Abstract
Methanotrophs can express a cytoplasmic (soluble) methane monooxygenase (sMMO) or membrane-bound (particulate) methane monooxygenase (pMMO). Expression of these MMOs is strongly regulated by the availability of copper. Many methanotrophs have been found to synthesize a novel compound, methanobactin (Mb), that is responsible for the uptake of copper, and methanobactin produced by Methylosinus trichosporium OB3b plays a key role in controlling expression of MMO genes in this strain. As all known forms of methanobactin are structurally similar, it was hypothesized that methanobactin from one methanotroph may alter gene expression in another. When Methylosinus trichosporium OB3b was grown in the presence of 1 μM CuCl2, expression of mmoX, encoding a subunit of the hydroxylase component of sMMO, was very low. mmoX expression increased, however, when methanobactin from Methylocystis sp. strain SB2 (SB2-Mb) was added, as did whole-cell sMMO activity, but there was no significant change in the amount of copper associated with M. trichosporium OB3b. If M. trichosporium OB3b was grown in the absence of CuCl2, the mmoX expression level was high but decreased by several orders of magnitude if copper prebound to SB2-Mb (Cu-SB2-Mb) was added, and biomass-associated copper was increased. Exposure of Methylosinus trichosporium OB3b to SB2-Mb had no effect on expression of mbnA, encoding the polypeptide precursor of methanobactin in either the presence or absence of CuCl2. mbnA expression, however, was reduced when Cu-SB2-Mb was added in both the absence and presence of CuCl2. These data suggest that methanobactin acts as a general signaling molecule in methanotrophs and that methanobactin “piracy” may be commonplace.
INTRODUCTION
Methanotrophs are distinguished from other microorganisms by their ability to utilize methane as a sole carbon and energy source yet are phylogenetically and physiologically diverse. Microbial methane oxidation can be coupled to a variety of terminal electron acceptors, including oxygen, sulfate, nitrate, and nitrite (1–4). The aerobic methanotrophs are typically mesophilic and group phylogenetically within the Gammaproteobacteria and Alphaproteobacteria (1). Thermo- and meso-acidophilic aerobic methanotrophs, however, that grow at pH <3 and at optimal temperatures ranging from 35°C to greater than 50°C have also been discovered in the phylum Verrucomicrobia (5–9). Further, novel oxygenic methanotrophs that couple methane oxidation to nitrite reduction have been reported, e.g., “Candidatus Methylomirabilis oxyfera” that generates oxygen from a unique denitrification pathway, which is then used for methane oxidation (2). Aerobic methanotrophs are found in many environments, e.g., freshwater and marine sediments, bogs, forest, and agricultural soils, among other locations (1, 2, 5–11).
These microorganisms have been extensively studied for many different reasons, including the fact that they play a key role in the global carbon cycle. All aerobic methanotrophs employ the enzyme methane monooxygenase (MMO) to convert methane to methanol in the first step of methane oxidation to CO2. One form of the enzyme, the particulate methane monooxygenase (pMMO), is found in most known aerobic methanotrophs and is located in the cytoplasmic membrane (1). Another form, the soluble methane monooxygenase (sMMO), is found in some aerobic methanotrophs and is located in the cytoplasm (1).
Aerobic proteobacterial methanotrophs are sensitive to copper, and this is a key factor regulating the expression of the genes encoding sMMO and pMMO as well as the activity of these enzymes. For the aerobic proteobacterial methanotrophs that can express both forms of MMO, sMMO is expressed only under conditions of copper deficiency, and various copper concentrations have a strong effect on the expression and activity of pMMO (1, 12–16).
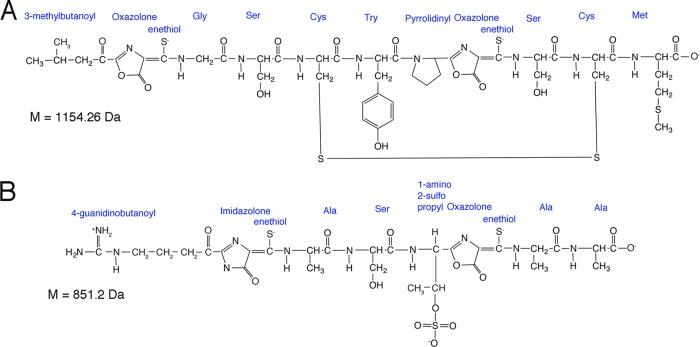
There are multiple mechanisms by which aerobic proteobacterial methanotrophs collect copper, including a membrane-bound copper binding protein, MopE (∼66 kDa), as well as a truncated form of MopE (∼46 kDa), termed MopE*, that is secreted into the growth medium (17–19). This mechanism to date, however, has been characterized only from Methylococcus capsulatus Bath. Instead, many proteobacterial methanotrophs secrete a chalkophore, or copper-binding compound (chalko is Greek for copper), called methanobactin (Mb), for copper uptake. The first form of methanobactin characterized was from Methylosinus trichosporium OB3b, and it was found to be a small modified polypeptide of 1,154 Da that utilizes two oxazolone rings, each associated with an enethiol group for copper binding (Fig. 1A) (20–22). More recently, methanobactins from four other methanotrophs have been characterized, and these methanobactins are all small (825 to 914 Da) and have two heterocyclic rings (one of which is an oxazolone ring and the other of which is either an imidazolone or pyrazinedione ring) with associated enethiol groups (23, 24). All methanobactins examined to date have very high copper affinities. For example, the measured copper affinity for methanobactin from M. trichosporium OB3b ranges from 1018 to 1058 M−1 (25–27), while methanobactin from Methylocystis sp. strain SB2 is reported to have a copper affinity of ∼1026 M−1 (28). Further, copper binding is quite rapid; i.e., the initial binding rate of Cu2+ to the first oxazolone ring for methanobactin from M. trichosporium OB3b is greater than 640 s−1, followed by a coordination rate of 121 s−1 to the second oxazolone ring (25). For methanobactin from Methylocystis sp. strain SB2, the coordination rates of Cu2+ to both rings are greater than 2,000 s−1 (29).
FIG 1.
Primary structures of methanobactin from M. trichosporium OB3b (A) and Methylocystis sp. strain SB2 (B). M, mass.
Recent studies have shown that methanobactin influences expression of the two forms of MMO; i.e., it forms part of the “copper switch.” Specifically, if purified methanobactin from M. trichosporium OB3b is added to cultures of M. trichosporium OB3b, increased expression of mmoX, encoding the α-subunit of the hydroxylase component of sMMO, is observed (30, 31).
Given this finding, it was hypothesized that as methanobactin is secreted into the surrounding growth environment and as the known forms of methanobactin have significant structural similarity, methanobactin from one methanotroph may alter gene expression in another. To that end, we investigated the effect of the addition of methanobactin from Methylocystis sp. strain SB2 (Fig. 1B) to M. trichosporium OB3b on the expression of genes encoding polypeptides of pMMO and sMMO and on methanobactin synthesis. That is, we wished to determine whether methanobactins act as signaling molecules in methanotrophs to regulate gene expression.
MATERIALS AND METHODS
Growth conditions and isolation of methanobactin.
Methylosinus trichosporium OB3b was grown in nitrate minimal salts (NMS) medium (32) at 30°C without any added copper or with 1 μM copper as CuCl2. The background copper concentration in standard NMS medium was 0.03 ± 0.01 μM, as determined using atomic absorption spectroscopy (33). Methanobactin from Methylocystis sp. strain SB2 (SB2-Mb) was isolated using procedures outlined by Bandow et al. (34). The purity of SB2-Mb was determined to be 98.2% ± 0.3% by high-performance liquid chromatography (HPLC) as described earlier (24). Copper-SB2-Mb complexes (Cu-SB2-Mb) were made as reported previously (33). Briefly, Cu-SB2-Mb was prepared by adding equimolar amounts of CuCl2 and SB2-Mb to create a 5 mM stock solution. Cu-SB2-Mb was freshly prepared at 30°C under constant mixing at 200 rpm in the dark for 1 h before use. The concentration of copper not bound to SB2-Mb in Cu-SB2-Mb solutions was tested by monitoring the copper concentration in the flowthrough fraction of samples passed through Sep-Pak cartridges as previously described (34). Copper concentrations in the flowthrough fractions were then determined by atomic absorption spectroscopy (33). The concentration of copper not associated with SB2-Mb in stock Cu-Mb-SB2 solutions was below detection, i.e., <1 nM. The stability of Cu-SB2-Mb at 30°C in NMS medium was monitored by changes in the UV-visible light absorption spectra and by the presence of unbound copper. Following a 3-day incubation period, 0.30% ± 0.01% sample loss was observed.
SB2-Mb or Cu-SB2-Mb was added to NMS medium at either 5 or 50 μM as the amount of methanobactin found in the spent medium typically ranges from 4 to 50 μM (1). Glass side-arm flasks (250 ml; each with 50 ml of NMS medium) for each condition were capped with butyl rubber stoppers. Methane was added at a methane-to-air ratio of 1:2. Cultures were grown at 30°C and shaken at 200 rpm for 3 days until the late exponential phase was reached. The optical density at 600 nm (OD600) was measured in a Genesys 20 Visible spectrophotometer (Spectronic Unicam, Waltham, MA) at 3- to 6-h intervals until the late exponential phase was reached. All conditions had at least duplicate biological samples and most commonly triplicate samples.
RNA extraction and quantitative reverse transcriptase PCR (RT-qPCR).
M. trichosporium OB3b cultures grown under various amounts of copper, SB2-Mb, and Cu-SB2-Mb were harvested after the addition of stop solution at a 9:1 (culture-to-stop solution) ratio. The stop solution consisted of 5% buffer-equilibrated phenol (pH 7.3) in ethanol. A 25-ml mixture of cell culture and stop solution was then centrifuged at 5,000 × g for 10 min at 4°C to collect biomass. RNA was extracted as described earlier (30). Any possible DNA contamination was examined for all RNA extractions via PCR amplification of the 16S rRNA gene, with each RNA sample treated with DNase until it was free of DNA contamination. A NanoDrop ND1000 instrument (NanoDrop Technologies, Inc., Wilmington, DE) was used to determine the concentration of purified RNA. Reverse transcription was performed on DNA-free RNA samples (500 ng) to synthesize cDNA using a Superscript III reverse transcriptase kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
RT-qPCR analyses were performed to quantify the relative expression of pmoA, mmoX, and mbnA genes in M. trichosporium OB3b grown with various amounts of copper, SB2-Mb, and Cu-SB2-Mb. Previously designed gene-specific primers (33) used for the amplifications included 5′-TTCTGGGGCTGGACCTAYTTC-3′ and 5′-CCGACAGCAGCAGGATGATG-3′ for pmoA, 5′-TCAACACCGATCTSAACAACG-3′ and 5′-TCCAGATTCCRCCCCAATCC-3′ for mmoX, 5′-TGGAAACTCCCTTAGGAGGAA-3′ and 5′-CTGCACGGATAGCACGAAC-3′ for mbnA, and 5′-GCAGAACCTTACCAGCTTTTGAC-3′ and 5′-CCCTTGCGGGAAGGAAGTC-3′ for 16S rRNA. The specificities of these primers were verified by gel electrophoresis and sequencing of PCR products. Quantitative PCR (qPCR) amplifications were performed in 96-well reaction PCR plates using Mx3000P qPCR systems (Stratagene, La Jolla, CA). The final reaction volume (20 μl) contained 0.8 μl of cDNA, 1× Brilliant III SYBR green qPCR Mastermix (Agilent Technologies, Santa Clara, CA), 15 nM 6-carboxy-X-rhodamine (ROX) dye, 0.5 μM (each) forward and reverse primer, and sterile water (Ambion Life Technologies, Grand Island, NY). The Mx3000P PCR program involved 40 cycles of denaturation (95°C for 30 s), annealing (58°C for 20 s), and extension (68°C for 30 s) after an initial denaturation at 95°C for 10 min. After the qPCR cycles, samples were subjected to melting curve analysis with temperatures ranging from 55°C to 95°C to confirm the specificity of qPCR products. MxPro software (Stratagene, La Jolla, CA) was used to import the threshold amplification cycle values. Relative gene expression levels were calculated from these threshold cycle (CT) values and the comparative CT method (2−ΔΔCT) (35), with 16S rRNA as the housekeeping gene (33).
Metal measurements.
Copper associated with biomass of M. trichosporium OB3b grown in the presence of various amounts of CuCl2, SB2-Mb, and Cu-SB2-Mb was determined as described previously (33). Briefly, cultures were centrifuged at 5,000 × g for 10 min at 4°C. Cell pellets were then resuspended in 1 ml of fresh NMS medium before storage at −80°C. These suspensions were then acidified in 1 ml of 70% HNO3 (vol/vol) and incubated for 2 h at 95°C. Copper associated with biomass was subsequently analyzed using inductively coupled plasma mass spectrometry (PerkinElmer, Waltham, MA).
Naphthalene assay for sMMO activity.
sMMO activity in M. trichosporium OB3b was assayed using a modified version of the naphthalene assay (36, 37). Briefly, growth was monitored by measuring the optical density at 600 nm (OD600) using a Milton Roy Spectronic 20D spectrophotometer (Milton Roy Company, Warminster, PA). Cultures were grown to an optical density of between 0.3 and 0.4, and triplicate samples of 3 ml each were put in 10-ml serum vials with several flakes of naphthalene, capped with Teflon-coated butyl rubber stoppers, and sealed. Cultures were incubated for 2 h at 200 rpm and 30°C. The cell suspension was then centrifuged for 5 min at 6,300 × g. A total of 130 μl of freshly prepared 4.21 mM tetrazotized o-dianisidine was then placed in a 1.5-ml cuvette with 1.3 ml of the culture supernatant, and the absorbance at 528 nm was monitored immediately using a Genesys 20 Visible spectrophotometer (Spectronic Unicam, Waltham, MA).
Statistical analyses.
One-way analyses of variance (ANOVA) were performed to determine any significant differences in the responses of M. trichosporium OB3b to various amounts of SB2-Mb or Cu-SB2-Mb in the presence or absence of CuCl2 as well as unpaired, two-tailed Student's t tests, assuming equal variance between groups when only two data sets were compared.
RESULTS
Growth and gene expression in M. trichosporium OB3b incubated with various concentrations of copper, methanobactin, and copper-methanobactin from Methylocystis sp. strain SB2.
In the absence of CuCl2, no significant trend in mmoX expression (encoding the α-subunit of the hydroxylase component of sMMO) was observed when various amounts of SB2-Mb were added (Fig. 2A) (ANOVA, P = 0.36). If 1 μM CuCl2 was provided, mmoX expression significantly decreased by more than 4 orders of magnitude compared to values when no CuCl2 was added (Student's t test, P = 2.3 × 10−5). Expression of mmoX, however, increased by more than 3 orders of magnitude with increasing amounts of SB2-Mb (ANOVA, P = 2 × 10−6).
FIG 2.
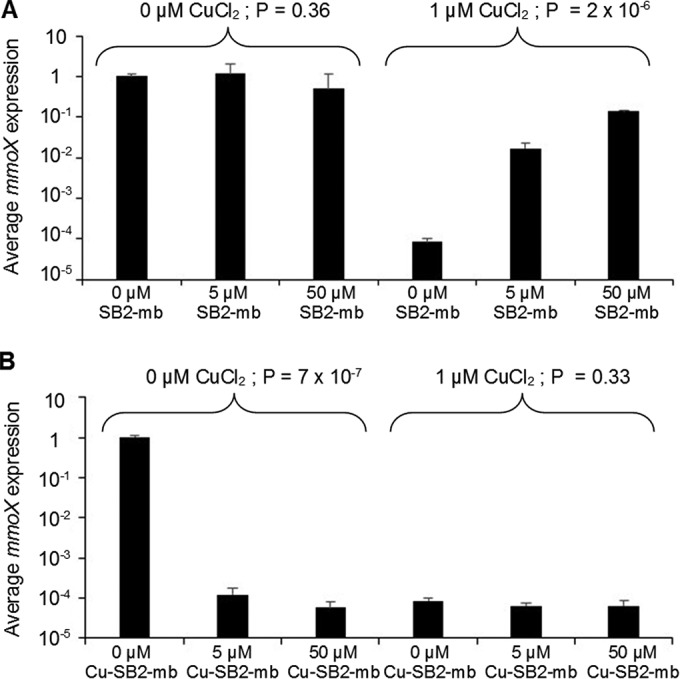
RT-qPCR of mmoX in M. trichosporium OB3b grown in either the absence or presence of 1 μM CuCl2 and various amounts of methanobactin from Methylocystis sp. strain SB2 (A) or copper-SB2 methanobactin complexes (B). Indicated P values are from ANOVA.
The addition of SB2-Mb had some effect on pmoA expression (encoding the 26-kDa subunit of pMMO) in the absence of CuCl2, with overall expression increasing ∼60% as SB2-Mb was increased (Fig. 3A) (ANOVA, P = 0.019). Further, expression of pmoA was ∼2.7-fold greater in the presence of CuCl2 than in its absence (Student's t test, P = 5.6 × 10−6), but pmoA expression did not change significantly in the presence of CuCl2 with the simultaneous addition of SB2-Mb (ANOVA, P = 0.08).
FIG 3.
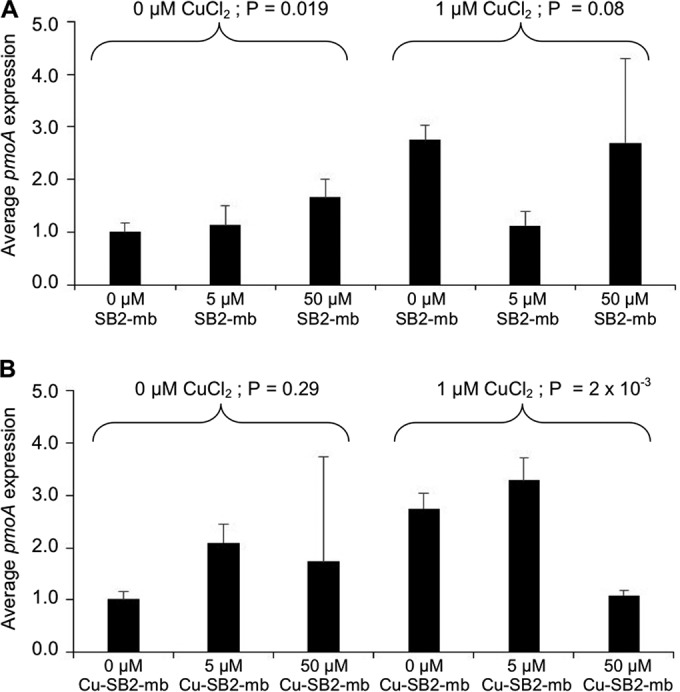
RT-qPCR of pmoA in M. trichosporium OB3b grown in either the absence or presence of 1 μM CuCl2 and various amounts of methanobactin from Methylocystis sp. strain SB2 (A) or copper-SB2 methanobactin complexes (B). Indicated P values are from ANOVA.
Finally, expression of mbnA (encoding the polypeptide precursor of methanobactin) in M. trichosporium OB3b was not found to vary significantly in the absence of CuCl2 in response to the addition of SB2-Mb (Fig. 4A) (ANOVA, P = 0.22). Further, the addition of 1 μM CuCl2 reduced mbnA expression ∼5-fold compared to the level in the absence of CuCl2 (Student's t test, P = 2 × 10−3). No significant change in mbnA expression was observed in response to various amounts of SB2-Mb when it was added in conjunction with 1 μM CuCl2 (ANOVA, P = 0.60).
FIG 4.
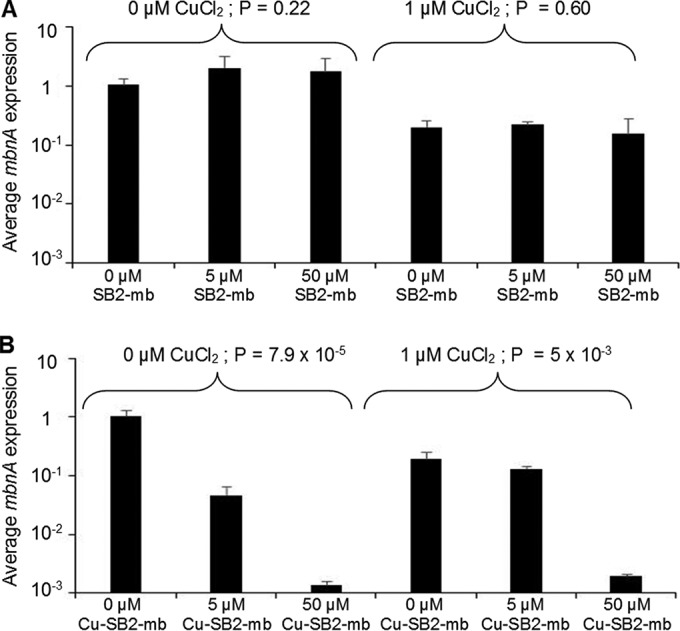
RT-qPCR of mbnA in M. trichosporium OB3b grown in either the absence or presence of 1 μM CuCl2 and various amounts of methanobactin from Methylocystis sp. strain SB2 (A) or copper-SB2 methanobactin complexes (B). Indicated P values are from ANOVA.
A different response was observed when SB2-Mb preincubated with copper (Cu-SB2-Mb) was added to cultures of M. trichosporium OB3b. In the absence of CuCl2, when Cu-SB2-Mb was added, mmoX expression dropped by approximately 4 orders of magnitude compared to when no Cu-SB2-Mb was added (Fig. 2B) (ANOVA, P = 7 × 10−7). Expression of mmoX in the presence of 1 μM CuCl2 was consistently low and did not change significantly with the addition of Cu-SB2-Mb (ANOVA, P = 0.33).
pmoA expression was less dependent on the presence of Cu-SB2-Mb. An ∼1.5- to 2-fold increase in pmoA expression was found in the absence of any CuCl2 when Cu-SB2-Mb was also added, but such a change was not significant (Fig. 3B) (ANOVA, P = 0.29). In the presence of 1 μM CuCl2, pmoA expression decreased by ∼3-fold when Cu-SB2-Mb was increased to 50 μM (ANOVA, P = 2 × 10−3).
Expression of mbnA was affected by the addition of Cu-SB2-Mb in both the presence and absence of CuCl2 (Fig. 4B). In the absence of CuCl2, mbnA expression decreased by ∼20- or 700-fold when either 5 or 50 μM Cu-SB2-Mb was added, respectively (ANOVA, P = 7.9 × 10−5). In the presence of 1 μM CuCl2, mbnA expression decreased only when 50 μM Cu-SB2-Mb was added (by ∼100-fold), and overall the addition of Cu-SB2-Mb in the presence of 1 μM CuCl2 had a significant effect (ANOVA, P = 5 × 10−3).
Impact of various amounts of copper, methanobactin, and copper-methanobactin from Methylocystis sp. strain SB2 on MMO activity in M. trichosporium OB3b.
Figure 5 summarizes the effect of SB2-Mb on sMMO activity in M. trichosporium OB3b, as determined using a naphthalene assay (36, 37). The addition of up to 50 μM SB2-Mb to M. trichosporium OB3b grown in the absence of CuCl2 had no apparent effect on sMMO activity (ANOVA, P = 0.4). If SB2-Mb was added in the presence of 1 μM CuCl2, naphthalene oxidation significantly increased ∼6-fold (ANOVA, P = 2.1 × 10−6).
FIG 5.
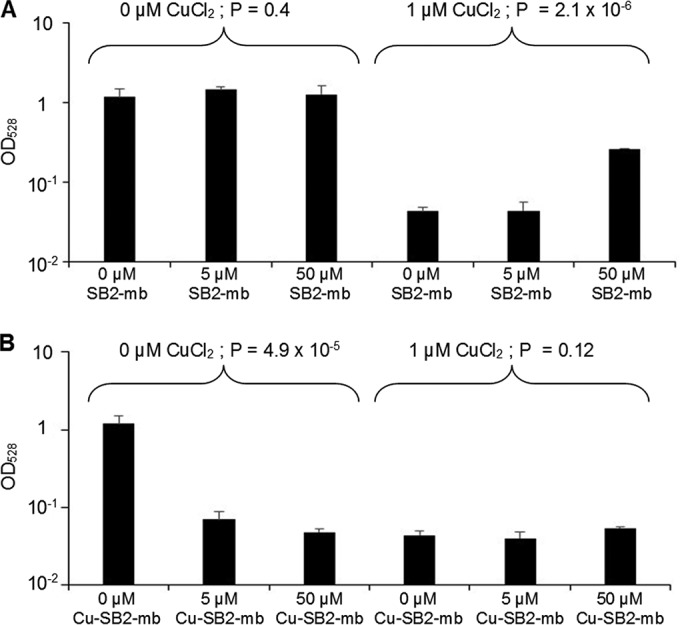
sMMO oxidation of naphthalene as indicated by changes in the OD528 in M. trichosporium OB3b grown in either the absence or presence of 1 μM CuCl2 and various amounts of methanobactin from Methylocystis sp. strain SB2 (A) or copper-SB2 methanobactin complexes (B). Indicated P values are from ANOVA.
When Cu-SB2-Mb was added to cultures of M. trichosporium OB3b grown in the absence of CuCl2, naphthalene oxidation decreased ∼20-fold, (ANOVA, P = 4.9 × 10−5). In the presence of copper, little sMMO activity was observed, and there was no significant effect of Cu-SB2-Mb on sMMO activity in M. trichosporium OB3b when Cu-SB2-Mb was added in conjunction with 1 μM CuCl2 (ANOVA, P = 0.12).
Uptake of copper by M. trichosporium OB3b in the presence of methanobactin and copper-methanobactin from Methylocystis sp. strain SB2.
To determine any effect of the addition of SB2-Mb on the ability of M. trichosporium OB3b to sequester copper, biomass-associated copper was also measured. The addition SB2-Mb to M. trichosporium OB3b grown in the absence of CuCl2 increased copper by ∼2.2-fold, from 0.01 μg of copper · mg protein−1 to 0.025 μg of copper · mg protein−1 (Fig. 6A) (ANOVA, P = 0.01). More copper was associated with biomass when 1 μM CuCl2 was added, i.e., 0.08 μg of copper · mg protein−1, and such an increase was significant compared to copper measured when CuCl2 was not added (Student's t test, P = 2.7 × 10−4). The addition of SB2-Mb, however, did not alter the amount of copper associated with M. trichosporium OB3b cultures grown in the presence of CuCl2 (ANOVA, P = 0.38).
FIG 6.
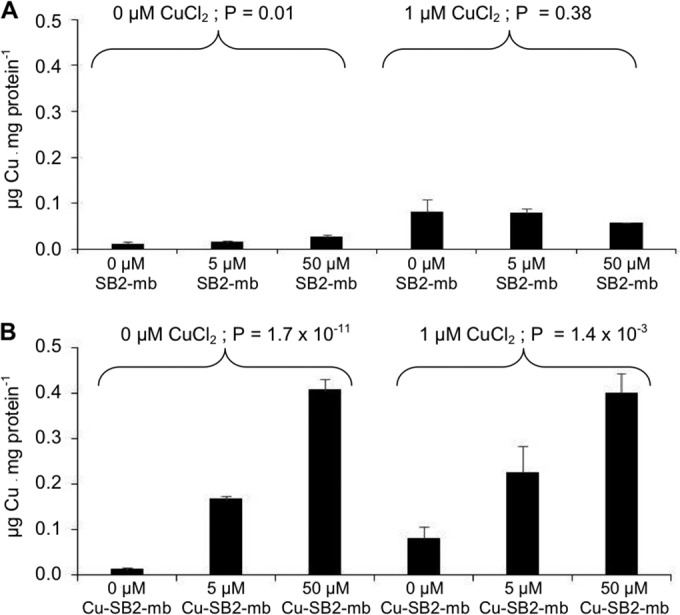
Copper associated with the biomass of M. trichosporium OB3b grown in either the absence or presence of 1 μM CuCl2 and various amounts of methanobactin from Methylocystis sp. strain SB2 (A) or copper-SB2 methanobactin complexes (B). Indicated P values are from ANOVA.
Cell-associated copper, however, significantly increased with increasing amounts of Cu-SB2-Mb in both the presence and absence of CuCl2 (Fig. 5B) (ANOVA, P = 1.7 × 10−11 and P = 1.4 × 10−3 for M. trichosporium OB3b grown in the absence and presence of CuCl2, respectively). It is also interesting that approximately the same amount of cell-associated copper was found when either 5 or 50 μM Cu-SB2-Mb was added in absence or presence of CuCl2. It was found earlier that M. trichosporium OB3b methanobactin cannot remove copper already bound to SB2-Mb and vice versa (28). Given this and the findings reported here (e.g., copper associated with biomass increased and mmoX and mbnA expression decreased with increasing amounts of Cu-SB2-Mb), it appears that M. trichosporium OB3b was able to take up Cu-SB2-Mb complexes and that such uptake then caused changes in the expression of specific genes.
DISCUSSION
It is well-known that methanotrophic metabolism is strongly affected by copper and that methanotrophs utilize multiple mechanisms for copper uptake (1, 17–24). Further, it has been shown that in M. trichosporium OB3b, methanobactin plays a significant role in controlling gene expression. Specifically, in both the M. trichosporium OB3b wild type and an M. trichosporium OB3b mutant where the mbnA gene was disrupted, the exogenous addition of methanobactin from M. trichosporium OB3b increased expression of mmoX encoding the α-subunit of the hydroxylase component of the soluble methane monooxygenase (30, 31). Given the significant structural similarity between the known forms of methanobactin, the role of methanobactin in controlling gene expression in at least one methanotroph, and the observation that methanobactin is secreted into the growth environment, it was hypothesized that methanobactin may also act as a signaling molecule.
The addition of SB2-Mb increased both mmoX expression and whole-cell activity of sMMO in the presence of 1 μM CuCl2 (Fig. 2A and 5A). SB2-Mb, however, had a much lower impact on either pmoA or mbnA expression in the absence or presence of CuCl2 (Fig. 3A and 4A), suggesting that methanobactin selectively controls gene expression in M. trichosporium OB3b. It should be stressed, however, that copper-methanobactin complexes appear to more broadly regulate gene expression in M. trichosporium OB3b. When Cu-SB2-Mb was added, mmoX expression was consistently low both in the presence and absence of CuCl2, indicating that Cu-SB2-Mb complexes could control expression of the mmo operon. Further, mbnA expression was found to be dependent on the presence of Cu-SB2-Mb; i.e., mbnA expression decreased with increasing amounts of Cu-SB2-Mb, suggesting that copper-methanobactin complexes can control methanobactin expression, possibly through a FecIRA-like system, as speculated earlier (38). Finally, Cu-SB2-Mb appeared to have relatively little effect on pmoA expression, suggesting that the control of MMO expression by methanobactin primarily targets regulation of the mmo operon.
The data presented here thus indicate that methanobactin caused differential gene expression in a methanotroph that did not produce it, and therefore methanobactin appears to be a signaling molecule. Signaling molecules are defined as substances used to either monitor cell density (i.e., quorum sensing) or the particular environmental niche a microbe inhabits to then induce specific changes in gene expression throughout the population (39). It should be noted, however, that a signaling molecule must satisfy four criteria (40): (i) the production of the signaling molecule must occur either during specific stages of growth, under definitive physiological conditions, or in response to environmental changes; (ii) the signaling molecule must accumulate extracellularly and be recognized by a specific receptor; (iii) after accumulation, the signaling molecule must induce a concerted response after some threshold concentration is reached; and (iv) the response must be more than changes required to metabolize or detoxify the signaling molecule. Methanobactin meets these requirements as follows: (i) methanobactin synthesis is tightly controlled by the availability of copper (1); (ii) methanobactin is secreted into the growth medium (21–24, 34, 41), and although the specific receptor by which methanobactin is recognized has yet to be definitively determined, it is speculated that methanobactin is recognized by a TonB-dependent transporter that is encoded upstream of the gene encoding the precursor polypeptide of methanobactin (30, 38); (iii) as shown here and in previous work (30, 31), after accumulation, methanobactin induced significant changes in expression of genes encoding sMMO polypeptides; and (iv) these genes are not the basis by which methanobactin is metabolized or detoxified [it should be noted that the biotic mechanism(s) by which methanobactin is degraded is still unknown].
To date, at least 11 general families of signaling molecules in bacteria have been identified (39, 42–50): (i) the N-acylhomoserine lactone family that is used in Gram-negative bacteria; (ii) the autoinducer oligopeptide family that is found typically in Gram-positive bacteria and also in some Gram-negative bacteria; (iii) the autoinducer-2 group, which is formed from the precursor compound 4,5-dihydroxy-2,3-pentanedione and is found in both Gram-negative and Gram-positive bacteria; (iv) the CAI-1 family, which is derived from α-hydroxyketones and found in Gram-negative bacteria; (v) the diffusible signal factor family which is derived from fatty acids and is mainly found in Gram-negative bacteria; (vi) the diketopiperazine or cyclic dipeptide family found in Gram-negative bacteria; (vii) indole found in Escherichia coli; (viii) the Pseudomonas quinolone signal (PQS) found in P. aeruginosa; (ix) an integrated quorum-sensing (IQS) signal, 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde, also found in P. aeruginosa; (x) the γ-butyrolactone family found in Streptomyces; and (xi) the family of ComX pheromones, i.e., modified peptides found in Gram-positive bacteria. Of these general families, it appears that methanobactin is most similar to ComX pheromones, but to the best of the authors' knowledge, this is the first report of any Gram-negative bacteria utilizing a modified polypeptide as a signaling molecule.
The finding that the addition of methanobactin from Methylocystis sp. strain SB2 affected specific gene expression and whole-cell activity in M. trichosporium OB3b is remarkable. It is also noteworthy that copper associated with M. trichosporium OB3b cultures increased in the presence of Cu-SB2-Mb (Fig. 5B). These data suggest that methanobactin “piracy” may occur in methanotrophic communities, as speculated earlier (31). Although the number and diversity of functions ascribed to methanobactin may seem surprising, other metal-binding compounds have been found to have similar functions. For example, the siderophore bacillibactin has been shown to bind to specific promoter regions in Bacillus subtilis and amplify expression of specific genes in this strain (51). Further, the siderophore pyoverdin has been shown not only to bind iron but also to regulate production of several virulence factors in Pseudomonas aeruginosa and act as a signaling molecule (52). Finally, it has been well documented that many microorganisms perform siderophore piracy, where siderophores produced by one microbe are stolen by another to promote its growth (53, 54).
In conclusion, we report here that methanobactin, in addition to binding copper, also serves as an interspecies signaling molecule. At this time it is unknown if methanobactin also serves as a signaling molecule to other microorganisms (e.g., ammonia-oxidizing bacteria) or how widespread the use of modified polypeptides as signaling molecules might be in Gram-negative bacteria. Such issues clearly warrant further investigation.
ACKNOWLEDGMENT
This research was supported by the Office of Science (BER) and the Advanced Research Projects Administration—Energy, U.S. Department of Energy.
REFERENCES
- 1.Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol Rev 34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 2.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 3.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 4.Haroon MF, Hu S, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 5.Op den Camp H, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 6.Dunfield PF, Yuryev A, Senin P, Smironova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain MW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–883. doi: 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- 7.Islam T, Jensen S, Reigstad LJ, Larsen Ø, Birkeland NK. 2008. Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci U S A 105:300–304. doi: 10.1073/pnas.0704162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MS, Op den Camp HJ. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874–878. doi: 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 9.van Teeseling MCF, Pol A, Harhangi HR, van der Zwart S, Jetten MSM, Op den Camp HJM, van Niftrik L. 2014. Expanding the verrucomicorbial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl Environ Microbiol 80:6782–6791. doi: 10.1128/AEM.01838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anthony C. 1982. The biochemistry of methylotrophs. Academic Press, London, United Kingdom. [Google Scholar]
- 11.Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol Rev 60:439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lontoh S, Semrau JD. 1998. Methane and trichloroethylene degradation by Methylosinus trichosporium expressing particulate methane monooxygenase. Appl Environ Microbiol 64:1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanley SH, Prior SD, Leak DJ, Dalton H. 1983. Copper stress underlies the fundamental change in intracellular location of methane monooxygenase in methane-oxidising organisms: studies in batch and continuous cultures. Biotechnol Lett 5:487–492. doi: 10.1007/BF00132233. [DOI] [Google Scholar]
- 14.DiSpirito AA, Gulledge J, Shiemke AK, Murrell JC, Lidstrom ME, Krema CL. 1992. Trichloroethylene oxidation by the membrane-associated methane monooxygenase in type I, type II, and type X methanotrophs. Biodegradation 2:151–164. doi: 10.1007/BF00124489. [DOI] [Google Scholar]
- 15.Choi D-W, Kunz R, Boyd ES, Semrau JD, Antholine WE, Han J-I, Zahn JA, Boyd JM, de la Mora A, DiSpirito AA. 2003. The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J Bacteriol 185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J-I, Semrau JD. 2004. Quantification of gene expression in methanotrophs by competitive reverse transcription-polymerase chain reaction. Environ Microbiol 6:388–399. doi: 10.1111/j.1462-2920.2004.00572.x. [DOI] [PubMed] [Google Scholar]
- 17.Karlsen OA, Berven FS, Stafford GP, Larsen Ø, Murrell JC, Jensen HB, Fjellbirkeland A. 2003. The surface-associated and secreted MopE protein of Methylococcus capsulatus (Bath) corresponds to changes in the concentration of copper in the growth medium. Appl Environ Microbiol 69:2386–2388. doi: 10.1128/AEM.69.4.2386-2388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helland R, Fjellbirkeland A, Karlsen OA, Ve T, Lillehaug JR, Jensen HB. 2008. An oxidized tryptophan facilitates copper binding in Methylococcus capsulatus-secreted protein MopE. J Biol Chem 283:13897–13904. doi: 10.1074/jbc.M800340200. [DOI] [PubMed] [Google Scholar]
- 19.Ve T, Mathisen K, Helland R, Karlsen OA, Fjellbirkeland A, Røhr ÅK, Andersson KK, Pedersen RB, Lillehaug JR, Jensen HB. 2012. The Methylococcus capsulatus (Bath) secreted protein, MopE* binds both reduced and oxidized copper. PLoS One 7:e43146. doi: 10.1371/journal.pone.0043146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behling LE, Hartsel SC, Lewis DE, DiSpirito AA, Masterson LR, Veglia G, Gallagher WH. 2008. NMR mass spectroscopy, and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. J Am Chem Soc 130:12604–12605. doi: 10.1021/ja804747d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HJ, Galeva N, Larive CK, Alterman M, Graham DW. 2005. Purification and physical-chemical properties of methanobactin: a chalkophore from Methylosinus trichosporium OB3b. Biochemistry 44:5140–5148. doi: 10.1021/bi047367r. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PMA. 2004. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science 305:1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 23.El Ghazouani A, Baslé A, Gray J, Graham DW, Firbank SJ, Dennison C. 2012. Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. Proc Natl Acad Sci U S A 109:8400–8404. doi: 10.1073/pnas.1112921109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krentz BD, Mulheron HJ, Semrau JD, DiSpirito AA, Bandow NL, Haft DH, Vuilleumier S, Murrell JC, McEllistrem MT, Hartsel SC, Gallagher WH. 2010. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry 49:10117–10130. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi DW, Zea CJ, Do YS, Semrau JD, Antholine WE, Hargrove MS, Pohl NL, Boyd ES, Geesey GG, Hartsel SC, Shafe PH, McEllistrem MT, Kisting CJ, Campbell D, Rao V, de la Mora AM, DiSpirito AA. 2006. Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry 45:1442–1453. doi: 10.1021/bi051815t. [DOI] [PubMed] [Google Scholar]
- 26.El Ghazouani A, Basle A, Firbank SJ, Knapp CW, Gray J, Graham DW, Dennison C. 2011. Copper-binding properties and structures of methanobactins from Methylosinus trichosporium OB3b. Inorg Chem 50:1378–1391. doi: 10.1021/ic101965j. [DOI] [PubMed] [Google Scholar]
- 27.Pesch M-L, Christl I, Hoffmann M, Kraemer SM, Kretzschmar R. 2012. Copper complexation of methanobactin isolated from Methylosinus trichosporium OB3b: pH-dependent speciation and modeling. J Inorg Biochem 116:55–62. doi: 10.1016/j.jinorgbio.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Bandow N, Gilles VS, Freesmeier B, Semrau JD, Krentz B, Gallagher W, McEllistrem MT, Hartsel SC, Choi DW, Hargrove MS, Heard TM, Chesner LM, Braunreiter KM, Cao BV, Gavitt MM, Hoopes JZ, Johnson JM, Polster EM, Schoenick BD, Umlauf AM, DiSpirito AA. 2012. Spectral and copper binding properties of methanobactin from the facultative methanotroph Methylocystis strain SB2. J Inorg Biochem 110:72–82. doi: 10.1016/j.jinorgbio.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Baral BS, Bandow NL, Vorobev A, Freemeier BC, Bergman BH, Herdendorf T, Fuentes N, Ellias L, Turpin E, Semrau JD, DiSpirito AA. 2014. Mercury binding by methanobactin from Methylocystis strain SB2. J Inorg Biochem 141:161–169. doi: 10.1016/j.jinorgbio.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Semrau JD, Jagadevan S, DiSpirito AA, Khalifa A, Scanlan J, Bergman BH, Freemeier BC, Baral BS, Bandow NS, Vorobev A, Haft DH, Vuilleumier S, Murrell JC. 2013. Methanobactin and MmoD work in concert to act as the “copper-switch” in methanotrophs. Environ Microbiol 15:3077–3086. doi: 10.1111/1462-2920.12150. [DOI] [PubMed] [Google Scholar]
- 31.Vorobev A, Jagadevan S, Baral BS, DiSpirito AA, Freemeier BC, Bergman BH, Bandow NL, Semrau JD. 2013. Detoxification of mercury by methanobactin from Methylosinus trichosporium OB3b. Appl Environ Microbiol 79:5918–5926. doi: 10.1128/AEM.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittenbury R, Phillips KC, Wilkinson JF. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol 61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 33.Kalidass B, Ul-Haque MF, Baral BS, DiSpirito AA, Semrau JD. 2015. Competition between metals for binding to methanobactin enables expression of soluble methane monooxygenase in the presence of copper. Appl Environ Microbiol 81:1024–2031. doi: 10.1128/AEM.03151-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandow NL, Gallagher WH, Behling L, Choi DW, Semrau JD, Hartsel SC, Gilles VS, DiSpirito AA. 2011. Isolation of methanobactin from the spent media of methane-oxidizing bacteria. Methods Enzymol 495:259–269. doi: 10.1016/B978-0-12-386905-0.00017-6. [DOI] [PubMed] [Google Scholar]
- 35.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 36.Brusseau GA, Tsien HC, Hanson RS, Wackett LP. 1990. Optimization of trichloroethylene oxidation by methanotrophs and the use of a colorimetric assay to detect soluble methane monooxygenase activity. Biodegradation 1:19–29. doi: 10.1007/BF00117048. [DOI] [PubMed] [Google Scholar]
- 37.Morton JD, Hayes KF, Semrau JD. 2000. Effect of copper speciation on whole-cell soluble methane monooxygenase activity in Methylosinus trichosporium OB3b. Appl Environ Microbiol 66:1730–1733. doi: 10.1128/AEM.66.4.1730-1733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenney GE, Rosenzweig AC. 2013. Genome mining for methanobactins. BMC Biol 11:17. doi: 10.1186/1741-7007-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan RP, Dow JM. 2008. Diffusible signals and interspecies communication in bacteria. Microbiology 154:1845–1858. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- 40.Winzer K, Hardie KR, Williams P. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr Opin Microbiol 5:216–222. doi: 10.1016/S1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- 41.DiSpirito AA, Zahn JA, Graham DW, Kim HJ, Larive CK, Derrick TS, Cox CD, Taylor A. 1998. Copper-binding compounds from Methylosinus trichosporium OB3b. J Bacteriol 180:3606–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chant EL, Summers DK. 2007. Indole signaling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol Microbiol 63:35–43. doi: 10.1111/j.1365-2958.2006.05481.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 44.Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. 2007. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318:652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- 45.Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, Chang C, Dong Y, Williams P, Zhang L-H. 2013. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol 9:339–343. doi: 10.1038/ncheMbio.1225. [DOI] [PubMed] [Google Scholar]
- 46.Okada M, Sato I, Cho SJ, Dubnau D, Sakagami Y. 2006. Chemical synthesis of ComX pheromone and related peptides containing isoprenoidal tryptophan residues. Tetrahedron 62:8907–8918. doi: 10.1016/j.tet.2006.06.074. [DOI] [Google Scholar]
- 47.Pesci EC, Milbank JBJ, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takano E. 2006. γ-Butyrolactones: Streptomyces signaling molecules regulating antibiotic production and differentiation. Curr Opin Microbiol 9:287–294. doi: 10.1016/j.mib.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Xavier KB, Miller ST, Lu W, Kim JH, Rabinowitz J, Pelczer I, Semmelhack MF, Bassler BL. 2007. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem Biol 2:128–136. doi: 10.1021/cb600444h. [DOI] [PubMed] [Google Scholar]
- 50.Yajima A. 2014. Recent progress in the chemistry and chemical biology of microbial signaling molecules: quorum-sensing pheromones and microbial hormones. Tetrahedron Lett 55:2773–2780. doi: 10.1016/j.tetlet.2014.03.051. [DOI] [Google Scholar]
- 51.Gaballa A, MacLellan S, Helmann JD. 2012. Transcription activation by the siderophore sensor Btr is mediated by ligand-dependent stimulation of promoter clearance. Nucleic Acids Res 40:3585–3595. doi: 10.1093/nar/gkr1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. 2010. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol 17:254–264. doi: 10.1016/j.cheMbiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traxler MF, Seyedsayamdost MR, Clardy J, Kolter R. 2012. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol Microbiol 86:628–644. doi: 10.1111/mmi.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]