Abstract
We sought to determine if the time, within a production day, that a cheese is manufactured has an influence on the microbial community present within that cheese. To facilitate this, 16S rRNA amplicon sequencing was used to elucidate the microbial community dynamics of brine-salted continental-type cheese in cheeses produced early and late in the production day. Differences in the microbial composition of the core and rind of the cheese were also investigated. Throughout ripening, it was apparent that cheeses produced late in the day had a more diverse microbial population than their early equivalents. Spatial variation between the cheese core and rind was also noted in that cheese rinds were initially found to have a more diverse microbial population but thereafter the opposite was the case. Interestingly, the genera Thermus, Pseudoalteromonas, and Bifidobacterium, not routinely associated with a continental-type cheese produced from pasteurized milk, were detected. The significance, if any, of the presence of these genera will require further attention. Ultimately, the use of high-throughput sequencing has facilitated a novel and detailed analysis of the temporal and spatial distribution of microbes in this complex cheese system and established that the period during a production cycle at which a cheese is manufactured can influence its microbial composition.
INTRODUCTION
Commercial cheeses produced with defined starter/adjunct strains often suffer from variations in cheese flavor profile and microbial content (1). This is thought to be primarily due to batch variations in milk quality and storage time as well as manufacturing practices (2) and the adventitious microbial populations present (3, 4). Indeed, in the latter case, aroma and taste defects, along with biogenic amine formation, mineral deposition (calcium lactate) issues, and irregular gas formation, are common defects associated with a variety of microorganisms (5).
Analysis of the bacterial composition of cheese has traditionally involved the use of culture-based techniques which, while effective for quantifying common starter or nonstarter bacteria as well as certain spoilage bacteria (Clostridium, Staphylococcus), do not always accurately reflect the total microbiota present (6, 7). PCR-based molecular profiling techniques targeting either particular populations or select taxonomic communities are also routinely used and have been extensively reviewed (8–10). PCR-based methods cannot, however, provide comprehensive coverage of total microbial populations.
The advent of high-throughput next-generation sequencing (NGS) has advanced the field of microbial ecology by providing a powerful means of analyzing dominant and subdominant populations and their dynamics in highly complex ecosystems (2). NGS has been applied extensively to a variety of environments, including the sea (11) and soil (12) as well as the gut (13). More recently, NGS of bacterial 16S rRNA amplicons has been used to characterize the microbial communities of a variety of fermented foods and beverages (14–20) as well as of raw milk and raw milk cheeses (21–26). Indeed, this approach has led to identification of a number of genera previously not associated with cheese ecosystems (Prevotella and Helcococcus) or with particular cheese types (Arthrobacter in goat's milk cheese). Microbial content has also been shown to vary with milk source, processing (raw or pasteurized), and addition of various ingredients (27). Ultimately, NGS platforms offer detection sensitivity that is significantly increased compared to more-traditional molecular methods with respect to the study of bacterial communities (2, 26, 28, 29). NGS-based approaches have also been used to profile communities present in production facilities, providing a unique insight into possible microbial reservoirs important for cheese sensory characteristics or for identifying potential biofilm-forming genera (2).
Both culture and molecular approaches have been used to better understand the spatial distribution of microbes in cheese. Microbial composition varies throughout the cheese block due to several factors, including salt, moisture, pH, and the availability of oxygen (30). The effect of salt is particularly important in brine-salted cheese varieties, as salt migrates to the core of the cheese over the course of the ripening process, affecting moisture levels and microbial growth (31). To date, the majority of studies examining the spatial distribution of microbial populations in cheese have relied on two methods. One involves nondestructive fluorescence microscopy, based on production of a gel cassette system (32) or the use of cryosectioning, followed by fluorescence in situ hybridization (FISH) using rRNA-targeted probes (33, 34). The second involves destructive sampling of selected regions of cheese followed by an assessment of the microbiota by culture-dependent and/or culture-independent methods (3, 30, 35–37). More recently, an NGS approach was used by Wolfe et al. to reveal both the microbial composition and the functional potential of 137 cheese rind communities. In that case, 16S rRNA gene and internal transcribed spacer (ITS) amplicon sequencing allowed characterization of microbial communities, while “shotgun” metagenomics permitted an in-depth analysis of pathways involved in flavor formation (38).
In this study, 16S rRNA amplicon sequencing was used to describe, from both a spatial perspective and a temporal perspective, the microbiota present in a brine-salted continental-type cheese produced within a single production day. This study built on results from a previous study which reported a significant interaction between time of day of manufacture and stage of ripening with respect to mean viable counts of nonstarter lactic acid bacteria (NSLAB) (P < 0.04), with the cheeses (n = 42) produced late in the day (in comparison to those produced early in the day or in the middle of the day of manufacture) having significantly higher mean viable NSLAB counts (39). We assessed whether production of the cheese earlier or later during the daily cheese-making cycle has an impact on the subsequent development of its bacterial community, investigated how these populations change throughout the ripening process, and examined variances in the microbial spatial distribution between the cheese core and rind. In each case, noteworthy variations in the microbial composition, resulting from differences in the production phase, the stage of ripening, or the part of the cheese being studied, were apparent.
MATERIALS AND METHODS
Cheese production, sampling, and nucleic acid extraction.
Four blocks of a semihard brine-salted continental-type cheese produced from pasteurized milk were sourced at 1 day postproduction. The blocks were produced in a single production day, from separate vats, and corresponded to early day (ED [morning sampling]; n = 2) and late day (LD [afternoon sampling]; n = 2) production, with 6 to 8 h separating ED and LD manufacture. Furthermore, two blocks were received from each respective vat. Cheeses were produced based on a Swiss-type model using the thermophilic starters Streptococcus thermophilus and Lactobacillus helveticus. Propionibacterium freudenreichii was added as an adjunct. Postproduction, cheeses were subjected to ripening at 10°C for 10 days prior to hot-room ripening (20°C) from day 10 to day 40. Cheeses were then stored at 6°C for the remainder of the ripening period.
Each individual block was sampled aseptically, using a cheese trier, at 4 stages: 1 day postproduction (TP1), 10 days postproduction (TP2), 40 days postproduction (TP3), and (after maturation) 64 days postproduction (TP4). Internal (core) and external (rind [1-cm3 segment]) regions of the cheese, at each time point, were also sampled. Cheese (1 g) was homogenized in 9 ml of a 2% trisodium citrate buffer (VWR, Dublin, Ireland). Enzymatic lysis treatment of homogenized cheese samples was conducted prior to DNA extraction and included treatment with lysozyme (1 mg/ml), mutanolysin (50 U/ml), and proteinase K (800 μg/ml) and incubation for 1 h at 55°C per the method of Quigley et al. (40). DNA was extracted using a PowerFood microbial DNA isolation kit (MoBio Laboratories Inc., Carlsbad, CA, USA). Grated samples from cheeses were analyzed for salt (41), moisture (42), and pH (43) at TP4.
PCR amplification of the microbial 16S rRNA gene.
Extracted DNA was amplified using universal primers targeting the V4 region of the bacterial 16S gene (239 nucleotides [nt]) (4, 27). Primers predicted to bind to 94.6% of all bacterial 16S genes consisted of a forward primer, F1 (5′-AYTGGGYDTAAAGNG), and a combination of four reverse primers, R1 (5′-TACCRGGGTHTCTAATCC), R2 (5′-TACCAGAGTATCTAATTC), R3 (5′-CTACDSRGGTMTCTAATC), and R4 (5′-TACNVGGGTATCTAATC) (RDP's Pyrosequencing Pipeline: http://pyro.cme.msu.edu/pyro/help.jsp). Primers also included a 19-mer sequence (GCCTGCCAGCCCGCTCAG) at the 5′ end to allow emulsion-based clonal amplification for the 454 pyrosequencing system. Identification of individual sequences from the pooled samples was achieved by incorporating molecular identifier tags between the primer sequence and the adaptamer.
PCRs were carried out in triplicate and contained 25 μl BioMix Red master mix (Bioline, London, United Kingdom), 1 μl of each primer (200 nmol liter−1), 5 μl of the DNA template (standardized to 100 ng DNA/sample), and nuclease-free water added to reach a final volume of 50 μl. PCR amplification was carried out using a G-Storm Thermal Cycler (Gene Technologies, United Kingdom). Amplification consisted of an initial denaturation at 94°C for 10 min followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1 min. This was followed by a final elongation step at 72°C for 2 min. PCR amplicons were cleaned using an AMPure XP purification system (Beckman Coulter, Takeley, United Kingdom). DNA quantity was assessed using Quant-It Picogreen double-stranded DNA (dsDNA) reagent (Invitrogen, USA) in accordance with manufacturer's guidelines and in conjunction with a NanoDrop 3300 Fluorospectrometer (Thermo-Fisher Scientific, Wilmington, MA, USA). Furthermore, DNA was standardized to equimolar concentrations prior to library preparation and sequencing.
High-throughput sequencing and bioinformatic analysis.
16S rRNA amplicons from the V4 region were sequenced on a Roche 454 FLX platform (Roche Diagnostics Ltd., West Sussex, United Kingdom) as previously described (17, 27) and according to protocols. Reads were quality filtered using the RDP sequencing pipeline (44). Reads with low-quality scores (below 40) and short lengths (less than 150 bp) and reads lacking exact matches with respect to primer sequence were discarded. Reads were clustered and aligned and chimeras removed also within the QIIME software package (45). All assigned operational taxonomic units (OTUs) were considered. A phylogenetic tree was generated using FastTree software, and alpha and beta diversities were subsequently calculated. Principal-coordinate analysis (PCoA), measuring dissimilarities at phylogenetic differences based on weighted/unweighted Unifrac analysis, were carried out using the QIIME suite of programs (45). Resultant PCoA plots were visualized with KiNG. Each trimmed FASTA sequence was assessed using the BLAST program (46) against the SILVA 16S database (version 1.06). The resultant BLAST program output was parsed using MEGAN (47). Bit scores were used for filtering the results prior to tree construction and summarization (absolute cutoff value, BLAST bit score of 86; relative cutoff value, 10% of top hit).
Nucleotide sequence accession number.
Reads were deposited in the SRA database under accession number PRJEB8181.
RESULTS
α-Diversity and β-diversity of microbial populations in early- and late-day production cheeses.
Blocks of a brine-salted continental-type cheese, manufactured early or late during a production cycle, were sampled at various stages throughout the ripening process. Post-DNA extraction, amplicons corresponding to the V4 region of the bacterial 16S rRNA gene were generated by PCR. These amplicons were then subjected to NGS, generating 294,853 reads. This corresponded to 87,156 reads for TP1, 97,045 reads for TP2, 62,248 reads for TP3, and 48,404 reads for TP4 (the full lists of reads and individual samples and associated bar graphs are located in Table S1 and Fig. S2 in the supplemental material). Species diversity (α-diversity) and richness were calculated for each time point as well as for the time of manufacture (early or late day) and the location (core or rind) from which the samples were collected. These are presented in Table 1. Chao1 values, reflective of operational taxonomic unit richness, ranged from 237.8 to 529.38, while the Shannon index values, used to measure overall sample diversity, ranged from 2.51 to 3.82. Analysis of these data revealed that α-diversity decreases throughout the ripening process. Cheeses produced early in the production day had a less diverse microbiota than those produced late in the production day. Diversity appeared greatest in the rinds of the samples at TP1, whereas, for all subsequent time points, core populations were more diverse. These observations held true regardless of whether the samples were from ED or LD manufacture. Rarefaction curves, used to determine species richness from sampling, were calculated at 97% similarity. These revealed that bacterial diversity was well represented, as data from samples are nearing positions parallel with the x axis (see Fig. S1 in the supplemental material).
TABLE 1.
Alpha diversity of continental-type cheeses segregated according to time of production day and spatial distribution (core or rind)
| Time or distribution of production or samplea | Chao1 value | Simpson value | Shannon index value | Phylogenetic diversity value | No. of observed OTUs |
|---|---|---|---|---|---|
| Time | |||||
| TP1 ED | 401.77 | 0.69 | 2.88 | 13.25 | 222.50 |
| TP2 ED | 328.80 | 0.65 | 2.62 | 11.13 | 198.25 |
| TP3 ED | 345.91 | 0.73 | 3.17 | 12.19 | 210.00 |
| TP4 ED | 304.11 | 0.66 | 2.63 | 9.72 | 165.25 |
| TP1 LD | 523.31 | 0.80 | 3.56 | 16.15 | 310.25 |
| TP2 LD | 478.63 | 0.75 | 3.29 | 14.58 | 292.75 |
| TP3 LD | 397.96 | 0.82 | 3.60 | 12.69 | 236.75 |
| TP4 LD | 357.94 | 0.78 | 3.34 | 12.46 | 215.33 |
| Distribution | |||||
| ED | |||||
| TP1 core | 372.24 | 0.67 | 2.80 | 12.18 | 194.00 |
| TP2 core | 294.59 | 0.62 | 2.51 | 11.37 | 182.00 |
| TP3 core | 417.14 | 0.72 | 3.16 | 13.00 | 238.50 |
| TP4 core | 370.37 | 0.61 | 2.56 | 11.65 | 183.50 |
| TP1 rind | 431.30 | 0.70 | 2.96 | 14.32 | 251.00 |
| TP2 rind | 363.00 | 0.67 | 2.72 | 10.89 | 214.50 |
| TP3 rind | 274.69 | 0.75 | 3.18 | 11.37 | 181.50 |
| TP4 rind | 237.84 | 0.71 | 2.70 | 7.78 | 147.00 |
| LD | |||||
| TP1 core | 517.23 | 0.80 | 3.52 | 14.80 | 290.00 |
| TP2 core | 471.02 | 0.75 | 3.32 | 14.86 | 295.00 |
| TP3 core | 412.17 | 0.83 | 3.60 | 12.71 | 244.50 |
| TP4 core | 405.02 | 0.83 | 3.82 | 15.40 | 241.00 |
| TP1 rind | 529.38 | 0.79 | 3.60 | 17.51 | 330.50 |
| TP2 rind | 486.25 | 0.76 | 3.26 | 14.29 | 290.50 |
| TP3 rind | 383.75 | 0.81 | 3.60 | 12.67 | 229.00 |
| TP4 rind | 334.40 | 0.76 | 3.10 | 10.99 | 202.50 |
ED, early day; LD, late day.
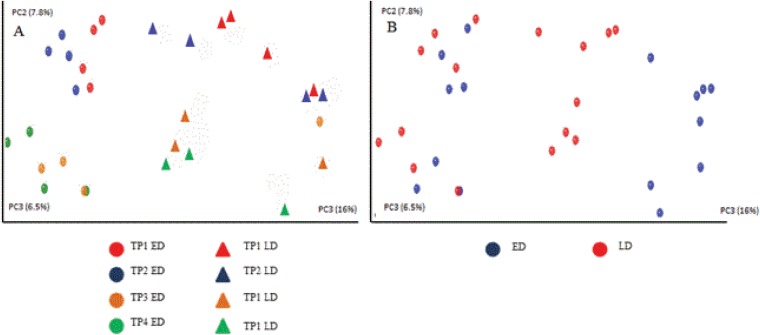
β-Diversity, based on the unweighted UniFrac matrix, and represented in the form of a PCoA plot, was used to determine whether samples grouped with respect to ripening point, time of manufacture (early or late), and collection from internal or external regions of the cheese (Fig. 1). Notably, samples from the same time point during the cheese-ripening process generally grouped together, with data points from TP1 and TP2 and from TP3 and TP4 also forming distinct clusters. In addition, samples clustered according to time of cheese production, with those produced early in the production day clustering together, away from a more diffuse cluster of data points corresponding to samples from cheeses manufactured later in the production cycle (Fig. 1A). Core and rind samples also formed distinct clusters. The distinction between the core and rind populations was more apparent in samples manufactured later in the production cycle (Fig. 1B).
FIG 1.
Principal-coordinate (PC) analysis of the β diversity (unweighted Unifrac) of cheese samples. (A) Coordinates reflect samples produced early and late in the production day (ED and LD, respectively) and are color coded to reflect the ripening phase of the cheese. (B) The same data are depicted, but in this instance, core and rind samples are distinguished.
Cheese composition.
Cheese pH, salt, and salt/moisture (S/M) levels were determined at TP4 for both ED and LD cheeses. Results were similar with respect to pH (5.39 ED and 5.45 LD), salt (0.59% ED and 0.57% LD), and salt/moisture (1.55% ED and 1.51% LD).
High-throughput sequencing reveals differences in microbial taxa between cheeses produced early and cheeses produced late in the production day.
Phylogenetic assignment of high-throughput sequence data revealed the presence of bacteria corresponding to 5 phyla: Firmicutes, Proteobacteria, Bacteroidetes, Deinococcus-Thermus, and Actinobacteria. As expected, the Firmicutes dominated throughout the study, representing 93.46% to 99.75% of reads in the ED samples. The percentages of the reads that corresponded to Firmicutes were lower in the LD samples and ranged from 72.26% to 85.56%. Deinococcus-Thermus was detected in both the ED and LD samples but at higher percentages in the LD samples. Less-dominant populations, corresponding to Actinobacteria and Proteobacteria, were also detected. Proteobacteria populations were highest at TP1 in both ED and LD samples.
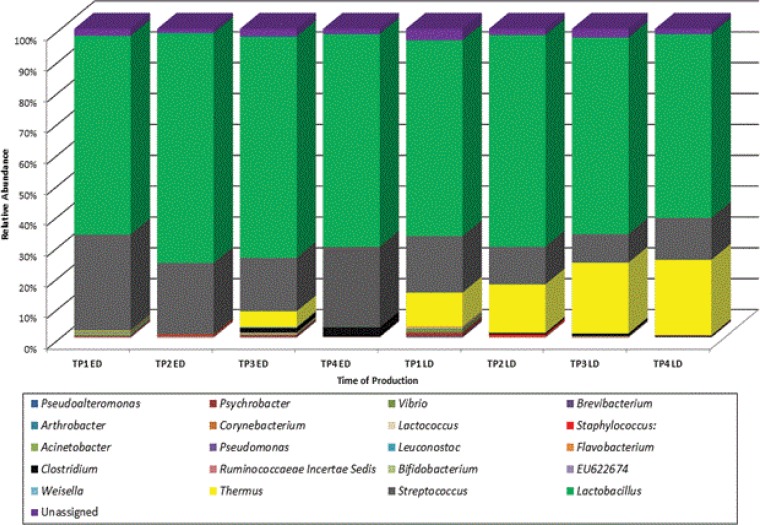
At the genus level, a number of differences were noted between cheese produced early and late in the production day (Fig. 2). Lactobacillus and Streptococcus populations dominated in both ED and LD samples throughout the study. Percentages of populations of Lactobacillus were similar in the ED and LD samples at TP1 (64.4% ED and 63.5% LD); thereafter, it was noticed that populations were consistently higher in the ED samples. Proportions of streptococci were greater in the ED samples (31.1%) than in the LD samples (18.3%), a trend that continued throughout the study. Thermus was detected in both ED and LD samples but in consistently greater proportions in the LD samples (0.1% to 5% in ED samples and 10.9% to 24.4% in LD samples).
FIG 2.
Relative abundances of bacteria at the genus level for a continental-type cheese produced early and late (ED and LD) in the production day. Results depicted are mean values of reads generated from individual core/rind samples from each respective cheese block.
Among the subdominant populations, there were a number of other notable observations. At TP1 and TP2, Acinetobacter and Pseudomonas were detected exclusively in the ED samples whereas Brevibacterium and Corynebacterium were detected only in the LD samples at TP1. Clostridium was identified at TP2 in both ED and LD samples and were consistently detected throughout the remainder of the study. In all instances, Clostridium was present at higher proportions in ED samples. Populations of Staphylococcus, a genus commonly associated with food spoilage, were detected in both ED and LD samples at TP2 only. Of the other subdominant populations detected, Vibrio, Lactococcus, and Psychrobacter were recurrently present in both ED and LD samples, while Pseudoalteromonas was present in ED and LD samples until TP4. A full list of both the dominant and subdominant genera present is located in Table S2 in the supplemental material.
Distribution of microbial communities present in the core and rind of a brine-salted continental-type cheese.
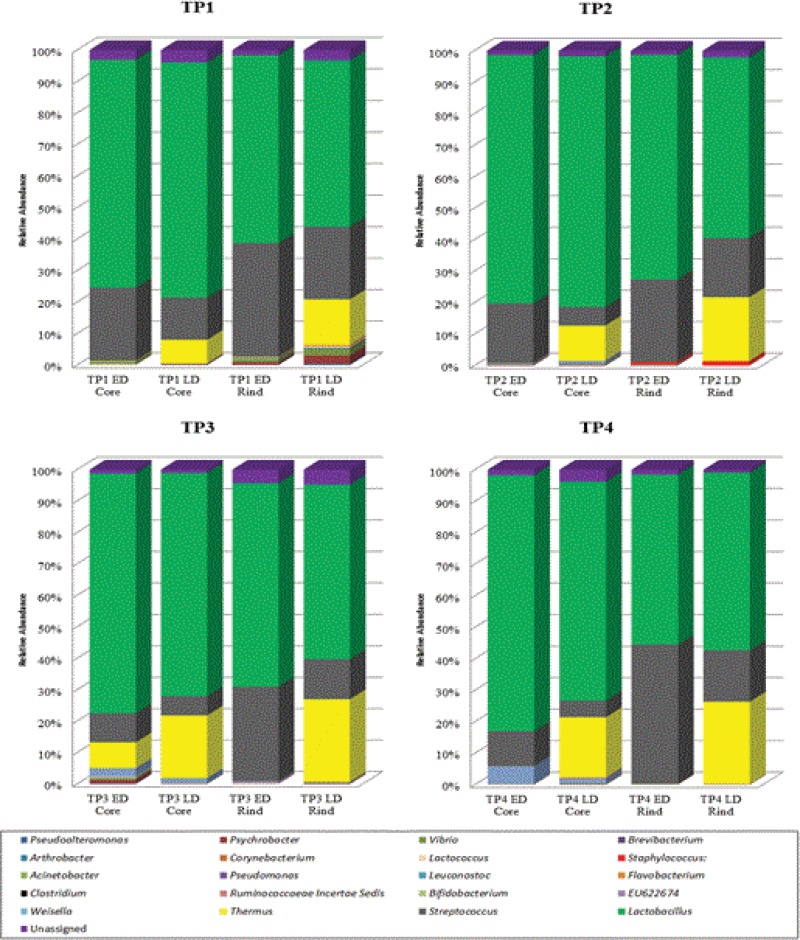
Although the majority of genera detected in this study were localized in both the core and the rind of the cheese sampled (Fig. 3 and 4), differences in proportions were noted. This is most obvious in examining the populations corresponding to the genus Lactobacillus, which were consistently higher in number in the core of the cheeses than in the rind throughout the ripening process. In contrast, Streptococcus populations were consistently higher in the respective rinds than in the core. Thermus populations were also noticeably higher in the rinds than the core. This difference was particularly apparent in the LD samples (i.e., the samples in which Thermus levels were highest). Populations that included Lactococcus, Vibrio, and Psychrobacter were consistently detected in both the core and the rind throughout the ripening process. Similarly, populations of Pseudomonas and Pseudoalteromonas were identified in the core and rind at the initial ripening stages but not at TP4. Of the other subdominant populations, clostridia, present in TP2, TP3, and TP4, were detected only in the respective cheese cores. Similarly, populations of Ruminococcaceae incertae sedis, Bifidobacterium, and Arthrobacter were sporadically detected in core regions only. Populations of Brevibacterium and Corynebacterium, genera commonly associated with surface-ripened cheeses, were located in the rind, as were populations of Staphylococcus and Weisella. A full list of both dominant and subdominant genera present is located in Table S3 in the supplemental material.
FIG 3.
Relative abundances of bacteria at the genus level for each TP according to the sample location (core or rind). Data presented are mean values of respective reads from individual cheese samples.
FIG 4.
Venn diagram depicting spatial differences in the microbial composition at each time point. Genera located in the intersecting region were detected in both the core and the rind, while those on the periphery were detected exclusively in the core or the rind.
DISCUSSION
In this study, NGS of 16S rRNA amplicons provided a detailed insight into the microbiota present in a brine-salted continental-type cheese produced with thermophilic starter bacteria. As expected, bacterial diversity was found to decrease throughout the ripening process. Interestingly, bacterial diversity in cheeses produced late in the day was determined to be greater than the diversity in those produced early in the production day. Differences in the microbial populations present in the respective cores and rinds were noted, while several genera not usually associated with cheese produced from pasteurized milk were also detected.
Microbial diversity (α-diversity) was greatest at TP1 (1 day postproduction) in both the samples produced early in the day and those produced late in the day. While the diversity may seem low in comparison to the diversity in gut or soil communities (12, 48), it is comparable to that seen in studies of similar cheese types (27). Cheeses that were produced later during the initial manufacturing day ultimately had a more diverse microbial population than their equivalents produced earlier in the day. This trend persisted throughout ripening, demonstrating, for the first time, that the time of day at which production occurs impacts not only the microbiota present in the final product but also the microbiota present throughout ripening. Greater diversity in terms of microbial populations present in LD cheeses may be due to an accumulating microbial load during the manufacturing process or may be a result of longer milk storage times. The significance of this phenomenon with respect to cheese quality will be the focus of further investigations.
Prior studies have described differences in the spatial distribution of microbial communities between the rind and core of several cheeses produced from both raw and pasteurized milk. Variation is likely due to the abiotic characteristics of the cheese, including O2, pH, salt, water activity (aw), redox potential, and temperature fluctuations (30, 49). In this study, the greater initial diversity in the rind may be due to the high cooking temperatures associated with some continental-type cheeses. Dependent on the block size, cheese cores may hold higher temperatures longer than the rind, consequently reducing microbial growth. Increased diversity in the rind, at TP1, may also have been due to the presence of halophiles (Vibrio and Pseudoalteromonas) associated with the salting process. Aerobic and aerotolerant microbes, including Streptococcus, Pseudoalteromonas, Psychrobacter, Vibrio, and Brevibacterium, were detected more often and at greater percentages in the cheese rind than in the core. This was likely due to the oxygen concentration present at or near the surface of the cheese in contrast to the more anaerobic core (35). Prior studies have shown that Gram-positive LAB are more likely to be distributed in the core than in the rind of smear-ripened and Swiss-type cheeses (Comté, Morbier, and Langres) (3). In agreement, we observed consistently higher proportions of Lactobacillus in the core than in the rind throughout ripening, possibly due to their preference for a microanaerobic environment. In contrast, streptococci, present in both the core and rinds throughout ripening, were found at higher percentages in the rind. In samples from TP2 to TP4, the cores of both ED and LD cheeses had higher microbial diversity than the rinds. This difference was particularly evident in the samples from late in the production day. Reduced diversity in the rind may have been due to several factors, including substrate competition and availability of O2 as well as pH/salt microgradients (49). Aerobic staphylococci were also identified in the rinds of both early- and late-day samples at TP2, in agreement with Maher and Murphy, who described rinds of smear-ripened cheeses as providing conditions that are complementary for the survival of spoilage microbes (50).
Species of Gram-negative bacteria, many of which would not generally be associated with a commercial cheese produced from pasteurized milk, were detected throughout this study. Thermus populations were detected throughout ripening and at higher percentages in the late-day samples (from 10.9% at TP1 to 24.4% at TP4). The presence of Thermus was confirmed by subsequent PCR using Thermus-specific primers (data not shown). This aerobic, marine-associated thermophilic and heterotrophic genus was originally isolated from alkaline hot springs in Yellowstone National Park (51, 52). As Thermus has previously been identified in two separate hot water systems, it is conceivable that this bacterium was introduced via a water source (52, 53). No negative health effects resulting from consumption of these cheeses have been reported, but further studies will be required to assess the effect Thermus has on cheese quality. Other Gram-negative genera detected include Pseudomonas, Pseudoalteromonas, Psychrobacter, Vibrio, and Flavobacterium. Vibrio and Pseudoalteromonas are marine-associated, halophilic genera and therefore may have gained access to the cheese via the brining process. While the significance of the presence of these populations is not yet clear, particularly at the levels present in the cheese, they may play a role in ripening (38, 49, 54). Psychrotrophic bacteria, including Psychrobacter and Pseudomonas, have previously been isolated from a variety of cheeses as well as raw milk and are particularly adapted to low-temperature milk storage conditions (49, 55, 56).
Many genera more commonly associated with artisanal and surface-ripened cheeses were detected. Brevibacterium and Corynebacterium were identified immediately postproduction and are associated with flavor and color development in smear-ripened cheese (22, 57–59). Arthrobacter, Weissella, and Acinetobacter, previously isolated from a variety of artisanal cheeses, were also identified, although their impact on cheese quality is unknown (60–67). The significance of the presence of gut-associated genera, including Bifidobacterium and Ruminococcaceae Incertae Sedis, is also unclear.
Clostridium was consistently identified in all time points aside from TP1. The percentages of clostridia present, with respect to samples from early in the production day, increased throughout ripening to 3.1% in TP4 ED cheeses. While the presence of Clostridium populations is a particular issue due its association with late gas production in various cheeses (5), no defects were noted, in this instance, at the time of sampling. Finally, Propionibacterium was not detected despite its addition as an adjunct. Further investigation of this revealed that Propionibacterium species are among the very few species that are not successfully amplified by the degenerate primers used in this study.
In conclusion, the use of high-throughput amplicon sequencing to profile the microbiota present in a brine-salted, continental-type cheese has revealed distinct differences in bacterial diversity, throughout ripening, between cheeses produced early and those produced late in the production day. As mentioned, the differences between ED and LD cheeses may have been due to increased microbial load and/or increased milk storage time between production runs; therefore, adapting these practices may result in a more microbiologically consistent product. Spatial variation due to environmental factors present in the core and rind was also found in this study. Furthermore, the presence of genera that would usually not be traditionally associated with this cheese type (Thermus, Bifidobacterium, Ruminococcaceae Incertae Sedis, Psychrobacter, and Pseudoalteromonas) was described. The significance of the presence of these genera requires further investigation.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Department of Agriculture, Food and the Marine of Ireland under the Food Institutional Research Measure through the Cheeseboard 2015 project. D. J. O'Sullivan is in receipt of a Teagasc Walsh Fellowship, grant number 2012205.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04054-14.
REFERENCES
- 1.Rehman S-U, McSweeney PLH, Banks JM, Brechany EY, Muir DD, Fox PF. 2000. Ripening of Cheddar cheese made from blends of raw and pasteurised milk. Int Dairy J 10:33–44. doi: 10.1016/S0958-6946(00)00024-8. [DOI] [Google Scholar]
- 2.Bokulich NA, Mills DA. 2013. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl Environ Microbiol 79:5214–5223. doi: 10.1128/AEM.00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogier J-C, Lafarge V, Girard V, Rault A, Maladen V, Gruss A, Leveau J-Y, Delacroix-Buchet A. 2004. Molecular fingerprinting of dairy microbial ecosystems by use of temporal temperature and denaturing gradient gel electrophoresis. Appl Environ Microbiol 70:5628–5643. doi: 10.1128/AEM.70.9.5628-5643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quigley L, McCarthy R, O'Sullivan O, Beresford TP, Fitzgerald GF, Ross RP, Stanton C, Cotter PD. 2013. The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J Dairy Sci 96:4928–4937. doi: 10.3168/jds.2013-6688. [DOI] [PubMed] [Google Scholar]
- 5.O'Sullivan DJ, Giblin L, McSweeney PL, Sheehan JJ, Cotter PD. 2013. Nucleic acid-based approaches to investigate microbial-related cheese quality defects. Front Microbiol 4:1. doi: 10.3389/fmicb.2013.00001.23346082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peláez C, Requena T. 2005. Exploiting the potential of bacteria in the cheese ecosystem. Int Dairy J 15:831–844. doi: 10.1016/j.idairyj.2004.12.001. [DOI] [Google Scholar]
- 7.Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM. 2001. Recent advances in cheese microbiology. Int Dairy J 11:259–274. doi: 10.1016/S0958-6946(01)00056-5. [DOI] [Google Scholar]
- 8.Jany J-L, Barbier G. 2008. Culture-independent methods for identifying microbial communities in cheese. Food Microbiol 25:839–848. doi: 10.1016/j.fm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Cocolin L, Alessandria V, Dolci P, Gorra R, Rantsiou K. 2013. Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int J Food Microbiol 167:29–43. doi: 10.1016/j.ijfoodmicro.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Bokulich NA, Mills DA. 2012. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep 45:377–389. doi: 10.5483/BMBRep.2012.45.7.148. [DOI] [PubMed] [Google Scholar]
- 11.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nacke H, Thurmer A, Wollherr A, Will C, Hodac L, Herold N, Schoning I, Schrumpf M, Daniel R. 2011. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6:e17000. doi: 10.1371/journal.pone.0017000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claesson MJ, O'Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O'Toole PW. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humblot C, Guyot J-P. 2009. Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearl millet slurries. Appl Environ Microbiol 75:4354–4361. doi: 10.1128/AEM.00451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam Y-D, Yi S-H, Lim S-I. 2012. Bacterial diversity of cheonggukjang, a traditional Korean fermented food, analyzed by barcoded pyrosequencing. Food Control 28:135–142. doi: 10.1016/j.foodcont.2012.04.028. [DOI] [Google Scholar]
- 16.Połka J, Rebecchi A, Pisacane V, Morelli L, Puglisi E. 2015. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol 46:342–356. doi: 10.1016/j.fm.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Marsh AJ, O'Sullivan O, Hill C, Ross RP, Cotter PD. 2013. Sequence-based analysis of the microbial composition of water kefir from multiple sources. FEMS Microbiol Lett 348:79–85. doi: 10.1111/1574-6968.12248. [DOI] [PubMed] [Google Scholar]
- 18.Marsh AJ, O'Sullivan O, Hill C, Ross RP, Cotter PD. 2014. Sequence-based analysis of the bacterial and fungal compositions of multiple kombucha (tea fungus) samples. Food Microbiol 38:171–178. doi: 10.1016/j.fm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Jung JY, Lee SH, Kim JM, Park MS, Bae J-W, Hahn Y, Madsen EL, Jeon CO. 2011. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl Environ Microbiol 77:2264–2274. doi: 10.1128/AEM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roh SW, Kim K-H, Nam Y-D, Chang H-W, Park E-J, Bae J-W. 2010. Investigation of archaeal and bacterial diversity in fermented seafood using barcoded pyrosequencing. ISME J 4:1–16. doi: 10.1038/ismej.2009.83.19587773. [DOI] [PubMed] [Google Scholar]
- 21.Masoud W, Vogensen FK, Lillevang S, Abu Al-Soud W, Sørensen SJ, Jakobsen M. 2012. The fate of indigenous microbiota, starter cultures, Escherichia coli, Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR. Int J Food Microbiol 153:192–202. doi: 10.1016/j.ijfoodmicro.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Quigley L, O'Sullivan O, Stanton C, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2013. The complex microbiota of raw milk. FEMS Microbiol Rev 37:664–698. doi: 10.1111/1574-6976.12030. [DOI] [PubMed] [Google Scholar]
- 23.Aldrete-Tapia A, Escobar-Ramírez MC, Tamplin ML, Hernández-Iturriaga M. 2014. High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Food Microbiol 44:136–141. doi: 10.1016/j.fm.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Delgado S, Rachid CTCC, Fernández E, Rychlik T, Alegría Á, Peixoto RS, Mayo B. 2013. Diversity of thermophilic bacteria in raw, pasteurized and selectively-cultured milk, as assessed by culturing, PCR-DGGE and pyrosequencing. Food Microbiol 36:103–111. doi: 10.1016/j.fm.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 25.Masoud W, Takamiya M, Vogensen FK, Lillevang S, Al-Soud WA, Sørensen SJ, Jakobsen M. 2011. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. Int Dairy J 21:142–148. doi: 10.1016/j.idairyj.2010.10.007. [DOI] [Google Scholar]
- 26.Alegría Á, Szczesny P, Mayo B, Bardowski J, Kowalczyk M. 2012. Biodiversity in oscypek, a traditional Polish cheese, determined by culture-dependent and -independent approaches. Appl Environ Microbiol 78:1890–1898. doi: 10.1128/AEM.06081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2012. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl Environ Microbiol 78:5717–5723. doi: 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ercolini D. 2013. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbiology and ecology. Appl Environ Microbiol 79:3148–3155. doi: 10.1128/AEM.00256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuka MM, Wallisch S, Engel M, Welzl G, Havranek J, Schloter M. 2013. Dynamics of bacterial communities during the ripening process of different Croatian cheese types derived from raw ewe's milk cheeses. PLoS One 8:e80734. doi: 10.1371/journal.pone.0080734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheehan A, O'Cuinn G, FitzGerald RJ, Wilkinson MG. 2009. Distribution of microbial flora, intracellular enzymes and compositional indices throughout a 12-kg Cheddar cheese block during ripening. Int Dairy J 19:321–329. doi: 10.1016/j.idairyj.2008.11.008. [DOI] [Google Scholar]
- 31.Guinee TP. 2004. Salting and the role of salt in cheese. Int J Dairy Technol 57:99–109. doi: 10.1111/j.1471-0307.2004.00145.x. [DOI] [Google Scholar]
- 32.Malakar PK, Brocklehurst TF, Mackie AR, Wilson PDG, Zwietering MH, van't Riet K. 2000. Microgradients in bacterial colonies: use of fluorescence ratio imaging, a non-invasive technique. Int J Food Microbiol 56:71–80. doi: 10.1016/S0168-1605(00)00222-1. [DOI] [PubMed] [Google Scholar]
- 33.Ercolini D, Hill PJ, Dodd CER. 2003. Development of a fluorescence in situ hybridization method for cheese using a 16S rRNA probe. J Microbiol Methods 52:267–271. doi: 10.1016/S0167-7012(02)00162-8. [DOI] [PubMed] [Google Scholar]
- 34.Fleurot I, Aigle M, Fleurot R, Darrigo C, Hennekinne J-A, Gruss A, Borezée-Durant E, Delacroix-Buchet A. 2014. Following pathogen development and gene expression in a food ecosystem: the case of a Staphylococcus aureus isolate in cheese. Appl Environ Microbiol 80:5106–5115. doi: 10.1128/AEM.01042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monfredini L, Settanni L, Poznanski E, Cavazza A, Franciosi E. 2012. The spatial distribution of bacteria in Grana-cheese during ripening. Syst Appl Microbiol 35:54–63. doi: 10.1016/j.syapm.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Fitzsimons NA, Cogan TM, Condon S, Beresford T. 2001. Spatial and temporal distribution of non-starter lactic acid bacteria in Cheddar cheese. J Appl Microbiol 90:600–608. doi: 10.1046/j.1365-2672.2001.01285.x. [DOI] [PubMed] [Google Scholar]
- 37.Gobbetti M, Burzigotti R, Smacchi E, Corsetti A, De Angelis M. 1997. Microbiology and biochemistry of gorgonzola cheese during ripening. Int Dairy J 7:519–529. doi: 10.1016/S0958-6946(97)00047-2. [DOI] [Google Scholar]
- 38.Wolfe BE, Button JE, Santarelli M, Dutton RJ. 2014. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 158:422–433. doi: 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daly DFM. 2014. Studies on factors relating to the development of defects in commercial cheese. Ph.D. thesis University College Cork, County Cork, Ireland. [Google Scholar]
- 40.Quigley L, O'Sullivan O, Beresford TP, Paul Ross R, Fitzgerald GF, Cotter PD. 2012. A comparison of methods used to extract bacterial DNA from raw milk and raw milk cheese. J Appl Microbiol 113:96–105. doi: 10.1111/j.1365-2672.2012.05294.x. [DOI] [PubMed] [Google Scholar]
- 41.International Dairy Federation (IDF). 1988. Cheese and processed cheese: determination of chloride content (potentiometric titration method). International standards 4a. International Dairy Federation, Brussels, Belgium. [Google Scholar]
- 42.IDF. 1982. Determination of total solids content (cheese and processed cheese). International standards 4a. International Dairy Federation, Brussels, Belgium. [Google Scholar]
- 43.British Standards Institute. 1976. British standard methods for chemical analysis of cheese: determination of pH value. British Standards Institute, London, England. [Google Scholar]
- 44.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res 17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stearns JC, Lynch MDJ, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. 25 November 2011, posting date Bacterial biogeography of the human digestive tract. Sci Rep doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montel M-C, Buchin S, Mallet A, Delbes-Paus C, Vuitton DA, Desmasures N, Berthier F. 2014. Traditional cheeses: rich and diverse microbiota with associated benefits. Int J Food Microbiol 177:136–154. doi: 10.1016/j.ijfoodmicro.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 50.Maher MM, Murphy PM. 2000. Microbiological changes during ripening in two Irish smear-ripened, farmhouse cheeses produced from raw milk. Irish J Agric Food Res 39:107–121. [Google Scholar]
- 51.Spanevello MD, Patel BKC. 2004. The phylogenetic diversity of Thermus and Meiothermus from microbial mats of an Australian subsurface aquifer runoff channel. FEMS Microbiol Ecol 50:63–73. doi: 10.1016/j.femsec.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Pask-Hughes R, Williams RAD. 1975. Extremely thermophilic Gram-negative bacteria from hot tap water. J Gen Microbiol 88:321–328. doi: 10.1099/00221287-88-2-321. [DOI] [PubMed] [Google Scholar]
- 53.Bagh LK, Albrechtsen H-J, Arvin E, Ovesen K. 2004. Distribution of bacteria in a domestic hot water system in a Danish apartment building. Water Res 38:225–235. doi: 10.1016/j.watres.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 54.Bleicher A, Neuhaus K, Scherer S. 2010. Vibrio casei sp. nov., isolated from the surfaces of two French red smear soft cheeses. Int J Syst Evol Microbiol 60:1745–1749. doi: 10.1099/ijs.0.016493-0. [DOI] [PubMed] [Google Scholar]
- 55.Delcenserie V, Taminiau B, Delhalle L, Nezer C, Doyen P, Crevecoeur S, Roussey D, Korsak N, Daube G. 2014. Microbiota characterization of a Belgian protected designation of origin cheese, Herve cheese, using metagenomic analysis. J Dairy Sci 97:6046–6056. doi: 10.3168/jds.2014-8225. [DOI] [PubMed] [Google Scholar]
- 56.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2011. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int J Food Microbiol 150:81–94. doi: 10.1016/j.ijfoodmicro.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Brennan NM, Brown R, Goodfellow M, Ward AC, Beresford TP, Simpson PJ, Fox PF, Cogan TM. 2001. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int J Syst Evol Microbiol 51(Pt 3):843–852. doi: 10.1099/00207713-51-3-843. [DOI] [PubMed] [Google Scholar]
- 58.Brennan NM, Ward AC, Beresford TP, Fox PF, Goodfellow M, Cogan TM. 2002. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl Environ Microbiol 68:820–830. doi: 10.1128/AEM.68.2.820-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mounier J, Rea MC, O'Connor PM, Fitzgerald GF, Cogan TM. 2007. Growth characteristics of Brevibacterium, Corynebacterium, Microbacterium, and Staphylococcus spp. isolated from surface-ripened cheese. Appl Environ Microbiol 73:7732–7739. doi: 10.1128/AEM.01260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Addis E, Fleet GH, Cox JM, Kolak D, Leung T. 2001. The growth, properties and interactions of yeasts and bacteria associated with the maturation of Camembert and blue-veined cheeses. Int J Food Microbiol 69:25–36. doi: 10.1016/S0168-1605(01)00569-4. [DOI] [PubMed] [Google Scholar]
- 61.Fuka MM, Engel M, Skelin A, Redzepović S, Schloter M. 2010. Bacterial communities associated with the production of artisanal Istrian cheese. Int J Food Microbiol 142:19–24. doi: 10.1016/j.ijfoodmicro.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 62.Irlinger F, Bimet F, Delettre J, Lefèvre M, Grimont PAD. 2005. Arthrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov., novel coryneform species isolated from the surfaces of cheeses. Int J Syst Evol Microbiol 55(Pt 1):457–462. doi: 10.1099/ijs.0.63125-0. [DOI] [PubMed] [Google Scholar]
- 63.Mounier J, Gelsomino R, Goerges S, Vancanneyt M, Vandemeulebroecke K, Hoste B, Scherer S, Swings J, Fitzgerald GF, Cogan TM. 2005. Surface microflora of four smear-ripened cheeses. Appl Environ Microbiol 71:6489–6500. doi: 10.1128/AEM.71.11.6489-6500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feurer C, Vallaeys T, Corrieu G, Irlinger F. 2004. Does smearing inoculum reflect the bacterial composition of the smear at the end of the ripening of a French soft, red-smear cheese? J Dairy Sci 87:3189–3197. doi: 10.3168/jds.S0022-0302(04)73454-2. [DOI] [PubMed] [Google Scholar]
- 65.Williams AG, Banks JM. 1997. Proteolytic and other hydrolytic enzyme activities in non-starter lactic acid bacteria (NSLAB) isolated from cheddar cheese manufactured in the United Kingdom. Int Dairy J 7:763–774. doi: 10.1016/S0958-6946(97)00092-7. [DOI] [Google Scholar]
- 66.Porcellato D, Østlie HM, Brede ME, Martinovic A, Skeie SB. 2013. Dynamics of starter, adjunct non-starter lactic acid bacteria and propionic acid bacteria in low-fat and full-fat Dutch-type cheese. Int Dairy J 33:104–111. doi: 10.1016/j.idairyj.2013.01.007. [DOI] [Google Scholar]
- 67.Litopoulou-Tzanetaki E, Tzanetakis N. 2011. Microbiological characteristics of Greek traditional cheeses. Small Rumin Res 101:17–32. doi: 10.1016/j.smallrumres.2011.09.022. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.