Abstract
This paper describes the transcriptional adaptations of nongrowing, retentostat cultures of Lactococcus lactis to starvation. Near-zero-growth cultures (μ = 0.0001 h−1) obtained by extended retentostat cultivation were exposed to starvation by termination of the medium supply for 24 h, followed by a recovery period of another 24 h by reinitiating the medium supply to the retentostat culture. During starvation, the viability of the culture was largely retained, and the expression of genes involved in transcription and translational machineries, cell division, and cell membrane energy metabolism was strongly repressed. Expression of these genes was largely recovered following the reinitiation of the medium supply. Starvation triggered the elevated expression of genes associated with synthesis of branched-chain amino acids, histidine, purine, and riboflavin. The expression of these biosynthesis genes was found to remain at an elevated level after reinitiation of the medium supply. In addition, starvation induced the complete gene set predicted to be involved in natural competence in L. lactis KF147, and the elevated expression of these genes was sustained during the subsequent recovery period, but our attempts to experimentally demonstrate natural transformation in these cells failed. Mining the starvation response gene set identified a conserved cis-acting element that resembles the lactococcal CodY motif in the upstream regions of genes associated with transcription and translational machineries, purine biosynthesis, and natural transformation in L. lactis, suggesting a role for CodY in the observed transcriptome adaptations to starvation in nongrowing cells.
INTRODUCTION
Lactococcus lactis has a long history of use in the manufacturing of fermented foods. In particular, dairy starter cultures containing L. lactis are important for primary acidification of cheese milk. In addition, L. lactis contributes to the texture and formation of flavor in cheese, which are key determinants of the product's sensory quality (1, 2). Next to these industrial food fermentation applications, L. lactis is frequently found in other environments, such as (decaying) plant material, especially when nutrients become available as a consequence of primary degradation of plant polymers by other microorganisms, including yeasts and fungi (3).
In nature, microbial populations mostly live in nutrient-limiting conditions due to low carbon and energy source availability (4). Under these environmental conditions, microbes either develop adaptive responses to high-affinity substrate acquisition or scavenge the ecosystem for alternative nutrients, resulting in a survival advantage (5). Similarly, in industrial fermentation applications, microbes may encounter restricted access to carbon sources for longer periods of time. For example, lactic acid bacteria (LAB) encounter long periods of low or no carbon availability during the ripening of dried sausages (6) and cheese (1), but they are able to survive under these conditions during months of ripening (6, 7, 8).
Physiological and molecular-level responses of microorganisms to growth stagnation as a consequence of nutrient starvation have been studied in batch cultures that proceed from the logarithmic phase of growth to the stationary phase of growth. An alternative design for the study of starvation in bacteria is a steady-state continuous cultivation in which the medium supply is turned off (9, 10). However, starvation induced by stationary phase in batch cultures and terminating the medium supply in steady-state continuous cultures of LAB encompass a major bottleneck that is generally not encountered in natural habitats. Both cultivation systems of LAB undergo continuous changes in culture conditions due to the increasing concentrations of fermentation end products (e.g., lactic acid) and eventually the accumulation of dead cells that emerge from stimulated cell death, which involves an autolysis process (11). Consequently, it is difficult to disentangle the effects elicited by nutrient depletion from those elicited by the secretion of fermentation end products and cell debris exposure. Also, it is unclear to what extent LAB responses to nutrient starvation and accumulation of end metabolites and dead cells contrast or overlap.
Retentostat cultivation provides an alternative way to study nutrient starvation in microorganisms. This cultivation setup is an adaptation of chemostat cultivation in which complete biomass retention in the fermentor is achieved by continuous removal of spent medium through a cross-flow filter effluent channel (12) (see Fig. S1 in the supplemental material). As a result of cell retention, the microbial biomass concentration increases over time while the carbon source supply remains constant, which results in a decreasing amount of carbon source availability for each individual cell (10, 13, 14). Prolonged retentostat cultivation leads to growth rates that approximate zero and induces a cellular physiology in which the metabolic energy is completely invested in maintenance-associated processes (10, 15), supporting high cell viability, and which is different from stationary-phase-induced starvation (10). Recent retentostat studies with L. lactis at a near-zero growth rate illustrated that lactococcal adaptation to these conditions includes the derepression of genes involved in alternative carbon source import as well as the repression of cell membrane metabolism (16). Some of these adaptations were also observed in batch cultures that entered the stationary phase of growth (17). In contrast to stationary-phase batch cultures, retentostat cultivation did not lead to repression of functions related to central metabolism, cell division, and macromolecule synthesis or the induction of autolytic processes and cell death. These findings illustrate the difference of these two approaches and illustrate that retentostat cultivation enables the study of nutrient starvation in nongrowing cells without the confounding effects of metabolite accumulation and the induction of autolysis and cell death.
The aim of this study was to disentangle genome-wide transcriptional responses in L. lactis to carbon starvation and carbon-limited conditions. To this end, L. lactis KF147, which was originally isolated from mung bean sprouts, was grown under anaerobic, carbon-limited retentostat conditions for 42 days (10). The near-zero-growth cultures obtained were exposed to starvation by terminating the medium supply for 24 h, followed by a recovery phase induced by restarting of the medium supply for another 24 h (see Fig. S1 in the supplemental material). During starvation, the expression of genes related to cell division, the transcription and translational machinery, and cell membrane energy metabolism were significantly decreased, whereas viability of the culture remained high. Subsequent reinitiation of the medium supply led to the recovery of initial expression levels of the translational machinery- and energy metabolism-associated genes. Strikingly, the gene repertoires predicted to be involved in natural transformation were highly induced during the starvation phase, which was sustained during the recovery period. The transcriptome adaptations were evaluated in the context of their regulation, where we identified a conserved cis-acting regulatory motif, and we propose a regulatory role for CodY in controlling expression of the translational machinery and natural competence. Notably, these findings suggest that CodY may play a dual role in the transcriptional adaptation to retentostat conditions (16) as well as the starvation adaptation studied here, which may involve different CodY coregulators.
MATERIALS AND METHODS
Bacterial isolates, media, and cultivation conditions.
Lactococcus lactis subsp. lactis strain KF147 originates from mung bean sprouts, and its genome sequence has been determined (3). Frozen stock cultures that had been stored at −80°C were used to inoculate 50 ml M17 broth (18) supplemented with 0.5% glucose (wt/vol) and grown overnight at 30°C, and these were used as precultures for retentostat cultivations (10). Overnight cultures were harvested by centrifugation (6,000 × g, 10 min, 4°C), washed twice with physiological salt solution (0.9% NaCl in water), and inoculated into chemically defined medium (19) containing 0.5% glucose (wt/vol) for anaerobic chemostat cultivation (dilution rate, 0.025 h−1; pH, 5.5; T, 30°C; stirring speed, 100 rpm; nitrogen flow, 15 ml min−1). Steady-state chemostat conditions were achieved after six volume changes, after which the fermentation was shifted to retentostat mode by removing the spent medium through a cross-flow filter (10).
Duplicate carbon-limited retentostat cultivations were run until (after 42 days) the estimated specific growth rate had decreased to approximately 0.0001 h−1 (10). At this time, the supply of fresh medium was switched off for 24 h, causing immediate carbon source starvation (starvation phase) (see Fig. S1 in the supplemental material), since the retentostat cultivation was already being performed under carbon-limiting conditions (10). After 24 h of starvation, the supply of fresh medium was switched on again, and retentostat cultivation was continued for another 24 h (recovery phase) (see Fig. S1). During these starvation and recovery phases, all other fermentation parameters were kept constant, including temperature (30°C), nitrogen flow (15 ml min−1), stirring speed (100 rpm), and pH (5.5) (10). Since removal of samples could potentially influence the fermentor conditions and the bacterial culture, sample volume and sampling frequency were minimized.
Biomass, substrate, and metabolite determination.
During the 24-h starvation period and subsequent retentostat recovery period of another 24 h, culture samples were taken at regular intervals to measure cell dry weight (CDW) and glucose and organic acid concentrations. CDW was measured using predried and preweighed membrane filters with a pore size of 0.45 μm (Merck Millipore, Darmstadt, Germany) as previously described (10). Concentrations of organic acids and residual glucose in the culture supernatant were determined by high-performance liquid chromatography as previously described (10, 20).
Cell viability and culturable cell estimation.
The viability of cells in the culture was assayed using the Live/Dead BacLight bacterial viability and counting kit (L34856; Molecular Probes Europe, Leiden, The Netherlands), according to the manufacturer's instructions, using the BD FACSAria II flow cytometer (BD Biosciences, San Jose, CA, USA). The kit contains two fluorescent dyes which intercalate with DNA: red fluorescent propidium iodide (PI) and green fluorescent SYTO9. While SYTO9 can diffuse into cells through intact cell membranes (viable cells), PI can penetrate cells only when cell membranes are permeabilized (dead or damaged cells). In addition, viability was also assessed by enumeration of CFU on GM17 agar plates, using serial dilutions of the cultures in triplicate (21).
RNA isolation and transcriptome analysis.
Microarray data used in this study have been merged into the data set described in reference 16 and represented as day 42 (the end of retentostat cultivation; 0 h of carbon starvation), 43 (after 24 h of starvation condition; 24 h), and 44 (after 24 h of reinitiated medium supply and recovery; 48 h). The microarray hybridization scheme for the transcriptome analyses of this experimental set-up consisted of a compound loop design with 7 arrays (see Fig. S6 in the supplemental material).
The gene expression intensities were compared and clustered using Short Time-series Expression Minor (STEM) (version 1.3.6; http://www.cs.cmu.edu/∼jernst/stem/) (22). The STEM clustering algorithm enriched with gene ontology (GO) terms was applied with the Bonferroni correction method, a maximum number of predefined, arbitrary model profiles of 8, and a minimum unit change in model profile between time points of 2 (ratio change of 1 in log2 scale). Profiles with statistically significant numbers of gene assignments were selected, and expression of significantly changed genes was visualized using heat maps generated by the MultiExperiment Viewer (MeV) program (http://www.tm4.org/mev.html) (23). The correlation of the transcriptome data at each time point in duplicate retentostat cultivations, starvation, and recovery procedures was calculated by Pearson correlation analysis and as hierarchical clustering using the MeV program.
DNA motif mining.
Significantly regulated genes that were identified by the significant model profile analyses of the STEM clustering were used to mine for transcription factor binding sites (TFBSs) in the genome of L. lactis KF147. Binding site searches were performed using the 300-bp upstream regions of the selected genes. To identify conserved upstream binding sites, the algorithm for fitting a mixture model by expectation maximization (MEME) was employed (24), using the parameters mod anr (unlimited number of motifs per upstream sequence) and revcomp (allowing motifs to be present on both plus and minus strands) and allowing a maximum of 3 motifs to be found per upstream region without any restriction on the total number of motifs to be found. The PePPER database was used as a source of literature-based regulon clusters (25).
Microarray data accession numbers.
The microarray data obtained in this study and the experimental procedures used to obtain them have been submitted to the NCBI Gene Expression Omnibus under accession numbers GPL17806 and GSE51494 (http://www.ncbi.nlm.nih.gov), respectively.
RESULTS AND DISCUSSION
Biomass accumulation and viability in retentostat-induced carbon starvation.
In previous studies, near-zero-growth cultivation of Lactococcus lactis KF147 was achieved using an anaerobic, carbon-limited retentostat setup (10). During retentostat cultivation, the carbon source was fed at a constant rate, while biomass accumulation increased, reaching a specific growth rate of approximately 0.0001 h−1 after 42 days of retentostat cultivation (10). Since there is an inverse relation between growth rate and doubling time, represented as growth rate = ln(2)/doubling time (26), this growth rate correlated with a culture doubling time of more than 260 days. At this stage, consequently, no carbon source was available for production of biomass, and the entire carbon source-derived energy was employed for maintenance-related processes in the cells (10). To contrast and compare the physiological and transcriptional responses of L. lactis KF147 at near-zero growth (retentostat) and under carbon starvation conditions, the medium supply was switched off after 42 days of retentostat cultivation, thereby exerting a virtually immediate carbon starvation. During the 24-h starvation period, biomass concentration, culture viability, and metabolite levels were determined at the starting point (stable retentostat condition; 0 h) and after 6, 14, and 24 h. Subsequently, the medium supply was switched on again (recovery phase), and the same parameters were determined after an additional 24 h of retentostat cultivation (48 h). In two independent experiments, optical density at 600 nm (OD600) (data not shown), the cell size estimated from flow cytometry analysis (data not shown), and the biomass concentration appeared constant during 24 h of starvation and subsequent 24 h of recovery (Fig. 1A). In parallel, viability of the culture was assessed using the enumeration of CFU (Fig. 1A) and Live/Dead staining linked with fluorescent-activated cell sorting (FACS) analysis (Fig. 1B). The CFU enumeration established only a very slight decrease of culture viability during the 24 h of starvation, which remained stable during the subsequent period of recovery (Fig. 1A). Previously, the viability assessment by discriminative staining and FACS analysis of L. lactis culture samples from the extended retentostat cultivation revealed a division into subpopulations of live, dead, and damaged cells. The latter state was tentatively considered a transition state that could no longer form a colony on solid medium but was still intact (10). The same approach in the present study confirmed the slight decrease of the live population (from approximately 76% to 69%) during the starvation period and its stable maintenance during the subsequent recovery period (Fig. 1B). In parallel, the initially present damaged cell population decreased during the starvation period from approximately 21% to 15%, whereas the population of dead cells increased from 4% to almost 17%, indicating that a substantial part of the damaged cells were dying during the starvation period. The observed damaged and dead population distributions remained stable during the recovery period (Fig. 1B).
FIG 1.
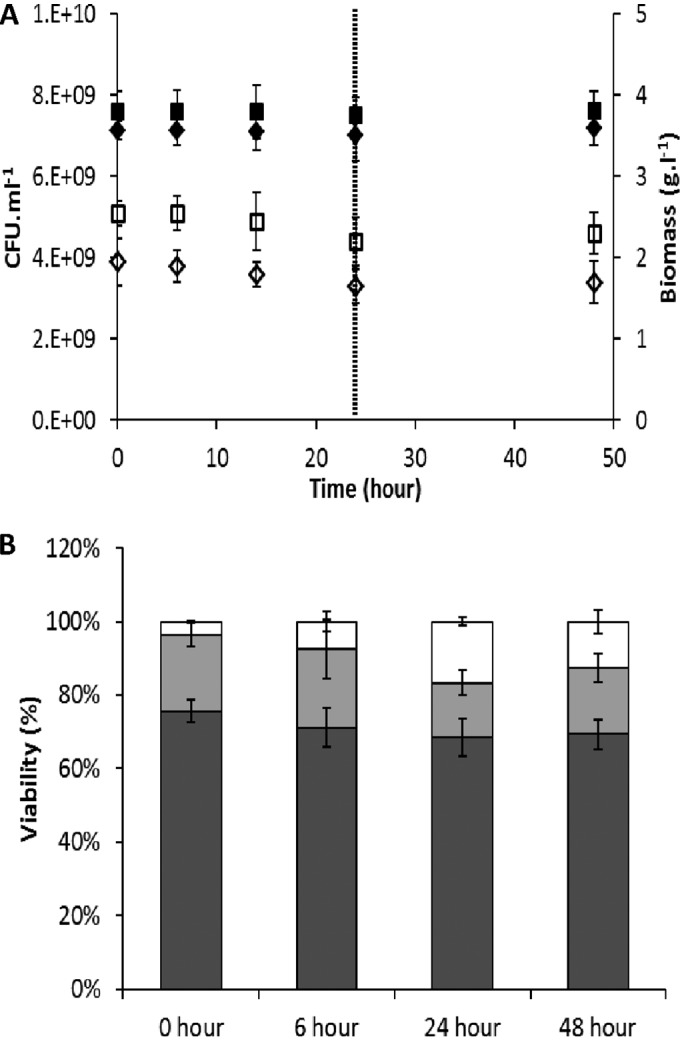
Biomass and viability assessment of L. lactis KF147 under extensive retentostat conditions (0 h), during medium starvation (up to 24 h), and during a subsequent recovery period in retentostat cultivation (48 h). (A) Biomass concentrations (g liter−1) of independent cultures 1 (closed diamonds) and 2 (closed squares) and CFU enumerations (CFU ml−1) of independent cultures 1 (open diamonds) and 2 (open squares). The dashed line indicates the end of the starvation period. Error bars indicate the standard deviations of triplicate measurements. (B) Biomass viability was assayed with a Live/Dead BacLight kit using FACS during the cultivations. This method enables discrimination of percentages of live (dark gray), damaged (light gray), and dead (white) cell populations. Error bars indicate the standard deviations for two independent cultivations.
Metabolic profiling in retentostat and starvation conditions.
The levels of the main fermentation metabolites lactate, formate, acetate, and ethanol were determined (see Fig. S2 in the supplemental material). Due to the extremely low growth rate of L. lactis at the end of retentostat cultivation, a mixed-acid fermentation was observed, with a relative metabolic distribution of 82% oriented toward formation of formate, acetate, and ethanol and 18% toward formation of lactate (see Fig. S2). The organic acid levels remained constant during the 24-h starvation and recovery periods in the current experiment, indicating that during the starvation period, L. lactis does not utilize its fermentation products in secondary fermentation processes that may generate additional energy (e.g., lactate conversion to acetate). Moreover, these data also illustrated that upon reinitiation of the medium supply (recovery phase), the culture executes the carbon flux distribution observed for the initial near-zero-growth retentostat culture (see Fig. S2). Notably, throughout the experiment, both carbon sources available in the medium (glucose and citrate) were completely consumed, and carbon balances calculated on basis of the major fermentation end products were consistently above 95% (data not shown).
Transcriptomic adaptations to starvation and recovery.
To investigate genome-level adaptation of near-zero-growth L. lactis KF147 to starvation conditions, transcriptome analyses were performed in samples derived from the two independent retentostat cultures. Samples were withdrawn before initiating starvation (0 h, i.e., after 42 days of retentostat cultivation), after 24 h of starvation, and after 24 h of subsequent reinitiated medium supply in retentostat cultivation mode (48 h; recovery phase). The initial analysis of the normalized transcriptome data sets included the comparative analysis of the duplicate intervention procedures, revealing that samples were strongly discriminated by their time point of sampling, while the samples taken at the same time point in the two independent cultivations were highly similar (see Fig. S3 in the supplemental material).
To identify gene expression changes during the course of the experiment, absolute expression levels of all genes (2,533 annotated genes in the L. lactis KF147 genome) were subjected to expression cluster analysis using STEM, which uses a process of statistical clustering of short-time-series data sets into precomposed patterns of time-dependent expression (22). STEM distributed the expression patterns into 8 time course model expression profiles, which were ordered on the basis of the number of genes assigned to the model profile. Moreover, STEM clustered 53% and 46% of the annotated L. lactis KF147 genes into 3 statistically significant model profiles (Table 1; also, see Fig. S4A and B in the supplemental material), using the data sets derived from the two independent cultivations, respectively. To explore specific transcriptional responses to starvation and recovery conditions, we focused on gene expression profiles with similar characteristics; i.e., model profile 3, characterized by repressed expression during the starvation period (24 h) and recovered expression during the recovery period (48 h), and model profiles 6 and 7, characterized by induction of gene expression during the starvation period (24 h), followed by, respectively, sustained high expression or dampening of expression during the recovery period (see Fig. S4A and B).
TABLE 1.
Gene ontology (GO) enrichment analysisa
| Model profile(s) | GO category | P value |
|---|---|---|
| 3 | Cellular macromolecule metabolic process | 9.2e−6 |
| Primary metabolic process | 7.6e−6 | |
| Protein metabolic process | 5.5e−6 | |
| Nucleic acid binding | 4.1e−6 | |
| Signal transducer activity | 8.3e−5 | |
| Ribosome | 6.8e−5 | |
| Gene expression | 6.5e−4 | |
| Cell division | 5.1e−4 | |
| Proton-transporting ATP synthase complex | 4.1e−4 | |
| 6 and 7 | Integral to membrane | 3.2e−4 |
| Membrane | 2.4e−4 | |
| Purine nucleoside monophosphate metabolic process | 2.2e−4 | |
| Amino acid transmembrane transporter activity | 2.5e−3 | |
| Heterocycle biosynthetic process | 1.7e−3 | |
| Organic acid transmembrane transporter activity | 1.4e−3 |
Shown are the GO enrichment results for the set of genes shown in Fig. S4 in the supplemental material. Enrichment is computed based on actual size enrichment.
Model profile 3 encompasses 328 and 360 genes in the two independent experiments, and 252 of those genes are shared in both experiments (see Fig. S5 in the supplemental material). Within these model profiles, the gene ontology (GO) terms that were strongly overrepresented included “cellular macromolecule metabolic process,” “protein metabolic process,” “ribosome,” “gene expression,” “proton-transporting ATP synthase complex,” and “cell division” (Table 1). Many transcriptional regulator encoding genes (bglR, glnR, hslR, nisRK, tcsKR, truA, uxaR, etc.) (Fig. 2A) and the complete membrane-associated ATP synthase-encoding operon (atpABCDEFH) were included in these model profiles (Fig. 2E). In addition, many genes involved in DNA replication (dcm, dnaAC, infC, lemA, papL, parC, rexB, topA, and xerC), genes encoding components of the RNA polymerase (rpoAEZ) (Fig. 2B), genes encoding several ribosomal proteins (rplIKOQU, rpmBEFGIHJ, and rpsJKLMO) (Fig. 2C), and cell division- and autolysis-associated genes (ddl, divIVA, engB, ftsAEWXZ, murDFGI, and acmAD) (Fig. 2D) were also assigned to model profile 3. These genes appear to be repressed in response to the carbon and energy starvation when the medium supply is shut down, but they recover their expression upon reinitiation of the medium supply. These observations indicate that L. lactis KF147 is adapted to starvation conditions through stringent responses, which suppress the transcription and translational machineries, DNA replication, and cell division. Notably, members of the acm gene family play prominent roles in stationary-phase-induced autolysis (27), but the N-acetyl muramidases (acmAD) assigned to profile 3 have also been reported to play a central role in cell wall repair and remodeling (28). The observation that only a marginal reduction of CFU enumeration is observed during these experiments implies that under these conditions the autolytic phenotype is not induced, which contrasts the typical induction of autolysis in stationary-phase cells obtained from batch cultures.
FIG 2.

Heat map of L. lactis KF147 genes differentially expressed (log2 scale; P ≤ 0.05) during medium starvation (24 h) and a recovery period in retentostat cultivation (48 h) compared to the initial retentostat culture (0 h) that are clustered in model profile 3 by the STEM module. Genes are grouped into statistically significant GO categories: (A) transcriptional regulators, (B) replication, (C) ribosome functions, (D) cell division and lysis, and (E) ATP synthases. In all panels, the genes are ordered on the basis of their expression changes from the highest to the lowest level. Gene expression patterns shown are derived from one of the experimental duplicates, and these expression patterns were highly similar between duplicates.
Remarkably, stringent responses have also been associated with the induction of stress response genes, including heat, acid, and cold shock genes (17), and starvation conditions during the stationary phase of batch cultures led to induction of stress responses in L. lactis (17, 29). These responses were not observed in the experiments performed here, which is likely explained by the fact that retentostat preculturing and near-zero-growth conditions had already led to strong induction of a panel of stress-related functions in the cultures that were used as the starting point in the current experiment (0 h). This retentostat-induced high-level expression of stress-related genes appeared to be sustained during the starvation and recovery periods of the current experiments. These findings imply that stress responses in L. lactis are important for maintenance of cellular function and viability under near-zero-growth conditions, as well as under starvation conditions, which may reflect the evolutionary advantage of stress robustness for bacteria that encounter conditions that do not permit growth and include (temporal) nutrient starvation (30, 31). Analogously, in industrial fermentation applications, increased stress robustness of lactococci may affect product characteristics by extending their viability, which may improve their contribution to aroma and flavor formation during product ripening or prolong shelf life of products that include probiotic bacteria (32).
Model profiles 6 and 7 were analyzed together because these profiles are both characterized by elevated expression levels during starvation (24 h), which either remained at a high level of expression (profile 7) or declined (profile 6) during the subsequent recovery period. Profiles 6 and 7 of the two replicate experiments collectively shared many genes, but their precise distribution between profiles 6 and 7 appeared to be inconsistent (see Fig. S4A and B and S5 in the supplemental material). These combined model profiles were significantly enriched for the functional categories related to “purine nucleoside monophosphate metabolic process,” “integral to membrane,” “organic acid transmembrane transporter activity,” “amino acid transmembrane transporter activity,” and “heterocycle biosynthetic process” (Table 1). In particular, genes and gene clusters encoding the enzymes for biosynthesis of histidine, branched-chain amino acids (valine, leucine, and isoleucine), purine, and riboflavin (hisACDGHBFIK, ilvBD-leuA, purCDEFHKLMNQ, and ribABH, respectively) were highly induced during starvation, and their high level of expression was sustained during the recovery period (Fig. 3A). In addition, the genes encoding metabolism of spermidine/putrescine (potA2B1B2C1C2D1) and gamma-aminobutyric acid (GABA) (yibEFG, ysjA, and yshA), transporters of metal ions (potassium [kdpBCE] and cobalt [corA and cbiQ]), and multidrug resistance transporters (blt, cydCD, ydiCD, ypgD, and sugE) were strongly induced during the starvation period (Fig. 3B).
FIG 3.
Heat map of L. lactis KF147 genes differentially expressed (log2 scale; P ≤ 0.05) during medium starvation (24 h) and a recovery period in retentostat cultivation (48 h) compared to the initial retentostat culture (0 h) that are clustered in model profiles 6 and 7 by the STEM module. Genes are grouped into statistically significant GO categories: (A) purine metabolic process, (B) membrane and transmembrane transporter activity, and (C) competence protein-related genes. In all panels, the genes are ordered on the basis of their expression changes from the highest to the lowest level. Gene expression patterns shown are derived from one of the experimental duplicates, and these expression patterns were highly similar between duplicates.
A remarkable molecular adaptation of L. lactis KF 147 to the carbon starvation and recovery regimen executed here was the enhanced transcription level of genes predicted to play a role in natural competence (coiA, comC, comEA, comEC, comFA, comGA, comGB, comGC, comGD, comGE, comGF, and comX) (Fig. 3C), a phenotype that to the best of our knowledge has not been observed in L. lactis strains to date (33). Natural competence enables cells to import exogenous genetic material to repair damaged endogenous genes, provide additional genetic diversity, or simply serve as a nutrient source (34). However, in the Gram-negative bacterium Shewanella oneidensis, it was demonstrated that extracellular DNA can also be used as a sole source of phosphorus, carbon, and energy under nutrient-limiting conditions (35). Furthermore, 8 genes involved in utilization of extracellular DNA as a carbon and energy source under carbon-limited conditions in Escherichia coli share significant homology with competence-associated com genes in Haemophilus influenzae and Neisseria gonorrhoeae (36). Previously, Redon and coworkers reported that five out of 13 genes related to a natural transformation pathway were induced during the stationary phase of growth of batch-cultured L. lactis IL1403 (17), whereas our experiments revealed the induction of all genes of L. lactis KF147 that are linked to competence. Induction of starvation in the high-density retentostat culture by the closing of the medium supply (0 h) is instant, which may explain the complete expression of pathways that could facilitate the import of exogenous DNA as a potential nutrient source. In this context, it is interesting that the Gram-positive bacterium L. lactis adapted to near-zero-growth conditions by extended retentostat cultivation (the state of the culture at 0 h), highly expressed the capacity to import alternative carbon sources, and was found to more rapidly utilize these carbon sources when they were provided to the cells (16). Apparently, starvation led to an expansion of this quest for alternative nutrients, including the machinery to import exogenous DNA.
Identification of a cis-acting DNA motif potentially involved in starvation and recovery gene regulation.
To identify candidate cis-acting DNA motifs that are potentially involved in controlling transcriptional adaptation to starvation and recovery, we searched for overrepresented DNA sequences in the upstream regions of genes that showed correlated expression. Remarkably, the previously identified highly conserved sequence element containing the palindromic 5′-CTGTCAG-3′ motif that resembles the CodY binding site (Fig. 4B) (16) was also identified in profiles 3, 6, and 7 in the present study (Fig. 4A). Analogous to our earlier findings (16), many of the identified motifs appeared in a duplicated head-to-tail orientation upstream of genes, which strengthens the palindromic nature of these regulatory motifs. The proposed CodY motif is present upstream of genes related to transcriptional and cell division machinery (ddl, murCDFG, mutM, papL, parC, rpmF, rexB, truA, and yacB), purine metabolic processing (guaC and purHL), and natural transformation protein (coiA, comEA, comEC, and comGC) (see Fig. S7 in the supplemental material). This result implies that the previously identified motif could play a prominent role in the regulation of the stringent response, purine metabolism, and natural competence in L. lactis KF147 in the adaptation to starvation.
FIG 4.
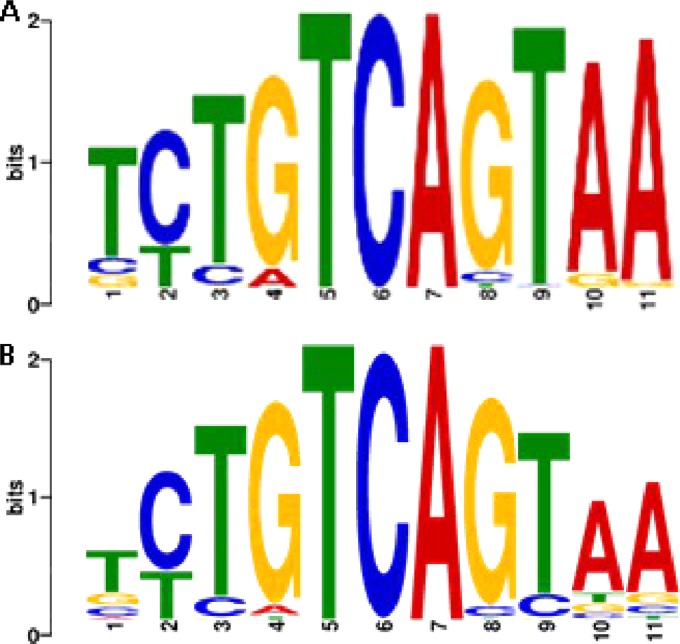
WebLogo visualization of the CodY motif in L. lactis KF147. (A) The postulated CodY upstream binding sequence found in front of the transcription and translational machinery, purine biosynthesis, and natural transformation related genes in L. lactis KF147. (B) The previously postulated CodY motif in L. lactis KF147 (16). The CTGTCAG palindrome sequence that forms the core of the motif is positioned from nucleotide 2 to 8. (Reprinted from reference 16.)
The lactococcal protein CodY has been reported to be a global regulator that contributes to the expression control of a variety of cellular functions, including macromolecular degradation, nutrient transport, and, amino acid and peptide metabolism (37, 38). CodY repression is alleviated in nutrient-limited environments, allowing the expression of many genes involved in adaptation to these restrictive conditions. In addition, several CodY-regulated genes are coregulated by additional mechanisms, allowing their fine-tuned modulation in response to other environmental conditions (38). In our earlier study, we proposed that the identified motif represents a high-affinity binding site for CodY in L. lactis KF147 and that CodY may orchestrate transcriptional adaptation of nitrogen- and carbon-metabolism in L. lactis KF147 at near-zero growth rates (16). In the present study, we expand the possible regulatory role of CodY to include the control of stringent response, purine metabolism, and natural transformation in nongrowing L. lactis KF147 upon its exposure to starvation conditions. Notably, some of the proposed CodY-mediated responses lead to downregulation of expression, while other genes are upregulated, implying that the CodY regulon may be subject to multiple mechanisms of dualistic and bimodal regulation. Since flavor formation in the ripening process of fermented food products, such as cheese, is linked with amino acid conversions that involve nitrogen metabolism-related enzymes (2), it is of importance to decipher regulation mechanisms and corresponding transcriptional regulators of nitrogen metabolism in the context of industrial applications of L. lactis in order to improve products' sensory characteristics.
Concluding remarks and perspective.
This paper presents the physiological and molecular responses of near-zero-growth cultures of L. lactis obtained by carbon source-limited retentostat cultivation to starvation elicited by closure of the medium supply. In addition, this study evaluates the recovery capacity of this starved culture upon reinitiation of the medium supply after 24 h of starvation.
Previous studies have shown that anaerobic retentostat cultivation of L. lactis mimics zero-growth conditions in which high culture viability is sustained and where metabolic energy is exclusively used for maintenance-related processes rather than cell growth-related processes (10). Notably, the maintenance-related adaptations included significant changes in both carbohydrate and nitrogen metabolisms, as well as the gene expression patterns that support expanded carbohydrate source flexibility (16). Contrary to the retentostat cultivation, the induction of starvation conditions strongly repressed typical growth-associated functions, including transcriptional, translational, DNA replication, and cell division machineries but also including the ATP synthase function. This response can be classified as a typical stringent response (17), which was not elicited under near-zero-growth conditions but was apparent only upon subsequent starvation. These observations are in clear contrast to the stringent response that is already significantly induced during retentostat cultivation of another Gram-positive bacterium, Bacillus subtilis (39), but also in cultures of Saccharomyces cerevisiae (40). Despite the starvation induced stringent response, the L. lactis culture viability remained relatively high, and the culture was capable of rapid recovery of metabolic activity upon reinitiation of the medium supply, which also led to a relief of the stringent response.
Intriguingly, the strong induction of the genes predicted to play a role in natural competence under starvation conditions may serve as an adaptation to sequester additional nutrients (i.e., exogenous DNA) rather than a system to expand the strain's genetic repertoire, although that option cannot be excluded. Our attempts to validate the proposed natural competence of the cells obtained from these cultures failed to prove that these cells could import and/or chromosomally integrate (linearized) plasmid DNA. To date, the functionality of the competence system and natural transformation in L. lactis have not been experimentally validated. Nevertheless, the observed activation of the complete competence gene set within the KF147 genome is intriguing and may offer possibilities to activate and study this relevant phenotype in lactococci.
Finally, mining of the transcriptome data sets for TFBSs identified a highly conserved motif, 5′-CTGTCAG-3′, which we previously detected as a potential regulatory motif involved in the adaptation to near-zero-growth conditions (16) and which is likely to represent the target site for CodY. This finding appears to indicate a dualistic and bimodal role of CodY-mediated regulation in response to changing environments, which may imply the involvement of additional coregulatory mechanisms to fine-tune these responses within the CodY regulon. Notably, structural analysis of the CodY of B. subtilis indicated that GTP acts as a ligand for this conserved regulator and that CodY may respond to (p)ppGpp levels during the stringent response (41, 42, 43). The proposed role of CodY in the stringent-response-related gene regulation patterns observed in this study supports an interaction between CodY and the alarmone (p)ppGpp, which may explain the dualistic and coordinated role of CodY in gene expression control during the adaptation to zero-growth conditions (retentostat) (16) and during the adaptation to starvation in which its interaction with (p)ppGpp may drive the stringent responses observed in the present study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Roelie Holleman (NIZO food research, Ede, The Netherlands) for technical assistance with HPLC. In addition, we thank our colleagues from the Industrial Microbiology Section, Delft University of Technology and Molecular Genetics Group, University of Groningen, in the joint zero-growth project group (Kluyver Centre, The Netherlands) for invaluable discussions.
This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03748-14.
REFERENCES
- 1.Smit G, Smit BA, Engels WJM. 2005. Flavour formation by lactic acid bacteria and biochemical flavor profiling of cheese products. FEMS Microbiol Rev 29:591–610. doi: 10.1016/j.fmrre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Smid EJ, Kleerebezem M. 2014. Production of aroma compounds in lactic fermentations. Annu Rev Food Sci Technol 5:313–326. doi: 10.1146/annurev-food-030713-092339. [DOI] [PubMed] [Google Scholar]
- 3.Siezen RJ, Bayjanov J, Renckens B, Wels M, van Hijum SAFT, Molenaar D, van Hylckama Vlieg JET. 2010. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J Bacteriol 192:2649–2650. doi: 10.1128/JB.00276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koch AL. 1971. The adaptive responses of Escherichia coli to a famine and feast existence. Adv Microb Physiol 6:147–217. doi: 10.1016/S0065-2911(08)60069-7. [DOI] [PubMed] [Google Scholar]
- 5.Egli T. 2010. How to live at very low substrate concentration. Water Res 44:4826–4837. doi: 10.1016/j.watres.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 6.Hugas M, Monfort JM. 1997. Bacterial starter cultures for meat fermentation. Food Chem 59:547–554. doi: 10.1016/S0308-8146(97)00005-8. [DOI] [Google Scholar]
- 7.Crow VL, Coolbear T, Gopal PK, Martley FG, McKay LL, Riepe H. 1995. The role of autolysis of lactic acid bacteria in the ripening of cheese. Int Dairy J 5:855–875. doi: 10.1016/0958-6946(95)00036-4. [DOI] [Google Scholar]
- 8.Erkus O, de Jager VCL, Spus M, van Alen-Boerrigter IJ, van Rijswijck IMH, Hazelwood L, Janssen PWM, van Hijum SAFT, Kleerebezem M. 2013. Multifactorial diversity sustains microbial community stability. ISME J 7:2126–2136. doi: 10.1038/ismej.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poolman B, Smid EJ, Veldkamp H, Konings WN. 1987. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J Bacteriol 169:1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ercan O, Smid EJ, Kleerebezem M. 2013. Quantitative physiology of Lactococcus lactis at extreme low-growth rates. Environ Microbiol 15:2319–2332. doi: 10.1111/1462-2920.12104. [DOI] [PubMed] [Google Scholar]
- 11.Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol 4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 12.van Verseveld HW, de Hollander JA, Frankena J, Braster M, Leeuwerik FJ, Stouthamer AH. 1986. Modeling of microbial substrate conversion, growth and product formation in a recycling fermenter. Antonie Van Leeuwenhoek 52:325–342. doi: 10.1007/BF00428644. [DOI] [PubMed] [Google Scholar]
- 13.Boender LGM, de Hulster EAF, van Maris AJA, Daran-Lapujade P, Pronk JT. 2009. Quantitative physiology of Saccharomyces cerevisiae at near-zero specific growth rates. Appl Environ Microbiol 75:5607–5614. doi: 10.1128/AEM.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goffin P, van de Bunt B, Giovane M, Leveau JHJ, Höppener-Ogawa S, Teusink B, Hugenholtz J. 2010. Understanding the physiology of Lactobacillus plantarum at zero growth. Mol Syst Biol 6:413. doi: 10.1038/msb.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Bodegom P. 2007. Microbial maintenance: a critical review on its quantification. Microbiol Ecol 53:513–523. doi: 10.1007/s00248-006-9049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ercan O, Wels M, Smid EJ, Kleerebezem M. 2015. Molecular and metabolic adaptations of Lactococcus lactis at near-zero growth rates. Appl Environ Microbiol 81:320–331. doi: 10.1128/AEM.02484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redon E, Loubiere P, Cocaign-Bousquet M. 2005. Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J Bacteriol 187:3589–3592. doi: 10.1128/JB.187.10.3589-3592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terzaghi BE, Sandine WE. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol 29:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poolman B, Konings WN. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol 170:700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugenholtz J, Starrenburg MJC. 1992. Diacetyl production by different strains of Lactococcus lactis subsp. lactis var. diacetylactis and Leuconostoc spp. Appl Microbiol Biotechnol 38:17–22. [Google Scholar]
- 21.Sieuwerts S, de Bok FAM, Mols E, de Vos WM, van Hylckama Vlieg JET. 2008. A simple and fast method for determining colony forming units. Lett Appl Microbiol 47:275–278. doi: 10.1111/j.1472-765X.2008.02417.x. [DOI] [PubMed] [Google Scholar]
- 22.Ernst J, Bar-Joseph Z. 2006. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. 2006. TM4 microarray software suite. Method Enzymol 411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 24.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 25.de Jong A, Pietersma H, Cordes M, Kuipers OP, Kok J. 2012. PePPER: a web server for prediction of prokaryote promoter elements and regulons. BMC Genomics 13:299. doi: 10.1186/1471-2164-13-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baty F, Delignette-Muller ML. 2004. Estimating the bacterial lag time: which model, which precision? Int J Food Microbiol 91:261–277. doi: 10.1016/j.ijfoodmicro.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Buist G, Karsens H, Nauta A, van Sinderen D, Venema G, Kok J. 1997. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl Environ Microbiol 63:2722–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steen A, Palumbo E, Deghorain M, Cocconcelli PS, Delcour J, Kuipers OP, Kok J, Buist G, Hols P. 2005. Autolysis of Lactococcus lactis is increased upon d-alanine depletion of peptidoglycan and lipoteichoic acids. J Bacteriol 187:114–124. doi: 10.1128/JB.187.1.114-124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartke A, Bouche S, Gansel X, Boutibonnes P, Auffray Y. 1994. Starvation-induced stress resistance in Lactococcus lactis subsp. lactis IL1403. Appl Environ Microbiol 60:3473–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schimel J, Balser T, Wallenstein M. 2007. Microbial stress-response physiology and its implications for ecosystem function. Ecology 88:1386–1394. doi: 10.1890/06-0219. [DOI] [PubMed] [Google Scholar]
- 31.Maharjan R, Nilsson S, Sung J, Haynes K, Beardmore RE, Hurst LD, Ferenci T, Gudelj I. 2013. The form of a trade-off determines the response to competition. Ecol Lett 16:1267–1276. doi: 10.1111/ele.12159. [DOI] [PubMed] [Google Scholar]
- 32.Smid EJ, Erkus O, Spus M, Wolkers-Rooijackers JC, Alexeeva S, Kleerebezem M. 2014. Functional implications of the microbial community structure of undefined mesophilic starter cultures. Microb Cell Fact 13(Suppl 1):S2. doi: 10.1186/1475-2859-13-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wydau S, Dervyn R, Anba J, Ehrlich SD, Maguin E. 2006. Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol Lett 257:32–42. doi: 10.1111/j.1574-6968.2006.00141.x. [DOI] [PubMed] [Google Scholar]
- 34.Johnsborg O, Eldholm V, Havarstein LS. 2007. Natural genetic transformation: prevalence, mechanisms, and function. Res Microbiol 158:767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Pinchuk GE, Amnons C, Culley DE, Li SMW, McLean JS, Romine MF, Nealson KH, Fredrickson JK, Beliaev AS. 2008. Utilization of DNA as a sole source of phosphorus, carbon, and energy by Shewanella spp.: ecological and physiological implications for dissimilatory metal reduction. Appl Environ Microbiol 74:1198–1208. doi: 10.1128/AEM.02026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palchevskiy V, Finkel SE. 2006. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol 188:3902–3910. doi: 10.1128/JB.01974-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.den Hengst CD, van Hijum SAFT, Geurts JMW, Nauta A, Kok J. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J Biol Chem 280:34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- 38.Guédon E, Sperandio B, Pons N, Ehrlich SD, Renault P. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151:3895–3909. doi: 10.1099/mic.0.28186-0. [DOI] [PubMed] [Google Scholar]
- 39.Overkamp W, Ercan O, Herber M, van Maris AJA, Kleerebezem M, Kuipers OP. 3 November 2014. Physiological and cell morphology adaptation of Bacillus subtilis at near-zero specific growth rates: a transcriptome analysis. Environ Microbiol doi: 10.1111/1462-2920.12676. [DOI] [PubMed] [Google Scholar]
- 40.Boender LGM, van Maris AJA, de Hulster EAF, Almering MJH, van der Klei IJ, Veenhuis M, de Winde JH, Pronk JT, Daran-Lapujade P. 2011. Cellular responses of Saccharomyces cerevisiae at near-zero growth rates: transcriptome analysis of anaerobic retentostat cultures. FEMS Yeast Res 11:603–620. doi: 10.1111/j.1567-1364.2011.00750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in Gram-positive bacteria. J Biol Chem 281:11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]
- 42.Wolz C, Geiger T, Gorke C. 2010. The synthesis and function of the alarmone (p)ppGpp in firmicutes. Int J Med Microbiol 300:142–147. doi: 10.1016/j.ijmm.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Geiger T, Wolz C. 2014. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int J Med Microbiol 304:150–155. doi: 10.1016/j.ijmm.2013.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



