ABSTRACT
LytM proteins belong to a family of bacterial metalloproteases. In Gram-negative bacteria, LytM factors are mainly reported to have a direct effect on cell division by influencing cleavage and remodeling of peptidoglycan. In this study, mining nontypeable Haemophilus influenzae (NTHI) genomes, three highly conserved open reading frames (ORFs) containing a LytM domain were identified, and the proteins encoded by the ORFs were named YebA, EnvC, and NlpD on the basis of their homology with the Escherichia coli proteins. Immunoblotting and confocal analysis showed that while NTHI NlpD is exposed on the bacterial surface, YebA and EnvC reside in the periplasm. NTHI ΔyebA and ΔnlpD deletion mutants revealed an aberrant division phenotype characterized by an altered cell architecture and extensive membrane blebbing. The morphology of the ΔenvC deletion mutant was identical to that of the wild-type strain, but it showed a drastic reduction of periplasmic proteins, including the chaperones HtrA, SurA, and Skp, and an accumulation of β-barrel-containing outer membrane proteins comprising the autotransporters Hap, IgA serine protease, and HMW2A, as observed by proteomic analysis. These data suggest that EnvC may influence the bacterial surface protein repertoire by facilitating the passage of the periplasmic chaperones through the peptidoglycan layer to the close vicinity of the inner face of the outer membrane. This hypothesis was further corroborated by the fact that an NTHI envC defective strain had an impaired capacity to adhere to epithelial cells and to form biofilm. Notably, this strain also showed a reduced serum resistance. These results suggest that LytM factors are not only important components of cell division but they may also influence NTHI physiology and pathogenesis by affecting membrane composition.
IMPORTANCE
Nontypeable Haemophilus influenzae (NTHI) is an opportunistic pathogen that colonizes the human nasopharynx and can cause serious infections in children (acute otitis media) and adults (chronic obstructive pulmonary disease). Several virulence factors are well studied, but the complete scenario of NTHI pathogenesis is still unclear. We identified and characterized three NTHI LytM factors homologous to the Escherichia coli LytM proteins. Although LytM factors are reported to play a crucial role in the cell division process, in NTHI they are also involved in other bacterial functions. In particular, YebA and NlpD are fundamental for membrane stability: indeed, their absence causes an increased release of outer membrane vesicles (OMVs). On the other hand, our data suggest that EnvC could directly or indirectly affect peptidoglycan permeability and consequently, bacterial periplasmic and outer membrane protein distribution. Interestingly, by modulating the surface composition of virulence determinants, EnvC also has an impact on NTHI pathogenesis.
INTRODUCTION
Bacterial metalloproteases are essential factors mainly involved in the hydrolysis of large polypeptide substrates for the recruitment of peptide nutrients (1), but they can also influence many other crucial aspects of bacterial life from metabolism to host colonization and from cell division to immune evasion (2–4). The fundamental role of metalloproteases in bacterial physiology prompted many researchers to select them as potential vaccine antigens and to test their protective capability in order to develop new vaccines (5–7).
Metalloproteases can be recognized by the presence of a short conserved signature sequence containing histidine and glutamate residues. The most common motif is HEXXH (zincins), but other motifs, such as HXXEH (inverzincins), HXXE (carboxypeptidase family), and HXH (e.g., lysostaphin-like), have also been reported (2). Lysostaphin-like metalloproteases (LytM proteins) belong to the M23 peptidase family and are found in bacteriophages and in Gram-positive and Gram-negative bacteria. The catalytic LytM domain (8) was identified for the first time in a secreted autolysin from Staphylococcus aureus (9). The physiological function of LytM-like proteins in Gram-positive bacteria has not been completely unraveled, even though they are known to cleave the pentaglycine bridges in peptidoglycan (PG) (10, 11). On the other hand, although not all the LytM proteins show a peptidase activity (12–14), their role in cell division and cell elongation has been established (15). In Escherichia coli, three LytM proteins (EnvC, NlpD, and YebA) are known to be involved in the cell division process (15). Strains lacking the divisome-associated LytM factors EnvC and NlpD show defects in cell splitting (15). It has been reported that these LytM proteins are potent and specific activators of PG hydrolysis by the amidases (16). YebA appears to play minor roles in cell separation, while it is likely to participate in other aspects of PG biogenesis (15). Instead, it maintains an active role in peptidoglycan cleavage to facilitate murein growth and enlargement (17). Consistent with this idea, both EnvC and NlpD are specifically recruited to the division site, whereas YebA shows a more dispersed peripheral localization pattern (15). LytM factors are also found in pathogenic bacteria, and they are involved not only in cell division but also in pathogenesis. In particular, HdpA from Helicobacter pylori has a role in the regulation of cell morphology and in colonization (18), NG1686 from Neisseria gonorrhoeae is important for resistance to hydrogen peroxide and polymorphonuclear neutrophil-mediated killing (19), while NlpD is essential for the development of bubonic and pneumonic plague in Yersinia pestis (7).
Although LytM metalloproteases are well characterized in E. coli and in other bacteria, not much information is available about their role in Haemophilus influenzae. In this study, through the screening of nontypeable H. influenzae (NTHI) genome sequences from public databases, we identified three open reading frames (ORFs) containing a LytM domain.
Detailed analysis on the role of the three newly identified LytM proteins was carried out in order to evaluate their contribution to cell division and pathogenesis of NTHI.
RESULTS
Three newly identified LytM factors in nontypeable Haemophilus influenzae.
By bioinformatic analysis, we identified three ORFs present in the NTHI 176 strain genome sequence that contain canonical features of LytM bacterial metalloproteases. The three proteins encoded by the ORFs were named YebA, EnvC, and NlpD on the basis of their homology with the E. coli proteins. Amino acid sequence analysis indicated that a catalytic LytM domain was localized at the C terminus in all three proteins, whereas a central LysM domain, recognized as responsible for binding to peptidoglycan (PG) (20), was present only in the YebA and NlpD proteins. Moreover, the presence of an N-terminal signal peptide sequence in each protein suggests a putative extracytoplasmic localization (Fig. 1, left). The genes were present in 23 publicly available NTHI genomes, and the related proteins were highly conserved at the amino acid sequence level (see Table S1 in the supplemental material). As expected, NTHI YebA, EnvC, and NlpD showed significant homology with a number of previously characterized LytM proteins expressed by other Gram-negative bacteria (Table S2). In particular, E. coli YebA, EnvC, and NlpD showed sequence identities of 49%, 40%, and 43% with the respective NTHI homologues. However, most of this similarity resides in the catalytic LytM domain (up to 79% amino acid identity [Fig. 1, right]). It is interesting that the typical M23 metalloprotease metal binding sites (HXXXD and HXH) were present only in YebA, whereas only few critical residues were conserved in NlpD and these motifs were completely absent in EnvC (Fig. 1, right).
FIG 1 .
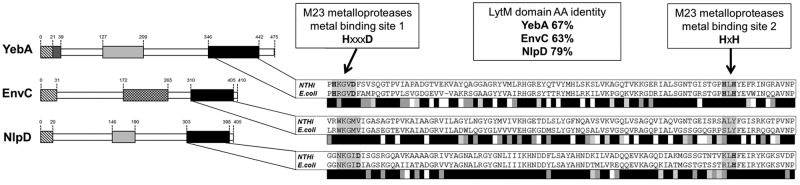
In silico analysis of NTHI LytM proteins. (Left) Domain organization of LytM proteins. LytM domains (black), LysM domains (light grey), transmembrane regions (dark grey), signal sequences (hatched boxes), and coiled coil regions (box with checkerboard pattern) are indicated. (Right) Alignments of LytM domains from NTHI and E. coli proteins are shown. Grey scale bars summarize results of LytM domain alignments of NTHI and E. coli proteins: black indicates positions that have fully conserved residues, dark grey indicates conservation between groups of strongly similar properties (scores of >0.5 in the Gonnet PAM 250 matrix), light grey indicates conservation between groups of weakly similar properties (score of <0.5 in the Gonnet PAM 250 matrix), and white indicates residues that were not conserved. AA, amino acid.
NTHI LytM proteins are localized in different cell compartments.
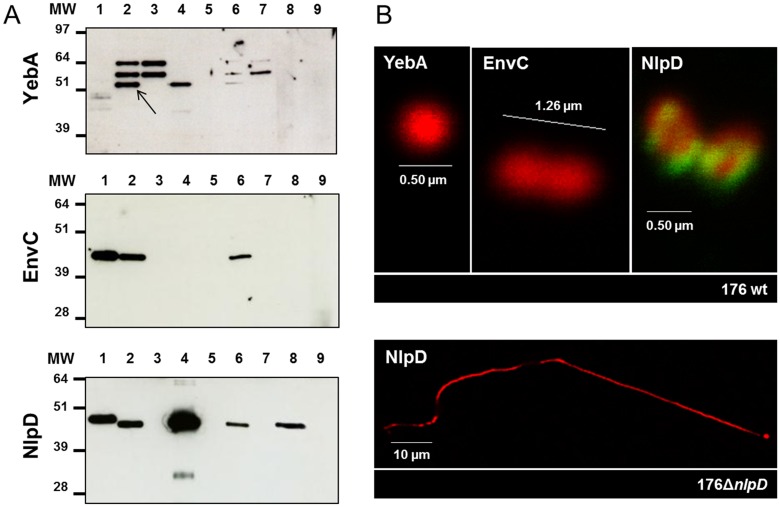
The presence of a typical leader peptide sequence suggests that NTHI LytM proteins are likely to be exported from the cytoplasm. In order to verify their expression and subcellular localization, periplasmic, outer membrane, and supernatant fractions from wild-type (wt) strain 176 and isogenic mutants generated by single-gene deletion were isolated. Cellular fractions were tested by immunoblotting using specific sera raised against each of the recombinant LytM proteins. As shown in Fig. 2A, YebA was mainly detected in the enriched outer membrane protein fraction and EnvC was mainly detected in the periplasmic fraction, while NlpD was present in both the membrane-associated and secreted fractions. No corresponding bands were detected in the respective knockout strains used as negative controls (Fig. 2A). The genuine nature of the different fractions was confirmed by testing the same samples with appropriate controls (see Fig. S1 in the supplemental material). Confocal immunofluorescence microscopy of bacteria stained with specific antibodies revealed no signal for YebA and EnvC proteins (Fig. 2B), indicating that the proteins are not exposed on the bacterial surface, while NlpD protein was clearly detected on the NTHI surface, and interestingly, mainly localized at specific foci, close to the division septum (Fig. 2B). No corresponding signal was detected in the nlpD knockout strain used as a negative control (Fig. 2B).
FIG 2 .
Expression and subcellular localization of NTHI LytM factors. (A) Western blot analyses on different cell compartment extracts were performed using specific antisera raised against YebA, EnvC, and NlpD. Bis-Tris 10% acrylamide gels were loaded as follows. Lanes: 1, recombinant protein; 2, whole-cell extract from wild-type (wt) strain 176; 3, whole-cell extract from knockout strain; 4, OMP from wt strain; 5, OMP from knockout strain; 6, periplasmic fraction from wt strain; 7, periplasmic fraction from knockout strain; 8, supernatant from wt strain; 9, supernatant from knockout strain. Antisera cross-react with other nonspecific bands that were not characterized. The black arrow indicates a specific signal for YebA. The positions of molecular weight (MW) markers (in thousands) are shown to the left of the gels. (B) Immunofluorescence microscopy analysis of wild-type strain 176 to detect surface localization of YebA, EnvC, and NlpD of LytM factors. Bacteria were stained red (antibody to the bacterium and a secondary fluorescent antibody), and LytM proteins were stained green (anti-LytM and a secondary fluorescent antibody). 176ΔnlpD mutant was used as a negative control to verify the absence of NlpD on the surface.
Deletion of yebA and nlpD genes leads to an aberrant cellular phenotype.
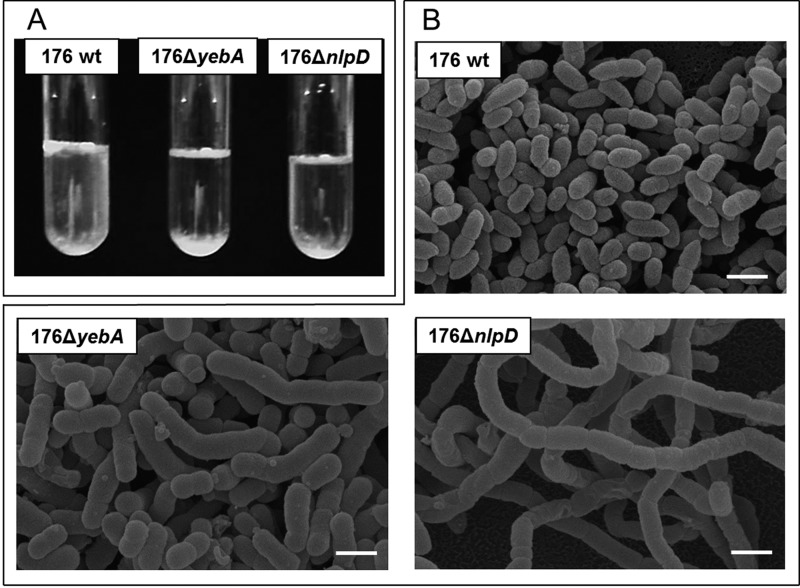
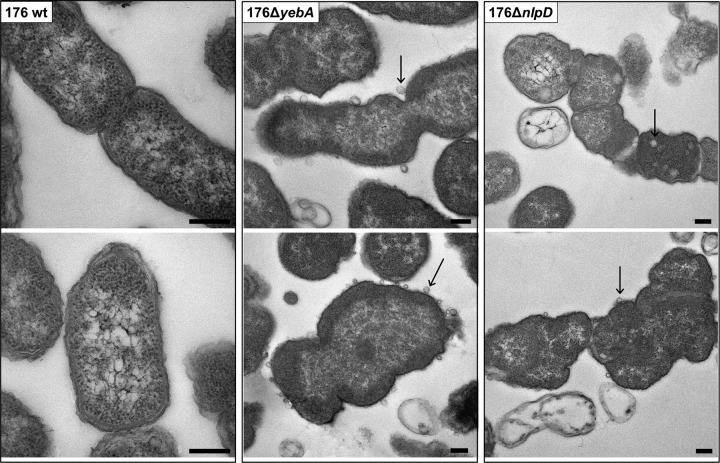
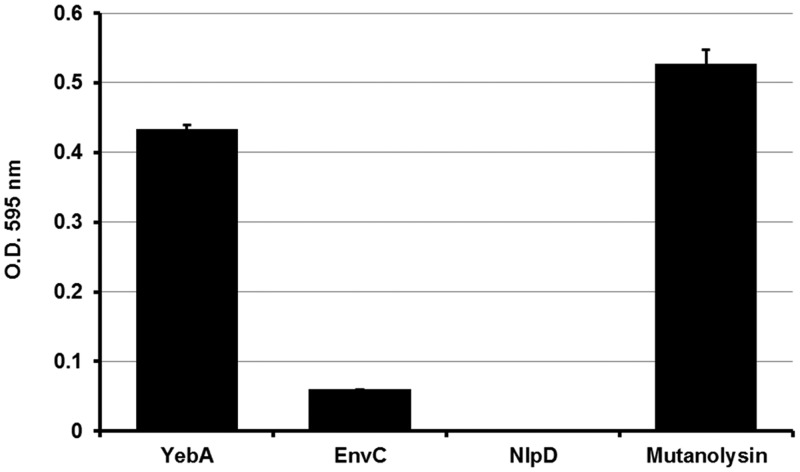
E. coli LytM proteins are known to have a crucial role in cell separation that may be maintained for the homologous proteins identified in NTHI. To address this hypothesis, we constructed deletion mutants for each of the three lytM genes. Although the absence of these genes did not significantly alter the bacterial growth rate (data not shown), bacterial clumping in liquid culture was observed for strains 176ΔyebA and 176ΔnlpD (Fig. 3A). Confocal and scanning electron microscopy confirmed that the observed phenotype was due to a failure in cell separation (Fig. 3B) associated with a diminished number of CFU when the two mutant strains were plated on chocolate agar (see Table S3 in the supplemental material). However, the morphology of these mutants was not identical. In 176ΔyebA bacteria, cells appeared roughly four times longer than the wild-type strain (4 µm) and bent in the central portion, while 176ΔnlpD bacteria formed very long chains (up to 100 µm), suggesting a different effect on cell division (Fig. 3B). In addition, as shown in Fig. 4, both mutant strains had defective septum formation and an extensive release of outer membrane vesicles (OMVs). In contrast, neither morphological differences nor overblebbing was observed for strain 176ΔenvC (see Fig. S2 in the supplemental material). LytM proteins have been reported to contribute to peptidoglycan cleavage, leading to the hypothesis that NTHI LytM factors might also share this function. The catalytic activity of these factors was tested in a peptidoglycan cleavage assay based on a previously reported protocol (14). As shown in Fig. 5, 4-h incubation of purified peptidoglycan from wild-type strain 176 with recombinant YebA, EnvC, and NlpD led to an increased release of peptidoglycan soluble fragments in the case of YebA, which was almost comparable to the mutanolysin (positive control). The PG cleavage activity was slightly detectable for EnvC and negative for NlpD. These data suggest that only YebA has a PG cleavage activity (Fig. 5).
FIG 3 .
Phenotypic characterization of NTHI wild-type strain 176 and lytM mutant strains. (A) Bacterial aggregation was evaluated growing the bacteria statically overnight at 37°C in liquid cultures. (B) Scanning electron microscopy of 176 wt, 176ΔyebA, and 176ΔnlpD strains. Bars, 1 µm.
FIG 4 .
Septum formation and OMV release in lytM mutants. Transmission electron microscopy of 176 wt strain and 176ΔyebA, and 176ΔnlpD mutant strains. Aberrant morphology and impaired septum formation are evident in the 176ΔyebA and 176ΔnlpD mutants. Arrows indicate OMVs that are released from the bacterial surface. Bars, 200 nm.
FIG 5 .
Contribution of LytM proteins to peptidoglycan cleavage. Results of peptidoglycan dye release assay are shown. Purified protein (4 µM) and mutanolysin (positive control) were incubated for 4 h at 37°C with remazol brilliant blue-stained peptidoglycan. The absorbance or optical density (O.D.) (595 nm) was measured after centrifugation to evaluate dye release.
EnvC influences periplasmic and outer membrane protein organization.
On the basis of the evidence that EnvC has a periplasmic localization, we hypothesized a role for this factor on periplasmic and outer membrane protein (OMP) distribution. To verify this hypothesis, naturally released OMVs (native OMVs) were purified from wild-type strain 176 and the 176ΔenvC mutant. Indeed, OMVs not only contain outer membrane proteins, but in the lumen, periplasmic elements belonging to the compartment localized between the peptidoglycan and the outer membrane are present (21, 22). As shown in Fig. 6A, comparative SDS-PAGE analysis of OMVs derived from wild-type strain 176 and 176ΔenvC strain revealed differences in the intensity of a few bands that were identified by peptide mass fingerprinting (Table 1). In particular, we found that the amount of HtrA, SurA, and OMP26 was reduced in OMVs from 176ΔenvC strain with respect to the wild-type strain. Interestingly, the H. influenzae OMP26 shares homology with the bacterial protein Skp, which along with HtrA and SurA forms a system of periplasmic chaperones essential for the proper assembly of OMP (23). Notably, Western blot analysis performed on cell fractions of both wild-type strain 176 and 176ΔenvC strain revealed that HtrA, SurA, and OMP26 were equally present in total extracts and periplasmic fractions, while their reduction in strain 176ΔenvC OMV preparation was confirmed (Fig. 6B). The outer membrane lipoprotein P6, which is found at a similar level in OMVs from both strains (Table 1), was used as a control in the Western blot (Fig. 6B). Taken together, these results suggest the possibility that in the 176ΔenvC mutant, these three periplasmic chaperones face difficulties in reaching the close vicinity of the inner layer of the outer membrane and consequently are not entrapped in the OMVs. OMVs from the 176ΔenvC strain were also characterized by an increase in OMP with a common β-barrel domain such as OMP1 and P5, as well as the autotransporters IgA protease, HxuA, Hap, and high-molecular-weight adhesin 2A (HMW2A) (Fig. 6A and Table 1). To assess whether the observed phenomenon could be extended to other periplasmic and β-barrel domain-containing proteins, a comparative label-free quantitative mass spectrometry based on the Hi3 method was applied (24). This analysis led to the identification of 60 proteins, including 28 predicted to be periplasmic proteins, 4 autotransporters, 8 OMPs, and 14 lipoproteins (see Table S4 in the supplemental material). All predicted periplasmic proteins, with the exception of NTHI1774, were less represented in the mutant, while all the autotransporters and the majority of β-barrel OMPs (6 out of 8) were more abundant (Fig. 6C and Table S4). Interestingly, a clear trend was not observed for lipoproteins. Of note, NlpD was identified in both OMVs (Fig. 6C). EnvC and YebA were not detected in the OMVs, suggesting that either the amounts are under the detection limit of the instrument or that, in the case of EnvC, it resides in the periplasmic space between the inner membrane and the peptidoglycan, and in the case of YebA, that it could be associated with the inner membrane.
FIG 6 .
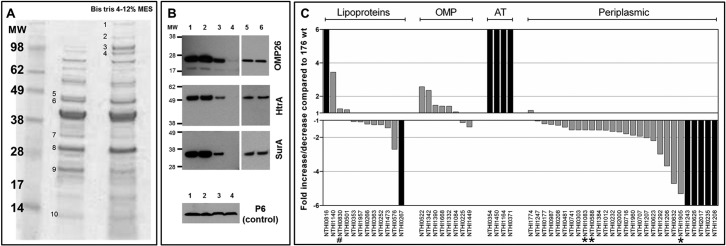
OMV analysis. (A) SDS-polyacrylamide gel was loaded with 10 µg of OMVs purified from 176 wt strain (middle lane) and 176ΔenvC mutant (right lane). Protein bands presenting the major staining intensity differences are numbered. The selected protein bands were identified by peptide mass fingerprinting (PMF) (Table 1). The positions of molecular weight (MW) markers (in thousands) are shown in the leftmost lane. MES, morpholineethanesulfonic acid. (B) Western blotting was performed to detect OMP26, HtrA, and SurA. Lanes: 1, whole-cell extract from wt strain; 2, whole-cell extract from knockout strain; 3, OMVs from wt strain; 4, OMVs from knockout strain; 5, periplasm from wt strain; 6, periplasm from knockout strain. Whole-cell extract was used as a control to verify the expression level of the three periplasmic chaperones. (C) Quantitative mass spectrometry analysis of OMVs shows differences in surface composition between the 176 wt strain and the 176ΔenvC mutant. For each protein, the fold increase/decrease in 176ΔenvC OMVs with respect to 176 wt (grey bars) is reported. Black bars indicate proteins that are present in only one of the two strains. The proteins identified were classified as lipoproteins, OMP, autotransporters (AT), and periplasmic proteins according to bioinformatic predictions. LipoP 1.0 server and PSORT were used to identify protein localization based on their signal peptide sequences. BOMP software was used to identify β-barrel domain and to confirm the outer membrane localization. In this analysis, all the proteins over the threshold of 0.1 ng were included, while proteins lacking an export signal sequence were excluded. The complete quantitative analysis is reported in Table S4 in the supplemental material. HtrA (NTHI1905), SurA (NTHI0588), and Omp26 (NTHI1083) (asterisks) and NlpD (NTHI0830) (pound sign) are indicated.
TABLE 1 .
Peptide mass fingerprinting identifications
| Band no.a | Protein(s) identifiedb | Class |
|---|---|---|
| 1 | NTHI1164 IgA-specific serine endopeptidase | Autotransporter |
| 2 | NTHI0354 adhesion and penetration protein Hap | Autotransporter |
| 3 | NTHI0782 hemoglobin-haptoglobin binding protein B | OMP |
| NTHI1450 HMW2A (high-molecular-weight adhesin 2) | Autotransporter | |
| 4 | NTHI0371 heme/hemopexin binding protein A | Autotransporter |
| NTHI1390 heme utilization protein | OMP | |
| 5 | NTHI1905 periplasmic serine protease HtrA* | Periplasmic |
| 6 | NTHI0522 OmpP1 long-chain fatty acid ABC transporter | OMP |
| 7 | NTHI0481 periplasmic chelated iron binding protein (protein F)* | Periplasmic |
| NTHI0588 survival protein SurA-like protein* | Periplasmic | |
| 8 | NTHI1332 OmpP5 (outer membrane protein P5) | OMP |
| 9 | NTHI1083 outer membrane protein 26* | Periplasmic |
| 10 | NTHI0353 hypothetical protein | Lipoprotein |
| NTHI0501 outer membrane protein P6 | Lipoprotein |
The band number was assigned as shown in the SDS-polyacrylamide gel in Fig. 6A. Bands 1 to 9 represented proteins with different staining intensities in the 176 wt and 176ΔenvC OMVs. Band 10 corresponded to proteins with similar staining intensities.
The proteins that are less abundant in the 176ΔenvC strain compared to the 176 wt strain are indicated by an asterisk.
Lack of EnvC expression affects NTHI adherence to epithelial cells, biofilm formation, and serum resistance.
The mislocalization of periplasmic chaperones, necessary for the maturation of the outer membrane proteins, could be responsible for the accumulation of these proteins in the 176ΔenvC OMVs in an unfolded/misfolded state. Moreover, the aberrant protein surface composition reported for the 176ΔenvC strain is likely to affect a number of cellular parameters crucial to NTHI pathogenesis and host colonization. In order to verify this hypothesis, we decided to test the 176ΔenvC mutant in different in vitro functional assays, focusing on the processes of adhesion, biofilm formation, and host immune evasion. Adhesion is an early step in NTHI persistence in the upper respiratory tract and precedes the formation of bacterial macrocommunities (25, 26). Both these phenomena were analyzed; in particular, we observed that the adherence of 176ΔenvC strain to Chang epithelial cells was severely impaired compared to the wild-type strain (Fig. 7A; see Fig. S3 in the supplemental material). In line with this result, we observed that the 176ΔenvC mutant strain showed reduced biomass formation compared to the wild-type strain in a classical in vitro crystal violet-based biofilm assay (Fig. 7B). Additionally, since the ability to evade the host defense is fundamental to the survival of NTHI during infection (25, 26), a serum resistance assay was performed using normal human sera (NHS) from healthy donors. The assay revealed that the 176ΔenvC strain was more susceptible than the wild-type strain 176 to NHS (Fig. 7C), confirming that surface alterations present in the mutant can also impair resistance to complement-mediated killing. Complementation of the envC gene expression fully restored both the adhesive and biofilm formation phenotypes, while the capacity to survive in the presence of NHS was only partially recovered (Fig. 7).
FIG 7 .
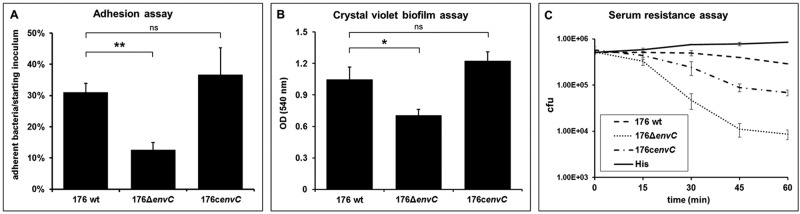
Defective pathogenicity of the 176ΔenvC mutant. (A) Chang cell monolayer was infected with an MOI of 100 of the different NTHI strains for 2 h. Quantification of the level of adhesion of 176ΔenvC and 176cenvC strains compared to the wild-type strain was calculated by counting the number of cell-associated CFU. Adherence is expressed as the percentage of adherent bacteria with respect to the starting inoculum. (B) Quantification of biofilm formation on plastic surface was performed after 24 h of static growth by crystal violet staining for wild-type strain 176 and 176ΔenvC mutant. Values are means plus standard deviations (error bars) of several experiments, each performed in triplicate. Values in panels A and B that are significantly different by analysis of variance (ANOVA) are indicated by a bar and asterisk as follows: *, P < 0.05; **, P < 0.01. Values that are not significantly different (ns) are also indicated. (C) Bacteria were incubated with 2% NHS from healthy patients, and bacterial survival was calculated by counting the number of CFU at different time points. Heat-inactivated serum (His) was used as a negative control.
DISCUSSION
Although LytM factors have been studied extensively in E. coli and proposed to contribute to bacterial physiology by affecting cell division (14–16), the relevance of such a protein family in H. influenzae physiology and pathogenesis has not been yet addressed. It is interesting that single-gene deletion of yebA and nlpD, two of the three newly identified lytM genes in NTHI revealed a crucial role in the cell division machinery showing immediately a substantial difference with respect to E. coli LytM factors, where only the triple knockout showed similar defects in cell division (15). In particular, the 176ΔyebA and 176ΔnlpD mutants showed not only aberrant cell morphology but also defects in septum formation and in outer membrane stability, as shown by the increased release of OMVs observed in the two isogenic mutants. Proteomic characterization for OMVs released by NTHI yebA and nlpD mutant strains is currently ongoing, and the results will be part of a follow-up study.
The contribution of NTHI NlpD and YebA in cell division appeared to be distinct. Indeed, the 176ΔnlpD knockout strain was able to duplicate but not to divide, while the 176ΔyebA mutant showed an intermediate phenotype with bacteria still able to split but with aberrant dimensions and an increased release of outer membrane vesicles, suggesting an influence of LytM proteins in membrane stability and peptidoglycan turnover. It is interesting that the ability of NTHI YebA to cleave peptidoglycan suggests a direct role of this protein in peptidoglycan biogenesis and rearrangement, as hypothesized for E. coli YebA (16, 17).
On the other hand, NTHI EnvC did not contribute to cell division. This observation was quite surprising considering the essential role played by E. coli EnvC in amidase activation (27). NTHI EnvC is localized in the periplasm, probably between the inner membrane and the peptidoglycan layer, and it is likely that the protein may express its function in this bacterial compartment. Indeed, NTHI EnvC could modulate the activity of other proteases involved in peptidoglycan cleavage, as reported for EnvC in E. coli (14, 27).
We hypothesized and discovered that NTHI EnvC affected periplasmic and outer membrane protein composition and localization, as the proteomic analysis of OMVs derived from the 176ΔenvC mutant evidenced a reduction in the amount of 27 out of 28 identified and quantified periplasmic proteins compared to the OMVs derived from the wild-type strain. This reduction was not observed for lipoproteins and for some OMPs, indicating for these proteins a mechanism of peptidoglycan translocation independent of EnvC.
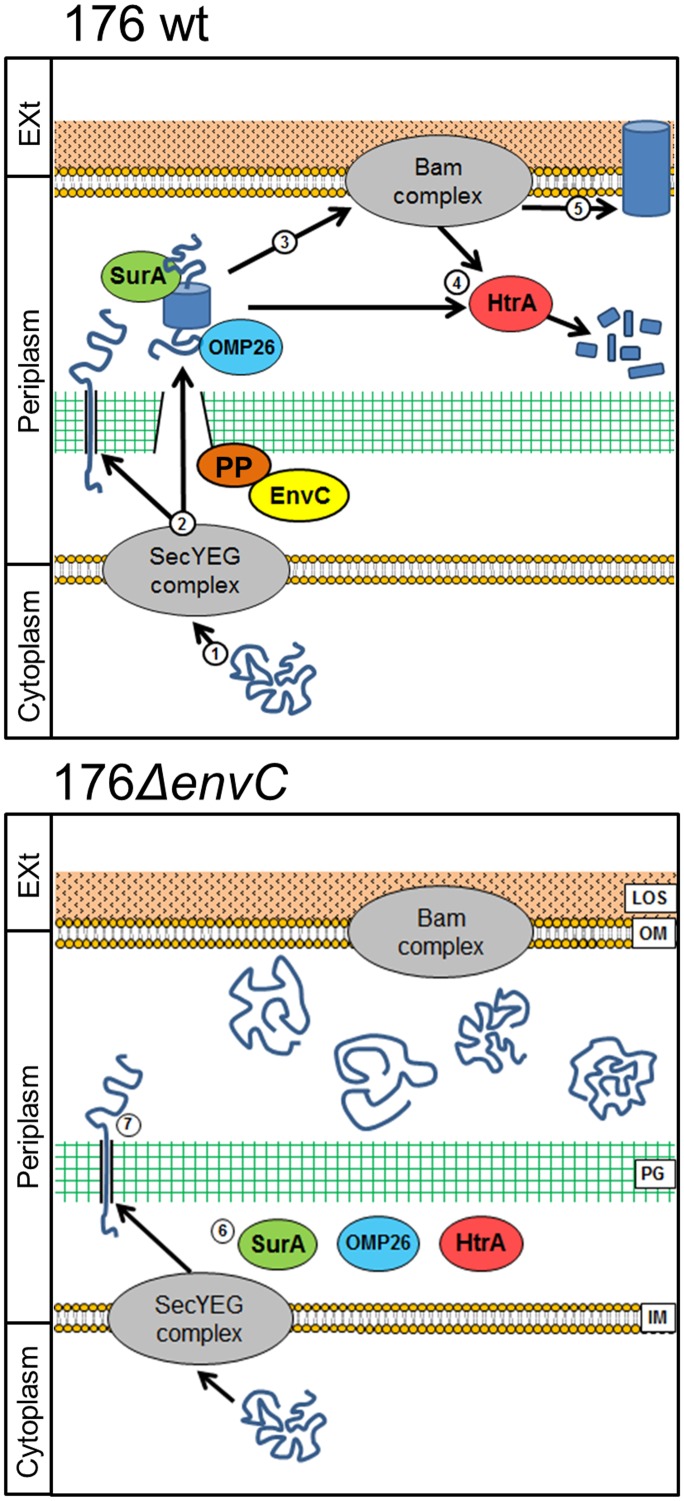
On the basis of the evidence reported in this study, we propose a plausible model of action of NTHI EnvC (Fig. 8), although the mechanism represented may be revised with extra data.
FIG 8 .
EnvC model of action. In the wild-type strain 176, unfolded proteins are exported from the cytoplasm through the secretion (SEC) system (step 1). The proteins reach the periplasm, and some of them pass through the peptidoglycan layer using a mechanism mediated by EnvC, by interacting with a potential partner (PP) able to cleave peptidoglycan (step 2). Periplasmic chaperones act to fold the outer membrane proteins and to protect them from degradation during passage in the periplasm (step 3). Misfolded proteins are degraded by HtrA (step 4), while properly folded proteins are inserted in the outer membrane (OM) (step 5). In the 176ΔenvC mutant, most of the periplasmic proteins fail to pass through peptidoglycan, particularly the periplasmic chaperones SurA, HtrA, and OMP26 (step 6). β-Barrel-containing OMPs and autotransporters instead, are able to pass through peptidoglycan (PG), but without the activity of periplasmic chaperones, they are accumulated and probably do not reach the outer membrane, causing defects in pathogenicity (step 7). EXt, exterior; LOS, lipooligosaccharide; IM, inner membrane.
Briefly, we postulate that as a consequence of the absence of EnvC, periplasmic chaperones, such as HtrA, SurA, and OMP26 (homologue to Skp), could not reach their site of action when in close vicinity to the outer membrane. This would lead to the impairment of both OMP and autotransporter maturation and to the degradation of the misfolded proteins (28–31). The accumulation of both OMPs and autotransporters observed in OMVs isolated from the 176ΔenvC mutant supports this interpretation, although further investigations are needed to understand the folding state and surface exposure of these proteins.
It has already been reported that the absence of just one of the periplasmic chaperones has a strong impact on the interaction with host cells for Gram-negative bacteria. In particular, the HtrA deletion in Campylobacter jejuni reduced bacterial binding to epithelial cells of 5 to 10 times more than the absence of any known adhesin, suggesting a pleiotropic effect (32), and a similar phenotype was also observed for the SurA deletion mutant in Yersinia pseudotuberculosis (33). Furthermore, in E. coli, the SurA knockout mutant showed a reduced ability to form biofilms (34), and the same phenomenon was observed for Listeria monocytogenes lacking HtrA (35). Last, an influence on resistance to human sera is reported for HtrA in Klebsiella pneumoniae (36) and for SurA in Yersinia species (33, 37). The alteration in OMV protein patterns prompted us to further investigate the influence of EnvC on NTHI pathogenesis. A confirmation of the effect of EnvC on NTHI pathogenicity was revealed by the defects of the 176ΔenvC strain in adhesion to epithelial cells and biofilm formation, probably due to the accumulation of nonfunctional proteins usually involved in adhesion to host tissues, such as Hap, HMW2, and P5 (38–40). Also, the decreased resistance to human sera could be derived from the defects in the functionality of surface determinants involved in this system like IgA protease and P5 (26, 41).
In conclusion, this study highlights the importance of LytM proteins in NTHI physiology, confirming their role in cell division, and shows their impact on periplasmic and outer membrane protein distribution and in pathogenesis.
MATERIALS AND METHODS
Bioinformatic analysis.
LytM proteins were analyzed using several online applications. Putative signal peptides were identified using SignalP (http://www.cbs.dtu.dk/services/SignalP/); Pfam (http://pfam.sanger.ac.uk/) was used to detect the presence of domains of known function, and SMART software (http://smart.embl-heidelberg.de/) was used to reconstruct the architectural structure of each protein. Homologues of nontypeable Haemophilus influenzae (NTHI) LytM factors were identified using BLASTP (http://blast.ncbi.nlm.nih.gov/). Identity percentages between different proteins were obtained comparing amino acid sequences with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and BLASTP. Proteins identified by mass spectrometry were further analyzed with DOLOP (http://www.mrc-lmb.cam.ac.uk/genomes/dolop/analysis.shtml) to detect lipoproteins, with PSORTb (http://www.psort.org/psortb/index.html) to predict localization, and with BOMP (http://services.cbu.uib.no/tools/bomp) to identify β-barrel domains.
Bacterial strains and growth conditions.
NTHI strain 176 was obtained from the middle ears of children as part of a Finnish otitis media cohort study. NTHI was cultivated on chocolate agar polivitex (bioMérieux) at 37°C with 5% CO2. Brain heart infusion (BHI) broth (Difco Laboratories) supplemented with 10 µg/ml of hemin (Fluka Biochemika) and 10 µg/ml nicotinamide adenine dinucleotide (NAD) (Sigma) was used as fluid growth medium. E. coli strains DH5α (Invitrogen), HK100 (42), and BL21(DE3) T1R (New England BioLabs [NEB]) were used for cloning and expression of LytM proteins.
Cell cultures.
Chang epithelial cells (ATCC CCL-20.2) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 25 mM HEPES, 15 mM l-glutamine, antibiotics, and 10% (vol/vol) heat-inactivated fetal calf serum (FCS) (Invitrogen Corporation) at 37°C with 5% CO2.
Generation of mutant strains.
NTHI isogenic mutants of yebA, envC, and nlpD genes (corresponding to NTHI0532, NTHI0915, and NTHI0830 in publicly available sequences of strain 86-028NP) were constructed by allelic replacement of each entire coding sequence with an erythromycin resistance cassette in strain 176. The genomic sequence of NTHI wild-type strain 176 was obtained using whole-genome sequencing (43), and it is reported in European Nucleotide Archive (ENA) (RR125021). Regions upstream and downstream of the three genes were amplified by PCR using the primers listed in Table S5 in the supplemental material and cloned in Stratagene pSC-A TOPO vector. The erythromycin resistance cassette was purified from plasmid pIM13. The plasmid constructs containing the region upstream of the three genes, antibiotic resistance cassette, and region downstream of the three genes were assembled. The plasmids obtained were linearized and used to transform NTHI strain 176 using M-IV medium as described previously (44). Isogenic mutants 176ΔyebA, 176ΔenvC, and 176ΔnlpD were confirmed by PCR, Western blotting, and locus sequencing. The 176ΔenvC complemented strain, 176cenvC, was constructed by replacing the erythromycin cassette present in the mutant with a construct containing the original envC gene and an adjacent chloramphenicol resistance cassette. The oligonucleotides used are listed in Table S5.
Peptidoglycan extraction and cleavage assay.
Peptidoglycan was purified from NTHI wild-type strain 176 by the method of Uehara et al. (15). One liter of exponential-phase culture was centrifuged, the pellet was resuspended in 20 ml of phosphate-buffered saline (PBS), boiled with 80 ml of 5% SDS for 30 min, and left overnight at room temperature (RT). Samples were ultracentrifuged for 1 h at 52,000 × g at RT and then washed with water several times to remove SDS. The peptidoglycan pellet was resuspended in 1 ml of PBS and incubated with 200 µg/ml amylase (catalog no. A6380; Sigma) overnight at 37°C. The sample was pelleted by ultracentrifugation at 200,000 × g for 15 min, washed with water three times, and resuspended in 1 ml of water. The dye release assay was performed staining purified peptidoglycan with remazol brilliant blue (catalog no. R8001; Sigma) (14) and then incubating it with 4 µM purified LytM proteins or mutanolysin (positive control) at 37°C for different incubation times. Dye release was quantified measuring the absorbance at 595 nm. Peptidoglycan incubated with PBS alone was used as a negative control (blank) and subtracted from all values.
NTHI native OMV purification and sample preparation for MS analysis.
Native outer membrane vesicles (OMVs) were isolated from wild-type and mutant strains, growing the bacteria overnight in 60-ml BHI cultures. The bacteria were then centrifuged, and supernatant fractions were filtered and left overnight at 4°C after the addition of protease inhibitor (Roche) and EDTA. The supernatant was ultracentrifuged for 3 h at 200,000 × g (maximum value), and the final pellet containing OMVs was resuspended in PBS. NTHI OMV proteins were precipitated with 10% (wt/vol) trichloroacetic acid and 0.04% (wt/vol) sodium deoxycholate and finally resuspended in 50 mM ammonium bicarbonate. Fifty micrograms of OMV proteins from wild-type and mutant strains were heated for 10 min at 100°C in 50 mM ammonium bicarbonate containing 0.1% RapiGest (Waters) and 5 mM Tris(2-carboxyethyl)phosphine (TCEP). Digestions were performed overnight at 37°C with 2.5 µg trypsin (Promega). Digestions were stopped with 0.1% (vol/vol) formic acid, desalted using OASIS cartridges (Waters) as described by the manufacturer, concentrated with a Centrivap concentrator (Labconco) to about 30 µl, and kept at −20°C until analysis by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) as reported in Text S1 in the supplemental material.
Infection of Chang epithelial cells: adhesion assays with NTHI strains.
Chang cell suspensions obtained from confluent monolayers were seeded at 3 × 105 cells per well in 24-well tissue culture plates (Nunc) and incubated for 24 h in an antibiotic-free medium. Overnight cultures of bacteria were washed once and resuspended in DMEM plus 1% inactivated fetal calf serum (FCSi) to a concentration of 3 × 107 bacteria ml−1 at a multiplicity of infection (MOI) of approximately 1:100. Aliquots of 1 ml of each strain were added to monolayer cultures and incubated for 3 h at 37°C in 5% CO2. Nonadherent bacteria were removed by washing the cultures three times with DMEM plus 1% FCSi and twice with PBS. The remaining bacteria were released by the addition of 1% saponin (Sigma) and incubation at 37°C for 15 min. Serially diluted bacteria were plated onto chocolate agar, and adhesion capability was quantified by counting the number of CFU.
Biofilm assay on plastic.
Bacteria from overnight cultures were diluted 1:100 and incubated statically in BHI on plastic 24-well plates at 37°C. After 24 h, the wells were gently washed once with 1 ml sterile PBS and then allowed to dry for 10 min. The bacteria were stained with 1 ml of filter-sterilized 0.2% crystal violet and incubated for 30 min at RT. Crystal violet was removed from the wells, followed by two washes with 1 ml PBS. The dye was extracted by adding 1 ml of 96% ethanol to each well and incubating for 30 min at RT. The absorbance was measured at 540 nm using a Tecan Infinite 200 PRO plate reader.
Serum resistance assay.
Bacteria were grown until early exponential phase and then diluted in Dulbecco’s PBS and split into a multiwell plate to have 104 CFU/ml. Normal human sera from healthy individuals were added to each sample to a final concentration of 2% (vol/vol). Heat-inactivated serum (30 min at 56°C) was also included as a negative control. Aliquots were spotted from each well at different time points onto chocolate agar plates to evaluate bacterial survival. Human sera were obtained from anonymous healthy donors (available from authorized blood banks). Informed consent was obtained before all blood donations. The study protocol was approved by the Novartis Research Center Ethical Committee and conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
SUPPLEMENTAL MATERIAL
Quality control of purified fractions. Subcompartmental fractions were analyzed by Western blotting using proper controls to assess their quality and exclude contamination. Lanes: 1, whole-cell extract from wild-type (wt) strain 176; 2, OMVs from wt strain; 3, OMPs from wt strain; 4, supernatant from wt strain, 5, periplasmic fraction from wt strain. Four proteins were selected as positive controls: protein D (OMP), HxuA (supernatant), cytochrome C-552 (periplasm), and RNA polymerase β (cytoplasm). Since cytochrome C-552 is also reported to be linked to the inner membrane (M. Doi, K. Takamiy, and M. Nishimura, Photosynth Res 4:49–60, 1983, doi:10.1007/BF00041800), the result suggests an absence of inner membrane protein contaminants in the OMP fraction. Download
Morphology of 176ΔenvC mutant. Scanning electron microscopy of wild-type strain 176 and 176ΔenvC mutant confirms the absence of defects in cell division. Bars, 1 µm. Download
Adhesion of 176ΔenvC mutant to epithelial cells. Immunofluorescence microscopy showing adhesion to Chang epithelial cells of wild-type strain 176 and 176ΔenvC mutant. The Chang cell monolayer was infected with the different NTHI strains for 2 h. Cells are stained in red (actin), blue (4′,6′-diamidino-2-phenylindole [DAPI]), and green (antibody against the whole bacterium). Download
Supplemental methods Download
Conservation level of LytM proteins in a panel of NTHI strains
Percentages of amino acid identity of NTHI LytM factors with bacterial homologues
CFU count of wild-type strain 176 and lytM mutants
Quantitative proteomic analysis
Oligonucleotides for the generation of NTHI mutants
ACKNOWLEDGMENTS
We thank R. M. Moxon (Oxford University) for helpful discussions, D. Hood (Oxford University) for providing NTHI strain 176 and for technical support, Danilo Gomes Moriel and Maria Scarselli for help with bioinformatic analysis, Sara Marchi for protein purification, Marco Tortoli for immunizing the mice, Annarita Taddei for electron microscopy images, Silvia Rossi Paccani for serum resistance assay assistance, Francesco Berti for peptidoglycan analysis, and Catherine Mallia for manuscript editing.
Footnotes
Citation Ercoli G, Tani C, Pezzicoli A, Vacca I, Martinelli M, Pecetta S, Petracca R, Rappuoli R, Pizza M, Norais N, Soriani M, Aricò B. 2015. LytM proteins play a crucial role in cell separation, outer membrane composition, and pathogenesis in nontypeable Haemophilus influenzae. mBio 6(2):e02575-14. doi:10.1128/mBio.02575-14.
REFERENCES
- 1.Rao MB, Tanksale AM, Ghatge MS, Deshpande VV. 1998. Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 62:597–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laarman AJ, Ruyken M, Malone CL, van Strijp JAG, Horswill AR, Rooijakkers SHM. 2011. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol 186:6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 3.Pruteanu M, Hyland NP, Clarke DJ, Kiely B, Shanahan F. 2011. Degradation of the extracellular matrix components by bacterial-derived metalloproteases: implications for inflammatory bowel diseases. Inflamm Bowel Dis 17:1189–1200. doi: 10.1002/ibd.21475. [DOI] [PubMed] [Google Scholar]
- 4.Miyoshi S, Shinoda S. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect 2:91–98. doi: 10.1016/S1286-4579(00)00280-X. [DOI] [PubMed] [Google Scholar]
- 5.Chen YC, Chang CC, Chang SY, Su JH. 2010. A recombinant metalloprotease antigen of Vibrio vulnificus elicits protective antibodies in a murine model. Lett Appl Microbiol 50:168–172. doi: 10.1111/j.1472-765X.2009.02771.x. [DOI] [PubMed] [Google Scholar]
- 6.Gong Y, Xu W, Cui Y, Zhang X, Yao R, Li D, Wang H, He Y, Cao J, Yin Y. 2011. Immunization with a ZmpB-based protein vaccine could protect against pneumococcal diseases in mice. Infect Immun 79:867–878. doi: 10.1128/IAI.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tidhar A, Flashner Y, Cohen S, Levi Y, Zauberman A, Gur D, Aftalion M, Elhanany E, Zvi A, Shafferman A, Mamroud E. 2009. The NlpD lipoprotein is a novel Yersinia pestis virulence factor essential for the development of plague. PLoS One 4:e7023. doi: 10.1371/journal.pone.0007023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawlings ND, Morton FR, Barrett AJ. 2006. MEROPS: the peptidase database. Nucleic Acids Res 34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler E, Safrin M, Olson JC, Ohman DE. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem 268:7503–7508. [PubMed] [Google Scholar]
- 10.Bochtler M, Odintsov SG, Marcyjaniak M, Sabala I. 2004. Similar active sites in lysostaphins and d-Ala-d-Ala metallopeptidases. Protein Sci 13:854–861. doi: 10.1110/ps.03515704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramadurai L, Jayaswal RK. 1997. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J Bacteriol 179:3625–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meisner J, Moran CP Jr. 2011. A LytM domain dictates the localization of proteins to the mother cell-forespore interface during bacterial endospore formation. J Bacteriol 193:591–598. doi: 10.1128/JB.01270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levdikov VM, Blagova EV, McFeat A, Fogg MJ, Wilson KS, Wilkinson AJ. 2012. Structure of components of an intercellular channel complex in sporulating Bacillus subtilis. Proc Natl Acad Sci U S A 109:5441–5445. doi: 10.1073/pnas.1120087109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang DC, Tan K, Joachimiak A, Bernhardt TG. 2012. A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Mol Microbiol 85:768–781. doi: 10.1111/j.1365-2958.2012.08138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uehara T, Dinh T, Bernhardt TG. 2009. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol 191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uehara T, Parzych KR, Dinh T, Bernhardt TG. 2010. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J 29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh SK, SaiSree L, Amrutha RN, Reddy M. 2012. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol 86:1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 18.Bonis M, Ecobichon C, Guadagnini S, Prévost MC, Boneca IG. 2010. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol 78:809–819. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- 19.Stohl EA, Chan YA, Hackett KT, Kohler PL, Dillard JP, Seifert HS. 2012. Neisseria gonorrhoeae virulence factor NG1686 is a bifunctional M23B family metallopeptidase that influences resistance to hydrogen peroxide and colony morphology. J Biol Chem 287:11222–11233. doi: 10.1074/jbc.M111.338830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 21.McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol 63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. 1998. On the origin of membrane vesicles in Gram-negative bacteria. FEMS Microbiol Lett 163:223–228. doi: 10.1111/j.1574-6968.1998.tb13049.x. [DOI] [PubMed] [Google Scholar]
- 23.Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- 24.Tani C, Stella M, Donnarumma D, Biagini M, Parente P, Vadi A, Magagnoli C, Costantino P, Rigat F, Norais N. 2014. Quantification by LC-MS(E) of outer membrane vesicle proteins of the Bexsero® vaccine. Vaccine 32:1273–1279. doi: 10.1016/j.vaccine.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Clementi CF, Murphy TF. 2011. Non-typeable Haemophilus influenzae invasion and persistence in the human respiratory tract. Front Cell Infect Microbiol 1:1. doi: 10.3389/fcimb.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foxwell AR, Kyd JM, Cripps AW. 1998. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol Mol Biol Rev 62:294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters NT, Morlot C, Yang DC, Uehara T, Vernet T, Bernhardt TG. 2013. Structure-function analysis of the LytM domain of EnvC, an activator of cell wall remodelling at the Escherichia coli division site. Mol Microbiol 89:690–701. doi: 10.1111/mmi.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazar SW, Kolter R. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol 178:1770–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz N, Kahne D, Silhavy TJ. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol 4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. 2009. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol 191:6571–6583. doi: 10.1128/JB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brøndsted L, Andersen MT, Parker M, Jørgensen K, Ingmer H. 2005. The HtrA protease of Campylobacter jejuni is required for heat and oxygen tolerance and for optimal interaction with human epithelial cells. Appl Environ Microbiol 71:3205–3212. doi: 10.1128/AEM.71.6.3205-3212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obi IR, Francis MS. 2013. Demarcating SurA activities required for outer membrane targeting of Yersinia pseudotuberculosis adhesins. Infect Immun 81:2296–2308. doi: 10.1128/IAI.01208-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niba ET, Naka Y, Nagase M, Mori H, Kitakawa M. 2007. A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res 14:237–246. doi: 10.1093/dnares/dsm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson RL, Brown LL, Kirkwood-Watts D, Warren TK, Lund SA, King DS, Jones KF, Hruby DE. 2006. Listeria monocytogenes 10403S HtrA is necessary for resistance to cellular stress and virulence. Infect Immun 74:765–768. doi: 10.1128/IAI.74.1.765-768.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cortés G, de Astorza B, Benedí VJ, Albertí S. 2002. Role of the htrA gene in Klebsiella pneumoniae virulence. Infect Immun 70:4772–4776. doi: 10.1128/IAI.70.9.4772-4776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biedzka-Sarek M, Venho R, Skurnik M. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect Immun 73:2232–2244. doi: 10.1128/IAI.73.4.2232-2244.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. Geme JW III, Falkow S, Barenkamp SJ. 1993. High-molecular-weight proteins of non-typable Haemophilus influenzae mediate attachment to human epithelial cells. Proc Natl Acad Sci U S A 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrixson DR, St. Geme JW. 1998. The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol Cell 2:841–850. doi: 10.1016/S1097-2765(00)80298-1. [DOI] [PubMed] [Google Scholar]
- 40.Reddy MS, Bernstein JM, Murphy TF, Faden HS. 1996. Binding between outer membrane proteins of nontypeable Haemophilus influenzae and human nasopharyngeal mucin. Infect Immun 64:1477–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosadini CV, Ram S, Akerley BJ. 2014. Outer membrane protein P5 is required for resistance of nontypeable Haemophilus influenzae to both the classical and alternative complement pathways. Infect Immun 82:640–649. doi: 10.1128/IAI.01224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klock HE, White A, Koesema E, Lesley SA. 2005. Methods and results for semi-automated cloning using integrated robotics. J Struct Funct Genomics 6:89–94. doi: 10.1007/s10969-005-3084-1. [DOI] [PubMed] [Google Scholar]
- 43.De Chiara M, Hood D, Muzzi A, Pickard DJ, Perkins T, Pizza M, Dougan G, Rappuoli R, Moxon ER, Soriani M, Donati C. 2014. Genome sequencing of disease and carriage isolates of non-typeable Haemophilus influenzae identifies discrete population structure. Proc Natl Acad Sci U S A 111:5439–5444. doi: 10.1073/pnas.1403353111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herriott RM, Meyer EM, Vogt M. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol 101:517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality control of purified fractions. Subcompartmental fractions were analyzed by Western blotting using proper controls to assess their quality and exclude contamination. Lanes: 1, whole-cell extract from wild-type (wt) strain 176; 2, OMVs from wt strain; 3, OMPs from wt strain; 4, supernatant from wt strain, 5, periplasmic fraction from wt strain. Four proteins were selected as positive controls: protein D (OMP), HxuA (supernatant), cytochrome C-552 (periplasm), and RNA polymerase β (cytoplasm). Since cytochrome C-552 is also reported to be linked to the inner membrane (M. Doi, K. Takamiy, and M. Nishimura, Photosynth Res 4:49–60, 1983, doi:10.1007/BF00041800), the result suggests an absence of inner membrane protein contaminants in the OMP fraction. Download
Morphology of 176ΔenvC mutant. Scanning electron microscopy of wild-type strain 176 and 176ΔenvC mutant confirms the absence of defects in cell division. Bars, 1 µm. Download
Adhesion of 176ΔenvC mutant to epithelial cells. Immunofluorescence microscopy showing adhesion to Chang epithelial cells of wild-type strain 176 and 176ΔenvC mutant. The Chang cell monolayer was infected with the different NTHI strains for 2 h. Cells are stained in red (actin), blue (4′,6′-diamidino-2-phenylindole [DAPI]), and green (antibody against the whole bacterium). Download
Supplemental methods Download
Conservation level of LytM proteins in a panel of NTHI strains
Percentages of amino acid identity of NTHI LytM factors with bacterial homologues
CFU count of wild-type strain 176 and lytM mutants
Quantitative proteomic analysis
Oligonucleotides for the generation of NTHI mutants