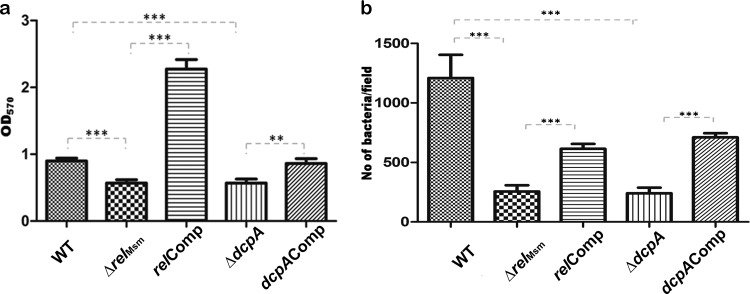
After the unexpected observation of the knockout strains showing resistance to antibiotics, we subsequently checked if biofilm formation was impaired in these strains. We observed that the pellicles formed on the surface of Sauton's medium by the ΔrelMsm and ΔdcpA strains were not as robust as those formed by the WT or the respective complemented strains (see Fig. S7 in the supplemental material). This observation was further confirmed by quantifying the biofilm using 1% crystal violet, as described in Materials and Methods. The amount of biofilm formed by the knockout strains was reduced by almost 40% compared with the WT (Fig. 2a) (P ≤ 0.001). As expected, biofilm formation was restored nearly to WT levels in the dcpAComp strain (P ≤ 0.001 compared with ΔdcpA), but the relComp strain consistently showed values that were even higher than those of the WT (Fig. 2a) (P ≤ 0.001 compared with the ΔrelMsm strain). We could not find any support in the literature for this observation, and the reasons are still unclear.
FIG 2.
(a) Quantification of biofilms using 1% crystal violet. The absorbance at 570 nm of crystal violet retained by the residual biofilm is shown. The error bars represent standard errors of the mean for two-tailed paired t tests. The experiment was repeated at least twice. The graph was plotted using GraphPad Prism 5. **, P ≤ 0.01, and ***, P ≤ 0.001. (b) Primary adherence assay on the polystyrene surface. The number of bacteria attached per field of view of the microscope were determined. Five randomly chosen fields of view were imaged and the bacteria counted. The error bars represent standard deviations for two-tailed paired t tests. The experiment was repeated at least twice. The graph was plotted using GraphPad Prism 5. ***, P ≤ 0.001.
Adherence to an abiotic surface is the first step of biofilm formation. Since the knockout strains are defective in biofilm formation, we anticipated that the number of cells attached per field of view in the microscope for the knockout strains would be less than for the WT or the complemented strains. A primary adherence assay revealed that this was indeed the case. The numbers of knockout cells attached per field of view in the microscope were significantly lower than those for the WT (P ≤ 0.001), and as expected, the ability to attach was regained by the complemented strains (P ≤ 0.001 compared with the respective complemented strains) (Fig. 2b). Overall, the primary adherence assay and biofilm quantification prove that the knockout strains are indeed defective in biofilm formation.
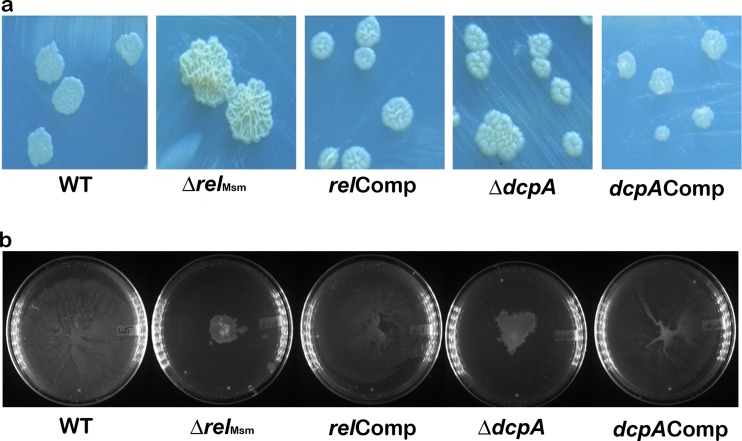
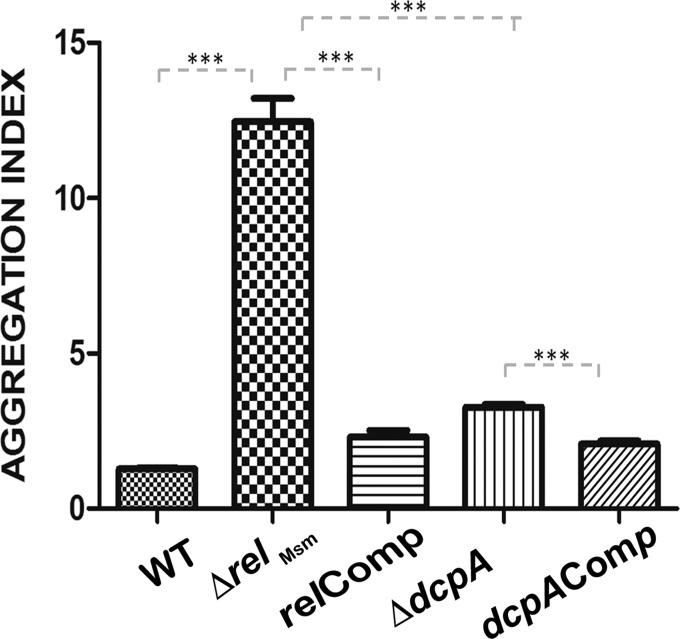
The ability to form biofilm is correlated with the amount of GPLs in the cell wall of M. smegmatis (40, 41). Hence, we hypothesized that the amounts of GPLs must be reduced in the knockout strains. Apart from biofilm formation, GPLs are responsible for macroscopic surface phenotypes, like colony morphology, sliding motility, and aggregation. Hence, we scored for these phenotypes, as well. We observed that the colonies of the ΔrelMsm and ΔdcpA strains were raised and dry, while those of the WT were flat, smooth, and glistening (Fig. 3a). This phenotype was rescued in the complemented relComp and dcpAComp strains (Fig. 3a). Similarly, sliding motility was impaired in the ΔrelMsm and ΔdcpA strains compared with the WT and the respective complemented strains (Fig. 3b). However, the colony morphology and sliding motility of the ΔrelMsm strain are more severely affected than those of the ΔdcpA strain. We also noticed that the liquid cultures of the knockout strains settled faster than those of the WT or the complemented strains. Upon quantifying this aggregation, we found that the aggregation indexes of the knockout strains were significantly higher than those of the WT (P ≤ 0.001) or the complemented strains (P ≤ 0.001) (Fig. 4).
FIG 3.
(a) Colony morphologies of strains on MB7H9 agar supplemented with 2% glucose. The plates were incubated at 37°C for 6 to 7 days. (b) Sliding motilities of strains on 0.3% agarose plates. The plates were incubated at 37°C for 4 days.
FIG 4.
Aggregation indexes of the strains under study. Both the ΔrelMsm and the ΔdcpA strains display significantly higher aggregation than the WT or the respective complemented strains, relComp and dcpAComp. The experiment was repeated twice with three technical replicates. The error bars represent standard errors of the mean of one-tailed unpaired t tests. The graph was plotted using GraphPad Prism 5. ***, P ≤ 0.001.
Amounts of GPLs and polar lipids are reduced in the ΔrelMsm and the ΔdcpA strains.
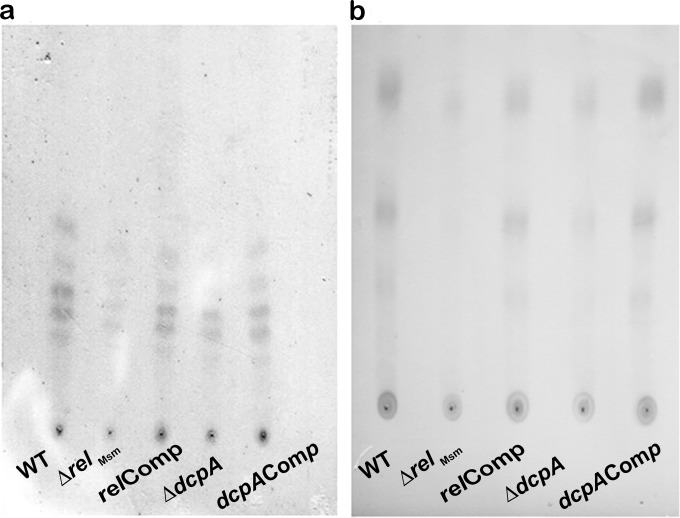
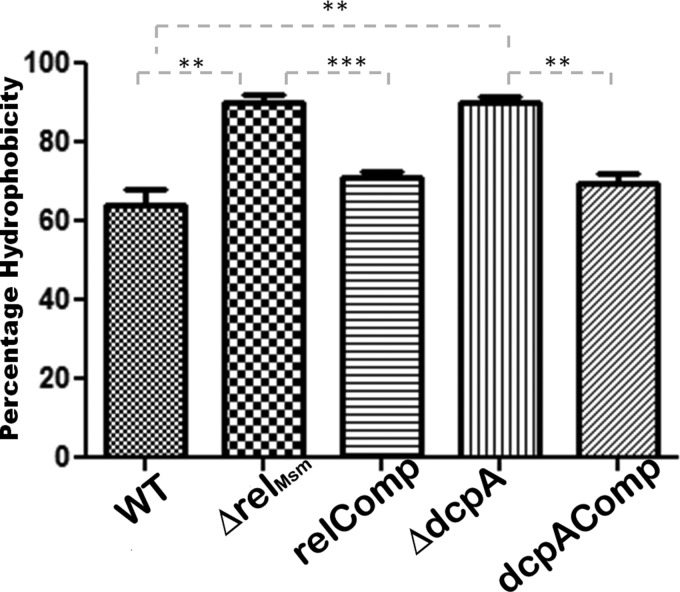
Reduction in biofilm formation and changes in the macroscopic surface properties of the knockout strains strongly suggested alterations in the lipid composition of the cell walls of the knockout strains. Hence, we isolated various cell wall fractions and analyzed them by TLC. We found that the amounts of glycopeptidolipids were significantly reduced in the knockout strains (Fig. 5a). We confirmed the identities of various GPL bands with matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (see Fig. S10 in the supplemental material). We also determined that the levels of mycolic acids were unaltered in the ΔrelMsm and ΔdcpA strains (see Fig. S8 in the supplemental material). Similarly, the levels of apolar lipids did not change in any of the strains (see Fig. S9 in the supplemental material). However, TLC analysis of the polar lipids revealed that the knockout strains had smaller amounts of polar lipids in their cell walls than the WT or the respective complemented strains (Fig. 5b). Reduced levels of polar lipids and unaltered levels of apolar lipids in the knockout strains led us to hypothesize that their cell walls may be more hydrophobic (Fig. 5b; see Fig. S9 in the supplemental material). A hydrophobicity assay done in p-xylene confirmed this prediction. Both the ΔrelMsm and ΔdcpA strains displayed significantly higher cell wall hydrophobicity than the WT (P ≤ 0.01) and the relComp (P ≤ 0.001) strains or the dcpAComp (P ≤ 0.001) strain (Fig. 6).
FIG 5.
Thin-layer chromatograms of glycopeptidolipids and polar lipids. (a) The thin-layer chromatogram was developed in chloroform-methanol (9:1), and GPLs were visualized by charring at 120°C after spraying with 20% ethanolic H2SO4. (b) The thin-layer chromatogram was developed in chloroform-methanol-water (60:30:6), and polar lipids were visualized by charring at 110°C after spraying with 5% ethanolic phosphomolybdic acid.
FIG 6.
Hydrophobicity assay in p-xylene. The experiment was done in triplicate and repeated twice. The graph was plotted using GraphPad Prism 5. **, P ≤ 0.01; ***, P ≤ 0.001.
DISCUSSION
In this study, we investigated whether M. smegmatis impaired in (p)ppGpp or c-di-GMP signaling would be sensitive to multiple antibiotics and defective in biofilm formation. We observed that the ΔrelMsm and ΔdcpA strains showed enhanced survival compared with the WT or the respective complemented strains following exposure to many antibiotics. The MIC values of the knockout strains, which were higher than those of the WT or the respective complemented strains, validated this observation. Thus, it seems that both the ΔrelMsm and ΔdcpA strains display multidrug resistance. We also found that the knockout strains were defective in biofilm formation and exhibited altered surface properties. These phenotypes directly correlate with the amount of GPLs present in the cell wall of M. smegmatis (40, 41). TLC analysis of various cell wall fractions revealed that the amounts of GPLs and polar lipids were reduced in the knockout strains. It has been shown previously that drug-resistant clinical isolates of the Mycobacterium avium complex have altered GPL profiles (42). The rough-colony variant of Mycobacterium abscessus, which lacks GPLs, is more virulent than the smooth morphotype (43). Since the absence or the reduction in the amount of GPLs or alteration in their profiles is linked with phenotypes such as drug resistance and virulence, we hypothesize that the reduced levels of GPLs in the cell walls of the ΔrelMsm and the ΔdcpA strains may possibly be responsible for their multidrug resistance. Such multidrug resistance in M. smegmatis was reported previously, where porins had been knocked out or when there was an upregulation of the efflux pumps (44, 45). Since (p)ppGpp and c-di-GMP are global regulators, the enhanced survival of the knockouts cannot be attributed only to the downregulation of porins or the upregulation of efflux pumps; however, this possibility cannot be ruled out. Similarly, the increased hydrophobicity of the knockout strains may hinder the uptake of some antibiotics, thus making them more tolerant of such antibiotics.
C-di-GMP regulates biofilm formation in many Gram-negative bacteria (14, 15). It was shown recently that it might also be involved in biofilm formation in the Gram-positive B. subtilis (46). The alarmone (p)ppGpp regulates biofilm formation, not only in Gram-negative bacteria, like Vibrio cholerae and E. coli, but also in Gram-positive bacteria, like Streptococcus mutans and Enterococcus faecalis (47–50). It was found that the relComp strain formed more biofilms than the WT strain; however, its primary adherence values were less than those of the WT. This discrepancy could be due to the different methods used to investigate the phenomenon of biofilm formation. The primary adherence assay involves imaging the cells that are attached to a surface without any staining. On the other hand, quantification involves staining the cells with crystal violet. The possibility that the relComp strain took up more stain cannot be ruled out. However, the molecular mechanism of the discrepancy between the primary adherence assay and the biofilm formation assay is not clear. Irrespective of this, our data suggest that both (p)ppGpp and c-di-GMP may be involved in the regulation of biofilm formation in the Gram-positive actinobacterium M. smegmatis.
Since both second messengers are global regulators, it is likely that they regulate some of the same phenotypes, and cell wall biosynthesis could be one of those phenotypes. The alarmone (p)ppGpp is involved in the regulation of fatty acids and lipopolysaccharide synthesis in E. coli (5, 51, 52). Recently, it was shown that c-di-GMP binds to a transcription factor, LtmA, that regulates the expression of genes involved in lipid transport in M. smegmatis (21). Since both the ΔrelMsm and the ΔdcpA strains have reduced levels of cell wall GPLs and polar lipids, we propose that both (p)ppGpp and c-di-GMP may be involved in the metabolism of GPLs and polar lipids in M. smegmatis.
Thus, our data suggest that (p)ppGpp and c-di-GMP regulate hitherto unreported phenotypes, like antibiotic resistance, biofilm formation, colony morphology, and sliding motility, by regulating GPL and polar lipid synthesis in M. smegmatis.
Supplementary Material
ACKNOWLEDGMENTS
K.R.G., S.K., and D.C. thank the Department of Biotechnology (DBT), Government of India, for funding this work. K.R.G. thanks the Council for Scientific and Industrial Research, Government of India, for a senior research fellowship. S.K. thanks the DBT for a research associateship.
We acknowledge Christina Stallings for providing the relComp strain. K.R.G. acknowledges Praveen Anand and Priyanka Baloni for their help in generating Fig. 1 using the R statistical analysis software. K.R.G. thanks N. V. Joshi for helpful discussions on t tests, Wareed Ahmad and Ujjwal Rathore for helpful discussions on the manuscript, and P. Anushya, M. Geetha, and Nandini Mani for improving the quality of the manuscript by proofreading it.
K.R.G. and D.C. designed the experiments. K.R.G. performed the experiments. S.K. helped K.R.G. with the PM experiments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03999-14.
REFERENCES
- 1.Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. 2013. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev 42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 2.Pesavento C, Hengge R. 2009. Bacterial nucleotide-based second messengers. Curr Opin Microbiol 12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Chatterji D, Ojha AK. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol 4:160–165. doi: 10.1016/S1369-5274(00)00182-X. [DOI] [PubMed] [Google Scholar]
- 4.Jain V, Kumar M, Chatterji D. 2006. ppGpp: stringent response and survival. J Microbiol 44:1–10. [PubMed] [Google Scholar]
- 5.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 6.Avarbock D, Salem J, Li LS, Wang ZM, Rubin H. 1999. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233:261–269. doi: 10.1016/S0378-1119(99)00114-6. [DOI] [PubMed] [Google Scholar]
- 7.Mathew R, Ojha AK, Karande AA, Chatterji D. 2004. Deletion of the rel gene in Mycobacterium smegmatis reduces its stationary phase survival without altering the cell-surface associated properties. Curr Sci 86:149–153. [Google Scholar]
- 8.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol 182:4889–4898. doi: 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE III. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A 100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC. 2010. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202:1397–1404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med 200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss LA, Stallings CL. 2013. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J Bacteriol 195:5629–5638. doi: 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murdeshwar MS, Chatterji D. 2012. MS_RHII-RSD, a dual-function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J Bacteriol 194:4003–4014. doi: 10.1128/JB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 15.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 17.Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol 61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bharati BK, Sharma IM, Kasetty S, Kumar M, Mukherjee R, Chatterji D. 2012. A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 158:1415–1427. doi: 10.1099/mic.0.053892-0. [DOI] [PubMed] [Google Scholar]
- 20.Sharma IM, Prakash S, Dhanaraman T, Chatterji D. 2014. Characterization of a dual-active enzyme, DcpA, involved in cyclic diguanosine monophosphate turnover in Mycobacterium smegmatis. Microbiology 160:2304–2318. doi: 10.1099/mic.0.080200-0. [DOI] [PubMed] [Google Scholar]
- 21.Li W, He ZG. 2012. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res 40:11292–11307. doi: 10.1093/nar/gks923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong Y, Zhou X, Fang H, Yu D, Li C, Sun B. 2013. Cyclic di-GMP mediates Mycobacterium tuberculosis dormancy and pathogenecity. Tuberculosis 93:625–634. doi: 10.1016/j.tube.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochner BR. 2003. New technologies to assess genotype-phenotype relationships. Nat Rev Genet 4:309–314. doi: 10.1038/nrg1046. [DOI] [PubMed] [Google Scholar]
- 25.Bochner BR, Gadzinski P, Panomitros E. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res 11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y, Asai K, Sadaie Y, Helmann JD. 2010. Transcriptomic and phenotypic characterization of a Bacillus subtilis strain without extracytoplasmic function sigma factors. J Bacteriol 192:5736–5745. doi: 10.1128/JB.00826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Eiff C, McNamara P, Becker K, Bates D, Lei XH, Ziman M, Bochner BR, Peters G, Proctor RA. 2006. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J Bacteriol 188:687–693. doi: 10.1128/JB.188.2.687-693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khatri B, Fielder M, Jones G, Newell W, Abu-Oun M, Wheeler PR. 2013. High throughput phenotypic analysis of Mycobacterium tuberculosis and Mycobacterium bovis strains' metabolism using Biolog phenotype microarrays. PLoS One 8:e52673. doi: 10.1371/journal.pone.0052673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 30.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. 2009. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathew R, Mukherjee R, Balachandar R, Chatterji D. 2006. Deletion of the rpoZ gene, encoding the omega subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology 152:1741–1750. doi: 10.1099/mic.0.28879-0. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. 1999. Genetic approaches to study of biofilms. Methods Enzymol 310:91–109. doi: 10.1016/S0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 34.Mohamed JA, Huang W, Nallapareddy SR, Teng F, Murray BE. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun 72:3658–3663. doi: 10.1128/IAI.72.6.3658-3663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez A, Torello S, Kolter R. 1999. Sliding motility in mycobacteria. J Bacteriol 181:7331–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deshayes C, Laval F, Montrozier H, Daffe M, Etienne G, Reyrat JM. 2005. A glycosyltransferase involved in biosynthesis of triglycosylated glycopeptidolipids in Mycobacterium smegmatis: impact on surface properties. J Bacteriol 187:7283–7291. doi: 10.1128/JB.187.21.7283-7291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khoo KH, Chatterjee D, Dell A, Morris HR, Brennan PJ, Draper P. 1996. Novel O-methylated terminal glucuronic acid characterizes the polar glycopeptidolipids of Mycobacterium habana strain TMC 5135. J Biol Chem 271:12333–12342. doi: 10.1074/jbc.271.21.12333. [DOI] [PubMed] [Google Scholar]
- 38.Parish T, Stoker NG. 1998. Mycobacteria protocols, vol 101 Humana Press, Totowa, NJ. [Google Scholar]
- 39.Mohamed JA, Teng F, Nallapareddy SR, Murray BE. 2006. Pleiotrophic effects of 2 Enterococcus faecalis sagA-like genes, salA and salB, which encode proteins that are antigenic during human infection, on biofilm formation and binding to collagen type i and fibronectin. J Infect Dis 193:231–240. doi: 10.1086/498871. [DOI] [PubMed] [Google Scholar]
- 40.Recht J, Martinez A, Torello S, Kolter R. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J Bacteriol 182:4348–4351. doi: 10.1128/JB.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schorey JS, Sweet L. 2008. The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis. Glycobiology 18:832–841. doi: 10.1093/glycob/cwn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khoo KH, Jarboe E, Barker A, Torrelles J, Kuo CW, Chatterjee D. 1999. Altered expression profile of the surface glycopeptidolipids in drug-resistant clinical isolates of Mycobacterium avium complex. J Biol Chem 274:9778–9785. doi: 10.1074/jbc.274.14.9778. [DOI] [PubMed] [Google Scholar]
- 43.Bernut A, Herrmann JL, Kissa K, Dubremetz JF, Gaillard JL, Lutfalla G, Kremer L. 2014. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci U S A 111:E943–E952. doi: 10.1073/pnas.1321390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephan J, Mailaender C, Etienne G, Daffe M, Niederweis M. 2004. Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob Agents Chemother 48:4163–4170. doi: 10.1128/AAC.48.11.4163-4170.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li XZ, Zhang L, Nikaido H. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob Agents Chemother 48:2415–2423. doi: 10.1128/AAC.48.7.2415-2423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Chai Y, Guo JH, Losick R. 2012. Evidence for cyclic di-GMP-mediated signaling in Bacillus subtilis. J Bacteriol 194:5080–5090. doi: 10.1128/JB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He H, Cooper JN, Mishra A, Raskin DM. 2012. Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol 194:2962–2972. doi: 10.1128/JB.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. 2009. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- 49.Lemos JA, Brown TA Jr, Burne RA. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect Immun 72:1431–1440. doi: 10.1128/IAI.72.3.1431-1440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chavez de Paz LE, Lemos JA, Wickstrom C, Sedgley CM. 2012. Role of (p)ppGpp in biofilm formation by Enterococcus faecalis. Appl Environ Microbiol 78:1627–1630. doi: 10.1128/AEM.07036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.My L, Rekoske B, Lemke JJ, Viala JP, Gourse RL, Bouveret E. 2013. Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. J Bacteriol 195:3784–3795. doi: 10.1128/JB.00384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schakermann M, Langklotz S, Narberhaus F. 2013. FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J Bacteriol 195:1912–1919. doi: 10.1128/JB.02134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.