Abstract
The traditional view of the dependency of subsurface environments on surface-derived allochthonous carbon inputs is challenged by increasing evidence for the role of lithoautotrophy in aquifer carbon flow. We linked information on autotrophy (Calvin-Benson-Bassham cycle) with that from total microbial community analysis in groundwater at two superimposed—upper and lower—limestone groundwater reservoirs (aquifers). Quantitative PCR revealed that up to 17% of the microbial population had the genetic potential to fix CO2 via the Calvin cycle, with abundances of cbbM and cbbL genes, encoding RubisCO (ribulose-1,5-bisphosphate carboxylase/oxygenase) forms I and II, ranging from 1.14 × 103 to 6 × 106 genes liter−1 over a 2-year period. The structure of the active microbial communities based on 16S rRNA transcripts differed between the two aquifers, with a larger fraction of heterotrophic, facultative anaerobic, soil-related groups in the oxygen-deficient upper aquifer. Most identified CO2-assimilating phylogenetic groups appeared to be involved in the oxidation of sulfur or nitrogen compounds and harbored both RubisCO forms I and II, allowing efficient CO2 fixation in environments with strong oxygen and CO2 fluctuations. The genera Sulfuricella and Nitrosomonas were represented by read fractions of up to 78 and 33%, respectively, within the cbbM and cbbL transcript pool and accounted for 5.6 and 3.8% of 16S rRNA sequence reads, respectively, in the lower aquifer. Our results indicate that a large fraction of bacteria in pristine limestone aquifers has the genetic potential for autotrophic CO2 fixation, with energy most likely provided by the oxidation of reduced sulfur and nitrogen compounds.
INTRODUCTION
Due to the lack of light-driven primary production, groundwater ecosystems were originally believed to be controlled by surface-derived allochthonous organic matter input (1–3) and to be dominated by heterotrophic prokaryotes adapted to nutrient limitation. However, there is increasing evidence of the important role of lithoautotrophy for carbon flow in aquifers (4–7). A large proportion of drinking water originates from groundwater resources (8), with karstic aquifers providing ∼25% of the drinking water sources on a global scale (9). Despite the crucial role of microbial activity in shaping groundwater geochemistry (10–12), the links between microbial diversity and function in groundwater ecosystems, especially with regard to chemolithoautotrophy, are still poorly understood (7). Recent studies suggest that microbial CO2 assimilation in aquifers could be fueled by energy conserved by nitrification, oxidation of ferrous iron and reduced sulfur compounds (6, 7), or oxidation of H2 or methane (13, 14). Oxidation of electron donors present as solid minerals such as pyrite can even yield highly reactive dissolved ions that might affect other minerals and dissolved ions in the aquifer, leading to changes to the makeup of rocks and groundwater.
Today, there are six known autotrophic CO2 fixation pathways (reviewed in reference 15): (i) the Calvin-Benson-Bassham cycle, (ii) the reductive tricarboxylic acid cycle, (iii) the reductive acetyl coenzyme A (acetyl-CoA) (Wood-Ljungdahl) pathway, (iv) the 3-hydroxypropionate cycle, (v) the 3-hydroxypropionate/4-hydroxybutyrate pathway, and (vi) the dicarboxylate/4-hydroxybutyrate cycle. The Calvin-Benson-Bassham cycle is quantitatively the most important mechanism of these pathways (15), and its key enzyme is ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), catalyzing the carboxylation of ribulose-1,5-bisphosphate to form two molecules of 3-phosphoglycerate. Four types of RubisCO are known to date. Bacteria utilize forms I and II (16, 17), which differ mainly with regard to their CO2 affinity and CO2/O2 substrate specificity (18). RubisCO form II is thought to be the evolutionarily older form of RubisCO, with a lower CO2/O2 specificity and, thus, a lower tolerance toward oxygen (19). The cbbL and cbbM genes, encoding the large subunit of RubisCO forms I and II, respectively, have been widely used to analyze the diversity of CO2-fixing chemolithoautotrophic bacteria in the environment. Such approaches revealed a wide distribution and diversity of RubisCO-encoding genes in soils (20–23) and groundwater (6, 7, 14, 24, 25). However, especially in aquifers, only a few studies investigated cbbM or cbbL genes at the transcript level or by quantitative approaches (7, 26).
Recent studies addressing the microbial diversity in aquifers or drinking water wells by 16S rRNA gene-targeted pyrosequencing allowed comprehensive insight into the composition of microbial communities (14, 27–29). However, those studies were DNA based and did not allow characterization of active aquifer microbial community members, providing only limited insight into the physiologic strategies that are dominant under the given environmental conditions. Microbial activities strongly depend on aquifer physicochemical properties but in turn may also contribute substantially to shaping these properties, i.e., by oxidizing inorganic compounds to dissolved ions as part of a chemolithoautotrophic life-style.
During regular groundwater sampling campaigns in the Hainich region in Thuringia, central Germany, we sampled two superimposed pristine limestone aquifers: an upper, oxygen-deficient aquifer and a lower, oxygen-rich aquifer at a maximum depth of 88 m. We aimed to assess the metabolically active total and RubisCO-encoding microbial communities by targeting 16S rRNA, cbbM, and cbbL transcripts, and we monitored the temporal dynamics of these three genes in the two aquifers over 2 years. A marker gene-targeted approach was preferred over a more comprehensive metatranscriptomics approach as the latter requires large amounts of RNA starting material, which are inherently difficult to obtain from pristine groundwater, and detection of less abundant microbial community members might fail. We hypothesized (i) that the metabolically active microbial communities differ strongly between the two aquifers, driven mainly by the differences in oxygen availability; (ii) that the energy-providing metabolisms coupled to RubisCO-based autotrophy, as suggested by the phylogenetic affiliation of RubisCO-encoding microorganisms, also differ between the two aquifers; and (iii) that, assuming a reduced input of surface-derived organic material with increasing depth, putative autotrophs constitute a larger fraction of the microbial population in the lower than in the upper aquifer.
MATERIALS AND METHODS
Study site and sampling of groundwater.
Groundwater samples were obtained from two superimposed limestone aquifers located in the Hainich region in northwest Thuringia. With a total size of 16,000 ha, the Hainich region is the largest connected deciduous forest in Germany, surrounded by agricultural areas of various management intensities. Groundwater was obtained from one well accessing an upper aquifer developed in the cycloides bank stratum (site H4-3 [12-m depth]) and two wells accessing a lower aquifer developed in a trochite limestone stratum (site H4-1 [48-m depth] and site H5-1 [88-m depth]). Sites were located on meadows (sites H4-1 and H4-3) and farmland (site H5-1), following a downhill slope with a distance of ∼1,560 m between site H4 and site H5.
Prior to sampling, groundwater was pumped with a groundwater sampling pump (MP1; Grundfoss, Erkrath, Germany) until up to three well volumes were discharged and physicochemical parameters were stabilized. Samples for chemical analysis were filtered through 0.2-μm-pore-size filters and stored at 4°C until analysis. For molecular analyses, samples were filtered through 0.2-μm Supor filters (Pall Corporation), with 5 to 6 liters of groundwater passing through one filter. By integrating over a large volume of groundwater, we expected to capture a representative fraction of the microbial community. The filtration procedure lasted between 40 min and 90 min per filter, and water samples were kept cold during filtration. We cannot completely rule out that temporal exposure of the samples to oxygen during filtration may have caused shifts within the transcript pools. After filtration, filters were transferred into sterile reaction tubes and frozen on dry ice within <1 min. Filters were stored at −80°C until nucleic acid extraction.
Physicochemical analyses.
Conductivity, pH, water temperature, oxygen concentration and saturation, and redox potential were measured electrometrically on-site with a flowthrough system using appropriate sensors (WTW, Weilheim, Germany). Dissolved CO2 and bicarbonate concentrations were determined by titration directly after sampling according to Deutsche Einheitsverfahren (DEV) methods (30). Nitrate, nitrite, and ammonium levels were determined according to methods described previously by Velghe and Claeys (31) and Grasshoff et al. (32). Concentrations of Fe(II) were measured by the phenanthroline method (33). Concentrations of sulfate were determined by ion chromatography (IC 20 system [Dionex, Sunnyvale, CA] equipped with an IonPac AS11-HC column and an IonPac AG11-HC precolumn), and total organic carbon (TOC) concentrations were determined by using a TOC analyzer (AnalytikJena, Jena, Germany).
Nucleic acid extraction, amplification, clone library construction, and pyrotag sequencing.
Genomic DNA and total RNA were isolated according to methods described previously by Church et al. (34), in combination with the DNA blood and tissue kit (Qiagen) and the RNeasy minikit (Qiagen). For each site, nucleic acids were extracted from one filter per time point. DNase treatment (Turbo DNase-free kit; Ambion, USA) and reverse transcription (RT) (Array Script reverse transcriptase; Ambion) was performed as previously described (35). Along with each sample, we prepared one control reaction mixture using the same RNA extract but without reverse transcriptase, which was subsequently PCR amplified to check for contamination of RNA extracts with genomic DNA.
Diversity analysis of both 16S rRNA and RubisCO-encoding genes was based on RNA only from groundwater samples obtained from the three wells at sites H4-3, H4-1, and H5-1 in November 2010 and April 2011. Analysis of transcripts of RubisCO-encoding cbbL genes was performed only for the samples obtained in April 2011. PCR amplification of Bacteria 16S rRNA fragments was performed by using primers 27F and 518R (36, 37), under the cycling conditions described in the supplemental material. Genes encoding RubisCO large subunit type IA (cbbL) and type II (cbbM) were amplified by using Hotstar Taq Mastermix (Qiagen) and primer pairs F-cbbL/R-cbbL and F-cbbM/R-cbbM, respectively, under cycling conditions reported previously (6). PCR products were checked by agarose gel electrophoresis and purified by using the NucleoSpin extract II kit (Macherey-Nagel, Germany) according to the manufacturer's instructions. For 16S rRNA and cbbM amplicons, barcode and linker sequences were added in a second PCR performed by GATC-biotech (Constance, Germany). Amplicons were sequenced with 454 pyrosequencing on a Roche GS FLX genome sequencer system by GATC-biotech. For analysis of cbbL transcript sequence diversity, we did not use next-generation sequencing but constructed clone libraries from RNA-based cbbL amplicons using the pGEM-T Easy vector and Escherichia coli JM109 chemically competent cells (Promega). The cloning approach was considered sufficient enough to assess the quantitatively dominant phyla potentially involved in CO2 fixation and expressing cbbL genes. In addition, one clone library from DNA-based cbbM amplicons was constructed to generate standards for quantitative PCR (qPCR). Plasmid vector inserts of transformed E. coli clones were commercially sequenced (Macrogen Inc., South Korea).
Sequence analysis of 16S rRNA, cbbM, and cbbL amplicons.
Pyrosequencing was performed with RNA-based 16S rRNA amplicons obtained from the three different wells sampled in November 2010 and April 2011. A total of 128,145 sequence reads were obtained for the 6 samples in total and were analyzed by using Mothur (38), along with SILVA bacterial reference alignment according to Schloss SOP (http://www.mothur.org/wiki/Schloss_SOP; see also the supplemental material) (39). For phylogenetic analysis of cbbL and cbbM transcripts, reference alignments were generated in ARB (40) (see the supplemental material). Nucleic acid sequences were transformed to deduced amino acid sequences for further phylogenetic analysis. Phylogenetic analysis of deduced CbbM and CbbL amino acid sequences of cultured representatives in ARB revealed that sequence similarities between closely related species inferred from their relative positions in the calculated phylogenetic trees ranged from ∼94 to 96%. Consequently, a 0.05 distance cutoff at the protein level was used for operational taxonomic unit (OTU) assignment of cbbM and cbbL sequences to allow for a good approximation of species-level distinctions. Analysis of RNA-based cbbM amplicons derived from pyrosequencing was performed by using Mothur, BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and ARB (see the supplemental material). Assignment to OTUs and analysis of shared OTUs between samples were also conducted with Mothur. Coverages of the clone libraries were calculated according to methods described previously by Singleton et al. (41). Closest relatives were determined based on a BLAST search of representative OTU sequences, phylogenetic affiliations inferred from protein-based neighbor-joining trees, and distances calculated in ARB based on the clustering of sequences together with cultured representatives.
Quantitative PCR.
Gene abundances of bacterial and archaeal 16S rRNA genes and of cbbL and cbbM genes were determined by quantitative PCR using Maxima SYBR green qPCR Mastermix (Fermentas) on an Mx3000P cycler (Agilent). 16S rRNA genes were amplified by using primer pair Bac8Fmod/Bac338EUB (42, 43), according to cycling conditions and standards described previously by Herrmann et al. (35). Functional genes were quantified by using primers F-cbbL/R-cbbL and F-cbbM/R-cbbM (6), as described in the supplemental material. Details on the estimation of relative fractions of the microbial population harboring RubisCO-encoding genes are provided in the supplemental material.
Statistical analysis.
Differences in chemical parameters or gene abundances between sites were analyzed by using the t test or Mann-Whitney U test. Correlations between gene abundances and chemical parameters over time were analyzed by using the Spearman rank correlation coefficient. All statistical calculations were done with SPSS, version 19.0.
Nucleotide sequence accession numbers.
Sequences obtained in this study were deposited in GenBank under accession numbers KF872279 to KF872456 (cbbL clone libraries) and KF872457 to KF872493 (cbbM clone library for qPCR standards). Pyrosequencing data for 16S rRNA and cbbM transcripts have been submitted to the European Nucleotide Archive under project number PRJEB5805 and sample accession numbers ERS420411 to ERS420422.
RESULTS
Biogeochemistry of the two aquifers.
Previous investigations showed that the two aquifers harbored comparable microbial population densities, as revealed by SYBR green II total cell counts (site H4-3, 1.1 × 107 cells liter−1 to 9.4 × 107 cells liter−1 groundwater; site H4-1, 2.7 × 106 cells liter−1 to 3.8 × 108 cells liter−1; site H5-1, 1.2 × 107 cells liter−1 to 3.7 × 108 cells liter−1) (44). Groundwater oxygen saturation and nitrate concentrations were significantly lower in the upper aquifer (well H4-3, 5% O2 and 6 μmol liter−1 nitrate) than in the lower aquifer (well H4-1, 37% O2 and 121 μmol liter−1 nitrate; well H5-1, 38% O2 and 137 μmol liter−1 nitrate). Maximum concentrations of sulfate (up to 3.20 mmol liter−1) were observed at site H5-1, whereas sulfate concentrations were much lower in both aquifers at site H4 (Table 1). Groundwater from both aquifers had high ion contents (measured as conductivity), high concentrations of bicarbonate and dissolved CO2, and a neutral pH slightly higher than 7 (Table 1). The TOC concentration was generally low but typical for pristine groundwater environments, ranging from 75 to 125 μmol liter−1. Dissolved concentrations of inorganic electron donors were rather low, with mean concentrations of NH4+ ranging from 4.6 to 9.0 μmol liter−1 (Table 1) and concentrations of Fe(II) usually being <5 μmol liter−1 (data not shown). When water was sampled for community analyses, physicochemical parameters were similar to the mean values over a 2-year period (Table 1).
TABLE 1.
Physicochemical parameters of the groundwater at site H4 with two wells and further downstream at site H5a
| Site and date | Mean conductivity (μS cm−1) ± SD | Mean pH ± SD | Mean oxygen saturation (%) ± SD | Mean dissolved CO2 concn (mmol liter−1) ± SD | Mean bicarbonate concn (mmol liter−1) ± SD | Mean TOC concn (μmol liter−1) ± SD | Mean NO3− concn (μmol liter−1) ± SD | Mean NH4+ concn (μmol liter−1) ± SD | Mean SO42− concn (mmol liter−1) ± SD |
|---|---|---|---|---|---|---|---|---|---|
| H4-3 | |||||||||
| 24-mo study period | 747 ± 27 | 7.2 ± 0.1 | 5.3 ± 9.9 | 0.46 ± 0.17 | 6.74 ± 0.63 | 94.9 ± 9.2 | 6.0 ± 6.8 | 7.8 ± 3.0 | 0.37 ± 0.1 |
| Nov 2010 | 645 | 7.3 | 1.0 | ND | ND | 112.4 | BD | 10.8 | 0.39 |
| Apr 2011 | 754 | 7.2 | 3.5 | 0.45 | 7.55 | 83.3 | BD | 5.5 | 0.41 |
| H4-1 | |||||||||
| 24-mo study period | 748 ± 50 | 7.3 ± 0.1 | 38.3 ± 15.0 | 0.39 ± 0.18 | 6.33 ± 0.79 | 74.1 ± 10.8 | 121.2 ± 51.7 | 9.0 ± 6.7 | 0.79 ± 0.32 |
| Nov 2010 | 656 | 7.4 | 39.6 | ND | ND | 68.3 | 142.6 | 11.4 | 0.76 |
| Apr 2011 | 776 | 7.3 | 62.8 | 0.39 | 5.81 | 81.6 | 160.0 | BD | 1.09 |
| H5-1 | |||||||||
| 24-mo study period | 1013 ± 72 | 7.1 ± 0.1 | 37.1 ± 9.1 | 0.44 ± 0.18 | 5.43 ± 0.56 | 79.1 ± 10.0 | 137.3 ± 22.2 | 4.6 ± 4.5 | 2.53 ± 0.34 |
| Nov 2010 | 842 | 7.2 | 40.7 | ND | ND | 87.4 | 186.8 | 2.9 | 2.09 |
| Apr 2011 | 1051 | 7.2 | 36.8 | 0.44 | 5.47 | 94.9 | 131.0 | BD | 2.64 |
Data are from site H4 with two wells (wells H4-3 [12-m depth] and H4-1 [48-m depth]) and further downstream at site H5 (well H5-1 [88-m depth]). Data are means ± standard deviations of results obtained during a 24-month study period. For each site, values are also given for the months when samples were taken for diversity analysis. Well H4-3 was in the upper cycloides bank aquifer, whereas wells H4-1 and H5-1 were located in the lower trochite limestone aquifer. Detection limits for NO3− and NH4+ were 1 μmol liter−1 and 0.5 μmol liter−1, respectively. ND, not determined; BD, below the limit of detection.
Diversity of active groundwater bacterial communities (16S rRNA).
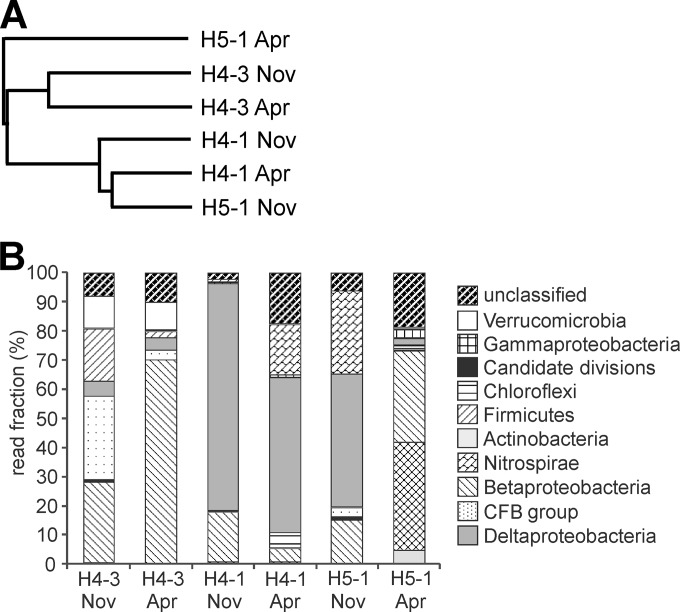
We analyzed sequences of bacterial 16S rRNA, cbbM, and cbbL gene transcripts to link RubisCO-based autotrophy to physiologies inferred from closely related described taxa within active groundwater microbial communities. 16S rRNA diversity was assessed by 454 pyrosequencing of RNA-based 16S rRNA amplicons for samples obtained in November 2010 and April 2011. For comparative analysis, we utilized the data set after normalization to the lowest number of reads found in sample H5-1 from April. More information is given in the supplemental material. Comparison of samples using Bray-Curtis indices revealed that samples from the upper aquifer (site H4-3) obtained at two different time points were most similar and that sample H5-1 from April, which had the highest level of diversity, was the most different from the other samples (Fig. 1A and Table 2).
FIG 1.
Results of 16S rRNA-targeted pyrosequencing. (A) Dendrogram based on Bray-Curtis similarity values; (B) phylogenetic affiliation at the phylum level. Only phyla represented by >1% of the sequence reads in at least one sample are shown. Candidate divisions include OD1, OP10, OP3, TG-1, TM7, WS3, BD1-5, and WCHB1-60. The CFB group includes members of the Cytophagales, Flavobacteria, and Bacteroidetes.
TABLE 2.
Overview of pyrosequencing data sets for 16S rRNA and cbbM transcripts after normalizationa
| Sample and mo | Gene | No. of sequences | Coverage | Inverse Simpson's index (1/D) | No. of observed OTUs | No. of estimated OTUs (chao) | No. of rare OTUs (singletons) | Shannon index |
|---|---|---|---|---|---|---|---|---|
| H4-3 Nov | 16S rRNA | 7,317 | 0.92 | 9.16 | 797 | 3,062 | 727 (610) | 3.36 |
| H4-3 Apr | 16S rRNA | 7,317 | 0.92 | 13.79 | 784 | 2,612 | 695 (571) | 3.74 |
| H4-1 Nov | 16S rRNA | 7,317 | 0.95 | 4.27 | 539 | 1,755 | 479 (395) | 2.65 |
| H4-1 Apr | 16S rRNA | 7,317 | 0.94 | 6.04 | 644 | 1,940 | 555 (453) | 3.19 |
| H5-1 Nov | 16S rRNA | 7,317 | 0.96 | 3.85 | 401 | 1,268 | 355 (289) | 2.28 |
| H5-1 Apr | 16S rRNA | 7,317 | 0.85 | 60.25 | 1,537 | 5,301 | 1,341 (1,115) | 5.46 |
| H4-3 Nov | cbbM | 4,830 | 0.99 | 10.21 | 97 | 250 | 64 (53) | 2.81 |
| H4-3 Apr | cbbM | 4,830 | 0.99 | 1.61 | 63 | 141 | 47 (38) | 0.97 |
| H4-1 Nov | cbbM | 4,830 | 0.99 | 7.78 | 95 | 195 | 58 (43) | 2.59 |
| H4-1 Apr | cbbM | 4,830 | 0.99 | 1.97 | 71 | 102 | 45 (30) | 1.41 |
| H5-1 Nov | cbbM | 4,830 | 0.99 | 5.95 | 93 | 159 | 59 (42) | 2.32 |
| H5-1 Apr | cbbM | 4,830 | 0.99 | 4.50 | 60 | 192 | 41 (33) | 1.97 |
Sequence distance cutoffs for OTU assignment were 0.03 (nucleic acid level) for 16S rRNA genes and 0.05 (protein level) for cbbM.
The majority of sequences (69% of total sequences) were related to members of the Proteobacteria, including Alpha-, Beta-, Gamma-, Delta-, and Epsilonproteobacteria (Fig. 1B). Members of the Deltaproteobacteria represented the highest number of sequence reads in almost all the samples obtained from the lower aquifer (46% of sequences from site H5-1 in November, 53% from site H4-1 in April, and 78% from site H4-1 in November) (Fig. 1B). Nitrospirae-affiliated phylotypes represented 29% and 17% of sequences in samples from the lower aquifer in November 2010 (site H5-1) and in April 2011 (site H4-1), respectively. In contrast, members of the Betaproteobacteria and the CFB (Cytophagales, Flavobacteria, and Bacteroidetes) group dominated the active community in the upper aquifer, together representing 56% and 73% of all sequences in November 2010 and April 2011, respectively (Fig. 1B).
The 25 most abundant OTUs accounted for ∼70% of all sequence reads. The highest number of overall sequence reads (21%) belonged to an OTU assigned to the Deltaproteobacteria with a sequence similarity to cultured microorganisms of <90%. This OTU accounted for a large fraction (37 to 44%) of sequence reads in the lower aquifer samples but was almost absent from the upper aquifer. It had 99% and 97% sequence similarities to environmental sequences from lake sediment (GenBank accession number AB661566 [45]) and groundwater (accession numbers KC606878 [46] and DQ407360 [47]), respectively. The second most abundant OTU (7% of total sequence reads) was related to Nitrospira moscoviensis (93.5% sequence identity). The two Nitrospira-related phylotypes found in our study were most closely related to environmental sequences obtained from beech forest soil (48) or from grassland soil (49). Abundant OTUs representing large fractions of sequences in samples from the upper aquifer (site H4-3) were related to the genera Pedobacter (up to 23.9%), Limnohabitans (up to 17%), Albidiferax (up to 16.1%), Opitutus (up to 10.1%), and Polaromonas (up to 8.6%); however, these groups represented <0.8% of total sequences in the lower aquifer (sites H4-1 and H5-1). Two OTUs closely related to Sulfuricella denitrificans (98% sequence identity) and Nitrosomonas sp. strain Is79A3 (98.4% sequence identity), groups known to fix CO2 via the Calvin-Benson-Bassham cycle (17, 50), were also detected in the 16S rRNA read pool from the lower aquifer, representing 0.8 to 5.6% (Sulfuricella) and 0.3 to 3.8% (Nitrosomonas) of total reads.
Diversity of the CO2-fixing microbial community based on cbbM and cbbL transcripts.
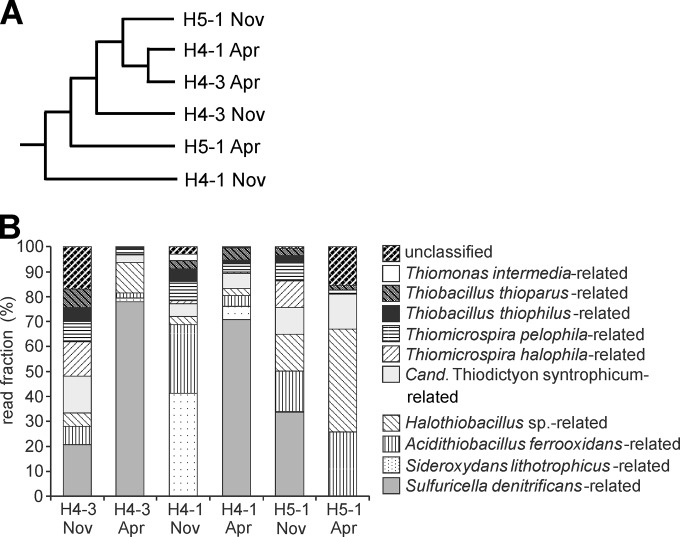
The composition of the microbial community transcribing cbbM genes was analyzed by 454 pyrosequencing for samples obtained in November 2010 and April 2011. Comparison of samples by using Bray-Curtis indices based on normalized data sets did not result in an obvious clustering pattern according to site or time (Fig. 2A). Only 7 of the 324 total OTUs were shared by communities of all samples; however, 4 of these OTUs accounted for ∼60% of all sequence reads and were detected at all three wells at both sampling times. The most abundant OTUs were related to Sulfuricella denitrificans (97.3% similarity at the protein level; 9,745 reads), Halothiobacillus sp. (94.6% similarity; 3,014 reads), Acidithiobacillus ferrooxidans (97.3% similarity; 2,593 reads), Sideroxydans lithotrophicus ES-1 (92.4 to 93.6% identity; 2,200 reads), and “Candidatus Thiodictyon syntrophicum” (98.7% identity; 2,005 reads) (Fig. 2B; see also Fig. S4 in the supplemental material). However, the relative fractions of sequence reads of each phylotype differed considerably between sampling sites or time points. While Sulfuricella-related sequence reads constituted >70% of all reads for samples from sites H4-3 and H4-1 in April 2011, this phylotype was represented by only a few reads in samples obtained from site H4-1 in November 2010 and from site H5-1 in April 2011 (Fig. 2B). Sequences related to Thiobacillus thioparus, Halothiobacillus neapolitanus, and Thiomicrospira pelophila usually formed only a small fraction of the cbbM transcript pool. Overall, the composition of the active cbbM-transcribing community did not show a pattern that would suggest that cbbM OTU patterns were related to the aquifer or sampling time point.
FIG 2.
Results of cbbM-targeted pyrosequencing, based on deduced cbbM protein sequences. (A) Dendrogram based on Bray-Curtis similarity values; (B) phylogenetic affiliation and relative frequency of the different sequence types. Only phylotypes represented by >1% of the sequence reads in at least one sample are shown. Closest relatives were determined based on BLAST results and distances calculated in ARB. Similarities of deduced cbbM protein sequences obtained in this study to cultivated strains are as follows: 90.4 to 97.3% for Sulfuricella denitrificans, 91.1 to 96.2% for Sideroxydans lithotrophicus ES-1, 90.5 to 97.3% for Acidithiobacillus ferrooxidans, 90.4 to 94.7% for Halothiobacillus sp., 91 to 98.7% for “Candidatus Thiodictyon,” 90.4 to 97.3% for Thiomicrospira halophila, 90.4 to 98.6% for Thiomicrospira pelophila, 90.5 to 94.6% for Thiobacillus thiophilus, 90.7 to 97.3% for Thiobacillus thioparus, and 90.4 to 97.3% for Thiomonas intermedia.
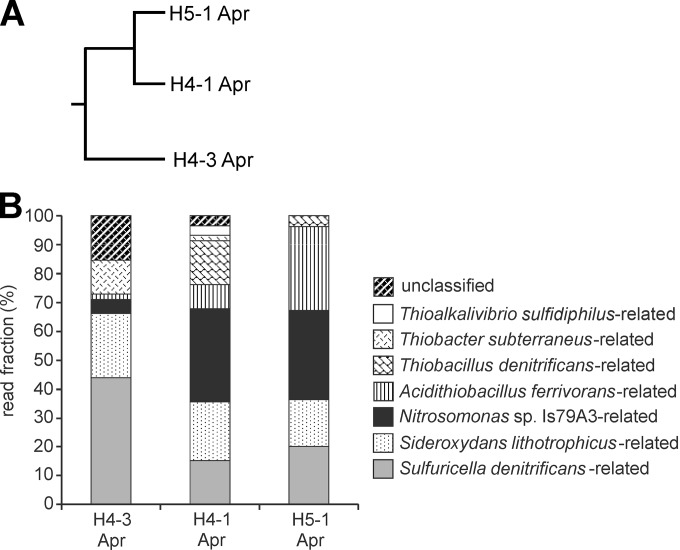
The active CO2-fixing microbial community harboring cbbL genes was analyzed by using a cloning approach targeting cbbL transcripts for one sampling time per well (April 2011). Using a 0.05 distance cutoff for deduced cbbL protein sequences, we detected 13 OTUs in total (see Fig. S5 in the supplemental material). Coverage ranged from 96.7 to 98.3% for 58 to 60 sequences per library (Table 3), indicating that the cloning approach sufficiently covered the major phylotypes. Similar to the cbbM-targeted approach, the 6 shared OTUs represented at least 70% of the sequences within each sample and 74% of total sequence reads. While the cbbL genotypes were dominated by sequences related to Sulfuricella denitrificans in the upper aquifer (site H4-3; ∼43%), the Sulfuricella-related fraction represented only 10 to 20% of sequences in the lower aquifer (Fig. 3B). In turn, ∼30% of genotypes in the lower aquifer were related to the ammonium-oxidizing bacterium Nitrosomonas sp. IsA73 (GenBank accession number YP_004694496), which was represented by only a few clones in the upper aquifer. This pattern was reflected by a higher level of similarity between samples obtained from the two wells of the lower aquifer (Fig. 3A). Sideroxydans lithotrophicus ES-1-related sequences formed a comparably large fraction at all three wells, while Acidithiobacillus ferrivorans-related sequences formed a larger fraction at site H5-1.
TABLE 3.
Overview of clone library-based analysis of cbbL transcriptsa
| Sample and mo | No. of sequences | Coverage (%) | Inverse Simpson's index (1/D) | No. of observed OTUs | No. of estimated OTUs (chao) | Shannon index |
|---|---|---|---|---|---|---|
| H4-3 Apr | 60 | 96.7 | 7.53 | 9 | 10 | 2.00 |
| H4-1 Apr | 60 | 98.3 | 8.12 | 11 | 11 | 2.16 |
| H5-1 Apr | 58 | 98.3 | 5.01 | 8 | 8 | 1.73 |
Sequence distance cutoffs for OTU assignment were 0.05 (protein level).
FIG 3.
Results of the cbbL cloning approach, based on deduced cbbL protein sequences. (A) Dendrogram based on Bray-Curtis similarity values; (B) phylogenetic affiliation and relative frequency of the different sequence types. Closest relatives were determined based on BLAST results and distances calculated in ARB. The upper aquifer included site H4-3, and the lower aquifer included sites H4-1 and H5-1. Similarities of deduced cbbL protein sequences obtained in this study to cultivated strains are as follows: 95.6 to 100% for Sulfuricella denitrificans, 95.5 to 98.3% for Sideroxydans lithotrophicus ES-1, 96.7 to 99.4% for Nitrosomonas sp. Is79A3, 94.4 to 96.1% for Acidithiobacillus ferrivorans, 92.2 to 96.7% for Thiobacillus denitrificans, 92.2 to 95.5% for Thiobacter subterraneus, and 91.6% for Thioalkalivibrio sulfidiphilus.
Temporal variation of the abundance of RubisCO-encoding and 16S rRNA genes.
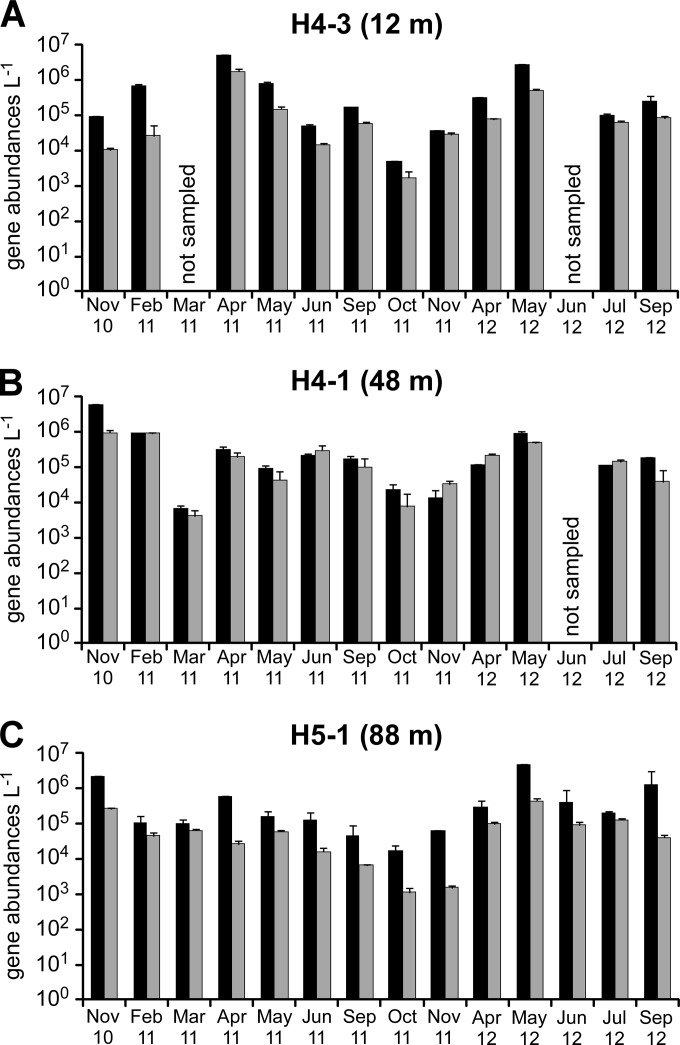
The abundance of potential CO2 fixers using RubisCO and the total prokaryotic population was approximated from analysis of RubisCO-encoding and 16S rRNA genes in genomic DNA extracted from groundwater. Numbers of cbbM genes, encoding RubisCO form II, ranged from 5.04 × 103 to 5.93 × 106 genes per liter of groundwater over a 2-year period, while numbers of cbbL genes, encoding RubisCO form IA, were between 2 and 8 times lower (ranging from 1.14 × 103 to 1.75 × 106 genes liter−1) (Fig. 4). Substantial temporal fluctuations of cbbL and cbbM gene abundances of 2 orders of magnitude at each site were observed but without significant differences in the abundances of these genes between the upper and lower aquifers or between the two wells of the lower aquifer. Groundwater bacterial 16S rRNA gene numbers, providing an approximation of the total microbial population, ranged from 3.11 × 105 to 2.69 × 108 genes liter−1 at the three different sites, as reported previously (44). Archaeal 16S rRNA genes were ∼2 orders of magnitude less abundant than bacterial 16S rRNA genes, and their numbers ranged from 1.3 × 103 to 4.6 × 105 genes liter−1 (44). Similar to the RubisCO-encoding genes, there were strong temporal fluctuations but no significant differences among the three sampling sites. Estimated cell numbers based on bacterial and archaeal 16S rRNA gene qPCR data were in the same range as the total cell counts performed on selected samples by Opitz et al. (44).
FIG 4.
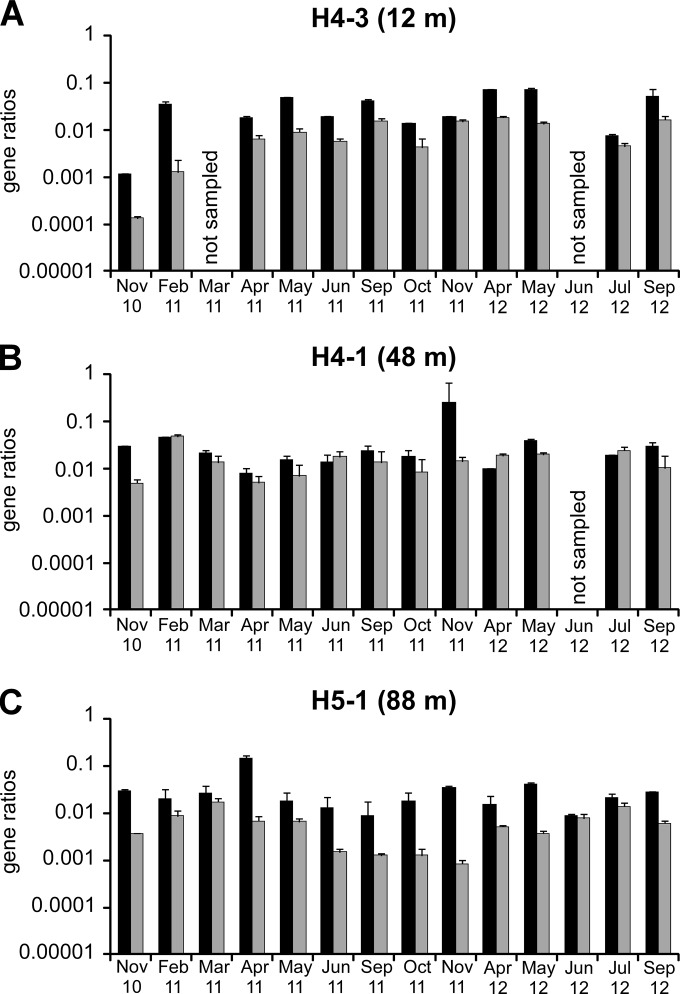
Abundance of cbbM (black) and cbbL (gray) genes per liter groundwater over a 2-year period. (A) Site H4-3 (upper aquifer; 12-m depth); (B) site H4-1 (lower aquifer; 48-m depth); (C) site H5-1 (lower aquifer; 88-m depth). Error bars represent the standard deviations of data from three replicate qPCRs.
For each well, cbbM and cbbL gene numbers were positively correlated with each other (Spearman rank correlation coefficients of 0.84 for site H4-3, 0.89 for site H4-1, and 0.72 for site H5-1; P < 0.01) and with bacterial 16S rRNA gene numbers (Spearman rank correlation coefficients of 0.89 for cbbM and 0.85 for cbbL; P < 0.001) over time. Based on 16S rRNA, cbbM, and cbbL gene numbers, the estimated fraction of organisms of the total community with the genetic potential to assimilate CO2 via the Calvin-Benson-Bassham cycle ranged from 0.5 to 14.4% in the upper aquifer (site H4-3) and from 2.1 to 17.3% in the lower aquifer (sites H4-1 and H5-1).
Temporally, the abundances of cbbM and cbbL genes showed maxima in spring and early summer, were low in autumn (Fig. 4), and thus followed the same patterns as those previously observed for bacterial 16S rRNA genes for the same samples (44). cbbM-to-16S rRNA gene ratios exhibited variations of usually <1 order of magnitude in both aquifers over time (Fig. 5), indicating that CO2-fixing microorganisms harboring cbbM genes made up a rather stable fraction of the total microbial community. However, cbbL-to-16S rRNA gene ratios were subject to substantial temporal variations in the lower aquifer at site H5-1, with minima from June to November 2011 (Fig. 5), suggesting stronger changes in the CO2-fixing community over time than in upstream well H4-1 or the upper aquifer (site H4-3). cbbM and cbbL gene abundances did not show significant correlations with oxygen saturation or CO2 concentrations over time, except for site H5-1 of the lower aquifer, where cbbM gene abundances were negatively correlated with oxygen saturation (Spearman rank correlation coefficient of −0.55; P < 0.05) and both cbbM and cbbL gene abundances were positively correlated with CO2 concentrations (Spearman rank correlation coefficients of 0.60 and 0.70, respectively; P < 0.05).
FIG 5.
Relative numbers of cbbM (black) and cbbL (gray) genes compared to bacterial and archaeal 16S rRNA gene numbers over a 2-year period. (A) Site H4-3 (upper aquifer; 12-m depth); (B) site H4-1 (lower aquifer; 48-m depth); (C) site H5-1 (lower aquifer; 88-m depth). Error bars represent the standard deviations of data from three replicate qPCRs.
Transcript-to-gene ratios of bacterial 16S rRNA, cbbM, and cbbL at selected time points were analyzed by qPCR (see Fig. S6 in the supplemental material). While transcript-to-gene ratios were usually between 1 and 10 for bacterial 16S rRNA, maximum estimates of transcript-to-gene ratios for cbbM and cbbL ranged from 0.38 to 0.01, corresponding to <1 × 105 cbbM or cbbL transcripts per liter groundwater.
DISCUSSION
Structure of active bacterial communities.
Factors that affect microbial diversity and abundance in groundwater ecosystems were previously linked to the interplay between organic contaminants, nutrients, and hydrogeology (8, 51–53). In addition, seasonal hydrological dynamics, e.g., resulting from snowmelt, may have strong effects on groundwater microbial community composition (54). In the system of two superimposed limestone aquifers investigated in this study, both aquifers were characterized as pristine, with low concentrations of organic carbon and nutrients and the same general lithology but with strong differences in oxygen concentrations. In contrast to our expectations, we did not observe a decrease in the concentration of TOC with increasing depth. This might be because the input of TOC is similarly low for both aquifers. Alternatively, assuming a higher input of TOC into the upper than in the lower aquifer, higher TOC turnover rates in the upper aquifer might result in the observed equally low TOC concentrations in both aquifers. The latter would be supported by the lower oxygen availability in the upper aquifer. Oxygen availability and distance to surface soils were likely key drivers of the observed differences between the compositions of the active microbial communities in the upper and the lower aquifers. In support of our hypothesis, we found little overlap in the active bacterial communities between the two aquifers. Members of the Betaproteobacteria and Cytophagales-Flavobacteria-Bacteroidetes group dominated the metabolically active communities in the upper aquifer. The dominance of Betaproteobacteria in groundwater agrees with data from previous reports (29, 55–57). Sequences related to facultative or obligate anaerobes, e.g., the genera Albidiferax, Limnohabitans, and Opitutus, together represented 26.2 to 30.5% of the sequences reads in the total active community in the upper aquifer, which reflects the oxygen-deficient conditions of this aquifer. Metabolically active communities in the upper aquifer also harbored a larger fraction of soil-related genera, e.g., Pedobacter, Opitutus, Albidiferax, and Polaromonas (58–61), belonging to the families Sphingobacteriaceae, Opitutaceae, and Comamonadaceae. In fact, a recent study of microbial communities in seepage water of agricultural soil revealed that representatives of these bacterial families originating from the rhizosphere and rhizoplane were subject to vertical migration (62). Consequently, large fractions of the bacterial community in the upper aquifer might actually originate from surface soils and could be preferentially transported vertically to the groundwater by event-driven transport upon heavy rainfalls or snowmelt (62, 63).
The metabolically active communities in the lower, oxygen-rich aquifer showed a very different structure and harbored large fractions of Deltaproteobacteria and Nitrospirae. The large proportions of Nitrospirae are similar to those reported for alpine karstic spring water (64) and granitic aquifers with low oxygen availability (65). Deltaproteobacteria are occasionally reported in groundwater environments (66) but in most cases represent sulfate reducers under conditions of low oxygen availability (46). Due to the low level of sequence similarity of the most abundant Deltaproteobacteria-related OTU found in our study, we cannot infer the potential metabolism of these organisms. The second most abundant OTU was related to Nitrospira moscoviensis, with 93.5% sequence identity, which agrees with data from previous reports of the presence of Nitrospira-related phylotypes in water of an alpine radioactive thermal spring (67), suggesting a role in nitrification. Moreover, the distribution pattern of this OTU, with a much higher relative abundance in the lower aquifer at higher nitrate concentrations than in the upper aquifer, would also support its role in nitrification.
Despite the strong differences in the compositions of the metabolically active microbial communities, total microbial abundances were not affected by the differences in physicochemistry between the two aquifers, as both aquifers harbored comparable microbial population densities. In fact, the microbial abundance estimated by 16S rRNA gene qPCR previously reported for the same samples (44) was comparable to those in limestone spring water (64) and two alpine deep granitic groundwater sites (65) but was 1 to 2 orders of magnitude lower than those in other granitic aquifers (57, 68).
Chemolithoautotrophy linked to sulfur and nitrogen cycling.
Given the differences in oxygen availability and microbial community composition between the two aquifers, we expected to find that the active microbial communities transcribing cbbM or cbbL genes would differ strongly between the two aquifers. In fact, there was a clear separation of the two aquifers at the level of cbbL but not cbbM transcripts. The lack of clustering at the level of cbbM transcripts could be linked to the fact that most of the dominant cbbM phylotypes belonged to microaerophilic or facultative anaerobic microorganisms able to thrive under the conditions of both aquifers. In addition, the composition of the CO2-fixing microbial community transcribing cbbM genes varied strongly between the November 2010 and April 2011 samples, suggesting substantial changes over time. The reasons for these changes remain speculative and could be linked to the availability of anoxic microsites within the karstic aquifer system.
Most of the RubisCO-expressing phyla detected in our study harbored both RubisCO forms I and II, which agrees with previous findings from groundwater environments (25). The existence of both form I and form II RubisCO in one organism has been explained by the different kinetic properties of the enzymes, allowing efficient CO2 fixation in environments with strong fluctuations in the levels of oxygen and CO2 (6). RubisCO form I is capable of selectively fixing CO2 regardless of the presence of oxygen and therefore is better adapted to lower CO2 concentrations and/or oxic conditions (19, 69, 70). The simultaneous expression of both cbbL and cbbM by some organisms in groundwater with oxygen saturation of 5 to 63% observed here is in contrast to findings of Alfreider and coworkers (7), who did not detect cbbM transcripts in oxygen-amended groundwater, but agrees with data from previous reports of cultivation-based studies (71, 72).
Most of the cbbL and cbbM phylotypes detected in this study were closely related to the genera Sulfuricella, Sideroxydans, and Acidithiobacillus, pointing to a strong link between autotrophy and the oxidation of reduced sulfur compounds, as previously reported for groundwater environments (6, 7, 25, 26). Since representatives of the genera Sideroxydans and Acidithiobacillus are capable of oxidizing Fe(II), chemolithoautotrophy in the two aquifers could also be coupled to iron oxidation. However, given the low concentrations of Fe(II) in the groundwater of both aquifers along with the prevailing oxic conditions in the lower aquifer, the availability of Fe(II) is likely too low to support a considerable contribution of iron oxidation to autotrophy. The active RubisCO-based autotrophic communities in both aquifers appeared to be dominated by a phylotype closely related to the strictly autotrophic, facultatively aerobic betaproteobacterium Sulfuricella denitrificans (>97% similarity at the protein level). The dominance of S. denitrificans may be explained by its versatile lithotrophic metabolism, as it is able to oxidize elemental sulfur and thiosulfate to sulfate aerobically or coupled to denitrification under anoxic conditions (50). A fraction of 16S rRNA reads related to S. denitrificans in the lower aquifer samples of 0.4 to 5.6% underlined the importance of this metabolic group within the total active microbial community. Oxidative weathering of pyrite, which is known to be associated with trochite limestone in this area, is likely to provide the energy for CO2 fixation by sulfide, thiosulfate, or iron oxidizers, contributing to the microbially mediated changes to the makeup of rock and water in this aquifer system.
Aquifer-driven clustering of cbbL sequences was linked primarily to the distribution of reads related to ammonia-oxidizing bacteria (AOB), specifically Nitrosomonas sp. strain IsA73, known to harbor only RubisCO form I. The distribution of these strict aerobic microorganisms was obviously influenced by oxygen availability, since this genus was represented by a much larger cbbL read fraction in the oxygen-rich lower aquifer. In line with this, 16S rRNA reads related to Nitrosomonas sp. IsA73 accounted for 0.3 to 3.8% of the total 16S rRNA sequence reads from the lower aquifer but only <0.1% of total 16S rRNA sequence reads of the upper aquifer. These findings agree with data for qPCR targeting amoA genes encoding ammonia monooxygenase of AOB, which were 1 to 2 orders of magnitude more abundant in the lower than in the upper aquifer (44). Moreover, phylogenetic analysis based on amoA sequences revealed that AOB communities were dominated by phylotypes closely related to Nitrosomonas ureae (44), which is closely related to Nitrosomonas sp. IsA73, the dominant AOB detected in the active microbial communities in this study. Nitrification activity was also detectable in the oxygen-rich lower aquifer at 0.6 nmol NO3 liter−1 h−1 (44), which is slightly higher than what has been reported for the Atlantic Ocean (73). Thus, these findings strongly indicate that nitrification plays a role in autotrophic CO2 fixation in the lower aquifer.
The larger fraction of Nitrospira-related 16S rRNA sequence reads in the lower aquifer suggested that nitrite oxidizers related to Nitrospira sp. could also contribute to carbon autotrophy, using the reverse tricarboxylic acid (rTCA) cycle as the CO2 fixation pathway (74). Moreover, recently reported results showed that thaumarchaeal ammonia oxidizers, which fix CO2 via the 3-hydroxypropionate/4-hydroxybutyrate cycle pathway (75, 76), represented up to 65% of the archaeal population in the lower aquifer (44). Future work employing metagenomics and metatranscriptomics approaches will allow us to integrate these alternative CO2 fixation pathways in a more comprehensive picture of chemolithoautotrophy in the Hainich limestone aquifers, especially with regard to the potential contribution of nitrite-oxidizing bacteria and ammonia-oxidizing Archaea to nitrification-based carbon autotrophy.
Estimated fraction of autotrophs within the total microbial population.
Quantitative PCR targeting the cbbM and cbbL genes clearly demonstrated that a considerable fraction of the total microbial population had the genetic potential to fix CO2 via the Calvin-Benson-Bassham cycle in both aquifers. Assuming a decreasing influence of surface-derived input of organic matter with increasing depth, we expected to find a greater genetic potential for autotrophy in the lower aquifer. However, in contrast to our hypotheses, we did not observe higher concentrations of TOC in the upper aquifer, nor did the abundances of RubisCO-encoding genes point to a larger fraction of autotrophs utilizing the Calvin-Benson-Bassham cycle within the total microbial population in the lower aquifer. Across the two aquifers and across all time points, the estimated fraction of putative CO2-fixing bacteria within the total groundwater microbial population ranged from <1 to ∼17%, which is >1 order of magnitude larger than fractions estimated for other aquifers (26), estuary sediments (77), or paddy soils (23). We cannot rule out that a certain fraction of the CO2-fixing microbial population using the Calvin-Benson-Bassham cycle was overlooked, since we did not target cbbL genes and transcripts encoding RubisCO form IC, which has often been associated with facultative autotrophs (17). However, the low transcript-to-gene ratios observed for cbbL and cbbM suggest that only a small fraction of the CO2-fixing microbial community was actually expressing their genes, indicating that rates of carbon fixation via the Calvin-Benson-Bassham cycle in the groundwater might have been low at these two time points. The genetic potential of the two aquifers to fix CO2 via the Calvin-Benson-Bassham cycle may still be underestimated, since we did not target microbial communities attached to the rock surfaces of the aquifer, which have been shown to differ from suspended communities in their composition and metabolic capacities (78, 79).
In conclusion, our results demonstrated that pristine limestone aquifers harbor diverse and dynamic microbial communities with a considerable fraction of bacteria that are capable of autotrophic CO2 fixation via the Calvin-Benson-Bassham cycle. Oxygen availability and distance to surface soils likely played key roles in shaping the structure of the active microbial communities, with a larger fraction of potentially soil-derived, heterotrophic microbial groups in the upper than in the lower aquifer. Most CO2-fixing bacterial groups identified harbored both RubisCO forms I and II, indicating the potential for CO2-fixing populations to adjust to fluctuating O2 and CO2 concentrations. Overall, our findings clearly point to a high genetic potential for chemolithoautotrophy in pristine limestone aquifers. Combined transcript analysis of 16S rRNA and RubisCO-encoding genes suggested that facultative aerobic thiosulfate oxidizers and strict aerobic ammonia oxidizers are involved in carbon fixation in the lower oxygen-rich aquifer. Further evidence of a role of ammonia oxidizers was provided by a parallel study confirming active nitrification and the presence of ammonia oxidizers based on amoA genes. Future studies are needed to address alternative CO2 fixation pathways, such as the rTCA cycle and the 3-hydroxypropionate/4-hydroxybutyrate pathway, to further elucidate the role of nitrite oxidizers as well as ammonia-oxidizing Archaea in carbon autotrophy in these limestone aquifers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Erika Köhnke and Holger Hartmann for their technical support with sampling. We acknowledge Patricia Lange for help with sample preparation and qPCR analyses. We acknowledge Steffen Kolb for helpful comments on the manuscript.
This study is part of the research project AquaDiv@Jena funded by the ProExcellence Initiative of the federal state of Thuringia, Germany. Additional financial support was provided by a grant from the Friedrich Schiller University Jena (ProChance Programmlinie A1) to M.H. and by CRC 1076 AquaDiva funded by the Deutsche Forschungsgemeinschaft.
We state that we have no conflict of interest. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03269-14.
REFERENCES
- 1.Culver DC. 1985. Trophic relationships in aquatic environments. Stygologia 1:43–53. [Google Scholar]
- 2.Baker MA, Valett HM, Dahm CN. 2000. Organic carbon supply and metabolism in a shallow groundwater ecosystem. Ecology 81:3133–3148. doi: 10.1890/0012-9658(2000)081[3133:OCSAMI]2.0.CO;2. [DOI] [Google Scholar]
- 3.Foulquier A, Simon L, Gilbert F, Fourel F, Malard F, Mermillod-Blondin F. 2010. Relative influences of DOC flux and subterranean fauna on microbial abundance and activity in aquifer sediments: new insights from 13C-tracer experiments. Freshw Biol 55:1560–1576. doi: 10.1111/j.1365-2427.2010.02385.x. [DOI] [Google Scholar]
- 4.Pedersen K, Ekendahl S. 1992. Assimilation of CO2 and introduced organic compounds by bacterial communities in groundwater from southeastern Sweden deep crystalline bedrock. Microb Ecol 23:1–14. doi: 10.1007/BF00165903. [DOI] [PubMed] [Google Scholar]
- 5.Stevens TO, McKinley MP. 1995. Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270:450–454. doi: 10.1126/science.270.5235.450. [DOI] [Google Scholar]
- 6.Alfreider A, Vogt C, Hoffmann D, Babel W. 2003. Diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from groundwater and aquifer microorganisms. Microb Ecol 45:317–328. doi: 10.1007/s00248-003-2004-9. [DOI] [PubMed] [Google Scholar]
- 7.Alfreider A, Schirmer M, Vogt C. 2012. Diversity and expression of different forms of RubisCO genes in polluted groundwater under different redox conditions. FEMS Microbiol Ecol 79:649–660. doi: 10.1111/j.1574-6941.2011.01246.x. [DOI] [PubMed] [Google Scholar]
- 8.Griebler C, Lueders T. 2009. Microbial biodiversity in groundwater ecosystems. Freshw Biol 54:649–677. doi: 10.1111/j.1365-2427.2008.02013.x. [DOI] [Google Scholar]
- 9.Ford DC, Williams PW. 2007. Karst geomorphology and hydrology, 2nd ed. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 10.Akob DM, Kusel K. 2011. Where microorganisms meet the rocks in the Earth's critical zone. Biogeosciences 8:3531–3543. doi: 10.5194/bg-8-3531-2011. [DOI] [Google Scholar]
- 11.Flynn TM, O'Loughlin EJ, Mishra B, DiChristina TJ, Kemner KM. 2014. Sulfur-mediated electron shuttling during bacterial iron reduction. Science 344:1039–1042. doi: 10.1126/science.1252066. [DOI] [PubMed] [Google Scholar]
- 12.Griebler C, Malard F, Lefébure T. 2014. Current developments in groundwater ecology—from biodiversity to ecosystem function and services. Curr Opin Biotechnol 27:159–167. doi: 10.1016/j.copbio.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen K. 2000. Exploration of deep intraterrestrial microbial life: current perspectives. FEMS Microbiol Lett 185:9–16. doi: 10.1111/j.1574-6968.2000.tb09033.x. [DOI] [PubMed] [Google Scholar]
- 14.Tiago I, Verissimo A. 2013. Microbial and functional diversity of a subterrestrial high pH groundwater associated to serpentinization. Environ Microbiol 15:1687–1706. doi: 10.1111/1462-2920.12034. [DOI] [PubMed] [Google Scholar]
- 15.Berg IA. 2011. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 71:576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badger MR, Bek EJ. 2008. Multiple Rubisco forms in Proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot 59:1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- 18.Tabita FR. 1999. Microbial ribulose bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res 60:1–28. doi: 10.1023/A:1006211417981. [DOI] [Google Scholar]
- 19.Tourova TP, Spiridonova EM. 2009. Phylogeny and evolution of the ribulose 1,5-bisphosphate carboxylase/oxygenase genes in prokaryotes. Mol Biol 43:713–728. doi: 10.1134/S0026893309050033. [DOI] [PubMed] [Google Scholar]
- 20.Selesi D, Schmid M, Hartmann A. 2005. Diversity of green-like and red-like ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes (cbbL) in differently managed agricultural soils. Appl Environ Microbiol 71:175–184. doi: 10.1128/AEM.71.1.175-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selesi D, Pattis I, Schmid M, Kandeler E, Hartmann A. 2007. Quantification of bacterial RubisCO genes in soils by cbbL targeted real-time PCR. J Microbiol Methods 69:497–503. doi: 10.1016/j.mimet.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Tolli J, King GM. 2005. Diversity and structure of bacterial chemolithotrophic communities in pine forest and agroecosystem soils. Appl Environ Microbiol 71:8411–8418. doi: 10.1128/AEM.71.12.8411-8418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao K-Q, Bao P, Bao Q-L, Jia Y, Huang F-Y, Su J-Q, Zhu Y-G. 2014. Quantitative analysis of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) large-subunit genes (cbbL) in typical paddy soils. FEMS Microbiol Ecol 87:89–101. doi: 10.1111/1574-6941.12193. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence JR, Hendry MJ, Wassenaar LI, Wolfaardt GM, Germida JJ, Greer CW. 2000. Distribution and biogeochemical importance of bacterial populations in a thick clay-rich aquitard system. Microb Ecol 40:273–291. doi: 10.1007/s002480000073. [DOI] [PubMed] [Google Scholar]
- 25.Alfreider A, Vogt C, Geiger-Kaiser M, Psenner R. 2009. Distribution and diversity of autotrophic bacteria in groundwater systems based on the analysis of RubisCO genotypes. Syst Appl Microbiol 32:140–150. doi: 10.1016/j.syapm.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Kellermann C, Selesi D, Lee N, Hügler M, Esperschütz J, Hartmann A, Griebler C. 2012. Microbial CO2 fixation potential in a tar-oil contaminated porous aquifer. FEMS Microbiol Ecol 81:172–187. doi: 10.1111/j.1574-6941.2012.01359.x. [DOI] [PubMed] [Google Scholar]
- 27.Lin XJ, McKinley J, Resch CT, Kaluzny R, Lauber CL, Frederickson J, Knight R, Konopka A. 2012. Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. ISME J 6:1665–1676. doi: 10.1038/ismej.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray CJ, Engel AS. 2013. Microbial diversity and impact on carbonate geochemistry across a changing geochemical gradient in a karst aquifer. ISME J 7:325–337. doi: 10.1038/ismej.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro-Noya YE, Suarez-Arriaga MC, Rojas-Valdez A, Montoya-Ciriaco NM, Gomez-Acata S, Fernandez-Luqueno F, Dendooven L. 2013. Pyrosequencing analysis of the bacterial community in drinking water wells. Microb Ecol 66:19–29. doi: 10.1007/s00248-013-0222-3. [DOI] [PubMed] [Google Scholar]
- 30.DEV. 1975. Deutsche Einheitsverfahren zur Wasser-, Abwasser- und Schlammuntersuchung. Physikalische, chemische, biologische und bakteriologische Verfahren. ed Fachgruppe Wasserchemie in der Gesellschaft Deutscher Chemiker in Gemeinschaft mit dem Normenausschuß Wasserwesen (NAW) im DIN Deutsches Institut für Normung e.V. VCH Verlagsgesellschaft, Weinheim, Germany. [Google Scholar]
- 31.Velghe N, Claeys A. 1985. Rapid spectrophotometric determination of nitrate in mineral waters with resorcinol. Analyst 110:313–314. doi: 10.1039/an9851000313. [DOI] [Google Scholar]
- 32.Grasshoff K, Ehrhardt M, Kremling K. 1983. Methods of seawater analysis, 2nd ed. Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 33.Kostka JE, Luther GW. 1994. Partitioning and speciation of solid-phase iron in salt-marsh sediments. Geochim Cosmochim Acta 58:1701–1710. doi: 10.1016/0016-7037(94)90531-2. [DOI] [Google Scholar]
- 34.Church MJ, Wai B, Karl DM, DeLong EF. 2010. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol 12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrmann M, Hädrich A, Küsel K. 2012. Predominance of thaumarchaeal ammonia oxidizer abundance and transcriptional activity in an acidic fen. Environ Microbiol 14:3013–3025. doi: 10.1111/j.1462-2920.2012.02882.x. [DOI] [PubMed] [Google Scholar]
- 36.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY. [Google Scholar]
- 37.Muyzer G, Dewaal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Kumar Y, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singleton DR, Furlong MA, Rathbun SL, Whitman WB. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol 67:4374–4376. doi: 10.1128/AEM.67.9.4374-4376.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 43.Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer KH, Wagner M. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol 68:5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opitz S, Küsel K, Spott O, Totsche KU, Herrmann M. 2014. Oxygen availability and distance to surface environments determine community composition and abundance of ammonia oxidizing prokaryotes in two superimposed pristine limestone aquifers in the Hainich region, Germany. FEMS Microbiol Ecol 90:39–53. doi: 10.1111/1574-6941.12370. [DOI] [PubMed] [Google Scholar]
- 45.Kojima H, Tsutsumi M, Ishikawa K, Iwata T, Mußmann M, Fukui M. 2012. Distribution of putative denitrifying methane oxidizing bacteria in sediment of a freshwater lake, Lake Biwa. Syst Appl Microbiol 35:233–238. doi: 10.1016/j.syapm.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 46.Flynn TM, Sanford RA, Ryu H, Bethke CM, Levine AD, Ashbolt NJ, Santo Domingo JW. 2013. Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol 13:146. doi: 10.1186/1471-2180-13-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fields MW, Bagwell CE, Carroll SL, Yan T, Liu X, Watson DB, Jardine PM, Criddle CS, Hazen TC, Zhou J. 2006. Phylogenetic and functional biomarkers as indicators of bacterial community responses to mixed-waste contamination. Environ Sci Technol 40:2601–2607. doi: 10.1021/es051748q. [DOI] [PubMed] [Google Scholar]
- 48.Pester M, Maixner F, Berry D, Rattei T, Koch H, Lücker S, Nowka B, Richter A, Spieck E, Lebedeva E, Loy A, Wagner M, Daims H. 2014. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol 16:3055–3071. doi: 10.1111/1462-2920.12300. [DOI] [PubMed] [Google Scholar]
- 49.Freitag TE, Chang L, Legg CD, Prosser J. 2005. Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils. Appl Environ Microbiol 71:8323–8334. doi: 10.1128/AEM.71.12.8323-8334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kojima H, Fukui M. 2010. Sulfuricella denitrificans gen. nov., sp. nov., a sulfur-oxidizing autotroph isolated from a freshwater lake. Int J Syst Evol Microbiol 60:2862–2866. doi: 10.1099/ijs.0.016980-0. [DOI] [PubMed] [Google Scholar]
- 51.Chapelle FH. 2000. The significance of microbial processes in hydrogeology and geochemistry. Hydrogeol J 8:41–46. doi: 10.1007/PL00010973. [DOI] [Google Scholar]
- 52.Christensen TH, Kjeldsen P, Bjerg PH, Jensen DL, Christensen JB, Braun A, Albrechtsen HJ, Heron G. 2001. Biogeochemistry of landfill leachate plumes. Appl Geochem 16:659–718. doi: 10.1016/S0883-2927(00)00082-2. [DOI] [Google Scholar]
- 53.Pearce AR, Rizzo DM, Mouser PJ. 2011. Subsurface characterization of groundwater contaminated by landfill leachate using microbial community profile data and a non parametric decision-making process. Water Resour Res 47:W06511. doi: 10.1029/2010WR009992. [DOI] [Google Scholar]
- 54.Zhou Y, Kellermann C, Griebler C. 2012. Spatio-temporal patterns of microbial communities in a hydrologically dynamic pristine aquifer. FEMS Microbiol Ecol 81:230–242. doi: 10.1111/j.1574-6941.2012.01371.x. [DOI] [PubMed] [Google Scholar]
- 55.Dojka MA, Hugenholtz P, Haack SK, Pace NR. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol 64:3869–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Detmers J, Strauss H, Bergmann U, Knittel K, Kuever J. 2004. FISH shows that Desulfotomaculum spp. are the dominating sulphate-reducing bacteria in a pristine aquifer. Microb Ecol 47:236–242. doi: 10.1007/s00248-004-9952-6. [DOI] [PubMed] [Google Scholar]
- 57.Bougon N, Aquilina L, Molénat J, Marie D, Delettre Y, Chancerel E, Vandenkoornhuyse P. 2012. Influence of depth and time on diversity of free-living microbial community in the variably saturated zone of a granitic aquifer. FEMS Microbiol Ecol 80:98–113. doi: 10.1111/j.1574-6941.2011.01273.x. [DOI] [PubMed] [Google Scholar]
- 58.Lee H-G, Kim S-G, Im W-T, Oh H-M, Lee S-T. 2009. Pedobacter composti sp. nov., isolated from compost. Int J Syst Evol Microbiol 59:345–349. doi: 10.1099/ijs.0.003061-0. [DOI] [PubMed] [Google Scholar]
- 59.Finneran KT, Johnson CV, Lovley DR. 2003. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int J Syst Evol Microbiol 53:669–673. doi: 10.1099/ijs.0.02298-0. [DOI] [PubMed] [Google Scholar]
- 60.Chin K-J, Liesack W, Janssen PH. 2001. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division “Verrucomicrobia” isolated from rice paddy soil. Int J Syst Evol Microbiol 51:1965–1968. doi: 10.1099/00207713-51-6-1965. [DOI] [PubMed] [Google Scholar]
- 61.Weon HY, Yoo S-H, Hong S-B, Kwon S-W, Stackebrandt E, Go S-J, Koo B-S. 2008. Polaromonas jejuensis sp. nov., isolated from soil in Korea. Int J Syst Evol Microbiol 58:1525–1528. doi: 10.1099/ijs.0.65529-0. [DOI] [PubMed] [Google Scholar]
- 62.Dibbern D, Schmalwasser A, Lueders T, Totsche KU. 2014. Selective transport of plant root-associated bacterial populations in agricultural soils upon snowmelt. Soil Biol Biochem 69:187–196. doi: 10.1016/j.soilbio.2013.10.040. [DOI] [Google Scholar]
- 63.Jaesche P, Totsche KU, Kögel-Knabner I. 2006. Transport and anaerobic biodegradation of propylene glycol in gravel-rich soil materials. J Contam Hydrol 85:271–286. doi: 10.1016/j.jconhyd.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 64.Farnleitner AH, Wilhartitz I, Ryzinska G, Kirschner AKT, Stadler H, Burtscher MM, Hornek R, Szewzyk U, Herndl G, Mach RL. 2005. Bacterial dynamics in spring water of alpine karst aquifers indicates the presence of stable autochthonous microbial endokarst communities. Environ Microbiol 7:1248–1259. doi: 10.1111/j.1462-2920.2005.00810.x. [DOI] [PubMed] [Google Scholar]
- 65.Konno U, Kouduka M, Komatsu DD, Ishii K, Fukuda A, Tsunogai U, Ito K, Suzuki Y. 2013. Novel microbial populations in deep granitic groundwater from Grimsel test site, Switzerland. Microb Ecol 65:626–637. doi: 10.1007/s00248-013-0184-5. [DOI] [PubMed] [Google Scholar]
- 66.Smith RJ, Jeffries TC, Roudnew B, Fitch AJ, Seymour JR, Delpin MW, Newton K, Brown MH, Mitchell JG. 2012. Metagenomic comparison of microbial communities inhabiting confined and unconfined aquifer systems. Environ Microbiol 14:240–253. doi: 10.1111/j.1462-2920.2011.02614.x. [DOI] [PubMed] [Google Scholar]
- 67.Weidler GW, Dornmayr-Pfaffenhuemer M, Gerbl FW, Heinen W, Stan-Lotter H. 2007. Communities of archaea and bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps, and evidence of ammonia-oxidizing crenarchaeota. Appl Environ Microbiol 73:259–270. doi: 10.1128/AEM.01570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pedersen K. 1997. Microbial life in deep granitic rocks. FEMS Microbiol Rev 20:399–414. doi: 10.1111/j.1574-6976.1997.tb00325.x. [DOI] [Google Scholar]
- 69.Jordan DB, Ogren WL. 1981. Species variation in the specificity of ribulose-1,5-bisphosphate carboxylase/oxygenase. Nature 291:513–515. doi: 10.1038/291513a0. [DOI] [Google Scholar]
- 70.Shively JM, van Keulen G, Meijer WG. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu Rev Microbiol 52:191–230. doi: 10.1146/annurev.micro.52.1.191. [DOI] [PubMed] [Google Scholar]
- 71.Jouanneau Y, Tabita FR. 1986. Independent regulation of synthesis of form I and form II ribulose bisphosphate carboxylase/oxygenase in Rhodopseudomonas sphaeroides. J Bacteriol 165:620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beller HR, Letain TE, Chakicherla A, Kane SR, Legler TC, Coleman MA. 2006. Whole-genome transcriptional analysis of chemolithoautotrophic thiosulfate oxidation by Thiobacillus denitrificans under aerobic versus denitrifying conditions. J Bacteriol 188:7005–7015. doi: 10.1128/JB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clark DR, Rees AP, Joint I. 2008. Ammonium regeneration and nitrification rates in the oligotrophic Atlantic Ocean: implications for new production estimates. Limnol Oceanogr 53:52–62. doi: 10.4319/lo.2008.53.1.0052. [DOI] [Google Scholar]
- 74.Hügler M, Sievert SM. 2011. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu Rev Mar Sci 3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 75.Berg IA, Kockelkorn D, Buckel W, Fuchs G. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 76.Auguet J-C, Borrego CM, Baneras L, Casamayor EO. 2008. Fingerprinting the genetic diversity of the biotin carboxylase gene (accC) in aquatic ecosystems as a potential marker for studies of carbon dioxide assimilation in the dark. Environ Microbiol 10:2527–2536. doi: 10.1111/j.1462-2920.2008.01677.x. [DOI] [PubMed] [Google Scholar]
- 77.Bräuer SL, Kranzler K, Goodson N, Murphy D, Simon HM, Baptista AM, Tebo BM. 2013. Dark carbon fixation in the Columbia River's estuarine turbidity maxima: molecular characterization of red-type cbbL genes and measurement of DIC uptake rates in response to added electron donors. Estuaries Coasts 36:1073–1083. doi: 10.1007/s12237-013-9603-6. [DOI] [Google Scholar]
- 78.Lehman RM, Colwell FS, Bala GA. 2001. Attached and unattached microbial communities in a simulated basalt aquifer under fracture- and porous-flow conditions. Appl Environ Microbiol 67:2799–2809. doi: 10.1128/AEM.67.6.2799-2809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Flynn TM, Sandford RA, Bethke CM. 2008. Attached and suspended microbial communities in a pristine confined aquifer. Water Resour Res 44:W07425. doi: 10.1029/2007WR006633. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.