Abstract
Small- and medium-size farms in the mid-Atlantic region of the United States use varied agricultural practices to produce leafy greens during spring and fall, but the impact of preharvest practices on food safety risk remains unclear. To assess farm-level risk factors, bacterial indicators, Salmonella enterica, and Shiga toxin-producing Escherichia coli (STEC) from 32 organic and conventional farms were analyzed. A total of 577 leafy greens, irrigation water, compost, field soil, and pond sediment samples were collected. Salmonella was recovered from 2.2% of leafy greens (n = 369) and 7.7% of sediment (n = 13) samples. There was an association between Salmonella recovery and growing season (fall versus spring) (P = 0.006) but not farming system (organic or conventional) (P = 0.920) or region (P = 0.991). No STEC was isolated. In all, 10% of samples were positive for E. coli: 6% of leafy greens, 18% of irrigation water, 10% of soil, 38% of sediment, and 27% of compost samples. Farming system was not a significant factor for levels of E. coli or aerobic mesophiles on leafy greens but was a significant factor for total coliforms (TC) (P < 0.001), with higher counts from organic farm samples. Growing season was a factor for aerobic mesophiles on leafy greens (P = 0.004), with higher levels in fall than in spring. Water source was a factor for all indicator bacteria (P < 0.001), and end-of-line groundwater had marginally higher TC counts than source samples (P = 0.059). Overall, the data suggest that seasonal events, weather conditions, and proximity of compost piles might be important factors contributing to microbial contamination on farms growing leafy greens.
INTRODUCTION
Increased awareness of the nutritional and economic benefits of eating fresh produce has caused global consumption to increase 4.5% from 1990 to 2004 (1), but field-grown foods such as vegetables and leafy greens (including lettuce, spinach, spring mix, and kale) can also serve as reservoirs of microorganisms, including bacteria, molds, and yeasts. Most of these microorganisms are not harmful and are part of the background microflora of the plant. However, human-pathogenic bacteria such as Salmonella, Listeria monocytogenes, Shigella spp., and Escherichia coli O157:H7 have been associated with foodborne outbreaks involving fresh produce (2). The ability of foodborne pathogens to colonize and persist as part of the plant microbiome as endophytes or epiphytes (reviewed in reference 3) represents a significant food safety risk, as fresh produce is often consumed raw without any processing “kill step.”
In the United States, estimates calculate approximately 4.9 million yearly incidents of food-related illnesses attributed to plant commodities, with leafy vegetables comprising 22.3% of these (4). Following the E. coli O157:H7 multistate outbreak in fall 2006, which was attributed to spinach (5), leafy greens have received significant attention from government, industry, and academic researchers. Other incidents have implicated leafy greens as a vehicle for E. coli O157:H7 transmission since the 2006 outbreak, including shredded lettuce (6), romaine lettuce (7), spinach and spring mix blend (8) and ready-to-eat salads (9).
Although the increase in foodborne disease linked to produce might be due to the increase in consumption of fresh produce or to changes in how fresh produce is processed and distributed, farm management and practices are still considered to play an important role (10). In the 2006 E. coli O157:H7 outbreak from spinach, colonization of livestock and feral swine with the implicated strain, together with harvesting practices, could have contributed to spinach contamination in the field (11). Preventing preharvest contamination is crucial, since remediation or elimination of contamination that occurs before harvest is difficult to achieve during the postharvest stage. E. coli O157:H7 can persist on leafy vegetables in the field (12), and leaf age (13, 14) and cultivar characteristics (such as leaf blade roughness) (15) have been shown to impact persistence. Another factor which likely contributes to foodborne outbreaks is use of contaminated irrigation water (16). New Jersey, one of the nation's leading producers of fresh market spinach, irrigates 19% of its cropland (which excludes pasture) (17), despite its average annual precipitation of 1,140 mm (18). This is in stark contrast to New York, which grows more than two dozen types of leafy greens, including spinach, and receives similar rainfall (average of 1,021 mm/year) but irrigates less than 2% of its cropland. The risk of using contaminated irrigation water is amplified in leafy greens production, as irrigation water is frequently applied via overhead sprinkler systems, and therefore water comes in direct contact with the edible portion of the crop, which is often consumed raw. Nonpathogenic E. coli strains have been shown to be consistently recovered from field-grown iceberg and romaine lettuce following overhead irrigation with contaminated water, but not with subsurface trickle (drip) or surface-applied furrow irrigation (19), and Salmonella has been shown to persist in the phyllosphere of greenhouse-grown parsley plants following overhead irrigation with contaminated water (20). In the mid-Atlantic (which consists of Delaware, Maryland, New Jersey, Pennsylvania, Virginia, and West Virginia [21]), surface water may be the main irrigation source available to growers (22). Bihn et al. reported that in New York more than half of surveyed fresh produce growers used surface water (23). Several studies have identified surface water as a predominant reservoir for Salmonella along the eastern coast of the United States (24–26), although Salmonella outbreaks associated with leafy greens are less frequently reported than those with other vegetables (27).
In the United States, the sales of organic foods have increased compared to those of conventional produce. Between 1997 and 2008, organic food sales quintupled from $3.6 billion to $21.1 billion, with produce and dairy accounting for over half of the total sales (28). However, organic food sales remain a small fraction of the market volume, about 3% of total food sales (28). The USDA Organic Rule implemented in 2002 included the acceptable production practices for foods marketed as organic, which largely limited the use of vegetable crop fertilizers to animal and plant wastes (29). As organic growers rely primarily on animal-derived fertilizer, it has been suggested that organically grown produce has a greater risk of pathogenic contamination than produce grown on conventional farms (30). Additionally, small- and medium-scale vegetable farms (defined as those less than 40 ha in size or with gross cash farm income of less than $999,999) (31, 32) differ from large-scale farms in their fertilization and cropping methods, harvesting and postharvest handling practices, and access to capital and labor resources. Small- and medium-size farms also tend to rely more on direct-to-consumer marketing channels, such as selling produce at local farmer's markets and through Community-Supported Agriculture programs (33). Within the mid-Atlantic states of Maryland, Delaware, and New Jersey, more than 90% (2,257 of 2,476) of fresh market vegetable farms are classified as small and medium scale, compared to 76% of farms in California (2,597 of 3,421) (31). Previous surveys report no difference in pathogen prevalence due to production scale (25) or between conventionally and organically grown produce (34, 35), although in the latter study, two “semiorganic” samples were contaminated with Salmonella (34). Our own work with tomatoes cultivated in the mid-Atlantic region found that farming system was not a significant factor for indicator bacteria or enteric pathogens (36). Data on leafy greens in the mid-Atlantic are still sparse, however. Due to differences in production scale, practices, and climate, data from other regions in the United States are not directly applicable to this region. The issue of whether organically grown leafy greens pose a greater risk for foodborne disease remains largely unresolved. Moreover, leafy greens are grown during two seasons in the mid-Atlantic, spring and fall, when the weather is wet and relatively mild, and the influence of the growing season on food safety risk is not known.
The objectives of this study were to address these data gaps by assessing pathogen prevalence and quantifying generic E. coli in leafy greens and their production areas in the mid-Atlantic region of the United States and to determine the influence of growing season, farming system, region, irrigation water source, and sampling time on food safety risk. Additionally, levels of indicator bacteria were used as a means to compare the influences of these factors on the microbiota of leafy greens and environmental samples from production areas. This survey focused on the prevalence of the pathogens Salmonella and Shiga toxin-producing E. coli (STEC) and the presence and enumeration of bacterial indicator organisms (generic E. coli, total coliforms, and aerobic mesophiles) on small- and medium-size farms growing leafy greens in the mid-Atlantic. Both organic and conventional farms in multiple states were included, together with a variety of leafy greens, irrigation water sources, and irrigation systems. This is the first extensive study to address microbial contamination of fresh leafy greens from small- and medium-size farms in the mid-Atlantic.
MATERIALS AND METHODS
Farmer recruitment.
Farms were recruited by personal invitation, through email, phone, or personal visit. In this study, farm invitation was based on several factors, including cultivation scale (small- or medium-size operations), geographical location (situated in the mid-Atlantic region), and willingness to provide samples and information about production practices. Cultivation scale can be measured in terms of acreage or annual sales, although both classification systems have limitations. For this study, small-size operations were those with less than four hectares of vegetable production (37) or gross cash farm incomes of less than $350,000 (32), while medium-size operations were those with less than 40 ha of leafy greens production (31) or gross cash farm incomes of less than $999,999 (32). No compensation was given to any participating farm or farmer. Information about on-farm production practices was obtained through email, phone, or in-person conversations with individual farmers prior to or following sample collection. Conventional and organic farms were included in the study. The term “organic” is a labeling term regulated by the USDA National Organic Program. In this paper, “organic farms” are defined as those which employed primarily or exclusively organic, agroecological, or sustainable practices, excluded or rarely used synthetic pesticides, and intentionally sought to sustain or improve soil quality. As with other agricultural research (34, 38–40), we are not referring to any particular certification criteria and include both certified and noncertified examples in our data. Farmers were recruited from four regions within three states: Maryland, Delaware, and New Jersey. In response to the diversity in regional agriculture and environmental and climatic conditions within Maryland, the state was split into two distinct sampling regions, central Maryland and Eastern Shore (east of the Chesapeake Bay). A total of 32 farms participated in this study, which consisted of 15 conventional and 17 organic operations (5 conventional and 4 organic farms in central Maryland, 1 conventional farm and 4 organic farms in Eastern Shore, 9 conventional and 4 organic farms in New Jersey, and 5 organic farms in Delaware).
Sample collection.
Samples were collected in fall 2012 and spring 2013. During the fall leafy greens growing season, participating farms were visited every 2 weeks from 24 September to 22 October; the final sampling trip was delayed until 13 November due to Hurricane Sandy (which made landfall on 29 October 2012). During the spring leafy greens growing season, participating farms were visited every week for 3 weeks from 13 May to 28 May. The following sample types were collected: foliage of leafy greenss; irrigation well, pond, or river water; pond or river sediment (based on whether a pond or river was used for irrigation); field soil; and compost (based on availability). Aseptic technique (use of latex gloves, disinfecting gloves with 70% ethanol between samples, and sterile bags) was employed during sampling. Each random leafy greens sample (approximately 25 g) consisted of “saleable” product (unsoiled internal leaves for head and cos lettuce and unsoiled leaves for loose leaf lettuce and other types of leafy greens) from random plants throughout the field collected in a serpentine pattern (random leafy greens samples [RLG]). Any leaves not touching the ground or plastic mulch could be included in an RLG. Targeted leafy greens samples (TLG) (approximately 25 g each) were not limited to “saleable” product and could include soiled and external leaves. A maximum of four leafy greens samples (2 RLG and 2 TLG) were aseptically collected into sterile Whirl-pak bags (Nasco, Fort Atkinson, WI) from each farm at each sampling trip. Preference was given to spinach during fall sampling and lettuce during spring sampling, though samples were collected based on what the participating farmer was growing for market. Irrigation well and pond and river water samples (approximately 1 liter) were collected from the source (well water tap and surface sample for pond or river) and from the end of the irrigation system (drip line or sprinkler). Well water taps and the open ends of the drip irrigation tubing and sprinkler heads were disinfected with 70% ethanol prior to sampling, and the well water taps and irrigation water systems were allowed to run freely for 1 min. Water layers were stirred before pond water collection, with efforts made not to disturb any sediment during the process. Two irrigation water samples (1 liter each) were collected into sterile Whirl-pak bags or Nalgene bottles (Thermo Scientific, Rochester, NY) from each farm at each sampling trip. Depending on availability, approximately 100 g each of field soil, pond and river sediment, and compost was collected into sterile Whirl-pak bags or Nalgene bottles from each farm, using sterile scoops (Fisher Scientific, Hampton, NH). Both field soil and compost samples consisted of mixed surface and subsurface layers. All environmental samples were placed in coolers with ice packs and transported to the lab for microbiological analysis within 24 h of collection. A total of 577 samples were collected (see Table S1 in the supplemental material). On-farm samples were collected by one team and analyzed in the laboratory by a different team, in an effort to maintain farmer confidentiality and minimize bias.
Enumeration of aerobic bacteria, generic E. coli, and total coliforms. (i) Leafy greens sample preparation.
Individual leafy greens samples were diluted at a 1:1 (wt/vol) ratio with buffered peptone water (BPW) (Becton, Dickinson and Company, Franklin Lakes, NJ) in sterile Whirl-pak filter bags (Nasco, Fort Atkinson, WI). Sample bags were palpated for 2 min at 250 rpm in a Stomacher 400 circulator (Seward Laboratory Systems Inc., Port Saint Lucie, FL) to homogenize leaf tissue. A dilution series (10−1 to 10−4) was prepared using the rinsate from each sample and 0.1% peptone water (PW) (Becton, Dickinson and Company, Franklin Lakes, NJ). A 1-ml aliquot of each dilution was then pipetted onto aerobic plate count (APC) Petrifilms (3M Global Headquarters, St. Paul, MN) for enumeration of aerobic mesophiles and onto E. coli/coliform Petrifilms (3M) (AOAC official method 991.14) for quantification of E. coli and total coliforms. Per the manufacturer recommendations, Petrifilms were incubated at 35°C for 24 h (for coliforms) or at 35°C for 48 h (for E. coli and APC) prior to counting. The limit of detection was 10 CFU/g of sample. The remaining BPW rinsate from each sample was incubated at 37°C for 24 h, followed by DNA extraction and PCR screening for Salmonella and STEC, as described in “Detection of Salmonella and STEC” below.
(ii) Field soil, compost, and sediment sample preparation.
For solid samples, 10 g of an individual sample was transferred to a sterile container and diluted 1:10 (wt/vol) with BPW. A dilution series (10−1 to 10−4) was then made with 0.1% PW, and aliquots of each dilution (1 ml each) were pipetted onto APC or E. coli/coliform 3M Petrifilms. The limit of detection was 10 CFU/g of sample. BPW enrichments were incubated for pathogen isolation as described in “Detection of Salmonella and STEC” below.
(iii) Irrigation water sample preparation.
Irrigation water samples were processed using the EPA standard membrane filtration protocol (EPA method 1604 [41]). First, 10-fold serial dilutions (ranging from 100 to 10−3 ml, depending on turbidity) were prepared. Then, aliquots of the original water sample (a 100-ml and a 250-ml aliquot) and of each dilution (a 10-ml aliquot) were passed through 0.45-μm mixed cellulose ester filters (Millipore, Billerica, MA) using a PALL filtration system (PALL Life Sciences, Ann Arbor, MI). The filters from the 100-ml and 10-ml aliquots were then aseptically removed, placed onto MI agar (Becton, Dickinson and Company, Franklin Lakes, NJ), and incubated at 37°C for 24 h, at which time blue colonies (E. coli count) were counted under ambient light conditions and fluorescent colonies (total coliform [TC] count) were counted under long-wavelength (365-nm) fluorescent light. The limit of detection was 0.01 CFU/ml of sample. The filter from the 250-ml aliquot was transferred to a sterile tube containing 40 ml BPW, vortexed, and incubated at 37°C for 24 h. Following incubation, this sample was analyzed for Salmonella and STEC using PCR, as described in “Detection of Salmonella and STEC” below.
Detection of Salmonella and STEC. (i) DNA extraction.
DNA extraction of preenrichment broths was accomplished by centrifuging 1-ml aliquots of each enriched sample at 16,000 × g for 3 min and then resuspending the pelleted cells in 100 μl of PrepMan Ultra sample preparation reagent (Applied Biosystems, Foster City, CA). The mixtures were then heated at 95°C for 10 min and centrifuged at 12,000 × g for 2 min, and 2 μl of each supernatant was removed for PCR amplification, as described below.
(ii) PCR amplification.
PCR amplification to detect the presence of Salmonella enterica was performed using species-specific primers and protocols. For Salmonella invA gene detection, we used a 25-μl PCR mixture containing 1× PCR buffer (Promega, Madison, WI), 0.2 mM each deoxynucleoside triphosphate (dNTP) (Life Technologies, Grand Island, NY), 1.5 mM MgCl2, 1 μM each primer (Integrated DNA Technologies [IDT], Coralville, IA), 0.25 μM probe, 1.5 U of GoTaq Hot Start polymerase (Promega), and 2 μl of DNA template (BPW suspension) (42). Quantitative PCR (qPCR) consisted of 45 cycles of denaturation at 95°C for 15 s followed by annealing and extension at 60°C for 30 s in an iCycler thermal cycler with the iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA). Fluorescence readings were acquired using the 6-carboxyfluorescein (FAM) channel, and the cycle threshold (CT) was obtained when the readings crossed 30 units. The limit of detection was 10 CFU/25 g or 10 CFU/25 ml with enrichment.
(iii) PCR amplification for STEC.
PCR amplification to detect the presence of Shiga toxin genes stx1 and stx2 was performed using published primers (43). Briefly, a 25-μl PCR amplification mixture containing 1× PCR buffer (Promega), 0.1 mM each dNTP (Life Technologies), 2.5 mM MgCl2, 0.5 μM each primer (Integrated DNA Technologies, Coralville, IA), 1.25 U of GoTaq Hot Start polymerase (Promega), and 2 μl of DNA template was amplified in a C1000 Touch thermal cycler (Bio-Rad). Cycling parameters consisted of 95°C for 10 min, followed by 30 cycles of denaturation at 94°C for 30 s, primer annealing at 58°C for 30 s, and extension at 72°C for 30 s and a final extension at 72°C for 5 min. PCR products were analyzed using an ethidium bromide-stained 1.5% agarose gel, observed under UV light, and documented with a Molecular Imager Gel Doc XR system (Bio-Rad). The limit of detection was 10 CFU/25 g or 10 CFU/25 ml with enrichment. E. coli O157:H7 EDL933 was used as a positive control in all PCRs.
Isolation of Salmonella and STEC.
To isolate Salmonella, 15 ml of tetrathionate (TT) Hajna broth (BD) with 1 ml of iodine solution was inoculated with 1 ml of BPW suspension from an invA PCR-positive sample and incubated at 37°C for 24 h. Then, a 10-μl aliquot was streaked onto XLT4 agar (BD), incubated at 37°C for 24 h, and evaluated for presumptive Salmonella colony growth. For STEC isolation, BPW suspensions were used following the colony hybridization method, as previously described (44).
Statistical analysis.
Data were analyzed using JMP version 10 (SAS Institute Inc., Cary, NC) for all bacterial counts. All microbial counts were log transformed to obtain a normal distribution and for statistical analyses. The data were examined both pooled and unpooled, based on growing season. Since indicator organism counts (APC, generic E. coli, and TC) for compost, sediment, and irrigation water samples were not significantly different between fall 2012 and spring 2013, data were pooled. For field soil samples, there were significant differences for all indicator organism counts based on growing season, so data were examined unpooled. For leafy greens samples, there were significant differences for APC between fall 2012 and spring 2013, so those data were examined unpooled. However, since generic E. coli and TC counts for leafy greens samples were not significantly different based on growing season, those data were pooled. Samples from individual farms were not analyzed separately. Prevalences of indicator organisms among produce, soil, and irrigation water samples from the different regions and farming systems were compared using standard and multiple chi-square tests. Statistically significant differences between sample types, region, sampling time, and farming system were determined using Student's t test. Regression analysis was used to examine correlations between indicator bacteria for a particular sample type. For all measures of association, P values of ≤0.05 were considered significant.
RESULTS
Microbiological survey of leafy greens and environmental samples.
A total of 577 samples were collected from the 32 farms sampled (see Table S1 in the supplemental material). Of the 577 samples collected, 140 were collected from central Maryland farms, 80 from Eastern Shore farms, 74 from Delaware farms, and 283 from New Jersey farms. A total of 369 leafy greens samples (191 grown using conventional practices and 178 using organic practices), 60 soil samples, 13 pond sediment samples, 11 compost samples, and 124 water samples were collected (see Table S1 in the supplemental material). Of the water samples, 93 were directly from the water source, including 66 from wells (groundwater), 23 from ponds, 2 from rivers, and 2 from the municipal water supply; the remaining 31 water samples were collected from the end of the irrigation system line. The majority of farms (26 of 32) used overhead sprinkler irrigation, with the remaining six farms using trickle (drip) irrigation.
Presence of pathogens on farms.
The prevalence of Shiga toxin genes was 0.3% (2/577) across all samples. One sample was positive for stx1, and one sample was positive for stx2. Both Shiga toxin gene-positive samples were collected from leafy greens samples (one from Delaware and the other from central Maryland). STEC was not isolated from either of the samples positive for Shiga toxin genes.
Out of 577 total samples, 4.2% (24/577) were qPCR positive for invA (Table 1). The samples comprised 15 leafy greens samples, six water samples, one compost sample, and two field soil samples. Seven farms had two or more samples that were PCR positive for invA. Growing season (fall 2012 versus spring 2013) was a significant factor for Salmonella PCR-positive leafy greens samples [χ2 (1) = 7.52, P = 0.006]. There was no significant relationship between invA presence and sample type [χ2 (4) = 1.70, P = 0.791], sampling time [χ2 (2) = 5.53, P = 0.063] or farming system [χ2 (1) = 2.09, P = 0.148] (numbers in parentheses indicate degrees of freedom). Salmonella was culture confirmed in nine of the samples positive for invA (Table 1). Eight of these positive samples were leafy greens samples collected in fall 2012 from both conventional and organic farming systems, split evenly between RLG and TLG, and each geographic region was represented (four from New Jersey, two from central Maryland, one from the Eastern Shore, and one from Delaware). The most common serotype was S. enterica serotype Mbandaka (5/8); S. Braenderup (2/8) and S. Newport (1/8) were also isolated from the leafy greens samples. A significant relationship between Salmonella presence and growing season was found [χ2 (1) = 7.54, P = 0.006]. The ninth sample from which Salmonella was isolated (S. Thompson) was a pond sediment sample collected in spring 2013 from an organic farm in New Jersey. The sediment sample was from a different New Jersey farm than the Salmonella-positive leafy greens samples from that region.
TABLE 1.
Salmonella-positive samples collected during fall 2012 and spring 2013a
| Sample no. | Date collected | Sample typeb | Region | Farming system | Farm no. |
Salmonella |
Serovar | |
|---|---|---|---|---|---|---|---|---|
| qPCR positive for invA | Culture confirmed | |||||||
| 1 | 24 September 2012 | Spinach | Delaware | Organic | DE3 | Yes | No | |
| 2 | 24 September 2012 | Field soil | Eastern Shore | Organic | ES1 | Yes | No | |
| 3 | 24 September 2012 | Bok choy | Central Maryland | Organic | MD8 | Yes | No | |
| 4 | 24 September 2012 | Spinach | New Jersey | Organic | NJ6 | Yes | No | |
| 5 | 1 October 2012 | Groundwater | New Jersey | Conventional | NJ10 | Yes | No | |
| 6 | 1 October 2012 | Groundwater | New Jersey | Conventional | NJ12 | Yes | No | |
| 7 | 9 October 2012 | Bok choy | Eastern Shore | Organic | ES1 | Yes | Yes | Newport |
| 8 | 9 October 2012 | Spinach | New Jersey | Organic | NJ7 | Yes | No | |
| 9 | 9 October 2012 | Spinach | New Jersey | Organic | NJ5 | Yes | No | |
| 10 | 9 October 2012 | Groundwater | New Jersey | Organic | NJ5 | Yes | No | |
| 11 | 15 October 2012 | Spinach | New Jersey | Conventional | NJ10 | Yes | Yes | Mbandaka |
| 12 | 15 October 2012 | Spinach | New Jersey | Conventional | NJ10 | Yes | Yes | Mbandaka |
| 13 | 15 October 2012 | Spinach | New Jersey | Conventional | NJ11 | Yes | Yes | Mbandaka |
| 14 | 15 October 2012 | Spinach | New Jersey | Conventional | NJ12 | Yes | Yes | Mbandaka |
| 15 | 15 October 2012 | Kale | Delaware | Organic | DE5 | Yes | Yes | Mbandaka |
| 16 | 22 October 2012 | Spinach | Central Maryland | Organic | MD2 | Yes | Yes | Braenderup |
| 17 | 22 October 2012 | Spinach | Central Maryland | Organic | MD8 | Yes | Yes | Braenderup |
| 18 | 13 May 2013 | Groundwater | New Jersey | Conventional | NJ11 | Yes | No | |
| 19 | 13 May 2013 | Pond water | New Jersey | Conventional | NJ14 | Yes | No | |
| 20 | 13 May 2013 | Field soil | New Jersey | Conventional | NJ15 | Yes | No | |
| 21 | 13 May 2013 | Pond water | New Jersey | Organic | NJ7 | Yes | No | |
| 22 | 20 May 2013 | Lettuce | Delaware | Organic | DE1 | Yes | No | |
| 23 | 20 May 2013 | Collards | Delaware | Organic | DE6 | Yes | No | |
| 24 | 20 May 2013 | Pond sediment | New Jersey | Organic | NJ7 | Yes | Yes | Thompson |
Total number of samples by region: 140 (central Maryland), 80 (Eastern Shore), 74 (Delaware), and 283 (New Jersey).
Leafy greens samples include both random and targeted; irrigation water samples include both source and end of line.
Indicator bacteria.
Indicator bacteria were isolated from environmental samples in each region and at each sampling date. Of the 577 total samples analyzed, 89.9% (519/577) were positive for APC, 10.2% (59/577) for generic E. coli, and 61.7% (356/577) for TC. Total samples were split almost evenly between farming systems (organic farms, n = 285; conventional farms, n = 292), though more than half (54.2%; 32/59) of samples positive for generic E. coli were collected from organic farms. As expected, sample type was a significant factor (P < 0.001) for all indicator bacteria. For organic farms, field soil samples had the highest average APC and TC counts, and compost samples had the highest generic E. coli counts (Table 2). For conventional farms, field soil samples had the highest average APC, and pond sediment samples had the highest average generic E. coli and TC counts. Compost was not available for sampling at any of the participating conventional farms. There was no significant relationship between the total number of positive samples and farming system for APC [χ2 (1) = 2.45, P = 0.118] or generic E. coli [χ2 (1) = 0.05, P = 0.826]. However, there was a significant relationship between the total number of positive samples and farming system for TC [χ2 (1) = 10.42, P = 0.001], with organic farms having more TC-positive samples (68.2%) than conventional farms (55.1%).
TABLE 2.
Average indicator bacterial counts by sample type and farming system
| Sample type | Organic farms |
Conventional farms |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of samples | Avg count, log CFU/g or log CFU/ml (% positive samples)a |
No. of samples | Avg count, log CFU/g or log CFU/ml (% positive samples) |
|||||
| APC | E. coli | Total coliforms | APC | E. coli | Total coliforms | |||
| Leafy greens | 178 | 5.71 a* (99) | 0.10 b (7) | 2.04 b* (71†) | 191 | 5.37 a* (99) | 0.12 b (6) | 1.30 a* (51†) |
| Irrigation water | 67 | 0.74 c (64) | 0.10 b (12) | 0.52 c (57) | 57 | 0.62 c (51) | 0.13 a (25) | 0.58 b (47) |
| Field soil | 26 | 5.89 a (96) | 0.48 a* (23†) | 3.00 a* (85) | 34 | 5.67 a (100) | 0.00 a* (0†) | 1.71 a* (71) |
| Compost | 11 | 5.70 ab (91) | 0.83 a (27) | 2.59 ab (91) | 0 | — | — | — |
| Pond Sediment | 3 | 4.07 b (100) | 0.54 ab (67) | 1.31 abc (67) | 10 | 4.11 b (100) | 0.16 a (30) | 2.00 a (80) |
Data for sample types having different letters were significantly different (P < 0.05) within the column. *, statistically significant difference between indicator bacterial counts of organic and conventional samples for a given sample type. †, statistically significant difference between percent positive for organic and conventional samples for a given sample type. —, no samples of that type were collected from that farming system. Data were log transformed prior to statistical analysis.
(i) Leafy greens samples.
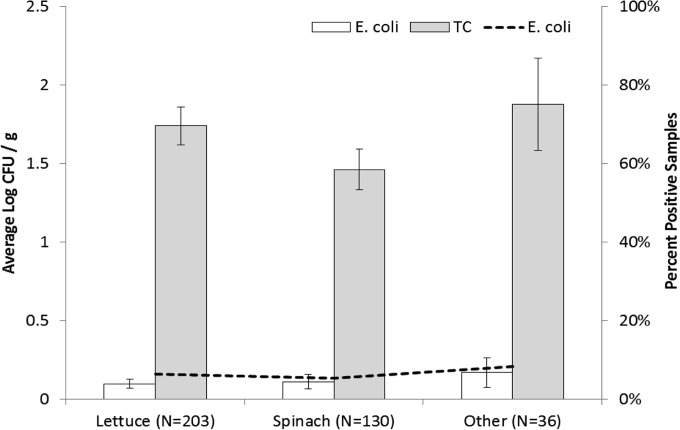
Of the 369 leafy greens samples analyzed, 98.9% (365/369) were positive for APC, 6.2% (23/369) for generic E. coli, and 61.0% (225/369) for TC. Growing season was not a significant factor for generic E. coli (P = 0.750) or TC (P = 0.163) levels, so data for leafy greens for those indicator organisms were pooled. The variety of leafy greens (spinach, lettuce, or other, which included bok choy and beet greens) was not a significant factor for generic E. coli (P = 0.693) or TC (P = 0.222) counts, and the percentage of samples positive for generic E. coli ranged from 5.4% (7/130) with spinach to 8.3% (3/36) with other (P = 0.803) (Fig. 1). Farming system was not a significant factor for generic E. coli levels (P = 0.646), and the number of positive leafy greens samples was not significantly different [χ2 (1) = 0.15, P = 0.697] between organic and conventional farms (6.7% [12/178] and 5.8% [11/191], respectively) (Table 2). However, farming system was a significant factor for TC (P < 0.001), and samples collected from organic farms had higher counts (average, 2.04 log CFU/g) than those from conventional farms (average, 1.30 log CFU/g) (Table 2). Although more TC-positive samples were collected from organic farms (71.3%) than from conventional farms (51.3%), they were not significantly different (P = 0.052). Region was not a significant factor for generic E. coli levels (P = 0.250); however, region was a significant factor for TC (P < 0.001), with central Maryland having lower average counts. Sampling time was a significant factor for both generic E. coli (P = 0.003) and TC (P = 0.003) counts, with sampling time 3 having counts similar to (generic E. coli data; average, 0.04 log CFU/g) or lower than (TC data; average, 1.26 log CFU/g) those at sampling time 1 (average, 0.06 log CFU/g and 1.81 log CFU/g, respectively).
FIG 1.
Average indicator bacterial counts (log CFU/g) (bars) and percentages of positive samples (lines) for different leafy greens by variety. Each variety consisted of random and targeted samples. “Other” includes bok choy, Swiss chard, kale, and beet greens. The dashed line indicates the percentage of positive samples for generic E. coli. Variety of leafy greens (spinach, lettuce, or other) was not a significant factor for any indicator bacteria. Data were log transformed prior to statistical analysis. Standard error bars shown.
Growing season was a significant factor for APC (P = 0.004) (fall 2012 samples had higher average counts), so data for that indicator were examined unpooled. For produce samples collected in fall 2012, leafy green variety (P = 0.259) and farming system (P = 0.236) were not significant factors for APC. However, sampling time (P < 0.001) and region (P = 0.004) were significant factors, with sampling time 3 and central Maryland having lower average counts, respectively. For produce samples collected in spring 2013, neither leafy green variety (P = 0.071), farming system (P = 0.078), nor region (P = 0.269) was a significant factor for APC. However, sampling time (P < 0.001) was a significant factor, and again, sampling time 3 had the lowest APC (average, 4.52 log CFU/g) compared to sampling times 1 (average, 5.41 log CFU/g) and 2 (average, 6.02 log CFU/g).
Bacterial counts on leafy greens were also assessed by TLG versus RLG. The majority of TLG (42.0%; 63/150) were selected because the plants had soiled leaves or the leaves were touching the ground or plastic mulch. Other common reasons for TLG selection included plants below power lines or on the edge of the field (16.0%; 24/150), diseased or unhealthy plants (8.7%; 13/150), plants near signs of animal intrusion (such as tracks, feathers, or fur) or contamination (such as scat) (8.0%; 12/104), plants in flooded areas of the field (4.7%; 7/150), plants with noticeable insect damage (4.7%; 7/150), plants near compost piles (4.0%; 6/150), or other (such as close proximity to weeds) (12.0%; 18/104). Categories of TLG were based on potential risk factors identified and/or reviewed in previous publications (10, 45–47). Type of leafy greens sample (TLG versus RLG) was not a significant factor for APC levels in fall 2012 (P = 0.864) or spring 2013 (P = 0.176) or for generic E. coli (data pooled; P = 0.084), although TLG had a higher rate of positive samples (9.3%; 14/150) than RLG (4.1%; 9/219). However, type of leafy greens sample was a significant factor for TC levels (data pooled; P = 0.027) with TLG having higher counts (average, 1.88 log CFU/g) than RLG (average, 1.50 log CFU/g) (Table 3). The highest average TC counts originated from leafy greens near compost piles (3.89 log CFU/g) and with noticeable insect damage (3.32 log CFU/g).
TABLE 3.
Average indicator bacterial counts for various targeted leafy greens samples
| Reason for TLGa | n | Avg count (log CFU/g) ± SDb |
||
|---|---|---|---|---|
| APC | Generic E. coli | Total coliforms | ||
| Soiled leavesc | 63 | 5.59 ± 1.01 | 0.12 ± 0.48 | 1.54 ± 1.45 b |
| Below power lines or on edge of field | 24 | 5.47 ± 1.64 | 0.08 ± 0.28 | 1.80 ± 1.40 ab |
| Diseased plants | 13 | 4.63 ± 1.48 | 0.23 ± 0.60 | 2.60 ± 1.77 ab |
| Signs of animalsd | 12 | 5.61 ± 0.57 | 0.22 ± 0.78 | 1.07 ± 1.42 b |
| In flooded areas | 7 | 6.70 ± 1.36 | 0.29 ± 0.76 | 2.36 ± 2.19 ab |
| Insect damage | 7 | 6.18 ± 1.00 | 0.35 ± 0.94 | 3.32 ± 2.40 ab |
| Near compost piles | 6 | 4.77 ± 2.57 | 0.35 ± 0.86 | 3.89 ± 0.49 a |
| Othere | 18 | 6.32 ± 0.85 | 0.14 ± 0.41 | 1.82 ± 1.72 ab |
| RLG | 219 | 5.50 ± 1.28 | 0.07 ± 0.41 | 1.50 ± 1.61 |
TLGs consisted of samples collected from a targeted area within the field, as specified; random leafy greens (RLG) were included for comparison.
Different letters indicate significant differences (P < 0.05) among various targeted leafy greens within the column. Reason for TLG was not significant for APC (P = 0.076) or generic E. coli (P = 0.852). Data were log transformed prior to statistical analysis.
Also includes plants with leaves touching the ground or plastic mulch.
Includes signs of animal intrusion (such as tracks or fur) and contamination (such as scat).
Includes close proximity to weeds and volunteer leafy green plants.
(ii) Irrigation water samples.
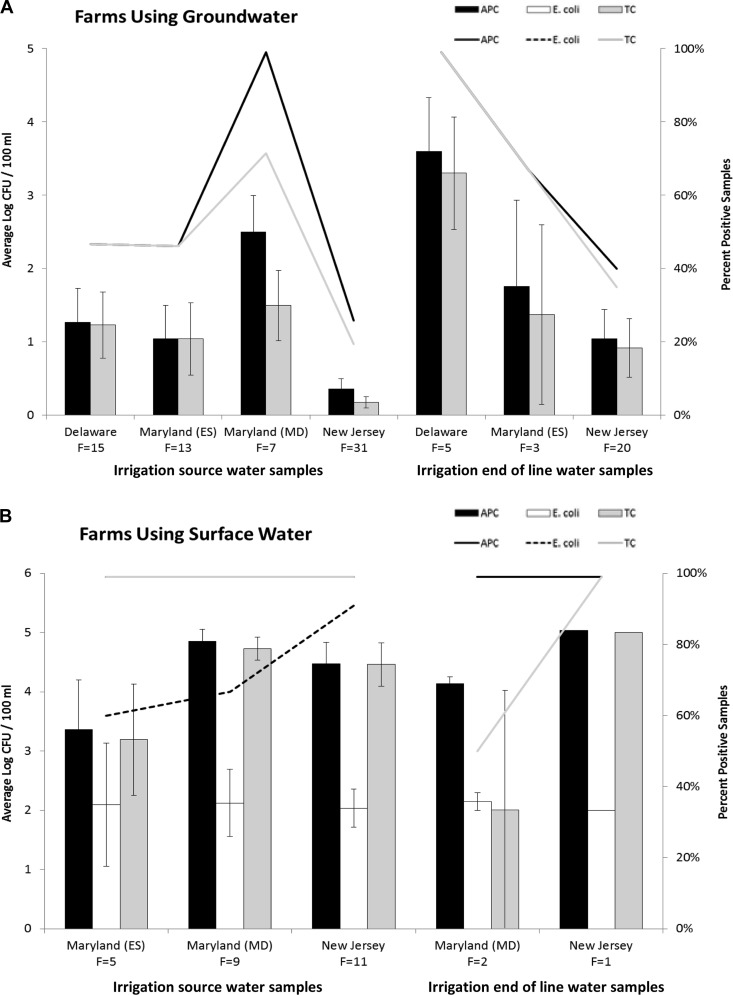
Of the 124 irrigation water samples analyzed, the majority consisted of groundwater (77.4%; 96/124), and the remaining 28 samples (22.6%) consisted of surface water (ponds, streams, and creek sources). Since U.S. municipal water supplies are chlorinated, the two municipal water samples collected from central Maryland were excluded from further data analysis. Source of irrigation water was a significant factor for all indicator bacteria (P < 0.001), and groundwater samples had lower APC, generic E. coli, and TC counts and percent positive samples than surface water samples (Fig. 2). Of the groundwater samples, 45.7% (43/94) were positive for APC, 0.0% (0/94) were positive for generic E. coli, and 40.4% (38/94) were positive for TC. Of the surface water samples, 100.0% (28/28) were positive for APC, 78.6% (22/28) were positive for generic E. coli, and 96.4% (27/28) were positive for TC. End-of-line groundwater samples were not significantly different from source groundwater samples for APC (P = 0.079) and TC (P = 0.059) levels; however, a weak effect was observed in the latter (significant at a P value of <0.1), with higher TC counts observed in end-of-line samples (1.39 log CFU/100 ml versus 0.73 log CFU/100 ml) (Fig. 2A). End-of-line samples also constituted a smaller proportion of the APC- and TC-positive groundwater samples (15/43 and 14/38, respectively) than the source samples (28/43 and 24/38). End-of-line surface water samples were not significantly different from source surface water samples in terms of APC (P = 0.943), generic E. coli (P = 0.982), or TC (P = 0.163) levels (Fig. 2B). Region was a significant factor for APC (P = 0.003) and TC (P = 0.009) in groundwater samples, with New Jersey having lower (average, 0.63 log CFU/100 ml and 0.47 log CFU/100 ml, respectively) counts than all other regions (Table 4). Sampling time was not a significant factor for any indicator bacteria for groundwater or surface water samples.
FIG 2.
(A) Average indicator bacterial counts (log CFU/100 ml) (bars) and percentages of positive samples (lines) for groundwater irrigation. (B) Average indicator bacterial counts (log CFU/100 ml) (bars) and percentages of positive samples (lines) for surface water irrigation. Groundwater included shallow and deep wells; surface water included ponds, creeks, and streams. Irrigation water samples consisted of source and end-of-line samples. F, number of individual farms represented by region. CFU per 100 ml of aerobic mesophilic bacteria (APC), generic E. coli, and total coliforms (TC) were averaged across source or end-of-line samples from farms using groundwater in Delaware (n = 15 and n = 5), eastern Maryland (ES) (n = 13 and n = 3), central Maryland (MD) (n = 7, source only), and New Jersey (n = 31 and n = 20) and across source or end-of-line samples from farms using surface water in eastern Maryland (n = 5, source only), central Maryland (n = 9 and n = 2), and New Jersey (n = 1 and n = 1). No Delaware farms using surface water participated in this study. Source of irrigation water was a significant factor (P < 0.001) for all indicator bacteria. No generic E. coli was detected in any of the groundwater samples. Data were log transformed prior to statistical analysis. Standard error bars shown.
TABLE 4.
Average aerobic mesophilic bacterial and total coliform counts in groundwater samples by regiona
| Region | Sample type | n | Avg count (log CFU/100 ml) ± SDb |
|
|---|---|---|---|---|
| Aerobic mesophilic bacteria | Total coliforms | |||
| Delaware | Source | 15 | 1.27 ± 1.75 | 1.23 ± 1.75 |
| End of line | 5 | 3.59 ± 0.74 | 3.30 ± 1.71 | |
| Total | 20 | 1.84 ± 2.00 a | 1.75 ± 1.93 a | |
| Central Maryland | Source | 7 | 2.50 ± 1.31 | 1.50 ± 1.27 |
| End of line | 0 | — | — | |
| Total | 7 | 2.50 ± 1.31 a | 1.50 ± 1.27 ab | |
| Eastern Shore | Source | 13 | 1.04 ± 1.63 | 1.04 ± 1.77 |
| End of line | 3 | 1.76 ± 2.03 | 1.37 ± 2.12 | |
| Total | 16 | 1.18 ± 1.66 ab | 1.10 ± 1.77 ab | |
| New Jersey | Source | 31 | 0.36 ± 0.78 | 0.18 ± 0.44 |
| End of line | 20 | 1.04 ± 1.04 | 0.92 ± 1.78 | |
| Total | 51 | 0.63 ± 1.30 b | 0.47 ± 1.20 b | |
Groundwater samples included shallow and deep wells; municipal water samples are omitted from this table. No generic E. coli was detected in any of the groundwater samples.
Data for regions having different letters were significantly different (P < 0.05) within the column. Data were log transformed prior to statistical analysis. —, no end-of-line groundwater samples were collected from that region.
(iii) Field soil, compost, and sediment samples.
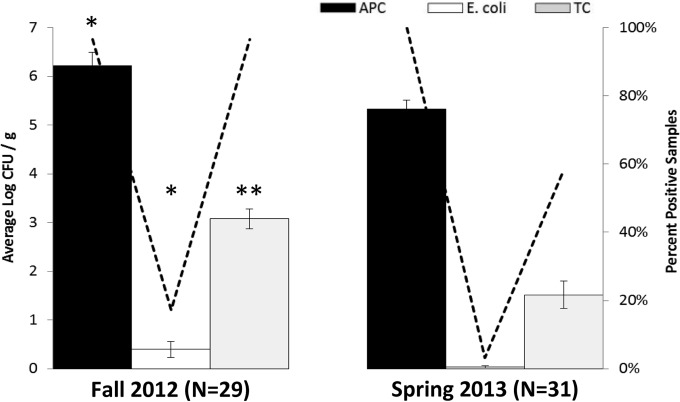
Of the 60 field soil samples analyzed, 98.3% (59/60) were positive for APC, 10.0% (6/60) for generic E. coli, and 76.7% (46/60) for TC. Growing season was a significant factor for APC (P = 0.006), generic E. coli (P = 0.031), and TC (P < 0.001), with fall 2012 field soil samples having higher average counts and generally more positive samples for all three indicator organism (Fig. 3). Farming system was a significant factor for TC (P = 0.001) in fall 2012 samples, with field soil collected from organic farms having higher counts (average, 3.56 log CFU/g) than those collected from conventional farms (average, 2.28 log CFU/g). Sampling time was a significant factor for APC in spring 2013 (P < 0.001) samples. Region was not statistically significant for any of the indicator bacteria counts for fall 2012 or spring 2013 field soil samples.
FIG 3.
Average indicator bacterial counts (log CFU/g) (bars) and percentages of positive samples (lines) for field soil by growing season. Field soil samples consisted of mixed surface and subsurface layers. The dashed line indicates the percentage of positive samples for aerobic mesophilic bacteria (APC), generic E. coli, and total coliforms (TC). Growing season (fall 2012 versus spring 2013) was a significant factor (P < 0.05) for all indicator bacteria; asterisks indicate degree of significance (*, P < 0.05; **, P < 0.001) for a given indicator organism between the two growing seasons. Data were log transformed prior to statistical analysis. Standard error bars shown.
Of the 13 pond and river sediment samples analyzed, 100% (13/13) were positive for APC, 38.5% (5/13) for generic E. coli, and 76.9% (10/13) for TC. The majority of sediment samples (69.2%; 9/13) were collected from New Jersey; the remaining samples were collected from central Maryland (3/13) and the Eastern Shore (1/13). No sediment samples were collected from Delaware.
Of the 11 compost samples analyzed, 90.9% (10/11) were positive for APC, 27.3% (3/11) for generic E. coli, and 90.9% (10/11) for TC. The majority of compost samples (54.5%; 6/11) were collected from New Jersey; the remaining samples were collected from central Maryland (2/11) and Delaware (3/11). No compost samples were collected from the Eastern Shore.
Associations among sample types.
There was a significant relationship between generic E. coli prevalence on leafy greens and sampling time [χ2 (2) = 8.45, P = 0.015] and type (TLG versus RLG) [χ2 (1) = 4.16, P = 0.041] but not between farming system [χ2 (1) = 0.15, P = 0.697], region [χ2 (3) = 6.04, P = 0.110], or variety (spinach, lettuce, or other) [χ2 (2) = 0.44, P = 0.802]. There was no association between the number of leafy greens samples that were culture confirmed for Salmonella and sampling time [χ2 (2) = 3.31, P = 0.191], region [χ2 (3) = 0.11, p = 0.991], or farming system [χ2 (1) = 0.01, p = 0.920]. However, there was an association between the number of leafy greens samples that were culture confirmed for Salmonella and growing season [χ2 (1) = 7.54, P = 0.006].
There was a significant relationship between generic E. coli prevalence in irrigation water and region [χ2 (3) = 13.11, P = 0.004] but not between sampling time [χ2 (2) = 0.41, P = 0.813] or irrigation water type (source versus end of line) [χ2 (1) = 1.96, P = 0.161]. APC was strongly correlated with TC count (P < 0.001, r2 = 0.882) for irrigation water samples but only moderately correlated with generic E. coli (P < 0.001, r2 = 0.395). TC counts were moderately correlated with E. coli counts (P < 0.001, R2 = 0.404).
For both fall 2012 and spring 2013 field soil samples, there was not a significant relationship between generic E. coli prevalence in soil and region [χ2 (3) = 2.34 (P = 0.505) and χ2 (3) = 0.30 (P = 0.960)], sampling time [χ2 (2) = 2.19 (P = 0.334) and χ2 (2) = 2.97 (P = 0.226)], or farming system [χ2 (1) = 3.69 (P = 0.055) and χ2 (1) = 0.36 (P = 0.549)]. However, TC counts in field soil were marginally correlated with APC counts (P = 0.098, r2 = 0.047) on leafy greens and were moderately correlated with TC counts (P = 0.002, r2 = 0.151) on leafy greens.
There was no association between any indicator bacterial counts in irrigation water and leafy greens or between irrigation water and field soil.
DISCUSSION
This report on the microbiological quality of leafy greens from small- and medium-size farms is, to our knowledge, the most extensive survey of its kind carried out in the mid-Atlantic region to date. Although STEC was not isolated, S. enterica was culture confirmed in 2.2% of leafy greens samples. S. Mbandaka was the most common serotype identified, and data from the Food-Borne Disease Outbreak Surveillance System indicate that among food-related illnesses caused by S. Mbandaka between 1998 and 2008, more than half were associated with plant commodities (48). Interestingly, we also isolated S. Braenderup from two of our leafy greens samples. Although this serotype is less common in foodborne outbreaks than S. Mbandaka and S. Newport, it has been associated with outbreaks involving iceberg lettuce in the United Kingdom (49). S. Newport has also caused tomato-associated outbreaks in the mid-Atlantic region (16) and has been previously isolated from other farms there (25). Only a limited number of studies have been conducted in the United States to investigate the prevalence of Salmonella and STEC on leafy greens at the farm level. Previous reports have indicated a very low level (<2%) of Salmonella contamination and largely an absence of STEC on preharvested produce, ranging from 0 to 0.3% (34, 50–53). The detection of 2.2% of Salmonella-positive leafy greens samples in this study appears to be higher than previously reported. Interestingly, all the Salmonella-positive leafy greens samples that were culture confirmed were collected in October 2012, and a significant association was found with growing season but not with farming system, region, or sampling time within a season. The recovery of Salmonella from leafy greens samples in the fall points to a significant contamination risk, possibly caused by a combination of factors. Specific seasonal events and potential pathogen sources other than those assessed could be contributing to the introduction into and persistence of foodborne pathogens in the farm ecosystem.
Although reported pathogen prevalence on produce samples tends to be low, pathogen prevalence in the agricultural environment varies. In a major leafy vegetable production region in central California, previous research has found the highest Salmonella contamination rate (7.1%) in water, followed by wildlife feces (4.3%) and soil/sediment (2.6%) (52). More recently, a two-and-a-half-year survey of farms growing leafy greens within that region found that 6.3% of water samples and 4.3% of sediment samples had detectable levels of Salmonella (54). Salmonella was also present in the majority (65%) of samples taken bimonthly at watershed sites in the vicinity of the central California leafy greens growing region (55), and in New York state, 9.2% of irrigation water samples and 2.2% of soil samples collected from fruit and vegetable farms were found to be Salmonella positive (26). In our study, only one pond sediment sample was culture confirmed for Salmonella. This low pathogen prevalence in the environment agrees with our previous survey on mid-Atlantic tomato farms performed in summer of 2012, where no Salmonella or STEC was isolated from 163 environmental samples from the same region (36).
Salmonella was isolated from spinach, kale, and bok choy on three separate sampling dates in October 2012, from organic and conventional farms geographically separated from each other in three different states. The diversity in type of leafy greens samples, on-farm production practices, and geography suggests that a factor with widespread influence, such as climate, may have played a role. Climate change and weather patterns are being recognized as drivers of transmission of human pathogens in ecosystems, as well as playing a role in pathogen survival and persistence in the environment (56, 57). In our study, most of the participating farms were impacted by Hurricane Sandy, the second most destructive tropical cyclone to hit the United States since 1900 (58). Hurricane Sandy made landfall in the mid-Atlantic on 30 October 2012, causing a catastrophic storm surge and dropping more than 8 in. of rain in Maryland, Delaware, and New Jersey as it traveled in a north-northeast direction (58). Consequently, we expected to see elevated levels of bacterial indicator organisms and human pathogens in our environmental samples collected following Hurricane Sandy, as a result of higher rainfall, damaging winds, flooding, and increased movement of wildlife across urban and agricultural landscapes. Previous research in North Carolina had found generic E. coli densities as high as 1.3 × 104 CFU/100 ml within the Pamlico Sound (an estuary which drains nearby low-lying agricultural fields) immediately following Hurricane Floyd (September 1999), as hurricane floodwaters largely replaced the estuary water (59). Over the next month as floodwaters receded, E. coli densities decreased at some collection sites but increased at others. Surprisingly, in our study, no pathogens were recovered and levels of bacterial indicator organisms and enteric pathogens were not elevated following Hurricane Sandy, despite heavy rainfall and tidal flooding. It is possible that crops in fields that receive heavy rain, but do not flood, could experience a washing effect of phyllospheric microbiota, thus explaining the absence of spikes in indicator counts following the storm. Hurricane Sandy also merged with an Arctic cold front prior to making landfall (60), and the resulting temperature fluctuations and near-freezing conditions along the East Coast (61) likely hindered bacterial growth.
In this study, sampling time was identified as a key factor affecting the carriage of bacterial indicators in produce and soil samples, as evidenced by the fact that samples collected during fall sampling time 3 (occurred between late October and early November 2012) tended to yield lower TC counts and/or APC levels and spring sampling time 3 lower APC levels. Ailes et al. observed higher concentrations of TC and higher APC on collards and spinach harvested in fall compared to winter (62), which agrees with our data from sampling time 3 during an unseasonably cold season (60). The dynamic changes in microbiological quality of produce and soil samples are attributable to weather conditions (such as temperature, precipitation patterns, humidity, and UV light) and farm practices and management (such as irrigation use, chemical inputs, and soil amendments), which all vary by season. Increased temperature and rainfall were indicated as the most important factors contributing to the persistence, survival, and spread of E. coli O157:H7 and Salmonella in produce and reservoirs, such as manure, compost, soil, and surface water (57). In a study examining factors affecting the occurrence of E. coli O157:H7 contamination in irrigation ponds, the variation among samples in E. coli O157:H7 levels, bacterial diversity, and culturable bacteria (including fecal coliforms) was associated with season (63). In this study, Salmonella was detected in leafy greens only in October, prior to Hurricane Sandy, and not during any other sampling time. This may have been due to seasonal differences in microbial diversity within the leafy greens phyllosphere community (64), impact of leaf age on Salmonella growth rates (13, 65), or local weather events prior to sampling. In the United States, 2012 was also the warmest year on record (66), and previous work on cilantro has shown that Salmonella can achieve a high population density on foliage at temperatures of 30°C (67). The warm year could have favored survival of Salmonella in the agro-environment, although no Salmonella was detected prior to October in this study. Bacterial populations in the phyllosphere frequently fluctuate, and subpopulations of the indicator bacteria we quantified may largely reflect the environmental conditions present at the time and place of sampling (reviewed in reference 68). Unfortunately, sampling time integrates multiple factors which require rigorous testing to identify, making the development of management practices that minimize the risk of enteric pathogen contamination of produce very challenging.
We also found that organic farms yielded more leafy greens and field soil samples positive for TC than conventional farms, and TC levels were significantly higher on those sample types from organic farms. Several studies have compared the prevalence of various indicator microorganisms on organic and conventional vegetables at the farm or retail level. Maffei and colleagues reported that some organic produce varieties such as lettuce, chicory, catalogna, and collard greens available in retail markets in Brazil had greater microbial counts than conventional items (69), which is similar to what Bohaychuk et al. found in Canadian farmers' markets (70). Assessment of the microbial quality of fresh produce collected from farms in the United States (34) and Spain (71) showed that organic products had significantly higher TC counts and a higher percentage of generic E. coli-positive samples. In this study, however, higher TC counts in leafy greens samples did not coincide with increases in generic E. coli or pathogen recovery (Table 2). Seeing that the adequacy of TC counts as predictors of foodborne pathogen occurrence in surface waters has been questioned as a result of weak or no correlations with enteric pathogens (72, 73), the higher TC concentrations from leafy greens obtained in this study do not support discrepancies in food safety risk as a result of organic versus conventional farming practices, since they did not correlate with higher pathogen isolation rates or generic E. coli counts. In spite of the limited value of using TC counts to assess food safety risks, this measure does indicate that farming system can impact the microbiological status of leafy greens production, which has implications for sustainable agriculture systems.
Leafy greens are easily contaminated due to large leaf surface areas and leaf topographical features, which facilitate the attachment or entrapment of microorganisms (15, 74). Although wrapper leaves that could collect more debris from the air or the ground are typically removed from head and cos lettuce before market, all leaves are marketed in many leafy green varieties such as baby spinach and baby leaf lettuce. In our study, significantly higher TC counts were obtained from leafy greens samples targeted for their proximity to compost heaps than from samples having soiled leaves or close to areas where signs of animal intrusion were present. A number of organic farms we visited performed composting of animal or plant waste on site, in close proximity to leafy greens production areas. Publications on the transmission of aerosolized enteric pathogens are sparse; however, strains of E. coli and S. enterica have been recovered at distances up to 125 m downwind from liquid hog manure application (75). Interestingly, the predominant bacterial species in aerosols may be different from those within fresh or dry manure, as was shown recently with cow manure at dairy farms (76) and beef cattle feedlots (77). Although compost piles at participating farms were separated from vegetable growing areas, maintenance (static or turned, on concrete slab or bare ground, etc.), barriers (covered or not, etc.), and environmental conditions varied, and growers were not asked to adjust their practices during the study. In this study, no confirmatory tests were conducted to track the source of TC on leaves. Further data are needed to assess risks associated with aerosolization of microorganisms and the impact of field placement and proximal surroundings and activities, including not only composting and compost storage but also animal operations and application of manure amendments to adjacent fields.
Splash-mediated soil dispersal was a potential contamination route for human-pathogenic bacteria onto fresh produce (78, 79), and the transfer of foodborne pathogens to external produce surfaces via contact with contaminated soil has been reported (80, 81). However, evidence also supports the notion that selection for phyllospheric microbial community members is largely plant regulated and less influenced by environmental factors such as water type (82) or airborne migration (83), which might explain why soiled leaves did not harbor significantly higher indicator bacterial counts. In any case, field surroundings and activities close to harvest are of concern in small- to medium-size farms, which tend to have larger production areas exposed to such risk factors. Establishment of buffer zones (a vegetative buffer zone or nonagricultural lands such as riparian forest plantings, wetlands, or grasslands) between the agricultural field(s) and potential environmental pathogen reservoirs has been demonstrated as an effective way to reduce pathogen prevalence in crops (26), but this may not be feasible for small farms with limited production acreage.
Irrigation water contaminated with high levels of enteric bacteria, viruses, protozoa, or helminths can result in increased frequency of pathogen isolations from harvested produce (63, 84, 85). The use of groundwater as an irrigation water source is common and preferred in the mid-Atlantic region, as groundwater is considered to have microbiological quality superior to that of surface water. In this study, a greater proportion of the groundwater samples positive for APC and TC were collected from the source, suggesting microbial intrusion into the well. Routine inspection and maintenance of the well casing, cap, and seals are therefore recommended to ensure structural integrity, and surface runoff should be directed away from wellheads. A marginal increase of 0.7 log CFU/100 ml in TC levels was also detected at the ends of irrigation lines delivering water to crops when groundwater was used, although the effect was weak and not significant at a P value of <0.05. This observation was also made in a previous study conducted during the summer on tomato fields in the same region, where a stronger and statistically significant effect was observed (P < 0.001) (34). This increase in microbial load in groundwater moving through irrigation lines might also be season dependent, with a more pronounced effect in the warmer summer months. Water may become contaminated by runoff from nearby livestock and poultry operations or from excessive land application of manure. Runoff may transport pathogens from the original field site of manure application to water bodies serving as irrigation sources (86, 87). Although the impact might be transient, irrigation methods (such as overhead sprinkler, surface-applied furrow, or subsurface trickle [drip] irrigation) can influence the microbiological quality of produce (88, 89). Trickle (drip) irrigation is considered to have a lower risk for contaminating produce since water does not contact the edible parts of produce; previous research has shown internalization and persistence of E. coli O157:H7 in lettuce leaves following spray irrigation with contaminated water (90). In contrast, a study using groundwater or surface water for pesticide mixing and application to tomatoes revealed no significant impacts to tomato fruit microbial communities (82).
In this study, the majority of participating farms (26 of 32) used overhead irrigation for leafy greens; however, no association was found between indicator bacterial counts in irrigation water and bacterial counts on leafy greens or between irrigation water and field soil. Regardless, practices that increase the potential risk of pathogen contamination should be reduced or eliminated. The establishment of pathogens in biofilms attached to the inside of irrigation lines is a potential risk that could lead to irrigation water contamination. Testing irrigation water at the end of irrigation lines, rather than at the source, is a simple measure that would identify any problems within irrigation lines. These findings continue to support the need for establishment of irrigation line maintenance guidelines.
Taking the findings together, this current study generated baseline microbiological data for small- and medium-scale farms growing leafy greens in the mid-Atlantic region. The presence of Salmonella in preharvest leafy greens within a specific time frame in the fall points to potential risks associated with growing season which are yet unidentified. Since leafy greens in the mid-Atlantic region are grown mainly in the spring and autumn, seasonal factors that interact with climate, such as field placement and proximal surroundings or activities might be the most important determinants contributing to microbiological inputs with food safety risk implications on small- and medium-size farms. Future work should assess these risks in a commodity-specific manner and at the local and regional scales.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Specialty Crop Research Initiative grant 2011-51181-30767 from the U.S. Department of Agriculture's National Institute of Food and Agriculture (USDA-NIFA).
The sponsor played no role in the study design and implementation. Any opinions, findings, and conclusions expressed in this material are those of the authors and do not necessarily reflect the views of the USDA-NIFA.
We thank all the farmers who participated in this study. We thank J. Nicci Coffie, Nazleen Khan, Louisa Martinez, Marie Pham, Anna Wallis, and Michael Newell for their assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00051-15.
REFERENCES
- 1.European Commission. 2007. Agricultural commodity markets: past developments—fruits and vegetables. European Commission Directorate-General for Agriculture and Rural Development, Directorate G. Economic analyses and evaluation and G.5 agricultural trade policy analysis. http://ec.europa.eu/agriculture/analysis/tradepol/worldmarkets/fruitveg/072007_en.pdf.
- 2.Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J Food Prot 67:2342–2353. [DOI] [PubMed] [Google Scholar]
- 3.Critzer FJ, Doyle MP. 2010. Microbial ecology of foodborne pathogens associated with produce. Curr Opin Biotechnol 21:125–130. doi: 10.1016/j.copbio.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities using outbreak data, United States, 1998-2008. Emerging Infect Dis 19:407–415. doi: 10.3201/eid1903.111866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. 6 October 2006. Update on multistate outbreak of E. coli O157:H7 infections from fresh spinach. http://www.cdc.gov/ecoli/2006/september/updates/100606.htm.
- 6.Sodha SV, Lynch M, Wannemuehler K, Leeper M, Malavet M, Schaffzin J, Chen T, Langer A, Glenshaw M, Hoefer D, Dumas N, Lind L, Iwamoto M, Ayers T, Nguyen T, Biggerstaff M, Olson C, Sheth A, Braden C. 2011. Multistate outbreak of Escherichia coli O157:H7 infections associated with a national fast-food chain, 2006: a study incorporating epidemiological and food source traceback results. Epidemiol Infect 139:309–316. doi: 10.1017/S0950268810000920. [DOI] [PubMed] [Google Scholar]
- 7.CDC. 2011. Multistate outbreak of E. coli O157:H7 infections linked to romaine lettuce (final update). http://www.cdc.gov/ecoli/2011/ecoliO157/romainelettuce/032312/index.html.
- 8.CDC. 2012. Multistate outbreak of Shiga toxin-producing Escherichia coli O157:H7 infections linked to organic spinach and spring mix blend (final update). http://www.cdc.gov/ecoli/2012/O157H7-11-12/index.html.
- 9.CDC. 2013. Multistate outbreak of Shiga toxin-producing Escherichia coli O157:H7 infections linked to ready-to-eat salads (final update). http://www.cdc.gov/ecoli/2013/o157h7-11-13/index.html.
- 10.Park S, Szonyl B, Gautam R, Nightingale K, Anciso J, Ivanek R. 2012. Risk factors for microbial contamination in fruits and vegetables at the preharvest level: a systematic review. J Food Prot 75:2055–2081. doi: 10.4315/0362-028X.JFP-12-160. [DOI] [PubMed] [Google Scholar]
- 11.Jay MT, Cooley M, Carychao D, Wiscomb GW, Sweitzer RA, Crawford-Miksza L, Farrar JA, Lau DK, O'Connell J, Millington A, Asmundson RV, Atwill ER, Mandrell RE. 2007. Escherichia coli O157:H7 in feral swine near spinach fields and cattle, central California coast. Emerging Infect Dis 13:1908–1911. doi: 10.3201/eid1312.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam M, Doyle MP, Phatak SC, Millner P, Jiang X. 2004. Presence of enterohemorrhagic Escherichia coli O157:H7 in soil and on leaf lettuce and parsley grown in fields treated with contaminated manure composts or irrigation water. J Food Prot 67:1365–1370. [DOI] [PubMed] [Google Scholar]
- 13.Brandl MT, Amundson R. 2008. Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl Environ Microbiol 74:2298–2306. doi: 10.1128/AEM.02459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pu S, Beaulieu JC, Prinyawiwatkul W, Ge B. 2009. Effects of plant maturity and growth media bacterial inoculum level on the surface contamination and internalization of Escherichia coli O157:H7 in growing spinach leaves. J Food Prot 72:2313–2320. [DOI] [PubMed] [Google Scholar]
- 15.Macarisin D, Patel J, Bauchan G, Giron JA, Ravishankar R. 2013. Effect of spinach cultivar and bacterial adherence factors on survival of Escherichia coli O157:H7 on spinach leaves. J Food Prot 76:1829–1837. doi: 10.4315/0362-028X.JFP-12-556. [DOI] [PubMed] [Google Scholar]
- 16.Greene SK, Daly ER, Talbot EA, Demma LJ, Holzbauer S, Patel NJ, Hill TA, Walderhaug MO, Hoekstra RM, Lynch MF, Painter JA. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol Infect 136:157–165. doi: 10.1017/S095026880700859X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.USDA ERS 2014. State fact sheets. http://www.ers.usda.gov/data-products/state-fact-sheets.aspx.
- 18.National Oceanic and Atmospheric Administration. 2013. National Climatic Data Center: climatological rankings. http://www.ncdc.noaa.gov/temp-and-precip/climatological-rankings/.
- 19.Fonseca JM, Fallon SD, Sanchez CA, Nolte KD. 2011. Escherichia coli survival in lettuce fields following its introduction through different irrigation systems. J Appl Microbiol 110:893–902. doi: 10.1111/j.1365-2672.2011.04942.x. [DOI] [PubMed] [Google Scholar]
- 20.Kisluk G, Yaron S. 2012. Presence and persistence of Salmonella enterica serotype Typhimurium in the phyllosphere and rhizosphere of spray-irrigated parsley. Appl Environ Microbiol 78:4030–4036. doi: 10.1128/AEM.00087-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ragland E, Tropp D. 2009. USDA national farmers market manager survey, 2006. USDA AMS, Washington DC. http://dx.doi.org/10.9752/MS037.05-2009.
- 22.Maryland Department of the Environment. 2006. Maryland source water assessment factsheet. http://www.mde.state.md.us/programs/water/water_supply/source_water_assessment_program/pages/programs/waterprograms/water_supply/sourcewaterassessment/factsheet.aspx.
- 23.Bihn EA, Smart CD, Hoepting CA, Worobo RW. 2013. Use of surface water in the production of fresh fruits and vegetables: a survey of fresh produce growers and their water management practices. Food Prot Trends 33:307–314. [Google Scholar]
- 24.Haley BJ, Cole DJ, Lipp EK. 2009. Distribution, diversity, and seasonality of waterborne Salmonellae in a rural watershed. Appl Environ Microbiol 75:1248–1255. doi: 10.1128/AEM.01648-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Micallef SA, Rosenberg Goldstein RE, George A, Kleinfelter L, Boyer MS, McLaughlin CR, Estrin A, Ewing L, Jean-Gilles Beaubrun J, Hanes DE, Kothary MH, Tall BD, Razeq JH, Joseph SW, Sapkota AR. 2012. Occurrence and antibiotic resistance of multiple Salmonella serotypes recovered from water, sediment and soil on mid-Atlantic tomato farms. Environ Res 114:31–39. doi: 10.1016/j.envres.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Strawn LK, Fortes ED, Bihn EA, Nightingale KK, Gröhn YT, Worobo RW, Wiedmann M, Bergholz PW. 2013. Landscape and meteorological factors affecting prevalence of three foodborne pathogens in fruit and vegetable farms. Appl Environ Microbiol 79:588–600. doi: 10.1128/AEM.02491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanning IB, Nutt JD, Ricke SC. 2009. Salmonellosis outbreaks in the United States due to fresh produce: Sources and potential intervention measures. Foodborne Pathog Dis 6:635–648. doi: 10.1089/fpd.2008.0232. [DOI] [PubMed] [Google Scholar]
- 28.Greene C, Dimitri C, Biing-Hwan L, McBridge W, Oberholtzer L, Smith T. 2009. Emerging issues in the U.S. organic industry. USDA ERS. EIB-55. USDA, Washington, DC: http://www.ers.usda.gov/media/155923/eib55_1_.pdf. [Google Scholar]
- 29.USDA AMS. 2000. National Organic Program: final rule. Docket TMD-00-02-FR. http://www.ams.usda.gov/AMSv1.0/getfile?dDocName=STELPRDC5087165.
- 30.Stephenson J. 1997. Public health experts take aim at a moving target: foodborne infections. JAMA 277:97–98. doi: 10.1001/jama.277.2.97,10.1001/jama.1997.03540260011004. [DOI] [PubMed] [Google Scholar]
- 31.USDA. 2009. Census of agriculture, 2007: United States summary and state data. Report AC-07-A-51. USDA, Washington DC. [Google Scholar]
- 32.Hoppe RA, MacDonald JM. 2013. Updating the ERS farm typology. USDA ERS. EIB-110. USDA, Washington, DC: http://www.ers.usda.gov/media/1070858/eib110.pdf. [Google Scholar]
- 33.Low SA, Vogel S. 2011. Direct and intermediated marketing of local foods in the United States. USDA ERS. ERR-128. USDA, Washington, DC: http://www.ers.usda.gov/media/138324/err128_2_.pdf. [Google Scholar]
- 34.Mukherjee A, Speh D, Dyck E, Diez-Gonzalez F. 2004. Preharvest evaluation of coliforms, Escherichia coli, Salmonella, and Escherichia coli O157:H7 in organic and conventional produce grown by Minnesota farmers. J Food Prot 67:894–900. [DOI] [PubMed] [Google Scholar]
- 35.Phillips CA, Harrison MA. 2005. Comparison of the microflora on organically and conventionally grown spring mix from a California processor. J Food Prot 68:1143–1146. [DOI] [PubMed] [Google Scholar]
- 36.Pagadala S, Marine SC, Micallef SA, Wang F, Pahl DM, Melendez MV, Kline WL, Oni RA, Walsh CS, Everts KL, Buchanan RL. 13 December 2014. Assessment of region, farming system, irrigation source and sampling time as food safety risk factors for tomatoes. Int J Food Microbiol doi: 10.1016/j.ijfoodmicro.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Newton DJ. 2014. Working the land with 10 acres: small acreage farming in the United States. USDA ERS. EIB-123. USDA, Washington, DC: http://www.ers.usda.gov/media/1391684/eib123_summary.pdf. [Google Scholar]
- 38.Kremen A, Greene C, Hanson J. 2004. Organic produce, price premiums, and eco-labeling in U.S. farmers' markets. USDA ERS. VGS-301-01. USDA, Washington, DC: http://www.ers.usda.gov/media/269468/vgs30101_1_.pdf. [Google Scholar]
- 39.Badgley C, Moghtader J, Quintero E, Zakem E, Jahi Chappell M, Avilés-Vázquez K, Samulon A, Perfecto I. 2007. Organic agriculture and the global food supply. Renew Agric Food Syst 22:86–108. doi: 10.1017/S1742170507001640. [DOI] [Google Scholar]
- 40.Seufert V, Ramankutty N, Foley JA. 2012. Comparing the yields of organic and conventional agriculture. Nature 485:229–232. doi: 10.1038/nature11069. [DOI] [PubMed] [Google Scholar]
- 41.EPA. 2002. Method 1604: total coliforms and Escherichia coli in water by membrane filtration using simultaneous detection technique (MI medium). Report EPA 821-R-02-024. Office of Science and Technology, EPA, Washington DC. [Google Scholar]
- 42.Cheng CM, Lin W, Van KT, Phan L, Tran NN, Farmer D. 2008. Rapid detection of Salmonella in foods using real-time PCR. J Food Prot 71:2436–2441. [DOI] [PubMed] [Google Scholar]
- 43.Xia X, Meng J, McDermott PF, Ayers S, Blickenstaff K, Tran TT, Abbott J, Zheng J, Zhao S. 2010. Presence and characterization of Shiga toxin-producing Escherichia coli and other potentially diarrheagenic E. coli in retail meats. Appl Environ Microbiol 76:1709–1717. doi: 10.1128/AEM.01968-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ju W, Shen J, Li Y, Toro MA, Zhao S, Ayers S, Najjar MB, Meng J. 2012. Non-O157 Shiga toxin-producing Escherichia coli in retail ground beef and pork in the Washington D.C. area. Food Microbiol 32:371–377. doi: 10.1016/j.fm.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 45.Barker-Reid F, Harapas D, Engleitner S, Kreidl S, Holmes R, Faggian R. 2009. Persistence of Escherichia coli on injured iceberg lettuce in the field, overhead irrigated with contaminated water. J Food Prot 72:458–464. [DOI] [PubMed] [Google Scholar]
- 46.Talley JL, Wayadande AC, Wasala LP, Gerry AC, Fletcher J, DeSilva U, Gilliland SE. 2009. Association of Escherichia coli O157:H7 with filth flies (Muscidae and Calliphoridae) captured in leafy greens fields and experimental transmission of E. coli O157:H7 to spinach leaves by house flies (Diptera: Muscidae). J Food Prot 72:1547–1552. [DOI] [PubMed] [Google Scholar]
- 47.DeNiro JL. 2013. Airborne transport of foodborne pathogens from bovine manure to vegetable surfaces. Master's thesis The Ohio State University, Columbus, OH: https://etd.ohiolink.edu/ap/10?0::NO:10:P10_ACCESSION_NUM:osu1376925440. [Google Scholar]
- 48.Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. 2013. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998-2008. Emerg Infect Dis 19:1239–1244. doi: 10.3201/eid1908.121511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gajraj R, Pooransingh S, Hawker JI, Olowokure B. 2012. Multiple outbreaks of Salmonella braenderup associated with consumption of iceberg lettuce. Int J Environ Health Res 22:150–155. doi: 10.1080/09603123.2011.613114. [DOI] [PubMed] [Google Scholar]
- 50.Duffy EA, Lucia LM, Kells JM, Castillo A, Pillai SD, Acuff GR. 2005. Concentrations of Escherichia coli and genetic diversity and antibiotic resistance profiling of Salmonella isolated from irrigation water, packing shed equipment, and fresh produce in Texas. J Food Prot 68:70–79. [DOI] [PubMed] [Google Scholar]
- 51.Mukherjee A, Speh D, Jones AT, Buesing KM, Diez-Gonzalez F. 2006. Longitudinal microbiological survey of fresh produce grown by farmers in the upper midwest. J Food Prot 69:1928–1936. [DOI] [PubMed] [Google Scholar]
- 52.Gorski L, Parker CT, Liang A, Cooley MB, Jay-Russell MT, Gordus AG, Atwill ER, Mandrell RE. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl Environ Microbiol 77:2734–2748. doi: 10.1128/AEM.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooley MB, Jay-Russell M, Atwill ER, Carychao D, Nguyen K, Quiñones B, Patel R, Walker S, Swimley M, Pierre-Jerome E, Gordus AG, Mandrell RE. 2013. Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS One 8:e65716. doi: 10.1371/journal.pone.0065716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamin L, Atwill ER, Jay-Russell M, Cooley M, Carychao D, Gorski L, Mandrell RE. 2013. Occurrence of generic Escherichia coli, E coli O157 and Salmonella spp in water and sediment from leafy green produce farms and streams on the Central California coast. Int J Food Microbiol 165:65–76. doi: 10.1016/j.ijfoodmicro.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Cooley MB, Quiñones B, Oryang D, Mandrell RE, Gorski L. 2014. Prevalence of Shiga toxin-producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front Cell Infect Microbiol 4:30. doi: 10.3389/fcimb.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tirado MC, Clarke R, Jaykus LA, McQuatters-Gollop A, Frank JM. 2010. Climate change and food safety: a review. Food Res Int 43:1745–1765. doi: 10.1016/j.foodres.2010.07.003. [DOI] [Google Scholar]
- 57.Liu C, Hofstra N, Franz E. 2013. Impacts of climate change on the microbial safety of pre-harvest leafy green vegetables as indicated by Escherichia coli O157 and Salmonella spp. Int J Food Microbiol 163:119–128. doi: 10.1016/j.ijfoodmicro.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 58.Blake ES, Kimberlain TB, Berg RJ, Cangialosi JP, Beven JL II. 2013. Tropical cyclone report: Hurricane Sandy (AL 182012). National Weather Service, National Hurricane Center; http://www.nhc.noaa.gov/data/tcr/AL182012_Sandy.pdf. [Google Scholar]
- 59.Bales JD. 2003. Effects of Hurricane Floyd inland flooding, September-October 1999, on tributaries to Pamlico Sound, North Carolina. Estuaries 26:1319–1328. doi: 10.1007/BF02803634. [DOI] [Google Scholar]
- 60.Halverson JB, Rabenhorst T. 2013. Hurricane Sandy: the science and impacts of a superstorm. Weatherwise 66:14–23. doi: 10.1080/00431672.2013.762838. [DOI] [Google Scholar]
- 61.Kunz M, Mühr B, Kunz-Plapp T, Daniell JE, Khazai B, Wenzel F, Vannieuwenhuyse M, Comes T, Elmer F, Schröter K, Fohringer J, Münzberg T, Lucas C, Zschau J. 2013. Investigation of Superstorm Sandy 2012 in a multi-disciplinary approach. Nat Hazards Earth Syst Sci 13:2579–2598. doi: 10.5194/nhess-13-2579-2013. [DOI] [Google Scholar]
- 62.Ailes EC, Leon JS, Jaykus L-A, Johnston LM, Clayton HA, Blanding S, Kleinbaum DG, Backer LC, Moe CL. 2008. Microbial concentrations on fresh produce are affected by postharvest handling, importation, and season. J Food Prot 71:2389–2397. [DOI] [PubMed] [Google Scholar]
- 63.Gu G, Luo Z, Cevallos-Cevallos JM, Adams P, Vellidis G, Wright A, van Bruggen AH. 2013. Factors affecting the occurrence of Escherichia coli O157 contamination in irrigation ponds on produce farms in the Suwannee River watershed. Can J Microbiol 59:175–182. doi: 10.1139/cjm-2012-0599. [DOI] [PubMed] [Google Scholar]
- 64.Williams TR, Moyne A-L, Harris LJ, Marco ML. 2013. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS One 8:e68642. doi: 10.1371/journal.pone.0068642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kroupitski Y, Pinto R, Belausov E, Sela S. 2011. Distribution of Salmonella typhimurium in romaine lettuce leaves. Food Microbiol 28:990–997. doi: 10.1016/j.fm.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 66.Crouch J, Heim RR Jr, Hughes P, Fenimore C. 2013. Regional climates: North America. Bull Am Meteor Soc 94:S147–S152. [Google Scholar]
- 67.Brandl MT, Mandrell RE. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl Environ Microbiol 68:3614–3621. doi: 10.1128/AEM.68.7.3614-3621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whipps JM, Hand P, Pink D, Bending GD. 2008. Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol 105:1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- 69.Maffei DF, Silveira NF, Catanozi M. 2013. Microbiological quality of organic and conventional vegetables sold in Brazil. Food Control 29:226–230. doi: 10.1016/j.foodcont.2012.06.013. [DOI] [Google Scholar]
- 70.Bohaychuk VM, Bradbury RW, Dimock R, Fehr M, Gensler GE, King RK, Rieve R, Romero Barrios P. 2009. A microbiological survey of selected Alberta-grown fresh produce from farmers' markets in Alberta, Canada. J Food Prot 72:415–420. [DOI] [PubMed] [Google Scholar]
- 71.Oliveira M, Usall J, Vinas I, Anguera M, Gatius F, Abadias M. 2010. Microbiological quality of fresh lettuce from organic and conventional production. Food Microbiol 27:679–684. doi: 10.1016/j.fm.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 72.Economou V, Gousia P, Kansouzidou A, Sakkas H, Karanis P, Papadopoulou C. 2013. Prevalence, antimicrobial resistance and relation to indicator and pathogenic microorganisms of Salmonella enterica isolated from surface waters within an agricultural landscape. Int J Hyg Environ Health 216:435–444. doi: 10.1016/j.ijheh.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 73.McEgan R, Mootian G, Goodridge LD, Schaffner DW, Danyluk MD. 2013. Predicting Salmonella populations from biological, chemical, and physical indicators in Florida surface waters. Appl Environ Microbiol 79:4094–4105. doi: 10.1128/AEM.00777-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leben C. 1988. Relative humidity and the survival of epiphytic bacteria with buds and leaves of cucumber plants. Phytopathology 78:179–185. doi: 10.1094/Phyto-78-179. [DOI] [Google Scholar]
- 75.Hutchison ML, Avery SM, Monaghan JM. 2008. The air-borne distribution of zoonotic agents from livestock waste spreading and microbiological risk to fresh produce from contaminated irrigation sources. J Appl Microbiol 105:848–857. doi: 10.1111/j.1365-2672.2008.03811.x. [DOI] [PubMed] [Google Scholar]
- 76.Ravva SV, Sarreal CZ, Mandrell RE. 2011. Bacterial communities in aerosols and manure samples from two different dairies in Central and Sonoma Valleys of California. PLoS One 6:e17281. doi: 10.1371/journal.pone.0017281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Durso LM, Harhay GP, Smith TPL, Bono JL, DeSantis TZ, Clawson ML. 2011. Bacterial community analysis of beef cattle feedlots reveals that pen surface is distinct from feces. Foodborne Pathog Dis 8:647–649. doi: 10.1089/fpd.2010.0774. [DOI] [PubMed] [Google Scholar]
- 78.Girardin H, Morris CE, Albagnac C, Dreux N, Glaux C, Nguyen-The C. 2005. Behavior of the pathogen surrogates Listeria innocua and Clostridium sporogenes during production of parsley in fields fertilized with contaminated amendments. FEMS Microbiol Ecol 54:287–295. doi: 10.1016/j.femsec.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 79.Monaghan JM, Hutchison ML. 2012. Distribution and decline of human pathogenic bacteria in soil after application in irrigation water and the potential for soil-splash-mediated dispersal onto fresh produce. J Appl Microbiol 112:1007–1019. doi: 10.1111/j.1365-2672.2012.05269.x. [DOI] [PubMed] [Google Scholar]
- 80.Guo X, Chen J, Brackett RE, Beuchat LR. 2002. Survival of Salmonella on tomatoes stored at high relative humidity, in soil, and on tomatoes in contact with soil. J Food Prot 65:274–279. [DOI] [PubMed] [Google Scholar]
- 81.Ibekwe MA, Grieve CM, Papiernik SK, Yang C-H. 2009. Persistence of Escherichia coli O157:H7 on the rhizosphere and phyllosphere of lettuce. Lett Appl Microbiol 49:784–790. doi: 10.1111/j.1472-765X.2009.02745.x. [DOI] [PubMed] [Google Scholar]
- 82.Telias A, White JR, Pahl DM, Ottesen AR, Walsh CS. 2011. Bacterial community diversity and variation in spray water sources and the tomato fruit surface. BMC Microbiol 11:81. doi: 10.1186/1471-2180-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. 2014. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio 5(1):e00682-13. doi: 10.1128/mBio.00682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilkes G, Edge T, Gannonc V, Jokinen C, Lyautey E, Medeiros D, Neumann N, Ruecker N, Topp E, Lapen DR. 2009. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within an agricultural landscape. Water Res 43:2209–2223. doi: 10.1016/j.watres.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 85.Edge TA, El-Shaarawi A, Gannon V, Jokinen C, Kent R, Khan IU, Koning W, Lapen D, Miller J, Neumann N, Phillips R, Robertson W, Schreier H, Scott A, Shtepani I, Topp E, Wilkes G, van Bochove E. 2012. Investigation of an Escherichia coli environmental benchmark for waterborne pathogens in agricultural watersheds in Canada. J Environ Qual 41:21–30. doi: 10.2134/jeq2010.0253. [DOI] [PubMed] [Google Scholar]
- 86.Thurston-Enriquez JA, Gilley JE, Eghball B. 2005. Microbial quality of runoff following land application of cattle manure and swine slurry. J Water Health 3:151–171. [PubMed] [Google Scholar]
- 87.Chen Z, Jiang X. 2014. Microbiological safety of chicken litter or chicken litter-based organic fertilizer: a review. Agriculture 4:1–29. doi: 10.3390/agriculture4010001. [DOI] [Google Scholar]
- 88.Solomon EB, Potenski CJ, Matthews KR. 2002. Effect of irrigation method on transmission to and persistence of Escherichia coli O157:H7 on lettuce. J Food Prot 65:673–676. [DOI] [PubMed] [Google Scholar]
- 89.Song I, Stine SW, Choi CY, Gerba CP. 2006. Comparison of crop contamination by microorganisms during subsurface drip and furrow irrigation. J Environ Eng 132:1243–1248. doi: 10.1061/(ASCE)0733-9372(2006)132:10(1243). [DOI] [Google Scholar]
- 90.Erickson MC, Webb CC, Diaz-Perez JC, Carlos J, Phatak SC, Silvoy JJ, Davey L, Payton AS, Liao J, Ma L, Doyle MP. 2010. Surface and internalized Escherichia coli O157:H7 on field-grown spinach and lettuce treated with spray-contaminated irrigation water. J Food Prot 73:1023–1029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.