Abstract
Reinforcing microbial thermotolerance is a strategy to enable fermentation with flexible temperature settings and thereby to save cooling costs. Here, we report on adaptive laboratory evolution (ALE) of the amino acid-producing bacterium Corynebacterium glutamicum under thermal stress. After 65 days of serial passage of the transgenic strain GLY3, in which the glycolytic pathway is optimized for alanine production under oxygen deprivation, three strains adapted to supraoptimal temperatures were isolated, and all the mutations they acquired were identified by whole-genome resequencing. Of the 21 mutations common to the three strains, one large deletion and two missense mutations were found to promote growth of the parental strain under thermal stress. Additive effects on thermotolerance were observed among these mutations, and the combination of the deletion with the missense mutation on otsA, encoding a trehalose-6-phosphate synthase, allowed the parental strain to overcome the upper limit of growth temperature. Surprisingly, the three evolved strains acquired cross-tolerance for isobutanol, which turned out to be partly attributable to the genomic deletion associated with the enhanced thermotolerance. The deletion involved loss of two transgenes, pfk and pyk, encoding the glycolytic enzymes, in addition to six native genes, and elimination of the transgenes, but not the native genes, was shown to account for the positive effects on thermal and solvent stress tolerance, implying a link between energy-producing metabolism and bacterial stress tolerance. Overall, the present study provides evidence that ALE can be a powerful tool to refine the phenotype of C. glutamicum and to investigate the molecular bases of stress tolerance.
INTRODUCTION
A high-G+C, Gram-positive bacterium, Corynebacterium glutamicum, was first discovered in Japan as a natural producer of glutamic acid (1) and has been exploited for industrial production of amino acids for over 50 years. Microbes employed in industrial-scale fermentation encounter a variety of stresses (2, 3), including thermal stress, which stems from heat generated by metabolic activities of microbial catalysts. Careful control of temperature during fermentation processes is crucial to protect microbes from thermal stress and to achieve optimal productivity. Engineering microbes with improved tolerance for thermal stress enables fermentation with more flexible temperature requirements and thereby leads to reductions in cooling costs. Different approaches have been developed to improve bacterial stress tolerance (4–6). Among them, adaptive laboratory evolution (ALE) makes use of naturally occurring mutations to generate individuals better adapted to certain environments (7–9). ALE has been successfully applied to metabolic engineering, as well as evolutionary studies on different organisms (10–15). Studies on Escherichia coli provide evidence that ALE can be a powerful tool to ameliorate bacterial stress tolerance (16–20). Independent groups have reported on ALE of the bacterium under thermal stress, and a 3 to 4°C increase in the maximal growth temperature (Tmax) relative to the respective parental strains was achieved in all cases (21–23). Apart from E. coli, application of ALE to improve bacterial thermotolerance has been limited.
Cells adapted to a certain form of stress often acquire tolerance for other, seemingly unrelated forms of stress. The phenomenon, referred to as cross protection/tolerance, is widespread among diverse species of bacteria (17, 24–26). In a recent ALE study on E. coli, all the strains that evolved independently under osmotic, acidic, oxidative, or solvent stress acquired tolerance for one or some of the stresses they never encountered during ALE (e.g., the strain adapted to solvent stress showed improved tolerance for acidic and osmotic stress), highlighting that cross protection/tolerance is a ubiquitous phenomenon in the bacterium (17). Given that microbes, as biocatalysts confront multiple stresses, such as fluctuations in pH, aeration, and temperature, understanding the interplay and compatibility among diverse forms of stress is of critical importance to facilitate microbial strain optimization for industrial applications (17). However, our knowledge of the molecular bases of cross protection/tolerance is still far from complete.
Previously, we optimized the glycolytic pathway of C. glutamicum for alanine production under oxygen deprivation, engineering the strain GLY3, in which four genes encoding glycolytic enzymes are overexpressed and two genes associated with organic acid production are inactivated (27, 28). The strain was also employed for isobutanol production, achieving higher productivity than in the preceding works on C. glutamicum (29). In the present study, we carried out an ALE experiment on strain GLY3 under thermal stress. Three strains adapted to supraoptimal growth temperatures were isolated, and all the mutations acquired by these strains were identified. Unexpectedly, the evolved strains acquired cross-tolerance for isobutanol during ALE. Allelic-exchange experiments revealed that the genomic deletion and the resulting loss of the transgenes inserted into the parental genome partly account for the increased tolerance for thermal and solvent stress, hinting at a pivotal role of energy-providing metabolism in microbial stress tolerance. The study demonstrates for the first time that ALE can facilitate refining complex phenotypic traits, such as stress tolerance, in C. glutamicum.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The C. glutamicum strains used in this study are listed in Table 1. E. coli strains JM109 (TaKaRa Bio Inc., Japan) and JM110 (30) were used for genetic manipulations. C. glutamicum strains were routinely grown at 33°C in nutrient-rich medium (A medium) supplemented with 4% glucose (31). Some experiments were carried out with minimal medium (BT medium), the recipe for which is essentially the same as that for A medium, except that yeast extract and Casamino Acids are omitted (31). E. coli strains were grown at 37°C in Luria-Bertani medium (30). Where appropriate, media were supplemented with 10 μM isopropyl-β-d-thiogalactopyranoside (IPTG), 50 μg/ml kanamycin, and/or 1.5% agar.
TABLE 1.
C. glutamicum strains used in this study
| Strain | Relevant genotype | Background | Source and/or reference |
|---|---|---|---|
| R | Wild type | NAb | JCM 18229; 33 |
| GLY3 | Δppc ΔldhA gapAc pgi pyk pfk | R | 28 |
| trm2 | Δregion 1 Δregion 2; 134 point mutationsa | GLY3 | This study |
| tam44 | Δregion 1 Δregion 2; 130 point mutationsa | GLY3 | This study |
| tam45 | Δregion 1 Δregion 2; 132 point mutationsa | GLY3 | This study |
| GLY3dfy | Δpyk (transgene) Δpfk (transgene) | GLY3 | This study |
| GLY3dr1 | Δregion 1 | GLY3 | This study |
| GLY3dr2 | Δregion 2 | GLY3 | This study |
| GLY3_35G348D | CgR_0035::1043C→T | GLY3 | This study |
| GLY3_141V157A | CgR_0141::470A→G | GLY3 | This study |
| GLY3_HemCA192T | hemC::574G→A | GLY3 | This study |
| GLY3_GlmUE295K | glmU::883C→T | GLY3 | This study |
| GLY3_GlmUE295K_rev | glmU::883T→C | GLY3_GlmUE295K | This study |
| GLY3_1064A256V | CgR_1064::767G→A | GLY3 | This study |
| GLY3_MhpAM358I | mhpA::1074G→A | GLY3 | This study |
| GLY3_NucSR161H | nucS::482G→A | GLY3 | This study |
| GLY3_MraWP283L | mraW::848G→A | GLY3 | This study |
| GLY3_PknLT367A | pknL::1099A→G | GLY3 | This study |
| GLY3_2303P166L | CgR_2303::497C→T | GLY3 | This study |
| GLY3_2333Q189R | CgR_2333::566A→G | GLY3 | This study |
| GLY3_OtsAR328H | otsA::983G→A | GLY3 | This study |
| GLY3_OtsAR328H_rev | otsA::983A→G | GLY3_OtsAR328H | This study |
| GLY3_PknGD144G | pknG::431A→G | GLY3 | This study |
| GLY3_2975Q62R | CgR_2975::185A→G CgR_2976::161T→C | GLY3 | This study |
| GLY3_SecND103G | secN::308T→C | GLY3 | This study |
| GLY3_SecNR23H | secN::68C→T | GLY3 | This study |
| GLY3_HrcAA332T | hrcA::994C→T | GLY3 | This study |
| GLY3_HrcAS29P | hrcA::85A→G | GLY3 | This study |
| GLY3_PtsSS340F | ptsS::1019G→A | GLY3 | This study |
| GLY3_PtsSL161F | ptsS::481G→A | GLY3 | This study |
| GLY3_1015-1016 | g.1123184T→C | GLY3 | This study |
| GLY3_1377-1378 | g.1509663G→A | GLY3 | This study |
| GLY3_1414-1415 | g.1553806G→A | GLY3 | This study |
| GLY3_1462-1463 | g.1616056A→G | GLY3 | This study |
| GLY3_2959-2960 | g.3284814T→C | GLY3 | This study |
| tam45_35WT | CgR_0035::1043T→C | tam45 | This study |
| tam45_141WT | CgR_0141::470G→A | tam45 | This study |
| tam45_HemCWT | hemC::574A→G | tam45 | This study |
| tam45_GlmUWT | glmU::883T→C | tam45 | This study |
| tam45_1064WT | CgR_1064::767A→G | tam45 | This study |
| tam45_MhpAWT | mhpA::1074A→G | tam45 | This study |
| tam45_NucSWT | nucS::482A→G | tam45 | This study |
| tam45_MraWWT | mraW::848A→G | tam45 | This study |
| tam45_PknLWT | pknL::1099G→A | tam45 | This study |
| tam45_2303WT | CgR_2303::497T→C | tam45 | This study |
| tam45_2333WT | CgR_2333::566G→A | tam45 | This study |
| tam45_OtsAWT | otsA::983A→G | tam45 | This study |
| tam45_OtsAWT_rev | otsA::983G→A | tam45_OtsAWT | This study |
| tam45_PknGWT | pknG::431G→A | tam45 | This study |
| tam45_2975WT | CgR_2975::185G→A CgR_2976::161C→T | tam45 | This study |
| trm2_SecNWT | secN::308C→T | trm2 | This study |
| tam45_SecNR23H | secN::68T→C | tam45 | This study |
| tam45_HrcAWT | hrcA::994T→C | tam45 | This study |
| tam44_HrcAWT | hrcA::85G→A | tam44 | This study |
| tam45_PtsSWT | ptsS::1019A→G | tam45 | This study |
| tam44_PtsSWT | ptsS::481A→G | tam44 | This study |
| tam45_1015-1016 | g.1123184C→T | tam45 | This study |
| tam45_1377-1378 | g.1509663A→G | tam45 | This study |
| tam45_1414-1415 | g.1553806A→G | tam45 | This study |
| tam45_1462-1463 | g.1616056G→A | tam45 | This study |
| tam45_2959-2960 | g.3284814C→T | tam45 | This study |
| GLY3OG | glmU::883C→T otsA::983G→A | GLY3_OtsAR328H | This study |
| GLY3dr1O | Δregion 1 otsA::983G→A | GLY3dr1 | This study |
| GLY3dr1G | Δregion 1 glmU::883C→T | GLY3dr1 | This study |
| GLY3dr1OG | Δregion 1 glmU::883C→T otsA::983G→A | GLY3dr1O | This study |
| R_GlmUE295K | glmU::883C→T | R | This study |
| R_OtsAR328H | otsA::983G→A | R | This study |
| ROG | glmU::883C→T otsA::983G→A | R_OtsAR328H | This study |
See Table S3 in the supplemental material.
NA, not applicable.
Strain GLY3 has two copies of gapA.
Evaluation of tolerance for thermal and solvent stress.
For evaluation of growth at high temperatures, seed cultures were grown in 2.5 ml A medium at 33°C overnight, starting from glycerol stocks. The overnight cultures were diluted to an optical density at 610 nm (OD610) of 0.1 in 10 ml A medium and incubated with agitation (200 rpm) in BR-43 FL constant-temperature incubators (Taitec Corp., Japan). The actual temperature in each incubator was measured using an SST-100 PT digital thermometer (Sansyo Co., Japan), whose resolution is 0.01°C, and adjusted to the desired temperatures in advance of growth tests. Bacterial growth was monitored by measuring the OD610 for 24 h, and growth rates were calculated as follows based on the OD610 recorded at 2, 4, 6, and 8 h after starting the growth tests: growth rate = [ln(ODt2) − ln(ODt1)]/(t2 − t1), where ODt1 and ODt2 are OD610 values measured at time points t1 and t2. The Tmax of a strain was arbitrarily defined as the maximal temperature at which growth of the strain reaches an OD610 of ≧1.0 within 24 h and was determined for each strain by examining its growth at various temperatures ranging from 40 to 42°C. Growth tests were performed at least three times for each strain and at each temperature.
Tolerance for isobutanol was examined by evaluating the growth of each strain in A medium supplemented with 1.5 to 2.25% (vol/vol) isobutanol essentially in the same way as described above, except that the growth temperature was always set to 33°C.
Adaptive laboratory evolution.
Strain GLY3 (28) was aerobically grown in 10 ml A medium in a 25- by 100-mm glass test tube, and growth was monitored by measuring the optical density at 610 nm. The culture was serially passed every 12 h for the first 39 days and every 24 h from day 40. The volume transferred was adjusted so that the initial OD610 was 0.1. Every week, an aliquot of the culture was mixed with an equal volume of 50% glycerol and stored at −80°C. The incubation temperature was initially set to 38°C and gradually increased up to 41.5°C as bacterial adaptation proceeded. In cases where the bacterial population failed to adapt to increasing temperatures and stopped growing, the experiment was restarted from the glycerol stocks. On day 65, aliquots of the culture were spread over plates containing A medium and incubated at 40°C. After 24 h of incubation, 48 clones were isolated and evaluated on their growth at high temperatures. Three isolates, trm2, tam44, and tam45, showing vigorous growth at temperatures over 41°C were selected for further analyses.
DNA microarray analysis.
The strains GLY3, trm2, tam44, and tam45 were aerobically grown in 10 ml A medium at 33°C and shifted to 41°C when the OD610 reached approximately 7.0. The cultures were further incubated for 1 h. Samples for RNA extraction were collected just before transferring the cultures from 33 to 41°C and after 1 h of growth at 41°C. Total RNA was extracted from each sample with NucleoSpin RNA (TaKaRa Bio Inc., Japan), according to the manufacturer's recommendations. Preparation of fluorescently labeled cDNA, hybridization to microarrays, and data manipulations were carried out as described previously (32).
Identification of genomic deletions.
PCR amplifications of regions 1 and 2 were carried out with the primer pairs dR1F and dr1R, and dR2F and dr2R (Table 2), respectively, using genomic DNA of the wild type (WT), GLY3, trm2, tam44, and tam45 as the template. Reaction mixtures contained 0.5 U PrimeStar GXL DNA polymerase (TaKaRa Bio Inc., Japan), 1× buffer, 0.8 mM deoxynucleoside triphosphate (dNTP), 1 μM each primer, and 150 ng of genomic DNA in a final volume of 20 μl, and the reaction was run for 35 cycles: 98°C for 10 s, 55°C for 5 s, and 68°C for 10 min. Two microliters of each reaction mixture was separated on a 0.8% agarose gel, and PCR products were visualized with ethidium bromide staining. After desalting of the reaction mixtures with NucleoSpin Gel and PCR Cleanup (TaKaRa Bio Inc., Japan), sequencing of the PCR products was performed using a BigDye terminator v1.1 cycle-sequencing kit (Applied Biosystems) on a 3130xl genetic analyzer (Applied Biosystems). The DNA sequences of the PCR products were aligned using the program ATGC version 6 (Genetyx Corp., Japan), and the deletion endpoints of regions 1 and 2 were manually determined.
TABLE 2.
Primers used to validate mutations in evolved strains
| Name | Sequence (5′→3′) | Target | Size (bp) |
|---|---|---|---|
| dR1F | ATGGAACGTTTCCGAAGAGG | Δregion 1 | 5,296a |
| dR1R | TCAGACAGAGTCTTGGTAGC | ||
| dR2F | GGGCTCACTCGAGGGATCTTGACTG | Δregion 2 | 8,066a |
| dR2R | GGCTGTGTACACGTCAGTGAGTCTC | ||
| 0035_F | TCGGCTTGGATGGAAAACCT | CgR_0035::1043C→T | 385 |
| 0035_R | TCCAACCAAATAGACCAATGCTCG | ||
| 0141_F | TCGGGTGCGTAATCGGAAAC | CgR_0141::470A→G | 235 |
| 0141_R | GGGATGGTCGCCTAGAGTG | ||
| 0489_F | TCTGGCTGAGCTTCCAGAAG | hemC::574G→A | 308 |
| 0489_R | GTTGAGCGCGCGGACGGT | ||
| 1044_F | ACCGATGCCCGTGAACTC | glmU::883C→T | 394 |
| 1044_R | TTTCTACGAAGCCACCGAGC | ||
| 1064_F | TGGTTCTCATGCCCGGAAAC | CgR_1064::767G→A | 351 |
| 1064_R | GGCCCGAGAAGTCAAGGAAA | ||
| 1240_F | GTAGGGCTCTCATCGCAGG | mhpA::1074G→A | 262 |
| 1240_R | ATGCGCTCATCGCGCAAA | ||
| 1294_F | AACTTCGACCTCGGTGAGGA | nucS::482G→A | 388 |
| 1294_R | TGGACTCAATGCCACGAAGC | ||
| 2048_F | TTGCGCGTTGAGGTGAACAA | mraW::848G→A | 321 |
| 2048_R | GTCTCCTGAGTTGTTGGCGA | ||
| 2057_F | GTGCCGGTTAATTCCGCAG | pknL::1099A→G | 496 |
| 2057_R | GTTCAGCCACTGCCACGAAA | ||
| 2303_F | ACACGGAGGATGCTGTTTGG | CgR_2303::497C→T | 802 |
| 2303_R | ACGGGGCGTTTTCTTCTAGC | ||
| 2333_F | TACAACGGTTACCCTGCGGA | CgR_2333::566A→G | 365 |
| 2333_R | GTGTCCGTACCTAGCACAGG | ||
| 2531_F | GGGCGTTCCCGATCTCGATT | otsA::983G→A | 472 |
| 2531_R | AAAGCACCAGTGCCGTCG | ||
| 2654_F | CGTGGTGCTCAAAGGCATGA | pknG::431A→G | 107 |
| 2654_R | CCTTCACGATACCGGGGTG | ||
| 2975_F | CGTCCTTCAGGTCAGAACCA | CgR_2975::185A→G CgR_2976::161T→C | 156 |
| 2975_R | TTTCTCAGAAGCTTGGCCCT | ||
| 1704_2/44_F | TGAAATCCAAAAGCTGCAGGAC | secN::308T→C | 274 |
| 1704_2/44_R | ATTCTCAGGATGTCCGTCAGTG | ||
| 1704_45_F | TCTAGCCATTTTTATCGTTCCCACC | secN::68C→T | 110 |
| 1704_45_R | TGACAATGCGGTCGCCTG | ||
| 2163_2/44_F | ATCGCCGACATTTTGCTTAGTG | hrcA::85A→G | 291 |
| 2163_2/44_R | ATCAACAAAAAGGCGGTAACCC | ||
| 2163_45_F | CACTGGGCGGATTGGGG | hrcA::994C→T | 110 |
| 2163_45_R | CTATTCGCCAGCGAGCACA | ||
| 2547_2/44_F | GACATTTTCGTCCCGCTGATTC | ptsS::481G→A | 276 |
| 2547_2/44_R | GTTAACCAGGGTTGGGAACAC | ||
| 2547_45_F | AGATCAGCGGTGTTGCTGAG | ptsS::1019G→A | 797 |
| 2547_45_R | GAGTGCAATCAAAGCGCCAC | ||
| 1015_1016_F | ATCACGACCACCACTGTACC | Intergenic mutation between CgR_1015 and CgR_1016 | 404 |
| 1015_1016_R | CTCGGCATATCCACTGCCAC | ||
| 1377_1378_F | CTACATGAGCCCGTACATTT | Intergenic mutation between CgR_1377 and CgR_1378 | 358 |
| 1377_1378_R | GGTACTCCGCATCATGTATT | ||
| 1414_1415_F | AGTACATACAGCCGGAGCGA | Intergenic mutation between CgR_1414 and CgR_1415 | 541 |
| 1414_1415_R | TACTGCAGGTCCGATGAGGT | ||
| 1462_1463_F | TTCATCGATAGGGTGGGGCT | Intergenic mutation between CgR_1462 and CgR_1463 | 402 |
| 1462_1463_R | CGAGGGAAACTGCGATGACT | ||
| 2959_2960_F | ACACCACCTCTCTTTTCAACATGA | Intergenic mutation between CgR_2959 and CgR_2960 | 293 |
| 2959_2960_R | TGAGGGCGACTCCAAGTCATA |
PCR product size is shown for the WT.
Whole-genome resequencing.
Genomic-DNA samples were prepared from GLY3, trm2, tam44, and tam45 with a Nucleobond AXG column (TaKaRa Bio Inc., Japan), following the manufacturer's instructions. Genomic-DNA library construction, whole-genome resequencing, and data analyses were carried out by the Dragon Genomic Center (TaKaRa Bio Inc., Japan). The libraries were prepared with NEBNext DNA sample prep reagent set 1 (New England BioLabs Inc.), and resequencing was performed using the Illumina Genome Analyzer IIx platform (Illumina, San Diego, CA). Sequence data were analyzed in two different ways to identify mutations. In one method, the paired-end reads were mapped onto the reference genome sequence of C. glutamicum R (33) using the BWA software (34), followed by detection of point mutations with the SAM tools (35). In the other method, the paired-end reads were first assembled into contigs with the Edena program (36). The contigs were mapped onto the reference C. glutamicum R genome sequence, and mutations, including point mutations and insertions/deletions, were identified using the MUMmer software (37). Among the mutations common to trm2, tam44, and tam45, intergenic point mutations and intragenic point mutations considered to cause amino acid substitutions were confirmed by PCR amplification, followed by Sanger sequencing. All intragenic insertions/deletions were validated in the same way. Information on the primers used for this purpose is provided in Table 2.
Estimation of mutation rates.
A fluctuation assay was carried out to estimate the mutation rates of the strains R, GLY3, trm2, tam44, and tam45 (38, 39). Cells picked up from a single colony were suspended in 10 ml A medium, and 20 replicates of 200-μl aliquots were distributed into 96-well cell culture plates. The culture plates were incubated at 33°C with agitation (550 rpm) until saturation (12 to 14 h). Five microliters of each replicate was dropped in duplicate onto plates containing A medium with 2 μg/ml rifampin, and the rifampin-resistant colonies that appeared after 24 h of incubation at 33°C were counted. In parallel, each replicate was serially diluted and plated on nonselective A medium to estimate the total number of cells (Nt).
The median number of rifampin-resistant mutants (ϕ) was calculated for each strain, and the number of mutation events (m) was estimated using the Lea-Coulson method (40) or the Drake formula of the median (41). The former method [ϕ/m − ln(m) = 1.24] was applied to the strains R and GLY3, whose values of m were low (15 ≧ m ≧ 1.5), whereas the latter formula [ϕ/m − ln(m) = 0] was used to determine the m values of the three evolved strains, which had higher m values (m ≧ 30). With the estimated m and the median number of Nt, the mutation rate (μ) was calculated for each strain as follows: μ = m × [ln(2)/median Nt] (42).
Plasmid construction and bacterial transformation.
The plasmid constructs used in the present study are listed in Table 3, and information on the primers used to generate the plasmid constructs is found in Table 4. All PCRs were carried out with PrimeStar HS DNA polymerase (TaKaRa Bio Inc., Japan). For overexpression of pfk and pyk, pCRB12i, a vector for an IPTG-inducible expression system, was generated by introducing lacIq from pDW363 into the E. coli-C. glutamicum shuttle vector pCRB12 (43). The coding region of pyk was amplified from genomic DNA of C. glutamicum R with the primers pyk_F_NdeI and pyk_R_NdeI. The amplified fragment was cloned into the NdeI site of pCRB214, a vector carrying the tac promoter and the rrnB terminator (44). The resulting construct, designated pCRE555, was digested with BamHI, and the fragment containing the pyk gene with the tac promoter and rrnB terminator was cloned into the BamHI site of pCRB12i, yielding pCRE556. pCRE557 was generated by cloning pfk into pCRB214 in the same way as described for construction of pCRE555. The coding region of pfk with the tac promoter and rrnB terminator was amplified from pCRE557 with the primers Ptac_KpnI and rrnB_KpnI, and the amplified fragment was cloned into pCRE556 linearized with KpnI, generating pCRE558.
TABLE 3.
Plasmid constructs used in the present study
| Name | Description | Restriction site(s)a | Source or reference |
|---|---|---|---|
| pCRB214 | Kmr; cloning vector with Ptac and rrnBT1T2 derived from pCASE1 | NA | 44 |
| pCASE1 | Plasmid identified in Corynebacterium casei JCM12072 | NA | 69 |
| pCRB12 | Kmr; pCG1-based C. glutamicum-E. coli shuttle vector | NA | 43 |
| pCG1 | Plasmid identified in C. glutamicum ATCC 31808 | NA | 70 |
| pDW363 | Apr; source of lacIq | NA | NBRP (NIG, Japan) |
| pCRB12i | Kmr; IPTG-inducible vector derived from pCRB12 | NA | This study |
| pHSG298 | Kmr; α-lac multicloning site; E. coli cloning vector | NA | TaKaRa Bio, Inc. |
| pCRA725 | Kmr; pHSG298 with Ptac-sacR-sacB | NA | 45 |
| pCRE500 | GLY3 ntb 39536–41166 in pCRA725; plasmid for allelic replacement of CgR_0035 | XbaI/SalI | This study |
| pCRE501 | tam45 nt 39536–41166 in pCRA725; plasmid for allelic replacement of CgR_0035 | XbaI/SalI | This study |
| pCRE502 | GLY3 nt 157063–158683 in pCRA725; plasmid for allelic replacement of CgR_0141 | SalI | This study |
| pCRE503 | tam45 nt 157063–158683 in pCRA725; plasmid for allelic replacement of CgR_0141 | SalI | This study |
| pCRE504 | GLY3 nt 545101–546040 in pCRA725; plasmid for allelic replacement of hemC | XbaI/SalI | This study |
| pCRE505 | tam45 nt 545101–546040 in pCRA725; plasmid for allelic replacement of hemC | XbaI/SalI | This study |
| pCRE506 | GLY3 nt 1151531–1153602 in pCRA725; plasmid for allelic replacement of glmU | XbaI/SalI | This study |
| pCRE507 | tam45 nt 1151531–1153602 in pCRA725; plasmid for allelic replacement of glmU | XbaI/SalI | This study |
| pCRE508 | GLY3 nt 1179149–1180226 in pCRA725; plasmid for allelic replacement of CgR_1064 | SalI | This study |
| pCRE509 | tam45 nt 1179149–1180226 in pCRA725; plasmid for allelic replacement of CgR_1064 | SalI | This study |
| pCRE510 | GLY3 nt 1356603–1357635 in pCRA725; plasmid for allelic replacement of mhpA | SalI | This study |
| pCRE511 | tam45 nt 1356603–1357635 in pCRA725; plasmid for allelic replacement of mhpA | SalI | This study |
| pCRE512 | GLY3 nt 1423170–1424283 in pCRA725; plasmid for allelic replacement of nucS | SalI | This study |
| pCRE513 | tam45 nt 1423170–1424283 in pCRA725; plasmid for allelic replacement of nucS | SalI | This study |
| pCRE514 | GLY3 nt 1899141–1900182 in pCRA725; plasmid for allelic replacement of secN | SalI | This study |
| pCRE515 | tam45 nt 1899141–1900182 in pCRA725; plasmid for allelic replacement of secN | SalI | This study |
| pCRE516 | trm2 nt 1899141–1900182 in pCRA725; plasmid for allelic replacement of secN | SalI | This study |
| pCRE517 | GLY3 nt 2257572–2259106 in pCRA725; plasmid for allelic replacement of mraW | XbaI/SalI | This study |
| pCRE518 | tam45 nt 2257572–2259106 in pCRA725; plasmid for allelic replacement of mraW | XbaI/SalI | This study |
| pCRE519 | GLY3 nt 2267254–2268257 in pCRA725; plasmid for allelic replacement of pknL | SalI | This study |
| pCRE520 | tam45 nt 2267254–2268257 in pCRA725; plasmid for allelic replacement of pknL | SalI | This study |
| pCRE521 | GLY3 nt 2385305–2386847 in pCRA725; plasmid for allelic replacement of hrcA | SalI | This study |
| pCRE522 | trm2 nt 2385305–2386847 in pCRA725; plasmid for allelic replacement of hrcA | SalI | This study |
| pCRE523 | tam45 nt 2385305–2386847 in pCRA725; plasmid for allelic replacement of hrcA | SalI | This study |
| pCRE524 | GLY3 nt 2537845–2538846 in pCRA725; plasmid for allelic replacement of CgR_2303 | SalI | This study |
| pCRE525 | tam45 nt 2537845–2538846 in pCRA725; plasmid for allelic replacement of CgR_2303 | SalI | This study |
| pCRE526 | GLY3 nt 2566914–2567767 in pCRA725; plasmid for allelic replacement of CgR_2333 | XbaI/SalI | This study |
| pCRE527 | tam45 nt 2566914–2567767 in pCRA725; plasmid for allelic replacement of CgR_2333 | XbaI/SalI | This study |
| pCRE528 | GLY3 nt 2797092–2798776 in pCRA725; plasmid for allelic replacement of otsA | SacI/SalI | This study |
| pCRE529 | tam45 nt 2797092–2798776 in pCRA725; plasmid for allelic replacement of otsA | SacI/SalI | This study |
| pCRE530 | GLY3 nt 2812261–2814354 in pCRA725; plasmid for allelic replacement of ptsS | XbaI/SalI | This study |
| pCRE531 | trm2 nt 2812261–2814354 in pCRA725; plasmid for allelic replacement of ptsS | XbaI/SalI | This study |
| pCRE532 | tam45 nt 2812261–2814354 in pCRA725; plasmid for allelic replacement of ptsS | XbaI/SalI | This study |
| pCRE533 | GLY3 nt 2936034–2937632 in pCRA725; plasmid for allelic replacement of pknG | XbaI | This study |
| pCRE534 | tam45 nt 2936034–2937632 in pCRA725; plasmid for allelic replacement of pknG | XbaI | This study |
| pCRE535 | GLY3 nt 3297021–3298018 in pCRA725; plasmid for allelic replacement of CgR_2975 | XbaI/SalI | This study |
| pCRE536 | tam45 nt 3297021–3298018 in pCRA725; plasmid for allelic replacement of CgR_2975 | XbaI/SalI | This study |
| pCRE537 | GLY3 nt 1122184–1123706 in pCRA725; plasmid for allelic replacement of CgR_1015-1016 | XbaI/SalI | This study |
| pCRE538 | tam45 nt 1122184–1123706 in pCRA725; plasmid for allelic replacement of CgR_1015-1016 | XbaI/SalI | This study |
| pCRE539 | GLY3 nt 1509110–1510281 in pCRA725; plasmid for allelic replacement of CgR_1377-1378 | XbaI/SalI | This study |
| pCRE540 | tam45 nt 1509110–1510281 in pCRA725; plasmid for allelic replacement of CgR_1377-1378 | XbaI/SalI | This study |
| pCRE541 | GLY3 nt 1552995–1554627 in pCRA725; plasmid for allelic replacement of CgR_1414-1415 | SacI/SalI | This study |
| pCRE542 | tam45 nt 1552995–1554627 in pCRA725; plasmid for allelic replacement of CgR_1414-1415 | SacI/SalI | This study |
| pCRE543 | GLY3 nt 1614878–1617235 in pCRA725; plasmid for allelic replacement of CgR_1462-1463 | SacI/XbaI | This study |
| pCRE544 | tam45 nt 1614878–1617235 in pCRA725; plasmid for allelic replacement of CgR_1462-1463 | SacI/XbaI | This study |
| pCRE545 | GLY3 nt 3283826–3285778 in pCRA725; plasmid for allelic replacement of CgR_2959-2960 | XbaI/SalI | This study |
| pCRE546 | tam45 nt 3283826–3285778 in pCRA725; plasmid for allelic replacement of CgR_2959-2960 | XbaI/SalI | This study |
| pCRE552 | R nt 2556378–2572116 in pCRA725; plasmid for deletion of transgenes pfk and pyk | XbaI | This study |
| pCRE553 | R nt 791912–792959 fused to R nt 799755–800795 in pCRA725; plasmid for deletion of region 2 | SalI | This study |
| pCRE554 | R nt 2554655–2555773 fused to R nt 2560444–2561451 in pCRA725; plasmid for deletion of region 1 | SalI | This study |
| pCRE555 | Coding region of pyk cloned into pCRB214 | NdeI | This study |
| pCRE556 | Ptac-pyk-rrnBT1T2 cloned into pCRB12i | BamHI | This study |
| pCRE557 | Coding region of pfk cloned into pCRB214 | NdeI | This study |
| pCRE558 | Ptac-pfk-rrnBT1T2 cloned into pCRE556 | KpnI | This study |
Restriction site(s) used for cloning. NA, not applicable.
nt, nucleotides.
TABLE 4.
Primers used to generate plasmid constructs
| Name | Sequence (5′→3′) | Size (bp) | Plasmid constructa |
|---|---|---|---|
| pyk_F_NdeI | GGAATTCCATATGATGGGCGTGGATAGACGAAC | 1,434 | pCRE555 |
| pyk_R_NdeI | GGAATTCCATATGTTAGAGCTTTGCAATCCTTGTG | ||
| pfk_ F_NdeI | GGAATTCCATATGATGGAAGACATGCGAATTGCTAC | 1,041 | pCRE557 |
| pfk_ R_NdeI | GGAATTCCATATGCTATCCAAACATTGCCTGGG | ||
| Ptac_KpnI | CGGGGTACCGGCTGTGCAGGTCGTAAATC | 1,704 | pCRE558 |
| rrnB_KpnI | CGGGGTACCAAAAAGGCCATCCGTCAGGA | ||
| CglR2326-Fw5(SalI) | CTCTGTCGACTGATTTCATCGCGCTGGTTG | 1,119 | pCRE554 |
| CglR2326-Rv6 | GCATGGGCGACCTATGG | ||
| CglR2326-Fw6 | ATCGCTACTATTGTTTCCATAGGTCGCCCATGCGACCGCCTCATCCTCGT | 1,008 | |
| CglR2326-Rv5(SalI) | CTCTGTCGACGAATGAAACTTCTCACCCGTG | ||
| CglR0733-Fw1(SphI) | CTCTGCATGCACTGCGTAGGTCGATTTCC | 1,048 | pCRE553 |
| CglR0733-Rv2 | CTCCCATAGTGGTAGACG | ||
| CglR0733-Fw2 | CCAGGGGAGCCGTCTACCACTATGGGAGACTCCCTGAACTGGTCTTTTAG | 1,041 | |
| CglR0733-Rv1(SalI) | CTCTGTCGACCTTACTTCTTGTACCCGAGC | ||
| indel10- Fw4(XbaI) | CTCTTCTAGAACACACCTCATCACGAGTAG | 15,739 | pCRE552 |
| indel10-Rv2(XbaI) | CTCTTCTAGACCTCGTCAGTTGCTGCAGTT | ||
| 0035-XbaI-F | GCTCTAGAGGCACTGAGCTTTGCAGCTA | 1,631 | pCRE500 pCRE501 |
| 0035-SalI-R | ACGCGTCGACCCCCGCAACGCATCATTAAC | ||
| 0141-SalI-F | TATGTCGACATGCTCGAAGGCATCATCTACC | 1,600 | pCRE502 pCRE503 |
| 0141-SalI-R | TATGTCGACATCGAACCATCATCGTGTTGGA | ||
| 0489-XbaI-F | GCTCTAGAACTGCACATTGTGACCACCC | 940 | pCRE504 pCRE505 |
| 0489-SalI-R | ACGCGTCGACGATTTTTGCACCACAGGCCG | ||
| 1044-XbaI-F | GCTCTAGACCAAAAACGTCATGGCAGGC | 2,072 | pCRE506 pCRE507 |
| 1044-SalI-R | ACGCGTCGACGAAATCGCGTGCCGTCATTG | ||
| 1064-SalI-F | TATGTCGACATCTCGCTCATAGATTCCACGG | 1,100 | pCRE508 pCRE509 |
| 1064-SalI-R | TATGTCGACACTTGCCAGAAATCCCGCTGTA | ||
| 1240-SalI-F | TATGTCGACATACCTGCTGCTACTTGTTGTG | 1,000 | pCRE510 pCRE511 |
| 1240-SalI-R | TATGTCGACAAGATCTCAAGGAACTCGCTTC | ||
| 1294-SalI-F | TATGTCGACAAGGCATGATTCAGCTCATCGC | 1,100 | pCRE512 pCRE513 |
| 1294-SalI-R | TATGTCGACAAGCCACTGAACTGGATGACAC | ||
| 2048-XbaI-F | GCTCTAGAACTCGATGACAAAGGCAGGC | 2,559 | pCRE517 pCRE518 |
| 2048-SalI-R | ACGCGTCGACTCTCTAGCGGCTCTTCTGCT | ||
| 2057-SalI-F | TATGTCGACAGGACAATCAGATTGTGGGCAC | 1,000 | pCRE519 pCRE520 |
| 2057-SalI-R | TATGTCGACACGAACGGTGCTTAAGGATCGA | ||
| 2303-SalI-F | TATGTCGACACAATGACTCTCCGCATTGGTC | 1,000 | pCRE524 pCRE525 |
| 2303-SalI-R | TATGTCGACATTGTCACGTTGCTTTCAGCGC | ||
| 2333-XbaI-F | GCTCTAGAACTGGGGAAAGTGGGGAAATG | 854 | pCRE526 pCRE527 |
| 2333-SalI-R | ACGCGTCGACAGCGGCGCTGTATAAGAAGC | ||
| 2531-SacI-F | TGCGAGCTCGTTGCTAACCGTCTGCCAGT | 1,721 | pCRE528 pCRE529 |
| 2531-SalI-R | ACGCGTCGACAGACAAGGTCACATAATCGGCA | ||
| 2654-XbaI-F | GCTCTAGAACTGCAGTACATGCCTGAGGAA | 1,560 | pCRE533 pCRE534 |
| 2654-XbaI-R | GCTCTAGAATTGGCGGTATTGCTCGTCTTC | ||
| 2975-XbaI-F | GCTCTAGATGTTTATCCCGCCAGGAAGC | 997 | pCRE535 pCRE536 |
| 2975-SalI-R | ACGCGTCGACAGTGAACACCTCACCGTGGA | ||
| 1704-SalI-F | TATGTCGACATTGCTAACCGACTGCTTCGTC | 1,000 | pCRE514 pCRE515 pCRE516 |
| 1704-SalI-R | TATCGATCGATCAACCAGACAACCACGCTTG | ||
| 2163-SalI-F | TATGTCGACAACAGAGATGCTGACC | 1,500 | pCRE521 pCRE522 pCRE523 |
| 2163-SalI-R | TATGTCGACAACAATGCGGCGCTTA | ||
| 2547-XbaI-F | GCTCTAGACGCAAAATCCCTTGATCGGACA | 2,094 | pCRE530 pCRE531 pCRE532 |
| 2547-SalI-R | ACGCGTCGACTTATGGTGTTGCTGGCACCG | ||
| 1015-XbaI-F | GCTCTAGAACCGGCTATTTTACCTCTGC | 1,523 | pCRE537 pCRE538 |
| 1016-SalI-R | ACGCGTCGACTGTGTAGCGGAATG | ||
| 1377-XbaI-F | GCTCTAGATTAATTGTTCTCGTCGTTGC | 1,172 | pCRE539 pCRE540 |
| 1378-SalI-R | ACGCGTCGACATGTGATGGGCAACTAATCT | ||
| 1414-SacI-F | AGCGAGCTCTGGAAGCCACTAGCACTCAT | 1,633 | pCRE541 pCRE542 |
| 1415-SalI-R | ACGCGTCGACCAAGATATTCGACGAGGGCG | ||
| 1462-SacI-F | AGCGAGCTCCTTGACCGATGATCTGCACC | 2,358 | pCRE543 pCRE544 |
| 1463-XbaI-R | GCTCTAGAAGCCAGGTTGAAGTCGTACA | ||
| 2959-XbaI-F | GCTCTAGAGTTCGGCCCAATCCTGTTCT | 1,953 | pCRE545 pCRE546 |
| 2960-SalI-R | ACGCGTCGACTGGGATACGAGTACGCAGGT |
Described in Table 3.
For allelic-exchange experiments, the genomic region surrounding each mutation was amplified from genomic DNA of one of the three evolved strains or GLY3. Restriction sites were anchored to both ends of the PCR fragments during amplification, and the amplicons were cloned into appropriate restriction sites of a suicide vector, pCRA725, carrying the sacB gene (45). To prepare the constructs for deletion of region 1 (Δregion 1), region 2 (Δregion 2), and the transgenes pfk and pyk (Δpfk Δpyk), the 5′ and 3′ flanks of the targeted genomic regions were amplified, using genomic DNA of C. glutamicum strain R as a template. The amplified fragments were tethered to each other by PCR fusion and cloned into pCRA725. For each plasmid construct, absence of any unintended mutation was verified by Sanger sequencing in advance of transformation. Transformation of C. glutamicum by electroporation was performed as previously described (46). Chromosomal allelic replacements and gene deletions were carried out by a markerless system (45).
Statistical analyses.
Statistical analyses were carried out by one-way analysis of variance (ANOVA), followed by the Tukey honestly significant difference (HSD) test for pairwise comparisons.
Accession numbers.
The microarray data were deposited in Gene Expression Omnibus with accession number GSE64654. The complete sequence of pCRB12i is available in GenBank with accession number KP340794.
RESULTS
Adaptive laboratory evolution of C. glutamicum under thermal stress.
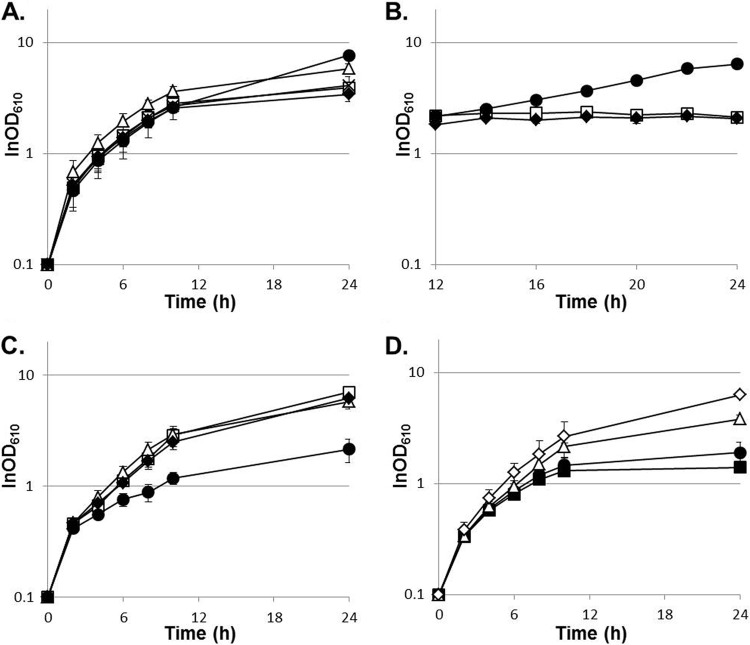
After 65 days of serial passage of C. glutamicum strain GLY3 under thermal stress, three single-colony isolates, trm2, tam44, and tam45, were derived from a single evolved population. No notable difference in cell shape or size was observed for the evolved strains compared to the parental strain (data not shown). Whether grown in nutrient-rich medium or in minimal medium, the evolved strains showed increased growth at supraoptimal temperatures compared to the parental strain. Indeed, these strains were able to grow at temperatures above the Tmax of GLY3 (Fig. 1 and Table 5). At 33°C, which is the optimal growth temperature for the WT strain, the maximal growth rates of the evolved strains in nutrient-rich medium were lower than that of GLY3 (P < 0.01) (Table 5), implying an evolutionary tradeoff as a result of adaptation to higher growth temperatures. These observations demonstrate that trm2, tam44, and tam45 have successfully adapted to supraoptimal growth temperatures.
FIG 1.
Evolved strains adapted to higher growth temperatures. Shown are growth curves of GLY3 (closed squares), trm2 (closed circles), tam44 (open triangles), and tam45 (open diamonds) with standard deviations (n = 3). (A) Strains grown in nutrient-rich medium at 33°C (solid lines) or 41°C (dotted lines). (B) Strains grown in minimal medium at 33°C (solid lines) or 38°C (dotted lines).
TABLE 5.
Growth properties of the parental and evolved strains
| Strain | Nutrient-rich medium |
Minimal medium |
||||
|---|---|---|---|---|---|---|
| Tmaxa (°C) | Maximal growth rate (h−1)b |
Tmax (°C) | Maximal growth rate (h−1)b |
|||
| 33°C | 41°C | 33°C | 38°C | |||
| GLY3 | 40.2 | 0.88 ± 0.041 | 0.14 ± 0.063 | 37 | 0.57 ± 0.050 | 0.26 ± 0.021 |
| trm2 | 41.5 | 0.72 ± 0.014c | 0.33 ± 0.041c | 40.5 | 0.55 ± 0.0099 | 0.38 ± 0.022c |
| tam44 | 41.2 | 0.66 ± 0.020c | 0.29 ± 0.025c | 40 | 0.55 ± 0.040 | 0.38 ± 0.035c |
| tam45 | 41.5 | 0.70 ± 0.056c | 0.33 ± 0.0091c | 40.5 | 0.55 ± 0.024 | 0.39 ± 0.013c |
At temperatures higher than the Tmax, growth of the strain never reaches an OD610 of 1.0 within 24 h.
Means ± standard deviations (n = 3).
A statistically significant difference was observed compared to GLY3 (P < 0.01).
Identification of a genomic deletion partly accounts for the increased thermotolerance of the evolved strains.
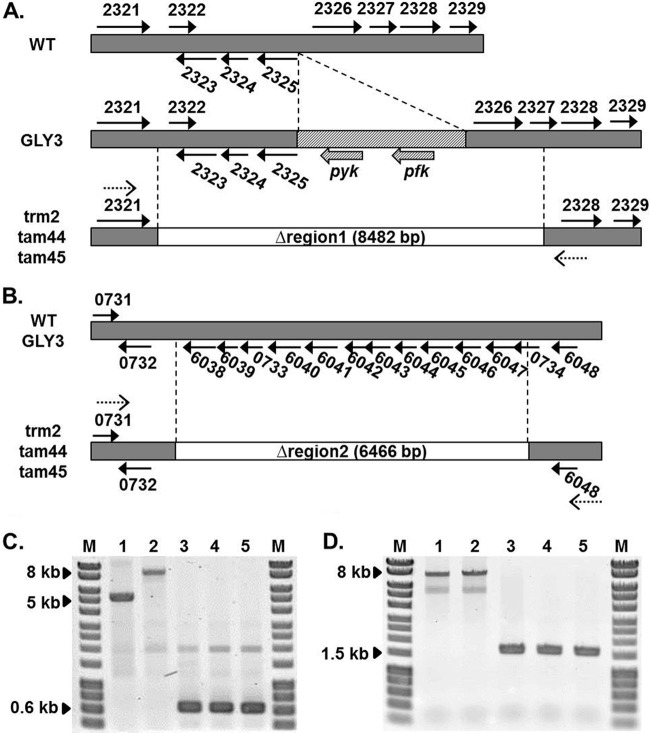
To gain insight into the molecular bases of the enhanced thermotolerance of the evolved strains, we compared the transcriptomes of the evolved strains with that of the parental strain using a DNA microarray. Expression of two clusters of genes located on two independent genomic regions, one spanning from CgR_2322 to CgR_2329 and the other from CgR_6038 to CgR_6048, was significantly reduced in the evolved strains compared to GLY3 (see Table S1 in the supplemental material). PCR amplification of CgR_2322 to CgR_2327 from the genomic DNA samples of the evolved strains generated much shorter products (626 bp) than those of the WT and GLY3 samples (Fig. 2). As GLY3 carries the two transgenes pfk and pyk, which encode a phosphofructokinase (PFK) and a pyruvate kinase (PYK), respectively, between CgR_2325 and CgR_2326 (28), larger PCR products were obtained for GLY3 (9,108 bp) than for the WT (5,296 bp). Sequencing of the PCR products from the evolved strains revealed that CgR_2322 to CgR_2326 and a portion of CgR_2327, including the transgenes pfk and pyk (8,482 bp, designated region 1) (see Table S2 in the supplemental material), are lost in these strains. Similarly, PCR amplification of CgR_6038 to CgR_6048 from the evolved strains yielded smaller products (1,600 bp) than the WT and strain GLY3 (8,066 bp) (Fig. 2), and sequencing of the PCR products indicated loss of CgR_6038 to CgR_6047, including CgR_0733 and a part of CgR_0734 (6,466 bp; designated region 2) (see Table S2 in the supplemental material), in trm2, tam44, and tam45. Sequencing of the GLY3 PCR products confirmed that regions 1 and 2 are intact in the parental strain. The genomic deletion events apparently took place in a common ancestor of the three evolved strains, as these regions were deleted from their genomes in exactly the same way.
FIG 2.
Deletion of regions 1 and 2 in the evolved strains. (A and B) Schematic diagrams of region 1 (A) and region 2 (B) are shown for the WT, GLY3, and the three evolved strains. The genomic regions lost in the evolved strains are shown as white rectangles. The arrows represent the genes in these regions, and the number next to each arrow represents the four-digit locus tag (CgR_nnnn). The transgenes introduced into GLY3 are shown as hatched rectangles and arrows. The dotted arrows indicate the positions of the primers used for PCR confirmation of the deletions shown in panels C and D. (C and D) PCR confirmation of Δregion 1 (C) and Δregion 2 (D). Reactions were carried out with the primer pairs dR1F and dR1R and dR2F and dR2R for regions 1 and 2, respectively, using genomic DNA of the WT (lanes 1), GLY3 (lanes 2), trm2 (lanes 3), tam44 (lanes 4), and tam45 (lanes 5) as templates. The PCR products were separated on a 0.8% agarose gel, and the images were taken with Typhoon Trio (GE Healthcare Life Sciences) after staining with ethidium bromide. Lanes M, HyperLadder I (Bioline).
To examine if these deletions account for the increased thermotolerance of the evolved strains, we deleted the same regions from the genome of the parental strain. Growth at high temperatures was unaffected in the Δregion 2 strain (data not shown). Deletion of region 1, in contrast, promoted growth of GLY3 under thermal stress. Although no significant improvement in the growth rate was observed, the Δregion 1 strain GLY3dr1 showed a prolonged growth phase compared to GLY3 (Fig. 3), and consequently, GLY3dr1 reached a higher cell density after 24 h of incubation at 40.2°C (P < 0.01; OD610 = 4.93 ± 0.30 for GLY3dr1 and 1.96 ± 0.16 for GLY3). No clear difference in growth was observed between GLY3dr1 and GLY3 at 33°C. As noted, GLY3 carries two transgenes in region 1. We tested how loss of these transgenes affects growth under thermal stress by deleting pfk and pyk from region 1 of the GLY3 genome. In terms of growth at high temperatures, strain GLY3dfy, in which only pfk and pyk were deleted, was indistinguishable from strain GLY3dr1 (Fig. 3). Plasmid-based overexpression of pfk and pyk reduced growth of GLY3dfy under thermal stress to the level of GLY3 (see Fig. S1 in the supplemental material). These results demonstrate that the enhanced thermotolerance of the evolved strains is partly attributable to deletion of region 1 and the resulting loss of the transgenes pfk and pyk.
FIG 3.
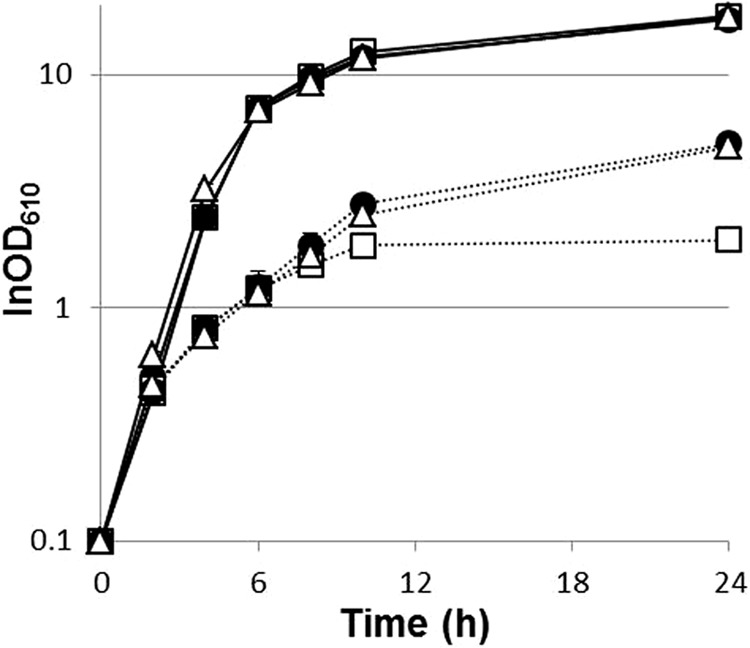
Transgenes pfk and pyk negatively affect growth of C. glutamicum under thermal stress. Shown are growth curves of different strains with standard deviations (n = 3). GLY3 (open squares), GLY3dr1 (open triangles; Δregion 1), and GLY3dfy (closed circles; Δpfk Δpyk) were grown in nutrient-rich medium at 33°C (solid lines) or 40.2°C (dotted lines).
Whole-genome resequencing identified all the mutations acquired by the evolved strains.
Under thermal stress, GLY3dr1 was not able to grow as vigorously as the evolved strains (data not shown), indicating that a mutation(s) other than the deletion of region 1 contributes to the enhanced thermotolerance of the evolved strains. In search of these mutations, we carried out whole-genome resequencing of the three evolved strains and the parental strain. The results of resequencing confirmed deletions of regions 1 and 2. In addition, the presence of 6 intragenic insertions and 14 intragenic deletions in the evolved strains was suggested (data not shown); however, reexamination by Sanger sequencing failed to detect these 20 mutations in the three evolved strains, indicating that the insertions/deletions were artifacts. Apart from the insertions/deletions, resequencing identified 295 point mutations, most of which (220 out of 295) are unique to one of the three evolved strains (see Table S3 in the supplemental material). Of the remaining 75 mutations, 26 were found in all three evolved strains, whereas 49 were detected only in trm2 and tam44. Except for the 26 mutations common to the three strains, tam45 does not share any mutation with trm2 or tam44, indicating that trm2 and tam44 are more closely related to each other than to tam45. A total of 261 mutations occurred in intragenic regions, while 34 were found in intergenic regions. Of the 261 intragenic mutations, 175 are nonsynonymous, and the rest are synonymous mutations.
The numbers of mutations acquired by each evolved strain during 65 days of serial passage, corresponding to ∼370 generations, were much higher than we expected from the earlier ALE studies on other bacterial species (22, 47). We speculated whether the evolved strains turned into mutators at some points in their evolution and carried out a fluctuation assay (38, 39) to estimate the mutation rates of the parental and evolved strains. Based on the frequencies of spontaneous mutations conferring resistance to the bactericidal antibiotic rifampin, the mutation rates of WT and GLY3 were estimated to be 4.4 × 10−9 and 3.5 × 10−9 mutations per cell per division, respectively. The deduced mutation rates of the evolved strains (trm2, 2.9 × 10−7; tam44, 2.0 × 10−7; tam45, 1.3 × 10−7 mutations per cell per division) were approximately 40- to 80-fold higher than that of the parental strain, indicating that the three evolved strains indeed acquired the mutator phenotype during ALE.
The missense mutations in glmU and otsA facilitate growth of the parental and wild-type strains under thermal stress.
All the mutations common to trm2, tam44, and tam45, except for the 7 synonymous mutations, were examined for their possible roles in adaptation to thermal stress (Table 6; see Table S4 in the supplemental material). The mutations in secN, hrcA, and ptsS were included in the analyses, as all three evolved strains carry missense mutations in these genes, although the positions of the mutations found for tam45 are different from those found for the other two strains (Table 6). Each mutation was introduced into the parental strain GLY3 or reverted to the wild-type allele in the evolved strain tam45, and the resulting transformants were characterized for their growth at high temperatures. Regarding the trm2/tam44-specific mutations in secN, hrcA, and ptsS, reversion experiments were carried out with trm2 or tam44.
TABLE 6.
Amino acid substitutions common to the evolved strains
| Position, nt [strain(s)] | Gene |
Protein | Mutation |
Thermotolerancea |
|||
|---|---|---|---|---|---|---|---|
| Locus tag | Name | mRNA | Protein | Introduction | Reversion | ||
| 40393 | CgR_0035 | Putative glycosyltransferase | 1043C→T | G348D | ± | ± | |
| 157961 | CgR_0141 | Transposase | 470A→G | V157A | ± | ± | |
| 545579 | CgR_0489 | hemC | Porphobilinogen deaminase | 574G→A | A192T | ± | ± |
| 1152246 | CgR_1044 | glmU | Bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase | 883C→T | E295K | + | ± |
| 1179478 | CgR_1064 | Putative esterase | 767G→A | A256V | ± | ± | |
| 1356940 | CgR_1240 | mhpA | 3-(3-Hydroxyphenyl)propionate hydroxylase | 1074G→A | M358I | ± | ± |
| 1423519 | CgR_1294 | nucS | Endonuclease | 482G→A | R161H | ± | ± |
| 2258272 | CgR_2048 | mraW | S-Adenosyl-methyltransferase | 848G→A | P283L | ± | ± |
| 2267850 | CgR_2057 | pknL | Serine/threonine protein kinase | 1099A→G | T367A | ± | ± |
| 2538467 | CgR_2303 | Putative ketosteroid isomerase | 497C→T | P166L | ± | ± | |
| 2567418 | CgR_2333 | Putative metal-dependent hydrolase | 566A→G | Q189R | ± | ± | |
| 2798011 | CgR_2531 | otsA | Trehalose-6-phosphate synthase | 983G→A | R328H | + | − |
| 2936712 | CgR_2654 | pknG | Serine/threonine protein kinase | 431A→G | D144G | ± | ± |
| 3297495 | CgR_2975 | Putative NUDIX hydrolase | 185A→G | Q62R | ± | ± | |
| CgR_2976 | Hypothetical protein | 161T→C | L54P | ||||
| 1899467 (trm2, tam44) | CgR_1704 | secNb | Preprotein translocase | 308T→C | D103G | ± | ± |
| 1899707 (tam45) | 68C→T | R23H | ± | ± | |||
| 2385598 (tam45) | CgR_2163 | hrcAb | Heat-inducible transcription repressor | 994C→T | A332T | ± | ± |
| 2386507 (trm2, tam44) | 85A→G | S29P | ± | ± | |||
| 2813228 (tam45) | CgR_2547 | ptsSb | PTS sucrose-specific transporter | 1019G→A | S340F | ± | ± |
| 2813766 (trm2, tam44) | 481G→A | L161F | ± | ± | |||
Mutations were introduced into GLY3 (Introduction) or replaced by the wild-type genes in the evolved strains (Reversion). ±, unchanged; +, increased; −, decreased.
Mutations in these genes occurred at different positions in tam45 than in the other two evolved strains.
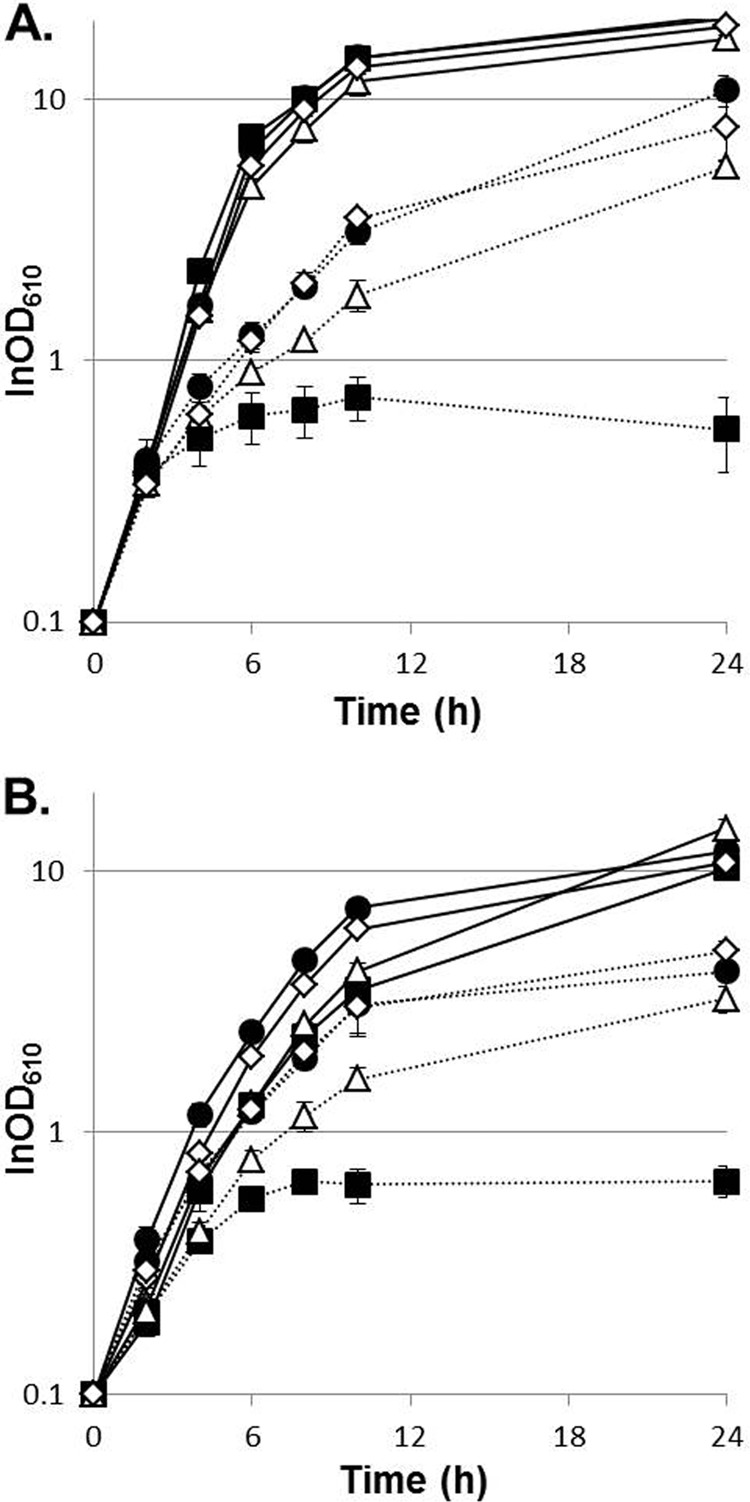
None of the 5 intergenic mutations affected thermotolerance, whether introduced into the parental strain or reverted in tam45 (data not shown). Among the 20 missense mutations examined, only two mutations, one in glmU and the other in otsA, facilitated growth of GLY3 at temperatures around 40°C (Fig. 4A). An increase in maximal cell density was observed for GLY3_GlmUE295K (OD610 = 7.7 ± 0.62) and GLY3_OtsAR328H (OD610 = 5.8 ± 0.57) compared to GLY3 (OD610 = 3.9 ± 0.29) after 24 h of incubation at 40°C (P < 0.01). When growth during the late growth phase was closely monitored, GLY3_GlmUE295K showed a prolonged growth phase compared to GLY3 (Fig. 4B), indicating that the GlmU::E295K substitution affects thermotolerance specifically during the late growth phase. In contrast, the difference in growth between GLY3_OtsAR328H and the WT was already evident after 10 h of incubation (P < 0.05; GLY3_OtsAR328H, OD610 = 3.7 ± 0.34; WT, OD610 = 2.8 ± 0.20), suggesting that the mutation in otsA influences exponential growth at supraoptimal temperatures. Reversion of each mutation in strain GLY3_GlmUE295K or GLY3_OtsAR328H restored GLY3-like growth (Fig. 4A and B). These mutations did not affect growth at 33°C (data not shown). When each of the 20 missense mutations was reverted to the wild-type allele, only the OtsA::R328H substitution was shown to be necessary for full thermotolerance of tam45 (Fig. 4C). Reversion of the mutant otsA markedly reduced the maximal growth rate of tam45 at 41.5°C (P < 0.01; tam45, 0.27 ± 0.044 h−1; tam45_OtsAWT, 0.18 ± 0.026 h−1). Reintroduction of the mutation into strain tam45_OtsAWT restored the tam45-like growth rate (0.24 ± 0.021 h−1). Despite the positive effect of the GlmU::E295K substitution observed for GLY3_GlmUE295K, reversion of the mutation in tam45 did not affect growth at high temperatures (0.29 ± 0.21 h−1) (Fig. 4C).
FIG 4.
The GlmU::E295K and OtsA::R328H substitutions facilitate growth of the wild-type and parental strains under thermal stress. Shown are growth curves of different strains with standard deviations (n = 4). (A and B) Growth in nutrient-rich medium at 40°C. Open squares, GLY3; closed circles, GLY3_GlmUE295K; open triangles, GLY3_OtsAR328H; closed diamonds, GLY3_GlmUE295K_rev; crosses, GLY3_OtsAR328H_rev. (C) Growth in nutrient-rich medium at 41.5°C. Open squares, tam45; open triangles, tam45_GlmUWT; closed circles, tam45_OtsAWT; closed diamonds, tam45_OtsAWT_rev. (D) Growth in nutrient-rich medium at 40.7°C. Closed squares, WT; closed circles, R_GlmUE295K; open triangles, R_OtsAR328H; open diamonds, ROG.
GLY3OG, which carries both the OtsA::R328H and GlmU::E295K substitutions, was able to grow at temperatures above the Tmax of the parental strain, whereas no improvement in the Tmax was observed for GLY3_GlmUE295K or GLY3_OtsAR328H (Table 7). An additive effect was also observed between the deletion of region 1 and the OtsA::R328H substitution, and strain GLY3dr1O carrying these two mutations was able to grow at 40.5°C. When grown at 40.2°C, GLY3dr1O achieved a higher maximal growth rate than GLY3dr1 (P < 0.01) (Table 7). No further improvement in the growth rate, maximal cell density, or Tmax was observed for GLY3dr1OG compared to GLY3OG or GLY3dr1O. As the combination of the Δregion 1, OtsA::R328H, and GlmU::E295K mutations failed to reproduce the growth phenotype of the evolved strains (Table 7; shown for tam45), there must be an additional mutation(s) accounting for the enhanced thermotolerance of the evolved strains.
TABLE 7.
Additive effects of Δregion 1, GlmU::E295K, and OtsA::R328H mutations on growth at high temperatures
| Strain | Genotype |
Maximal growth rate at 40.2°C (h−1)a | Maximal cell density (OD610; 40.2°C; 24 h)a | Maximal growth temp (°C) | ||
|---|---|---|---|---|---|---|
| Region 1 | glmU | otsA | ||||
| GLY3 | Present | WT | WT | 0.24 ± 0.027A | 2.5 ± 0.29A | 40.2 |
| GLY3_GlmUE295K | Present | Mutant | WT | 0.23 ± 0.021A | 3.1 ± 0.45A | 40.2 |
| GLY3_OtsAR328H | Present | WT | Mutant | 0.30 ± 0.021B | 3.7 ± 0.34AB | 40.2 |
| GLY3OG | Present | Mutant | Mutant | 0.28 ± 0.011AB | 6.6 ± 0.21CD | 40.5 |
| GLY3dr1 | Deleted | WT | WT | 0.23 ± 0.012A | 3.8 ± 0.57AB | 40.2 |
| GLY3dr1G | Deleted | Mutant | WT | 0.23 ± 0.016A | 5.1 ± 0.43B | 40.2 |
| GLY3dr1O | Deleted | WT | Mutant | 0.31 ± 0.015B | 5.8 ± 0.23BC | 40.5 |
| GLY3dr1OG | Deleted | Mutant | Mutant | 0.29 ± 0.010B | 6.6 ± 0.63CD | 40.5 |
| tam45 | Deleted | Mutant | Mutant | 0.45 ± 0.014C | 7.3 ± 0.40D | 41.5 |
Means ± standard deviations (n = 3). Different letters denote significant differences among the strains (P < 0.01).
Finally, we examined whether the OtsA::R328H and GlmU::E295K substitutions could enhance thermotolerance in the wild-type context. Introduction of the OtsA::R328H substitution promoted growth of the WT at 40.7°C (Fig. 4D), and a significant increase in maximal cell density was observed for R_OtsAR328H (OD610 = 3.8 ± 0.30) compared to the WT (P < 0.01; OD610 = 1.4 ± 0.10). The GlmU::E295K substitution by itself did not affect growth of the WT under thermal stress; nevertheless, a combination of the mutation with the OtsA::R328H substitution further improved the tolerance of R_OtsAR328H for thermal stress (Fig. 4D). Although no improvement in the Tmax was observed, strain ROG, carrying both substitutions, showed an increased growth rate (0.33 ± 0.029 h−1) compared to the WT (0.27 ± 0.017 h−1; P < 0.05) when grown at 40.7°C and achieved a maximal cell density (OD610 = 6.3 ± 0.29) higher than those of the WT and R_OtsAR328H (P < 0.01). These observations demonstrate that the positive effects of the OtsA::R328H and GlmU::E295K substitutions on thermotolerance are not specific to strain GLY3 and its descendants.
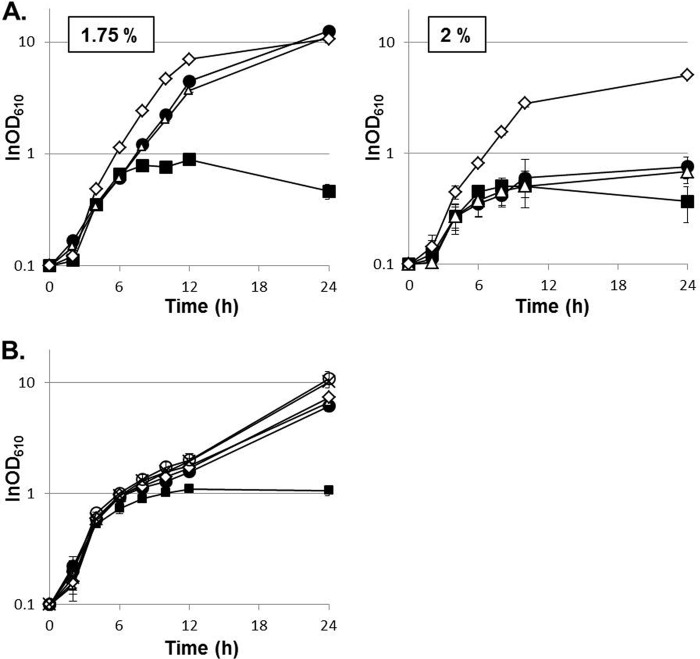
The evolved strains acquired cross-tolerance for isobutanol.
Through preliminary characterization of the evolved strains for their sensitivities to diverse stresses seemingly unrelated to thermal stress, we found that the evolved strains acquired cross-tolerance for isobutanol. In the presence of 1.75% (vol/vol) isobutanol, GLY3 stopped growing when the cell density reached an OD610 of approximately 1.0, whereas trm2, tam44, and tam45 showed vigorous growth, and their maximal cell densities attained an OD610 of >10 (Fig. 5A). Among the three evolved strains, tam45 was most tolerant of the solvent stress and was able to grow even in the presence of 2% (vol/vol) isobutanol (Fig. 5A).
FIG 5.
The evolved strains acquired cross-tolerance for isobutanol. Shown are growth curves of different strains with standard deviations (n = 3). (A) GLY3 (closed squares), trm2 (closed circles), tam44 (open triangles), and tam45 (open diamonds) were grown in nutrient-rich medium with 1.75 or 2% (vol/vol) isobutanol. (B) GLY3 (closed squares), GLY3dr1 (closed circles), GLY3dfy (open triangles), GLY3dr1G (open diamonds), GLY3dr1O (open circles), and GLY3dr1OG (crosses) were grown in nutrient-rich medium with 1.75% (vol/vol) isobutanol.
The three mutations positively affecting thermotolerance, namely, Δregion 1, OtsA::R328H, and GlmU::E295K, were evaluated for their influences on tolerance for isobutanol. When the two missense mutations were introduced into GLY3 independently or in combination, no improvement in solvent tolerance was observed (data not shown). Likewise, reversion of these mutations did not affect the growth of tam45 in the presence of isobutanol, indicating that the missense mutations in glmU and otsA play no role in refining isobutanol tolerance. Deletion of region 1, in contrast, was found to ameliorate the solvent tolerance of the parental strain (Fig. 5B). After 24 h of growth in medium containing 1.75% (vol/vol) isobutanol, the maximal cell density of GLY3dr1 (OD610 = 6.1 ± 0.63) was substantially higher than that of GLY3 (OD610 = 1.1 ± 0.11; P < 0.01). Growth of GLY3dfy was also improved to a similar extent, and no appreciable difference was observed between GLY3dr1 and GLY3dfy (Fig. 5B). Overexpression of pfk and pyk on a plasmid restored GLY3-like sensitivity to isobutanol to GLY3dfy (see Fig. S1 in the supplemental material). These results indicate that the enhanced solvent tolerance is attributable to loss of the transgenes and the resulting reduction in the pfk and pyk transcript levels. GLY3dr1G, GLY3dr1O, and GLY3dr1OG did not show any noticeable improvement in growth under solvent stress compared to GLY3dr1, further indicating that OtsA::R328H and GlmU::E295K are not associated with acquisition of isobutanol tolerance. As deletion of region 1 alone failed to enhance solvent tolerance to the level of the evolved strains, there must be an additional mutation(s) contributing to the increased isobutanol tolerance. These observations suggest shared and distinct genetic bases of adaptation of the evolved strains to thermal and solvent stress.
DISCUSSION
In the present study, we applied ALE to breeding thermotolerant strains of C. glutamicum. After approximately 2 months of serial passage, we were able to isolate three strains adapted to higher growth temperatures. In E. coli, mutational genetic adaptation to higher growth temperatures was found to occur within only 100 to 200 generations (48). Rapid adaptation was also observed for our case, and 65 days of serial passage, corresponding to ∼370 generations, improved the Tmax of C. glutamicum strain GLY3 from 40.2 to 41.5°C in the nutrient-rich medium and from 37 to 40.5°C in the minimal medium. In general, fitness gains tend to be greater in an early stage of ALE and decrease as an evolving population approaches an adaptive peak (49). It takes much longer for these small beneficial mutations than for mutations that provide large benefits to be fixed in a population, likely accounting for our failed attempts to further reinforce thermotolerance of the evolved strains through a few more months of serial passage. Longer-time-scale ALE experiments, which continued for almost 2 years, have been reported for E. coli (21, 22), and the improvements in Tmax in these studies were more significant than in the present study.
In the fluctuation assay of rifampin resistance, approximately 40- to 80-fold increases in mutation rates were observed for the three evolved strains compared to the parental strain. Mutators characterized by their increased mutation rates frequently emerge in the course of ALE experiments (22, 50). Because of high mutation rates, mutator phenotypes can facilitate ALE processes by accelerating the occurrence of beneficial mutations. On the other hand, they can complicate analyses of the genetic basis of adaptation, as not only beneficial but also neutral/deleterious mutation rates increase in mutators. In fact, our search for beneficial mutations among those common to trm2 and tam44 or unique to each strain has been frustrated so far by an overwhelming number of mutations, and perhaps by epistatic interactions among them. The pervasive presence of epistasis among beneficial mutations was suggested in the ALE study on E. coli (51), and a sign of epistasis was indeed observed in our study. The lack of reduction in thermotolerance in strain tam45_GlmUWT, in which the GlmU::E295K substitution was reverted, may be explained by the presence of another mutation that transforms the GlmU::E295K substitution to neutral. By focusing on the mutations common to the three evolved strains, we managed to identify three mutations associated with adaptation to higher growth temperatures, although not all the mutations were found to account for the enhanced thermotolerance of the evolved strains. The present work is the first demonstration that ALE can be a powerful tool to ameliorate the phenotype of C. glutamicum and to gain insights into molecular mechanisms for stress tolerance in the bacterium.
GLY3 carries an extra chromosomal copy of pfk and pyk under the control of the strong tac promoter (28), and these transgenes were shown to compromise growth under thermal and solvent stress. As GLY3 and GLY3dfy showed comparable growth under the optimal conditions (Fig. 3), stress tolerance, rather than aerobic growth in general, is affected by the transgenes. Cells under stress are considered to require an increased ATP supply to provide energy to protein and DNA repair machinery, like ATP-dependent chaperones. Genome-wide screening of the E. coli single-gene knockout library revealed that several genes associated with pyruvate metabolism and ATP synthesis are indispensable specifically for growth at high temperatures (52), and a proteomic analysis of the steady-state heat shock response of the bacterium implicated the components of glycolysis and the tricarboxylic acid (TCA) cycle, as well as ATP synthesis, in thermotolerance (53). Our observations are another implication of the link between energy-providing metabolism and microbial stress tolerance. In GLY3, the increased PFK and PYK activities may cause tuning of the central carbon metabolism to deteriorate in response to thermal or solvent stress, and consequently, the cells fail to meet the demand of the macromolecule repair systems for ATP.
Isobutanol is a promising alternative to fossil fuels, possessing several properties superior to those of ethanol, like higher energy density and lower hygroscopicity (54). One of the current obstacles to microbial production of this biofuel is its toxicity to cells, and intensive efforts are being made to engineer production strains with improved isobutanol tolerance (19, 55). Generally, the toxicity of biofuels is considered to stem from their intercalation into membrane lipid bilayers. The consequences of accumulation of the alcohols in cytoplasmic membranes include increased membrane permeability and fluidity, dissipation of proton motive force, and disruption of membrane protein functions (56). Thermal stress also affects the physical properties of cytoplasmic membranes (57), and in this respect, it may not be surprising to observe a link between microbial tolerances for thermal and solvent stress. Denaturation of cytoplasmic proteins is another common consequence of these stresses (56, 58). Trehalose, which facilitates protein folding and prevents aggregation of denatured proteins (59, 60), is widely recognized as a thermoprotectant (61–63). Indeed, we showed that the missense mutation in otsA, encoding the key enzyme for trehalose synthesis (64, 65), partly accounts for the improved thermotolerance of the evolved strains. Though we failed to connect the mutation in otsA with solvent tolerance, it is still worth testing whether trehalose contributes to bacterial tolerance for isobutanol. Glucose and nutrient transport can also be affected by biofuels, as shown for butanol, which inhibits glucose uptake by Clostridium acetobutylicum (66). Under the assumption that isobutanol interferes with glucose transport by C. glutamicum, overexpression of pfk and pyk, along with the solvent-mediated reduction in glucose uptake, may cause a critical imbalance in glycolytic flow. As discussed above, such imbalance in energy-providing metabolism can lead to a shortage in the energy supply to protein and DNA repair machineries and, consequently, retard growth.
In summary, we found one mutation that improves tolerance for both thermal and isobutanol stress and two mutations associated specifically with enhanced thermotolerance, suggesting overlap and divergence of underlying mechanisms for enhanced tolerance for two different types of stress. Microbes tolerant of multiple forms of stress are desired for industrial applications (67, 68). Progress in studies on cross protection/tolerance will promote the understanding of shared and distinct molecular bases of tolerance for different types of stress and may help develop a strategy for microbial strain optimization.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03973-14.
REFERENCES
- 1.Kinoshita S, Udaka S, Shimono M. 1957. Studies on the amino acid fermentation. I. Production of L-glutamic acid by various microorganisms. J Gen Appl Microbiol 3:193–205. [PubMed] [Google Scholar]
- 2.Gibson BR, Lawrence SJ, Leclaire JP, Powell CD, Smart KA. 2007. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev 31:535–569. doi: 10.1111/j.1574-6976.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- 3.Machielsen R, van Alen-Boerrigter IJ, Koole LA, Bongers RS, Kleerebezem M, Van Hylckama Vlieg JE. 2010. Indigenous and environmental modulation of frequencies of mutation in Lactobacillus plantarum. Appl Environ Microbiol 76:1587–1595. doi: 10.1128/AEM.02595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. 2006. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Li Y, Pei X, Yu L, Feng Y. 2007. Genome-shuffling improved acid tolerance and L-lactic acid volumetric productivity in Lactobacillus rhamnosus. J Biotechnol 129:510–515. doi: 10.1016/j.jbiotec.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Pan J, Wang J, Zhou Z, Yan Y, Zhang W, Lu W, Ping S, Dai Q, Yuan M, Feng B, Hou X, Zhang Y, Ma R, Liu T, Feng L, Wang L, Chen M, Lin M. 2009. IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLoS One 4:e4422. doi: 10.1371/journal.pone.0004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad TM, Lewis NE, Palsson BO. 2011. Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol 7:509. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portnoy VA, Bezdan D, Zengler K. 2011. Adaptive laboratory evolution—harnessing the power of biology for metabolic engineering. Curr Opin Biotechnol 22:590–594. doi: 10.1016/j.copbio.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Dragosits M, Mattanovich D. 2013. Adaptive laboratory evolution: principles and applications for biotechnology. Microb Cell Fact 12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng Y, Fong SS. 2011. Laboratory evolution and multi-platform genome re-sequencing of the cellulolytic actinobacterium Thermobifida fusca. J Biol Chem 286:39958–39966. doi: 10.1074/jbc.M111.239616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maughan H, Nicholson WL. 2011. Increased fitness and alteration of metabolic pathways during Bacillus subtilis evolution in the laboratory. Appl Environ Microbiol 77:4105–4118. doi: 10.1128/AEM.00374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S. 2010. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol 6:378. doi: 10.1038/msb.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cakar ZP, Seker UO, Tamerler C, Sonderegger M, Sauer U. 2005. Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae. FEMS Yeast Res 5:569–578. doi: 10.1016/j.femsyr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 14.de Crecy E, Jaronski S, Lyons B, Lyons TJ, Keyhani NO. 2009. Directed evolution of a filamentous fungus for thermotolerance. BMC Biotechnol 9:74. doi: 10.1186/1472-6750-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izutsu M, Zhou J, Sugiyama Y, Nishimura O, Aizu T, Toyoda A, Fujiyama A, Agata K, Fuse N. 2012. Genome features of “Dark-fly”, a Drosophila line reared long-term in a dark environment. PLoS One 7:e33288. doi: 10.1371/journal.pone.0033288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alcantara-Diaz D, Brena-Valle M, Serment-Guerrero J. 2004. Divergent adaptation of Escherichia coli to cyclic ultraviolet light exposures. Mutagenesis 19:349–354. doi: 10.1093/mutage/geh039. [DOI] [PubMed] [Google Scholar]
- 17.Dragosits M, Mozhayskiy V, Quinones-Soto S, Park J, Tagkopoulos I. 2013. Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol Syst Biol 9:643. doi: 10.1038/msb.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoebel DM, Hokamp K, Last MS, Dorman CJ. 2009. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet 5:e1000671. doi: 10.1371/journal.pgen.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atsumi S, Wu TY, Machado IM, Huang WC, Chen PY, Pellegrini M, Liao JC. 2010. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol 6:449. doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horinouchi T, Tamaoka K, Furusawa C, Ono N, Suzuki S, Hirasawa T, Yomo T, Shimizu H. 2010. Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics 11:579. doi: 10.1186/1471-2164-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph B, Gebendorfer KM, Buchner J, Winter J. 2010. Evolution of Escherichia coli for growth at high temperatures. J Biol Chem 285:19029–19034. doi: 10.1074/jbc.M110.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kishimoto T, Iijima L, Tatsumi M, Ono N, Oyake A, Hashimoto T, Matsuo M, Okubo M, Suzuki S, Mori K, Kashiwagi A, Furusawa C, Ying BW, Yomo T. 2010. Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet 6:e1001164. doi: 10.1371/journal.pgen.1001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaby IK, Lyons BJ, Wroclawska-Hughes E, Phillips GC, Pyle TP, Chamberlin SG, Benner SA, Lyons TJ, Crecy-Lagard VD, Crecy ED. 2012. Experimental evolution of a facultative thermophile from a mesophilic ancestor. Appl Environ Microbiol 78:144–155. doi: 10.1128/AEM.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leyer GJ, Johnson EA. 1993. Acid adaptation induces cross-protection against environmental stresses in Salmonella typhimurium. Appl Environ Microbiol 59:1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergholz TM, den Bakker HC, Fortes ED, Boor KJ, Wiedmann M. 2010. Salt stress phenotypes in Listeria monocytogenes vary by genetic lineage and temperature. Foodborne Pathog Dis 7:1537–1549. doi: 10.1089/fpd.2010.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canovas D, Fletcher SA, Hayashi M, Csonka LN. 2001. Role of trehalose in growth at high temperature of Salmonella enterica serovar Typhimurium. J Bacteriol 183:3365–3371. doi: 10.1128/JB.183.11.3365-3371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jojima T, Fujii M, Mori E, Inui M, Yukawa H. 2010. Engineering of sugar metabolism of Corynebacterium glutamicum for production of amino acid L-alanine under oxygen deprivation. Appl Microbiol Biotechnol 87:159–165. doi: 10.1007/s00253-010-2493-7. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto S, Gunji W, Suzuki H, Toda H, Suda M, Jojima T, Inui M, Yukawa H. 2012. Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl Environ Microbiol 78:4447–4457. doi: 10.1128/AEM.07998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto S, Suda M, Niimi S, Inui M, Yukawa H. 2013. Strain optimization for efficient isobutanol production using Corynebacterium glutamicum under oxygen deprivation. Biotechnol Bioeng 110:2938–2948. doi: 10.1002/bit.24961. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 31.Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196. doi: 10.1159/000079827. [DOI] [PubMed] [Google Scholar]
- 32.Inui M, Suda M, Okino S, Nonaka H, Puskas LG, Vertès AA, Yukawa H. 2007. Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504. doi: 10.1099/mic.0.2006/005587-0. [DOI] [PubMed] [Google Scholar]
- 33.Yukawa H, Omumasaba CA, Nonaka H, Kos P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M. 2007. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058. doi: 10.1099/mic.0.2006/003657-0. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez D, Francois P, Farinelli L, Osteras M, Schrenzel J. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res 18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosche WA, Foster PL. 2000. Determining mutation rates in bacterial populations. Methods 20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pope CF, O'Sullivan DM, McHugh TD, Gillespie SH. 2008. A practical guide to measuring mutation rates in antibiotic resistance. Antimicrob Agents Chemother 52:1209–1214. doi: 10.1128/AAC.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lea D, Coulson C. 1949. The distribution of the number of mutants in bacterial populations. Genetics 49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 41.Drake JW. 1991. Constant rate of spontaneous mutations in DNA based microbes. Proc Natl Acad Sci U S A 88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Armitage P. 1952. The statistical theory of bacterial populations subject to mutations. J R Stat Soc B 14:1–40. [Google Scholar]
- 43.Ozaki A, Katsumata R, Oka T, Furuya A. 1984. Functional expression of the genes of Escherichia coli in gram-positive Corynebacterium glutamicum. Mol Gen Genet 196:175–178. doi: 10.1007/BF00334113. [DOI] [PubMed] [Google Scholar]
- 44.Jojima T, Igari T, Gunji W, Suda M, Inui M, Yukawa H. 2012. Identification of a HAD superfamily phosphatase, HdpA, involved in 1,3-dihydroxyacetone production during sugar catabolism in Corynebacterium glutamicum. FEBS Lett 586:4228–4232. doi: 10.1016/j.febslet.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 45.Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol 8:243–254. doi: 10.1159/000086705. [DOI] [PubMed] [Google Scholar]
- 46.Vertès AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H. 1993. Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res Microbiol 144:181–185. doi: 10.1016/0923-2508(93)90043-2. [DOI] [PubMed] [Google Scholar]
- 47.Charusanti P, Fong NL, Nagarajan H, Pereira AR, Li HJ, Abate EA, Su Y, Gerwick WH, Palsson BO. 2012. Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction. PLoS One 7:e33727. doi: 10.1371/journal.pone.0033727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett AF, Dao KM, Lenski RE. 1990. Rapid evolution in response to high-temperature selection. Nature 346:79–81. doi: 10.1038/346079a0. [DOI] [PubMed] [Google Scholar]
- 49.Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 50.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 51.Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. 2012. The molecular diversity of adaptive convergence. Science 335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 52.Murata M, Fujimoto H, Nishimura K, Charoensuk K, Nagamitsu H, Raina S, Kosaka T, Oshima T, Ogasawara N, Yamada M. 2011. Molecular strategy for survival at a critical high temperature in Eschierichia coli. PLoS One 6:e20063. doi: 10.1371/journal.pone.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luders S, Fallet C, Franco-Lara E. 2009. Proteome analysis of the Escherichia coli heat shock response under steady-state conditions. Proteome Sci 7:36. doi: 10.1186/1477-5956-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Connor MR, Liao JC. 2009. Microbial production of advanced transportation fuels in non-natural hosts. Curr Opin Biotechnol 20:307–315. doi: 10.1016/j.copbio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Minty JJ, Lesnefsky AA, Lin F, Chen Y, Zaroff TA, Veloso AB, Xie B, McConnell CA, Ward RJ, Schwartz DR, Rouillard JM, Gao Y, Gulari E, Lin XN. 2011. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb Cell Fact 10:18. doi: 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicolaou SA, Gaida SM, Papoutsakis ET. 2010. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng 12:307–331. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Beney L, Gervais P. 2001. Influence of the fluidity of the membrane on the response of microorganisms to environmental stresses. Appl Microbiol Biotechnol 57:34–42. doi: 10.1007/s002530100754. [DOI] [PubMed] [Google Scholar]
- 58.Jaenicke R. 1991. Protein folding: local structures, domains, subunits, and assemblies. Biochemistry 30:3147–3161. doi: 10.1021/bi00227a001. [DOI] [PubMed] [Google Scholar]
- 59.Jain NK, Roy I. 2009. Effect of trehalose on protein structure. Protein Sci 18:24–36. doi: 10.1002/pro.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elbein AD, Pan YT, Pastuszak I, Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 61.Woodruff PJ, Carlson BL, Siridechadilok B, Pratt MR, Senaratne RH, Mougous JD, Riley LW, Williams SJ, Bertozzi CR. 2004. Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem 279:28835–28843. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- 62.Ells TC, Truelstrup Hansen L. 2011. Increased thermal and osmotic stress resistance in Listeria monocytogenes 568 grown in the presence of trehalose due to inactivation of the phosphotrehalase-encoding gene treA. Appl Environ Microbiol 77:6841–6851. doi: 10.1128/AEM.00757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reina-Bueno M, Argandona M, Salvador M, Rodriguez-Moya J, Iglesias-Guerra F, Csonka LN, Nieto JJ, Vargas C. 2012. Role of trehalose in salinity and temperature tolerance in the model halophilic bacterium Chromohalobacter salexigens. PLoS One 7:e33587. doi: 10.1371/journal.pone.0033587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolf A, Kramer R, Morbach S. 2003. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol Microbiol 49:1119–1134. doi: 10.1046/j.1365-2958.2003.03625.x. [DOI] [PubMed] [Google Scholar]
- 65.Tzvetkov M, Klopprogge C, Zelder O, Liebl W. 2003. Genetic dissection of trehalose biosynthesis in Corynebacterium glutamicum: inactivation of trehalose production leads to impaired growth and an altered cell wall lipid composition. Microbiology 149:1659–1673. doi: 10.1099/mic.0.26205-0. [DOI] [PubMed] [Google Scholar]
- 66.Bowles LK, Ellefson WL. 1985. Effects of butanol on Clostridium acetobutylicum. Appl Environ Microbiol 50:1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallace-Salinas V, Gorwa-Grauslund MF. 2013. Adaptive evolution of an industrial strain of Saccharomyces cerevisiae for combined tolerance to inhibitors and temperature. Biotechnol Biofuels 6:151. doi: 10.1186/1754-6834-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallström BM, Petranovic D, Nielsen J. 2014. Altered sterol composition renders yeast thermotolerant. Science 346:75–78. doi: 10.1126/science.1258137. [DOI] [PubMed] [Google Scholar]
- 69.Tsuchida Y, Kimura S, Suzuki N, Inui M, Yukawa H. 2009. Characterization of a new 2.4-kb plasmid of Corynebacterium casei and development of stable corynebacterial cloning vector. Appl Microbiol Biotechnol 81:1107–1115. doi: 10.1007/s00253-008-1746-1. [DOI] [PubMed] [Google Scholar]
- 70.Okibe N, Suzuki N, Inui M, Yukawa H. 2010. Antisense-RNA-mediated plasmid copy number control in pCG1-family plasmids, pCGR2 and pCG1, in Corynebacterium glutamicum. Microbiology 156:3609–3623. doi: 10.1099/mic.0.043745-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.