ABSTRACT
The bacterial flagellum is driven by a bidirectional rotary motor, which propels bacteria to swim through liquids or swarm over surfaces. While the functions of the major structural and regulatory components of the flagellum are known, the function of the well-conserved FliL protein is not. In Salmonella and Escherichia coli, the absence of FliL leads to a small defect in swimming but complete elimination of swarming. Here, we tracked single motors of these bacteria and found that absence of FliL decreases their speed as well as switching frequency. We demonstrate that FliL interacts strongly with itself, with the MS ring protein FliF, and with the stator proteins MotA and MotB and weakly with the rotor switch protein FliG. These and other experiments show that FliL increases motor output either by recruiting or stabilizing the stators or by increasing their efficiency and contributes additionally to torque generation at higher motor loads. The increased torque enabled by FliL explains why this protein is essential for swarming on an agar surface expected to offer increased resistance to bacterial movement.
IMPORTANCE
FliL is a well-conserved bacterial flagellar protein whose absence leads to a variety of motility defects, ranging from moderate to complete inhibition of swimming in some bacterial species, inhibition of swarming in others, structural defects that break the flagellar rod during swarming in E. coli and Salmonella, and failure to eject the flagellar filament during the developmental transition of a swimmer to a stalk cell in Caulobacter crescentus. Despite these many phenotypes, a specific function for FliL has remained elusive. Here, we established a central role for FliL at the Salmonella and E. coli motors, where it interacts with both rotor and stator proteins, increases motor output, and contributes to the normal rotational bias of the motor.
INTRODUCTION
Bacterial flagella promote motility by rotation of a helical flagellar filament and confer an advantage by facilitating chemotaxis toward nutrients or away from toxic chemicals (1–6). Flagella also promote swarming, where bacteria migrate as a group over a solid surface to colonize large swaths of territory, a behavior associated with increased pathogenic potential and resistance to antimicrobials (7–9).
The flagellum consists of a moving rotor and stationary stator (see Fig. S1 in the supplemental material). The rotor includes the cytoplasmic C ring (FliG, FliM, and FliN proteins) attached to the membrane MS ring (FliF protein), the periplasmic rod, and external hook. These three structures together comprise the basal body (BB), which is continuous with a long external helical flagellar filament. Stationary elements in the inner membrane include the MotA/MotB protein complexes, or stators, which conduct protons to power motor rotation. MotA interacts with FliG positioned at the top of the C ring (see Fig. S1). There are ~11 MotAB stators surrounding the BB (10–12). Stators are incorporated into the membrane independently of the BB and, until needed, are proposed to be held in an inactive form by a periplasmic region of MotB that serves as a plug preventing premature proton flow (13) (see Fig. S1). As protons travel through, protonation and deprotonation of an essential aspartate residue near the cytoplasmic end of each proton channel in MotB drive conformational changes in MotA that generate torque, causing MotA to move from one FliG subunit in the C-ring rotor to the next (14, 15). The C ring controls the switching between clockwise (CW) and counterclockwise (CCW) rotor directions in response to chemotaxis signals. Rotation is tightly coupled to proton motive force (Δp). Torque, or the force generated on the motor, is directly proportional to Δp and is transmitted through the basal structure to the hook and filament, enabling the whole structure to rotate in the membrane. Rotation speed varies inversely with viscosity, implying a constant torque. Escherichia coli motors spin at ~125 Hz when attached to a filament; when the viscous load is light, as when the filament is absent, motors rotate at 300 Hz or faster (2, 3, 16, 17).
In order to swarm, many bacteria increase flagellar numbers and/or secrete powerful extracellular surfactants, which are adaptations for increasing thrust and lowering surface tension, respectively; these adaptations assist in overcoming the resistive forces of surface friction, tension, and viscous drag (9). Salmonella and E. coli do not display either of these adaptations (18). In these bacteria, the FliL protein is essential for swarming (19) and likely increases torque without increasing flagellar numbers (18). In these and other bacteria, the fliL gene is either the first member of the fliLMNOPQR flagellar operon or present upstream of fliM/N (20); FliM and FliN are C-ring proteins (see Fig. S1 in the supplemental material). In other bacteria, fliL is proximal to motAB genes. The specific function of FliL has remained unknown, although it is clear that it must associate closely with the flagellar motor, because its absence abrogates swimming in Caulobacter, Pseudomonas, and Rhodobacter (21–23), decreases swimming speeds in Salmonella, E. coli, and Borrelia (19, 24), and abolishes swarming in Salmonella, E. coli, and Proteus (19, 25). Cryo-electron tomography (cryo-ET) studies in Borrelia place FliL between the stators and the rotor (24), and mutations that suppress the fliL swimming phenotype in Rhodobacter map to the MotB stator (23). FliL plays various other roles as well. For example, fliL mutations cause flagellar rod breakage in Salmonella and E. coli only during swarming (19) (see Fig. S2 in the supplemental material), influence positioning of the periplasmic flagellum in Borrelia (24), interfere with the sensory role of the motor during Proteus swarming (25, 26), and impede FliF degradation and subsequent swarmer-to-stalked-cell transition in Caulobacter (27, 28).
In this study, we established a role for FliL both in maximizing motor torque and in influencing motor bias and switching behavior. We show that FliL interacts with both stator proteins, strengthens their interaction with each other, increases their recovery/retention at the motor, increases the efficiency of proton flow through the motor, and improves motor function at high loads. We present a model for FliL location within the basal flagellar structure and discuss how its interaction with the rotor proteins might influence the structure and function of the flagellum.
RESULTS
The flagellar structures of E. coli and Salmonella are very similar, and all parts are interchangeable, including FliL. Indeed, FliL affects swimming and swarming in the same manner in both organisms (see Fig. S2 in the supplemental material). This work was initiated in Salmonella, where most of the experiments were conducted. Exceptions using E. coli proteins are noted with an “Ec” subscript. The single-motor experiments in Ficoll necessitated use of E. coli motors because their behavior was more robust in this viscous agent.
Single motors run at reduced speed and reverse less frequently in the absence of FliL.
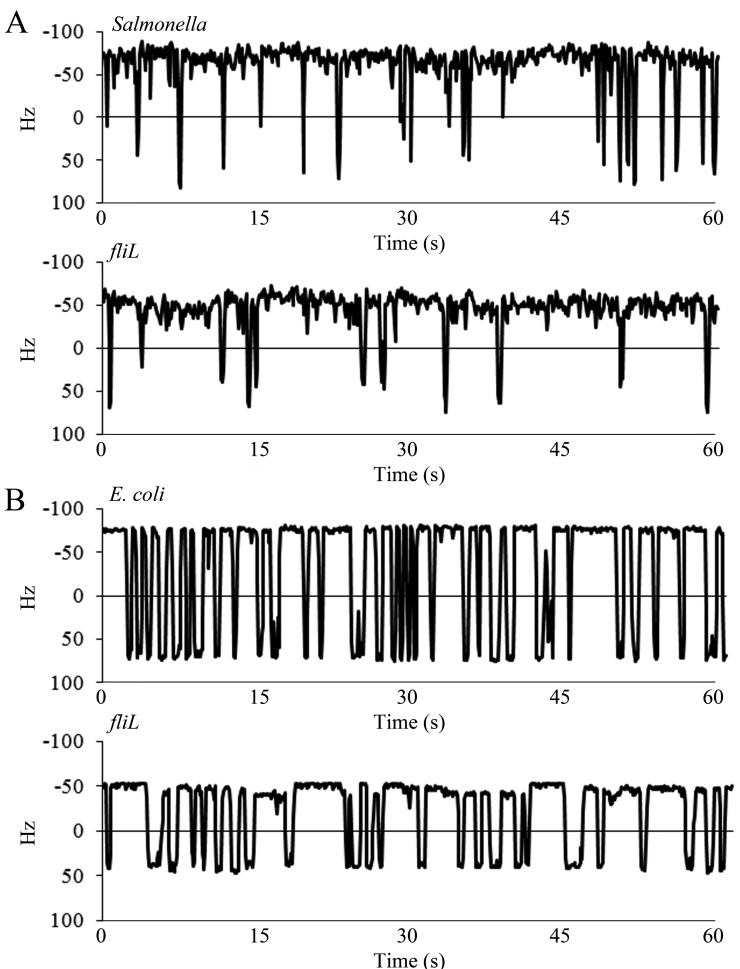
To determine whether FliL has a direct effect on the output of individual motors, the single-motor behavior of the wild type and isogenic fliL mutants was tracked by monitoring the rotation of 0.75-µm polystyrene beads attached to “sticky” flagellar filament stubs as described in Materials and Methods. Wild-type Salmonella motors had rotational speeds of 59 Hz ± 4 with 22 ± 3 reversals per minute (rvpm; mean ± standard deviation) (Fig. 1A). Its ΔfliL mutant derivative showed a reduction in both speed (47 ± 7 Hz) and reversals (15 ± 6 rvpm). Similar results were obtained with E. coli, where the wild-type speeds were 71 ± 4 Hz with 37 ± 4 rvpm, and isogenic ΔfliL motors rotated at 55 Hz ± 9 with 27 ± 8 rvpm (Fig. 1B). The lower speeds and slower switching behavior of wild-type Salmonella motors were confirmed in a different wild-type strain (SJW1103 fliCst), which was reported previously (29). The faster rotation of E. coli motors than Salmonella motors is surprising, given that E. coli is a less robust swimmer than Salmonella, as measured by migration in swim plates, an assay that reflects both swimming speeds and motor reversals (see Fig. S3 in the supplemental material).
FIG 1 .
Behavior of single motors of Salmonella (A) and E. coli (B) over a 60-s time frame. Rotation of motors of Salmonella (JP1107) and E. coli (MT02) was monitored by recording the motion of 0.75 µm polystyrene beads attached to sheared “sticky” filaments. Rotation speeds are expressed in Hz. The positive and negative values represent CW and CCW rotations, respectively. Switching (reversal in motor direction) occurs when the trace crosses zero. The profiles are representative of at least 20 individual motors.
An earlier single-motor assay, where a tethered flagellum drives the cell body, had not revealed a difference in rotation speeds of the Salmonella ΔfliL mutant and the wild type, even though bacterial swimming speeds were lower in the ΔfliL mutant (19). When we repeated the tethered-cell assay, however, we found that ΔfliL motors (UA74) displayed an average speed of 1.8 ± 0.65 Hz, with wild-type (14028) cells showing a speed of 2.4 ± 0.5 Hz; the lower motor speeds in this assay are due to the high load exerted on the filament, which drives the entire cell body. Thus, fliL motors run more slowly in this assay as well. The difference between this and the earlier measurements is likely due to an inherent instability of ΔfliL motors, as judged by the larger standard deviation in speeds compared to that in the wild type.
FliL interacts with both stator and rotor proteins.
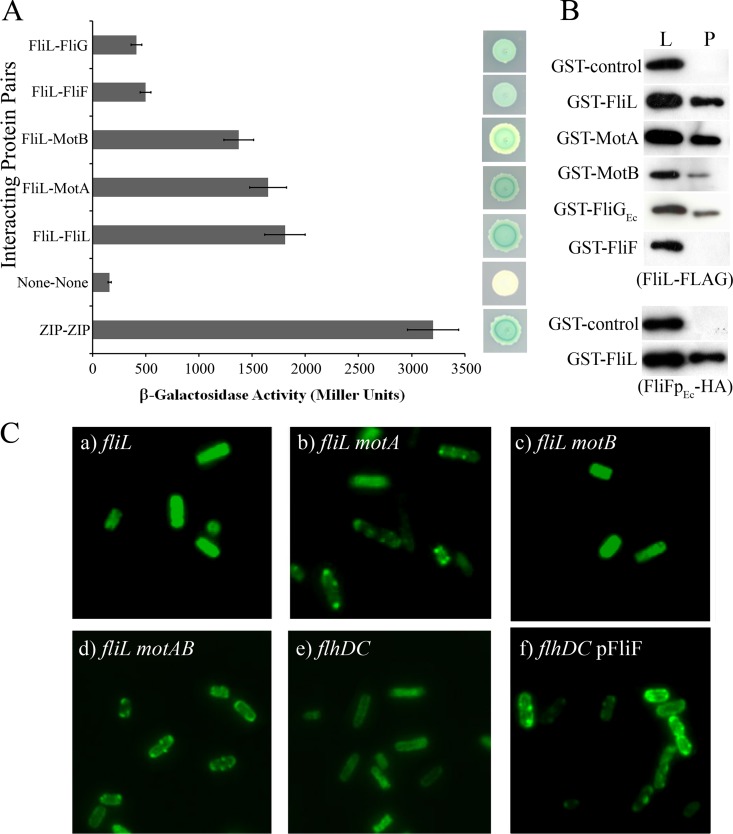
In Salmonella, FliL has a short segment in the cytoplasm (~11 N-terminal residues) and a single transmembrane domain, and the remainder of the 155-residue protein is in the periplasm (19). To determine if FliL interacts with specific rotor or stator components, two-hybrid and pulldown assays were carried out using Salmonella proteins as described in Materials and Methods (30, 31). In the former assay, FliL appeared to interact strongly with itself as well as with the stator proteins MotA and MotB and showed weak interactions with the MS ring protein FliF and the C-ring protein FliG, as measured by both quantitative β-galactosidase assays and qualitative colony color assays (Fig. 2A; ZIP is a positive control). No interaction was seen between FliL and the rod proteins (see Table S2 in the supplemental material). Positive controls using the known interacting partners MotA-MotB and MotA-FliG and negative controls using noninteracting proteins (e.g., stator-rod) were found to respond in an appropriate manner (see Table S2). The FliL interactions deduced by this assay were confirmed by pulldown assays. Here, N-terminal glutathione S-transferase (GST) fusions to prospective targets and FliL fused to the FLAG epitope were coexpressed pairwise from inducible plasmids in a strain where no other flagellar proteins were present. The detergent CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) was used to solubilize membrane proteins. FliL-FLAG was reproducibly coisolated or pulled down with the FliL-GST, MotA-GST, and MotB-GST fusions at levels significantly above those of GST-only controls, as determined by Western blotting (Fig. 2B). FliL association with FliGEc-GST was sporadic, showing positive results in only a minority of repeats (Fig. 2B), as was its association with FliF-GST. We explored the latter interaction further by coexpressing FliL-GST and a soluble periplasmic portion of FliF (FliFpEc) with a hemagglutinin (HA) epitope tag (the latter protein remained in the cytoplasm, i.e., was not exported to the periplasm). In these experiments FliFpEc-HA was reproducibly retrieved with FliL-GST (Fig. 2B, bottom). The in vivo localization patterns of FliL-GST are consistent with these data and show in addition that FliL can localize to the BB when only the MS ring is assembled (Fig. 2C).
FIG 2 .
Interaction of Salmonella FliL with itself and with rotor-stator components and localization of GFP-FliL at the BB in Salmonella. (A) Two-hybrid assays using the BACTH system, employing full-length proteins expressed from pKT25 or pUT18C, are described in Materials and Methods. Positive controls utilized proteins that dimerize via leucine zipper motifs (ZIP). Negative controls were the empty vectors (None). The strength of interactions between the indicated pairs of proteins was measured quantitatively by the Miller assay (left) as well as visually by colony color (right). The Miller values are averages from six independent biological repeats, each with three technical repeats. (B) Pulldown assays. GST fusions of indicated full-length proteins were coexpressed with either FliL-FLAG (upper set) or FliFpEc-HA (bottom set) in a ΔflhDC mutant strain (RP3098; i.e., no other flagellar proteins were present). Negative controls expressed GST alone. The detergent CHAPS was used to solubilize membrane proteins. Cell lysates were passed over Sepharose beads to trap the GST fusion protein and eluted with glutathione to determine if either FliL-FLAG or FliFpEc-HA was pulled down using appropriate antibodies to detect the epitope-tagged proteins. Lysate controls (L) show protein levels in samples prior to pulldown (P). FliFpEc, periplasmic domain of FliF from E. coli. Assays are representative of at least three independent experiments. All fusion proteins were expressed from plasmids (see Table S1 in the supplemental material). (C) All strains (a to f) expressed GFP-FliL from pJP02 and carried deletions in the indicated genes. The strains are all derived from UA74. The absence of fluorescent puncta when stators are present (a) is likely because the bulky GFP moiety prevents FliL incorporation into the assembled stators. When either MotA alone (b; JP350) or both MotA/MotB (d; JP393) were absent, a punctate pattern indicative of GFP-FliL localization at the BB was observed. The absence of puncta in the ΔmotB strain (c; JP452) is consistent with the report that MotA can localize to the BB by itself (64), where it would be expected to prevent GFP-FliL incorporation. Punctate patterns were eliminated in a ΔflhDC mutant (UA22) (no flagellar machinery present) (e) but restored if only FliF was expressed in this strain (pFliF; FliF oligomerizes into the MS ring by itself [1, 33]), suggesting that FliL can localize to the MS ring in the absence of other flagellar machinery (f).
In summary, three separate assays place FliL in close proximity to the stators and the MS ring. FliL also interacts strongly with itself. FliL interaction with the rotor switch protein FliG was weak but detectable.
FliL strengthens stator interactions and increases stator retention at the rotor.
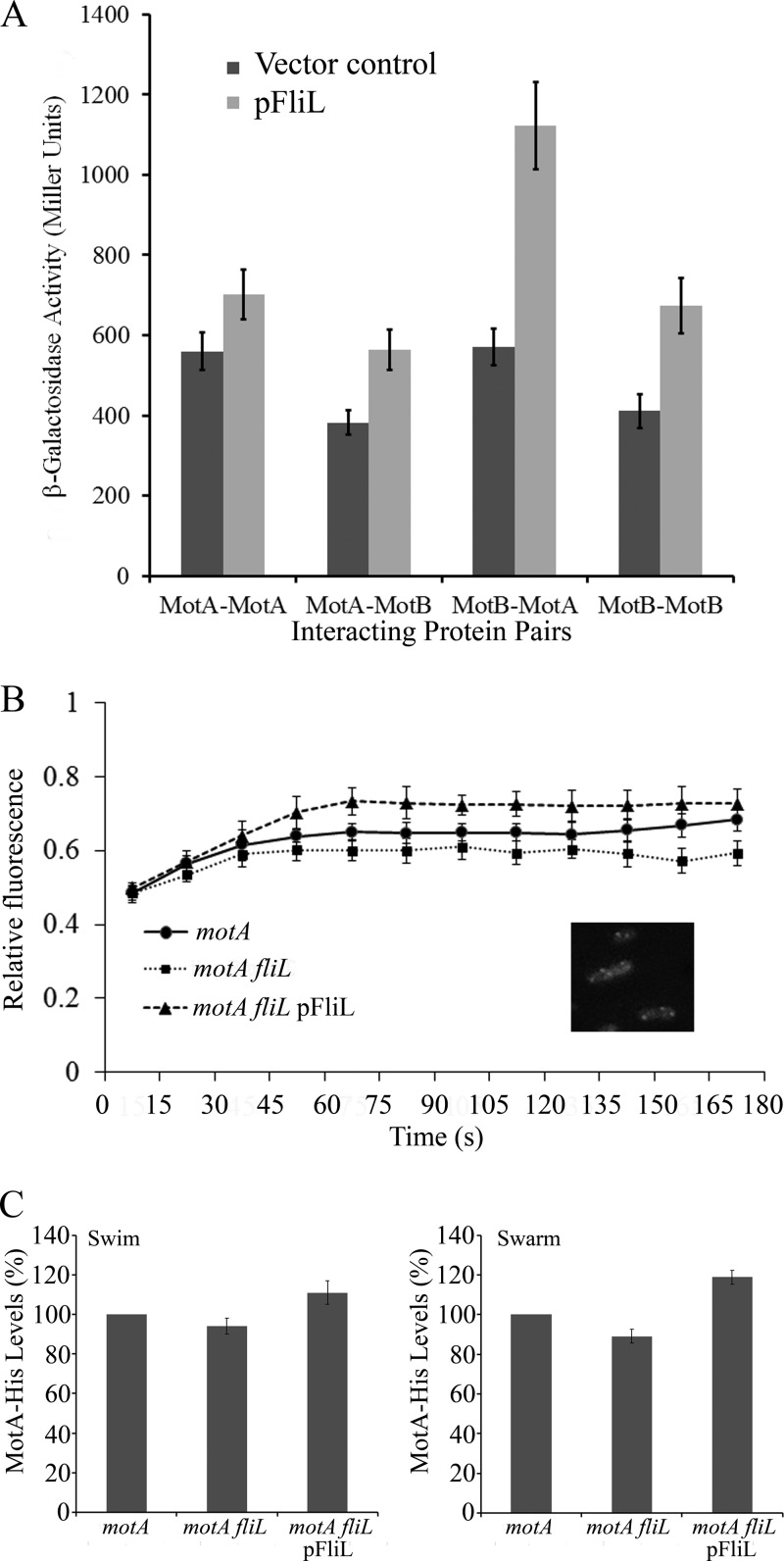
Each stator has a MotA4B2 stoichiometry (3, 32). Two MotA subunits and one MotB subunit are thought to form a single ion channel. With the finding that FliL interacts with both stator components, we investigated whether FliL was important in aiding the assembly or overall stability of the stator complex. To achieve this, we again employed the two-hybrid system and measured the effect of the presence of FliL on MotA-MotA, MotB-MotB, and MotA-MotB interacting pairs. In every combination, the presence of FliL (expressed on a third plasmid) increased the interaction of the stator proteins, with the largest effect on the MotA-MotB pair (Fig. 3A).
FIG 3 .
Influence of FliL on stator protein interaction and stator recovery at the BB in Salmonella. (A) Stator protein interaction in the absence (black bars) and presence (grey bars) of FliL was measured by two-hybrid assays as described for Fig. 2A. The vector control is empty pBAD33. MotA and MotB vectors were as in Fig. 2A. FliL was expressed from pJP134. (B) Kinetics of FRAP. Recovery of MotAEc-YFP (provided on plasmid pHL3) fluorescence after bleaching of puncta (inset) was monitored in the indicated Salmonella genetic backgrounds as described in Materials and Methods. Fluorescence intensity prior to bleaching is given a relative value of 1. Bleaching was controlled so as not to reduce the signal to 0, because recovery was poor otherwise. The data are a mean of 3 separate experiments and at least 9 individual puncta. (C) Western blots to monitor the recovery of functional MotA-His from isolated BBs in the indicated Salmonella strains under swimming or swarming conditions, with and without FliL, as well as with FliL overexpression (pFliL; pJP133) conditions as described in the supplemental material (representative blots are shown in Fig. S4). Data are the averages from 5 separate experiments. Error bars represent standard deviations from the mean.
In a second approach, we used FRAP (fluorescence recovery after photobleaching) to monitor the influence of FliL on the recycling or occupancy of the stator protein MotAEc conjugated to yellow fluorescent protein (MotAEc-YFP) at the BB in a Salmonella strain where the resident motA gene was deleted. MotAEc-YFP is functional for motility in E. coli (33), and we ascertained that it is so in Salmonella as well. Such experiments with MotB-GFP have shown that MotB turns over between the membrane pool and the motor, with a dwell time of ~30 s at the motor (34). To do this, we first located MotAEc-YFP at the motor by its punctate appearance (Fig. 3B, inset). The presence of 4 to 6 puncta per cell is consistent with the number of flagellar BBs. The puncta were bleached as described in Materials and Methods, and recovery of fluorescence at the bleached spot was monitored over a period of 3 min in a strain with or without FliL, as well as in a strain overexpressing FliL (Fig. 3B). Although the recovery was not complete, the data were highly reproducible. For the strain with FliL present (ΔmotA), we observed an initial fast recovery up to 45 s, followed by a plateau after 60 s. Final recovery (measured between 2 and 3 min) was 5 to 10% higher in the presence of FliL than its absence (ΔmotA ΔfliL). Overexpression of FliL in the ΔmotA strain showed that the initial fast phase of recovery was extended to ~70 s, with the final recovery being 5 to 10% higher than in the wild type. These data suggest that FliL promotes retention of the stators at the BB either by increasing their rate of incorporation or by slowing their turnover.
In a third approach, we used Western blots to monitor the recovery of functional MotA-His from isolated BBs under different conditions. The data show that BBs recovered from broth-grown cells (swimming conditions) had 6% less MotA in the ΔfliL mutant and 11% more in the FliL-overexpressing strain than in the wild type (Fig. 3C; also, see Fig. S4 in the supplemental material). Under swarming conditions, the BBs had 11% less MotA in the ΔfliL mutant and 19% more in the FliL overexpressing strain than in the wild type. Whole-cell controls were evaluated in each background and found to contain the same amounts of MotA, suggesting that FliL affects only the ability of MotA to associate with the BB (see Fig. S4). The swimming/swarming behavior of these strains either lacking or overexpressing FliL is shown in Fig. S5 in the supplemental material.
We conclude that FliL improves interaction between the stator proteins, as well as stator recovery at the BBs; there is a larger effect of FliL on the latter under swarming conditions.
Mutations in the “plug” region of the MotB stator alleviate motility-related fliL defects but not the rod breakage defect.
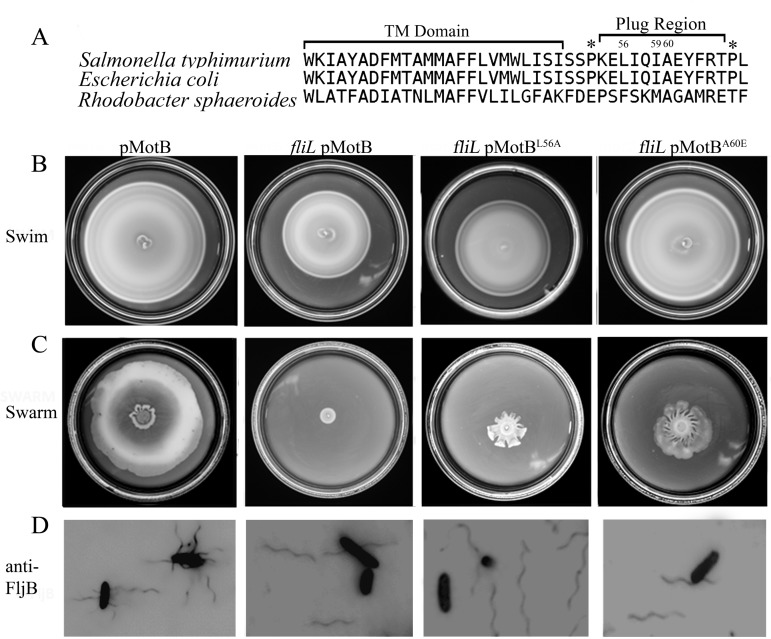
The plug region was originally identified as an eleven-residue periplasmic segment of E. coli MotB that prevents proton flow until the stators are fully assembled (Fig. 4A; also, see Fig. S1 in the supplemental material) (13). Mutations in the plug region led to proton leaks with concomitant growth defects in both E. coli (13) and Salmonella (35); growth impairment of the plug mutants is thought to be due to a premature proton leak (i.e., prior to docking with the rotor) through the unplugged mutant stators. In the course of this study, the swimming defect of a ΔfliL mutant in Rhodobacter sphaeroides was reported to be suppressed (to various degrees) by mutations in the plug region of MotB, implying a functional interplay between the two proteins, although a direct interaction was not detected (23).
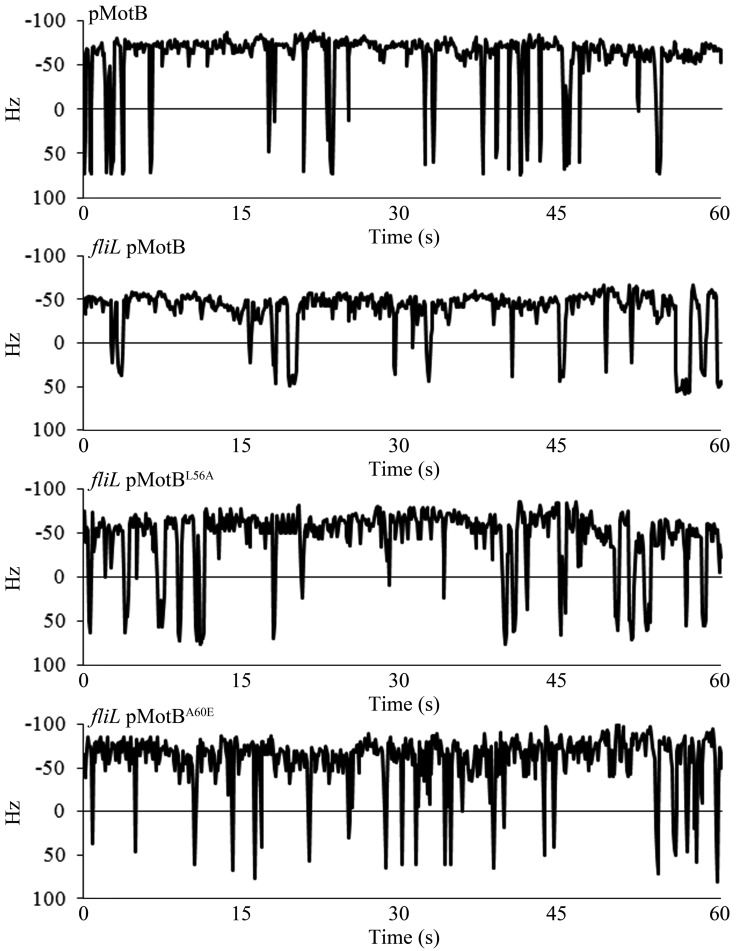
FIG 4 .
Mutations in the MotB plug suppress the motility defect of Salmonella fliL mutants but not their rod breakage phenotype. (A) The MotB plug region in Salmonella, E. coli, and Rhodobacter was aligned using T-Coffee (65). This region follows the transmembrane (TM) domain and is typically flanked by proline residues (*). The residues mutated in this study are indicated by numbers above the Salmonella sequence. L56 and A60 are equivalent to positions F63 and A67 in R. sphaeroides, respectively, substitutions at which suppressed the fliL motility defect (23), while substitutions at I59 resulted in growth defects in E. coli (13). (B and C) Two MotB plug mutations (L56A and A60E) expressed in a ΔfliL background (last two columns) suppress both swimming and swarming motility compared to the ΔfliL mutant alone (second column). The wild-type parent is shown in the first column. All strains have motB deleted on the chromosome (QW180) and are complemented by MotB from plasmid pJP74 or respective mutants from pJP114 (L56A) or pJP88 (A60E). (D) Cells picked from the edge of a swarm colony and stained with anti-flagellin (FljB) antibodies. The flagella are attached in the presence of FliL but detached in its absence, irrespective of the MotB plug mutation.
To test if mutations in the plug region of Salmonella MotB would suppress the ΔfliL motility defect as well, we engineered mutations equivalent to those isolated from E. coli as well as Rhodobacter into the MotB plug region in Salmonella (Fig. 4A; residues 56, 59 and 60) (23). As reported for E. coli, these mutations showed various growth defects in Salmonella as well (see Table S3 in the supplemental material). Interestingly, the growth defects were partially suppressed in the absence of FliL and exacerbated when FliL expression was increased, suggesting that FliL modulates the conformation of this region (see Table S3). These mutants also restored swimming motility to a ΔfliL mutant in a manner that tracked with the reported severity of their proton leak; i.e., mutants reported to have a larger proton leak (e.g., MotBA60E) (13) showed better motility than those with a less severe leak (MotBL56A) in the ΔfliL mutant (Fig. 4B). Restoration of swimming motility could be due to suppression of the growth defect; however, the observation that the more leaky stators showed higher motor speeds in the ΔfliL strain suggests that there is an additional effect of such stators on the motor (Fig. 5). The best suppressors of swimming motility (Fig. 4B) (MotBL56A and MotBA60E) in the ΔfliL mutant background showed partial rescue of swarming motility (~30 and 40% of wild-type values, respectively) (Fig. 4C). On swarm plates, the MotBL56A and MotBA60E suppressors appeared to be moving as well as the wild type when observed under the microscope, yet the swarm colony diameter was smaller than that of the wild type. When stained for the presence of flagella, the swarmer cells of the suppressors displayed broken flagella, similar to the ΔfliL mutant (Fig. 4D), likely accounting for the reduced swarm colony diameter.
FIG 5 .
Influence of FliL on single-motor behavior of MotB plug mutations in Salmonella. All strains are ΔmotB (QW180) and are complemented with either wild-type MotB (pJP74) or its indicated plug mutant derivatives (pJP114 and pJP88). Motor function was monitored by the bead assay as described for Fig. 1.
The behavior of single motors carrying the MotBL56A and MotBA60E mutant variants was consistent with the motility plate data (Fig. 5). A ΔmotB mutant expressing wild-type MotB from a plasmid averaged 58 ± 4 Hz (26 ± 3 rvpm), which dropped to 44 ± 6 Hz (15 ± 6 rvpm) when fliL was deleted, as observed for the wild-type--ΔfliL pair in Fig. 1. The plug mutations restored motor speeds and reversals to close to FliL+ levels: MotBL56A averaged 52 ± 4 Hz (21 ± 5 rvpm), and MotBL60A averaged 55 ± 5 Hz (21 ± 5 rvpm). Expression of these plasmids in a ΔmotB strain (i.e., otherwise wild type) increased motor output in this background as well (see Fig. S6 in the supplemental material): MotBL56A averaged 63 ± 4 Hz (24 ± 4 rvpm) and MotBL60A averaged 61 ± 4 Hz (28 ± 5 rvpm), slightly above the 58 Hz (26 ± 3 rvpm) seen when native MotB was expressed from a plasmid.
We conclude from these data that FliL has two separate functions, one contributing to motility and the other to rod stability. The motility function likely involves FliL interaction with the stators, and the rod stability function likely involves interaction with the MS ring. Restoration of normal motor speeds in a ΔfliL mutant when MotB is unplugged suggests that FliL normally favors the unplugged state and increases stator engagement with the rotor and/or that FliL increases the efficiency of proton flow through the motor.
FliL defects are exacerbated under conditions of high load.
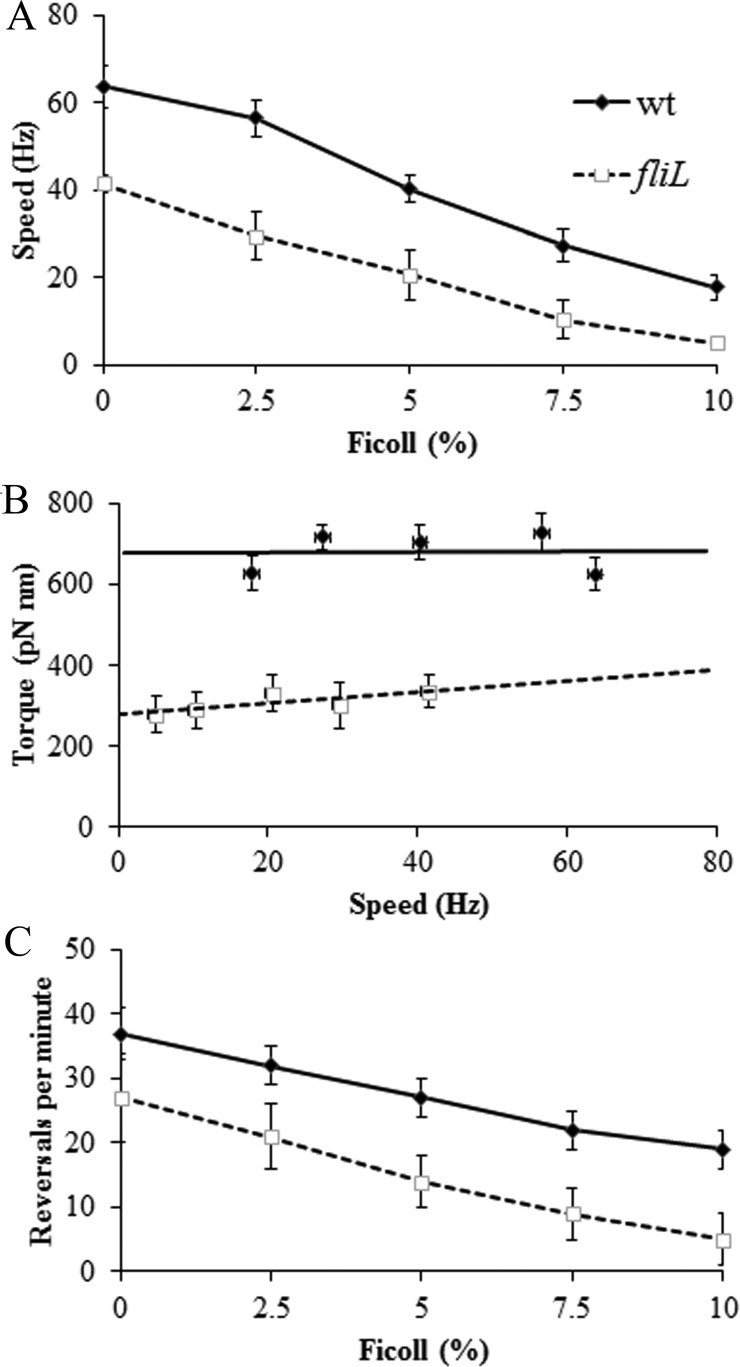
FliL is critically important to Salmonella and E. coli under swarming conditions (Fig. 4; also, see Fig. S2 in the supplemental material) (18, 19). While it is hard to know exactly what feature of the surface prevents swarming or causes the flagella to break, we expect that there is at least a higher viscous drag on the flagellum. To replicate this condition, external load on the motor was increased by using different concentrations of the viscous agent Ficoll (16). E. coli was selected for this experiment because Salmonella motors were difficult to work with in Ficoll. As expected, increasing Ficoll caused both wild-type and ΔfliL mutant motors of E. coli to decrease speed, with the ΔfliL mutant being slower at all Ficoll concentrations tested as measured by the bead assay (Fig. 6A). The power output of a motor is torque × angular velocity (2π × torque × rotational speed) (16). The torque or force required to spin the motor (with its attached bead) in a viscous medium is its rotational frictional drag coefficient × the angular velocity. When we estimated the torque values in the different Ficoll concentrations and plotted them versus speed as described previously (36), the torque for wild-type motors was constant over the experimental speed range, as observed earlier (16) (Fig. 6B). However, the fliL mutant produced much less torque at higher than lower loads. Switching behavior of wild-type and ΔfliL E. coli motors was also evaluated over the range of Ficoll concentrations. The finding that ΔfliL switches less frequently than the wild type (Fig. 1) was evident across all Ficoll concentrations (Fig. 6C).
FIG 6 .
Effect of viscosity on the behavior of fliL mutant motors of E. coli. (A) Motor speeds of wild-type E. coli (MT02) and its ΔfliL derivative (JP1297) monitored with increasing Ficoll concentrations as described for Fig. 1. (B) Torque versus speed plots of the data in panel A, derived as described in Materials and Methods. (C) Switching frequency of the motor, derived from data in panel A. At least 10 individual motors were monitored at each Ficoll concentration.
We conclude that FliL contributes to torque at all viscous loads tested and plays a critical role in torque generation at higher loads, because the motor produced less torque without FliL at these higher loads. This property of FliL is likely essential on a surface where the load is expected to be high.
DISCUSSION
The FliL protein is found across all bacterial genera and has diverse developmental and motility phenotypes associated with its loss. This study addresses the molecular basis of the motility phenotype in Salmonella and E. coli, which is likely to be shared with other bacteria.
Model for FliL location at the motor.
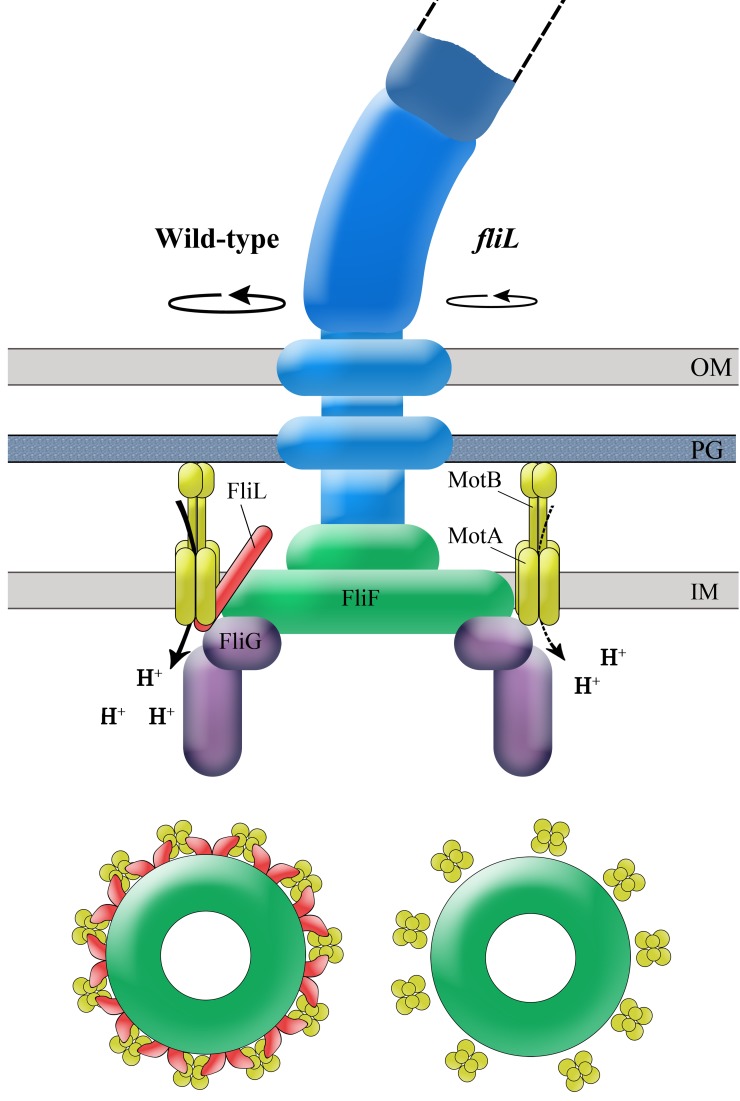
Based on the data presented in Fig. 1 to 5 and in Fig. S4 to S6 in the supplemental material, a model for FliL positioning at the motor is diagrammed in Fig. 7. The strong inter-FliL interactions suggest that FliL is at least a dimer, positioned at the motor so as to interact with both stators (MotA and MotB), the MS ring (FliF), and the C-ring switching apparatus (FliG), from where it contributes to both motor output and switching (Fig. 1). In a study using FRET assays, strong FliL-FliL and weak FliL-FliG interactions were also observed in E. coli (33). FliL-FliL interactions have been detected in R. sphaeroides as well, using two-hybrid assays (23). The location of FliL as depicted in Fig. 7 is likely to be similar in other bacteria, because two-hybrid analysis detected FliL-MotB interactions in Campylobacter jejuni (37), suppressors of the fliL motility defect in R. sphaeroides mapped to the plug region of MotB (23), and cryo-ET data in Borrelia burgdorferi place FliL near the stators (24).
FIG 7 .
Model showing the location and function of FliL at the motor. FliL is positioned to interact closely with both the stators and the MS ring (side view of the flagellum and cross view of the MS ring and stators). This arrangement stabilizes the stators and enhances proton flow/efficiency, delivering a higher torque, as depicted on the left side of the flagellum image. Interaction of FliL with FliG as well as MotA (which interacts with FliG) contributes to the switching behavior of the motor. The lower rotation speed in the absence of FliL is depicted on the right side of the flagellum image. Strong FliL-FliL interactions suggest that FliL is at least a dimer, as indicated in the cross view. Association of FliL with the MS ring protein FliF may reinforce the rod, helping it to withstand high external load during swarming. IM, inner membrane; PG, peptidoglycan layer; OM, outer membrane.
FliL and motor output.
A clear finding in this study is that in the absence of FliL, single motors of both Salmonella and E. coli rotate at lower speeds (Fig. 1). Earlier, measurement of bacterial free swimming speeds showed ~20% speed reduction in the Salmonella ΔfliL mutant (19), consistent with the reduced single-motor speeds measured in this study for fliL mutants of both Salmonella and E. coli. The drop in speed could be related to stator occupancy. The MotAB stator complexes can vary in number from 0 to 11, depending on the load on the motor (34, 38, 39). Without FliL, the stators may be unable to achieve maximum occupancy, as suggested by FRAP and Western blot data (Fig. 3B and C; also, see Fig. S4 in the supplemental material). (We note that simply overexpressing MotAB could not compensate for the swimming defect associated with the loss of FliL [data not shown], consistent with FliL's essential role in maximizing stator function [18].) Enhanced stator occupancy could increase motor speed by delivering more power. Additionally, the increased interaction strength between stator proteins in the presence of FliL (Fig. 3A) may increase the efficiency of proton flow. These results appear to be at odds with a recent study in E. coli by Lele et al., where the kinetics of stator incorporation into motors subjected to a sudden load increase was stated to be independent of FliL (39); the study did not report the actual speeds achieved in the absence of FliL. It is possible that FliL does not increase the rate of stator incorporation but does increase their dwell time at the motor.
Mutations in the plug region of Salmonella MotB, analogous to those in E. coli MotB that were reported to leak protons prematurely (13), were able to restore motor speeds to fliL mutant motors (Fig. 5). Because the unplugged conformation of MotB is normally attained when the stators dock with the rotor (see Fig. S1 in the supplemental material), these unplugged mutant stators might favor docking and produce higher motor speeds. Indeed, the increased stator retention at the motor observed in the presence of FliL (Fig. 3) could well be due to stabilization of the unplugged state by FliL. Alternatively, the higher motor speeds of these mutants could be due to increased proton flow through the leaky stators in the presence of FliL. Comparison of Fig. 5 and Fig. S6 in the supplemental material would favor this idea, because motor speed with the leaky stators is higher in the presence of FliL (see Fig. S6) than in its absence (Fig. 5).
In E. coli, the MotB plug mutations cause various degrees of growth defects because they are thought to open the proton channel before engaging with the rotor (13). That the absence of FliL suppresses the growth defect of equivalent Salmonella MotB mutants, while overexpression of FliL exacerbates their growth defect (Table S3), suggests that FliL likely modulates the MotB plug conformation prior to their engagement with the motor.
In wild-type E. coli motors, torque is approximately constant at heavy viscous loads and low speeds, where motor efficiency is high (2, 16) (Fig. 6B). A curious finding was that while absence of FliL reduces motor torque at all viscous loads tested, the torque reduction was more pronounced at higher viscous loads (Fig. 6B). If FliL has a rotor-stabilizing role (for example, rotor stiffening), FliL might act to resist certain deformations of the structure, and these deformations might have a role in signaling the presence of high load.
FliL and motor bias.
An unexpected property of FliL was its influence on motor bias in both Salmonella and E. coli; motors switched less frequently without FliL (Fig. 1 and 5). Since FliL interacts, albeit weakly, with the switch protein FliG in both bacteria (Fig. 2) (33), the altered bias could be directly related to abrogation of FliL-FliG contacts. Alternatively, FliL may influence switching indirectly by altering MotA-FliG contacts (Fig. 7); stator occupancy is known to affect motor bias (39–41). It has been suggested that the switch may respond to torque or sense motor speed by means of proton flux (40). FliL affects all these parameters. Another possibility is that FliL-FliF interactions are communicated to the switch protein FliG via FliF, on which FliG is directly mounted. This is an attractive possibility, given that FliL-FliF interactions are communicated to rod proteins, as discussed below.
Structural and sensory role of FliL.
Cells attempting to swarm in the absence of FliL break their flagella within the rod in Salmonella and E. coli (19) (see Fig. S2 in the supplemental material). We did not detect an interaction of FliL with rod proteins (see Table S2 in the supplemental material), suggesting that FliL stabilizes the rod indirectly, likely via FliF, to which the rod is connected. This conjecture is supported by the observation that a point mutation in Salmonella FliF exhibited a similar rod fracture phenotype (42). Interestingly, even though ΔfliL motor speeds and swim colony diameters were restored by MotB plug mutations, and even though cells swarmed vigorously, their flagella still broke during swarming (Fig. 4). These experiments reveal at least two separate FliL functions—one optimizing stator function and the other, a structural role, reinforcing the periplasmic segment of the BB.
In C. crescentus, a developmental signal communicated via cyclic di-GMP (c-di-GMP) to the flagellum elicits FliL-dependent FliF degradation, which results in ejection of the flagellum during transition from a motile to a sessile state; the flagellum is severed within the rod, similar to the swarming phenotype of Salmonella/E. coli, except in the presence of FliL (21, 27, 28, 43, 44). FliL is absolutely essential for motility in C. crescentus, as it is in R. sphaeroides, unlike its selective role only during swarming in Salmonella/E. coli. The C. crescentus FliL is 55 residues longer than the FliL in Salmonella/E. coli and clearly performs roles in addition to motility, such as sensing c-di-GMP cues. However, FliL-FliF interactions may be communicated to the rod in analogous ways in all three bacteria.
Flagellated bacteria are sensitive to a variety of environmental factors and are well known to use a chemotaxis system to transmit information about their chemical environment to the flagellar motor (2). Conversely, they use the flagellar apparatus to transmit information about the physical environment to various transcriptional and posttranscriptional pathways that ensure an appropriate developmental response. For example, swimming in viscous solutions or on surfaces triggers changes such as induction of lateral flagellar synthesis in Vibrio parahaemolyticus (45, 46) or upregulation of flagellar and virulence genes in Proteus mirabilis, responses that enhance swarming (47). In the former case, changes in lateral flagellar gene expression also occur by exposure to sodium channel blockers, indicating that they might be caused by interference with the function of the sodium ion-powered stators that drive the polar flagellum (48). In C. crescentus, surface adherence via retractile pili, which restricts rotation of the flagellum, induces secretion of holdfasts that attach bacteria to the surface (49). In Bacillus subtilis, inactivation of the stators induces the transcription of genes for the synthesis of a highly mucoid polymer that promotes biofilms (50, 51). While the flagellar motor is clearly implicated as a sensory device in all these bacteria, how such a device would work is not yet understood. It is likely that external mechanical stimuli and internal disengagement of stators (52), whether they alter proton flux, stator occupancy, or switch conformation, ultimately all manipulate the rotor-stator interface, relaying the signal to rotor- or stator-associated proteins. FliL, which interacts with both rotor and stator, may provide an important handle to probe this sensory device.
Concluding remarks.
The hallmark of a robust biological system is its ability to persist (show only subtle effects) in the face of perturbations, whether genetic or nongenetic. Often, the advantages of apparently dispensable parts are revealed only when populations face diverse environmental challenges. This is the case for FliL, whose effect on swimming is subtle but whose effect on swarming is drastic in E. coli and Salmonella. Our findings speak to both effects. With regard to the former, single-motor analysis shows that the “normal” motor bias has a hitherto-unsuspected FliL component to it. Given that small changes in bias have a huge impact on competitive fitness, as measured by chemotaxis, our results are expected to factor this additional parameter into understanding the robustness of the chemotactic response, the best-modeled signal transduction system (53, 54). Our results are also relevant to motor function in organisms where FliL is essential for swimming. With regard to swarming, our results show that FliL has two separable functions, one providing increased motor torque and the other stabilizing the rod.
MATERIALS AND METHODS
Only a partial set of materials and methods is included here; more standard ones can be found in the supplemental material. Strains and plasmids are listed in Table S1 in the supplemental material.
Measurement of single-flagellum motor rotation.
Single-flagellum motors were analyzed as described previously (36, 55). Salmonella strains used in these experiments carried a chromosomal replacement of fliC with fliCst (from SJW1103 fliCst) linked to tetRA and moved into a strain expressing only FliC (hin+C) (JP1107). FliC was rendered “sticky” by the deletion of codons 205 to 293 (56). E. coli strains used had FliC replaced by an unmarked fliCst variant (MT02; Armitage lab, Oxford, United Kingdom). Cells with sheared filaments were attached to a glass slide, and polystyrene beads were attached to the sticky filament stubs as described below. High-speed video of individual beads was captured, and bead rotation was used as an indicator of motor function.
Overnight cultures of Salmonella and E. coli were diluted 1:100 in fresh LB (5 ml) and grown at 30°C for 5 h. One milliliter of cells was centrifuged at 5,000 × g for 5 min, and the pellet was resuspended in 500 µl of potassium phosphate motility buffer (10 mM potassium phosphate buffer [pH 7.0], 0.1 mM EDTA [pH 7.0], 10 mM NaCl, 75 mM KCl). The suspension was subjected to syringe passage to shear filaments (36). Forty microliters of the cell suspension was loaded into the space between an 18- by 18-mm polylysine-treated coverslip and a 24- by 50-mm glass slide that were separated by double-sided tape and incubated at room temperature for 10 min to allow the cells to attach to the coverslip. The space between the coverslip and slide was gently washed 2 or 3 times with 40 µl of motility buffer to remove the remaining unattached cells. Cells were washed with 40 µl of a 1:50 dilution of polystyrene beads (Polysciences, Warrington, PA), 0.75 µm in diameter. The mixture was incubated at room temperature for 10 min to allow the beads to attach to the flagellar filaments. They were gently washed 3 times again with 40 µl of motility buffer to remove unattached beads. Ficoll 400 (Sigma) solutions in motility buffer, when used, were added after this final washing step with a further 15-min incubation at room temperature before bead capture. The rotational motions of the beads were observed by phase-contrast microscopy (BX53F; Olympus, Tokyo, Japan) and recorded with a high-speed charge-coupled device (CCD) camera (ICL-B0620M-KC0; Imperx, Boca Raton, FL) at 1,250 frames/s. Phase-contrast images of each bead were cropped to the proper pixel size (16 by 16 to 22 by 22 pixels) and converted to videos (.avi files) with XCAP image processing software (Epix Inc., Buffalo Grove, IL, USA). Contrast enhancing, to fix image brightness and reduce background noise, was performed using ImageJ software (http://rsb.info.nih.gov/ij/). Videos of each bead were processed using custom analytical programs within LabVIEW 2012 (National Instruments, Austin, TX), which were provided by Yuichi Inoue (Ishijima Lab, Tohoku University, Sendai, Japan). Program 1 fitted each video by a two-dimensional Gaussian function to determine the center of the bead (x,y coordinates). The data were stored as a text file (.txt) and used by Program 2 to calculate the angular (rotation) speed from the center of the bead. Torque output of motors was calculated as described previously (36).
The tethered-cell assay was carried out as described previously (57). Tethered bacteria were observed through phase-contrast microscopy (BX53F; Olympus, Tokyo, Japan) and recorded using Olympus cellSens software. Average rotation speed per minute was recorded from playback at reduced speed and converted to Hz for this report.
Two-hybrid screen of protein-protein interactions.
Interactions between Salmonella proteins of interest were screened using the BACTH bacterial two-hybrid system (Euromedex) as described by Karimova et al. (30, 31). In this assay, proteins of interest are genetically fused in various combinations to two fragments (T25 and T18) of the catalytic domain of Bordetella pertussis adenylate cyclase (AC) and coexpressed in an E. coli strain deficient in endogenous AC. Interaction of the two hybrid proteins brings the T25 and T18 fragments together, leading to cyclic AMP (cAMP) synthesis and in turn to transcriptional activation of a lacZ reporter system. The pKT25 and pUT18C plasmids were used with the host strain BTH101, with a deletion of the flagellar master regulon gene flhC (JP319) to prevent native expression of flagellar genes and assembly of flagellar structures. Positive controls utilized the plasmids pKT25-ZIP and pUT18C-ZIP, with each expressing fusion proteins that associate through dimerization of leucine zipper motifs expressed. Negative controls were empty vectors. Genes of interest were amplified from Salmonella enterica genomic DNA by PCR and fused in frame to the gene fragments in plasmids pKT25 and/or pUT18C. Plasmid pairs were cotransformed into the host strain and plated on LB agar supplemented with ampicillin, kanamycin, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 µg·ml−1), and IPTG (isopropyl-β-D-thiogalactopyranoside; 0.5 mM). Plates were incubated at 30°C for 48 h, with noninteracting proteins remaining white and potential interactants exhibiting a range of blue coloration. Levels of interaction were quantified in liquid cultures with the Miller assay for measuring β-galactosidase (58). Overnight cultures were subinoculated (1:100) and grown at 30°C to an optical density at 600 nm (OD600) of 0.5 to 0.7 in the presence of 0.5 mM IPTG plus appropriate antibiotics.
GST pulldown assays.
Protein pairs to be tested were coexpressed in an flhDC strain (RP3098) so that the two proteins of interest were the only flagellar proteins present (59). The plasmid pHT100 (59) was used to express the GST fusions, with FliLFLAG coexpressed from pTRC99a (pUA2) (19). All proteins tested in this assay were from Salmonella, except for FliFpEc, which was from E. coli (pKP553). Control experiments used empty pHT100 (GST only). The basic pulldown assay was performed as described elsewhere (60). Several of the tester proteins are in the membrane. Although the detergent dodecylphosphocholine (DPC) has been shown to be especially effective for solubilizing MotA/MotB complexes from E. coli membranes (61), we had better success with CHAPS [3-(3-cholamidopropyl)dimethylammonio-1-propanesulfonate] (15, 59, 62), which was added to the cells as follows. Plasmid-bearing cells were grown overnight at 30°C in 40 ml tryptone broth (TB) containing appropriate antibiotics and 400 µM IPTG, harvested, and resuspended in 1 ml resuspension buffer (phosphate-buffered saline [PBS]: 8 g NaCl liter−1, 0.2 g KCl, 1.44 g Na2HPO4, and 0.24 g KH2PO4 in 1 liter distilled H2O [adjusted to pH 7.4] with 5 mM EDTA, 0.2 mM PMSF [phenylmethanesulfonylfluoride], 0.1% CHAPS, and 10 mg·ml−1 lysozyme). After 1 h incubation on ice, cells were disrupted by sonication before centrifugation at 4°C (13,000 rpm, 40 min). The supernatant was retained, with a 50-µl aliquot taken to estimate protein levels and the remainder (~1 ml) mixed with 150 µl of glutathione Sepharose 4B (Pharmacia) that had been subjected to 3 washes with resuspension buffer (without lysozyme) and resuspended as a 50% slurry. Samples were incubated at room temperature for 2 h with gentle rotation to allow binding. Following incubation, Sepharose beads were collected by centrifugation (13,000 rpm, 30 s) and washed with PBS. This spin-and-wash step was repeated twice. Samples for pulldowns with FliG were washed with PBS containing 1% bovine serum albumin (BSA) and 0.1% Triton-X as detailed in reference 60. After a washing, the beads were incubated with 50 µl of elution buffer (50 mM reduced glutathione in 50 mM Tris-HCl [pH 8.0]) for 10 min at room temperature with gentle rotation to release the GST fusion protein and associated proteins. Beads were then pelleted and the supernatant was collected for analysis by SDS-PAGE and immunoblotting using appropriate antibodies, either anti-FLAG or anti-HA (Sigma).
FRAP analysis.
FRAP of MotAEc-YFP (pHL3; gift from Victor Sourjik [33]) was carried out in Salmonella strains grown at 30°C in TB supplemented with 10 µM IPTG and appropriate antibiotics to an OD600 of 0.5. Cells were harvested (5 min, 4,000 rpm) and washed once in tethering buffer (10 mM potassium phosphate, 0.1 mM EDTA, 1 mM l-methionine, 67 mM sodium chloride, 10 mM sodium lactate [pH 7]) before resuspension in tethering buffer and attachment to a polylysine-treated coverslip. Measurements were performed at room temperature using a Leica SP2 AOBS confocal microscope with a 63× oil objective and YFP emission channel (525 to 650 nm). Cells with similar levels of fluorescence were selected for bleaching experiments; a region of interest (ROI) containing a motor was bleached for 3 s at maximal power. Bleaching was controlled so as not to reduce the signal to 0, because recovery was poor otherwise. One prebleach image was taken and postbleach images captured every 15 s (up to 165 s) in 512-by-512 format with line averaging (2×) using Leica Confocal software, version 2.61. Captured images were subsequently analyzed using ImageJ software as described elsewhere (63). For comparison of multiple experiments with different bleaching depths and cluster intensities, the relative fluorescence intensity of the ROI in the image sequence was normalized to the relative ROI intensity prior to bleaching. Data are representative of least 9 separate ROI evaluated over 3 separate experiments (days).
SUPPLEMENTAL MATERIAL
The flagellar apparatus of Salmonella and E. coli. The entire structure from the cytoplasmic C ring to the external filament rotates (curved arrows). Stationary elements are MotAB complexes, which conduct protons and power the rotor, and the L and P rings, which act as bushings for the rod. MotAB insert and drift in the membrane until they encounter the basal MS ring structure, upon which the plugged proton channel opens. See the text for other descriptions. OM, outer membrane; PG, peptidoglycan; IM, inner membrane. Download
The effects of fliL on motility and rod fracture are similar in both Salmonella and E. coli. (A and B) Salmonella wild type (14028) and ΔfliL (UA74) and E. coli wild type (RP437) and ΔfliL (JP633a) were monitored for swimming and swarming motility in LB media by inoculating soft agar (0.3%) and swarm agar (0.6% supplemented with 0.5% glucose), respectively. Plates were incubated at 30°C (E. coli) or 37°C (Salmonella) for 6 h (swimming) or 8 h (swarming). (C) Cells were picked from swarm plates shown in the panel above and immune-stained for flagella as described in Text S1. Download
Salmonella displays more robust swimming motility than E. coli. Two different wild-type E. coli (top) and Salmonella (bottom) strains (see Table S1) were monitored for swimming motility by inoculating overnight cultures at the center of soft agar plates (LB media with 0.3% agar) and incubating them at 30°C for 6 h. This assay monitors both swimming speed and chemotactic proficiency. Download
Western blots to monitor the recovery of functional MotA-His under various conditions in Salmonella. Protein loading in whole cells and BBs was controlled for with antibodies to the periplasmic protein MBP and the BB protein FliM, respectively. Protein bands were quantified as described in the supplemental material. All strains were ΔmotA, with MotA-His provided from pJP45. BBs recovered under swimming conditions (broth-grown cells) showed 4% less MotA in the ΔfliL mutant (pJP530) and 10% more in the FliL-overexpressing strain (pJP133) than in the wild type (QW178); swim diameters of these strains followed a similar trend (see Fig. S5). BBs recovered under swarming conditions had 10% less MotA in the ΔfliL mutant and 18% more in the FliL overexpressing strain than in the wild type. The swarm diameter of the overexpression strain was also larger than that of the wild type (see Fig. S5). Blots are representative of 5 independent experiments. Download
Effect of absence or overexpression of FliL on swimming and swarming motility in Salmonella. Wild-type Salmonella (14028), an isogenic ΔfliL mutant (UA74), and a wild-type strain with pFliL (pJP133) were inoculated on swim (0.3% agar) and swarm (0.6%) plates as described in Text S1 and incubated for 6 h at 37°C. Download
MotB plug mutations increase speed of Salmonella wild-type motors. Single motors of ΔmotB strains (QW180) containing plasmids expressing the indicated MotB variants were monitored by bead assay as described for Fig. 1. Motors with wild-type MotB rotated at 58 Hz (26 ± 3 rvpm), MotBL56A averaged 63 Hz ± 4 (24 ± 4 rvpm), and MotBL60A averaged 61 Hz ± 4 (28 ± 5 rvpm). Download
(A) Strains and plasmids used in this study. (B) Extent of gene deletions in this study.
Two-hybrid analysis of basal body proteins.
Effect of FliL on growth defects associated with mutations in the MotB plug region.
Supplemental methods. Download
ACKNOWLEDGMENTS
We thank Judy Armitage, David Blair, Akihiko Ishijima, Yuichi Inoue, Michael Manson, and Victor Sourjik for strains, plasmids, and helpful discussion and Ryan Sullivan for help with data analysis. We also thank the anonymous reviewers of the original manuscript for useful insights and helpful suggestions.
This work was supported by National Institutes of Health grants GM57400 and GM112507 and in part by the Robert Welch Foundation, grant F-1351.
Footnotes
Citation Partridge JD, Nieto V, Harshey RM. 2015. A new player at the flagellar motor: FliL controls both motor output and bias. mBio 6(2):e02367-14. doi:10.1128/mBio.02367-14.
REFERENCES
- 1.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 2.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 3.Kojima S, Blair DF. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol 233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 4.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minamino T, Imada K, Namba K. 2008. Molecular motors of the bacterial flagella. Curr Opin Struct Biol 18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Harshey RM. 2011. New insights into the role and formation of flagella in Salmonella, p 163–186. In Powollik S (ed), Salmonella: from genome to function. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 7.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 8.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partridge JD, Harshey RM. 2013. Swarming: flexible roaming plans. J Bacteriol 195:909–918. doi: 10.1128/JB.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Block SM, Berg HC. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 11.Blair DF, Berg HC. 1988. Restoration of torque in defective flagellar motors. Science 242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 12.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosking ER, Vogt C, Bakker EP, Manson MD. 2006. The Escherichia coli MotAB proton channel unplugged. J Mol Biol 364:921–937. doi: 10.1016/j.jmb.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci U S A 95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Berg HC. 2000. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys J 78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan J, Berg HC. 2008. Resurrection of the flagellar rotary motor near zero load. Proc Natl Acad Sci U S A 105:1182–1185. doi: 10.1073/pnas.0711539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partridge JD, Harshey RM. 2013. More than motility: Salmonella flagella contribute to overriding friction and facilitating colony hydration during swarming. J Bacteriol 195:919–929. doi: 10.1128/JB.02064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Attmannspacher U, Scharf BE, Harshey RM. 2008. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol Microbiol 68:328–341. doi: 10.1111/j.1365-2958.2008.06170.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu R, Ochman H. 2007. Origins of flagellar gene operons and secondary flagellar systems. J Bacteriol 189:7098–7104. doi: 10.1128/JB.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenal U, White J, Shapiro L. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J Mol Biol 243:227–244. doi: 10.1006/jmbi.1994.1650. [DOI] [PubMed] [Google Scholar]
- 22.Segura A, Duque E, Hurtado A, Ramos JL. 2001. Mutations in genes involved in the flagellar export apparatus of the solvent-tolerant Pseudomonas putida DOT-T1E strain impair motility and lead to hypersensitivity to toluene shocks. J Bacteriol 183:4127–4133. doi: 10.1128/JB.183.14.4127-4133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suaste-Olmos F, Domenzain C, Mireles-Rodríguez JC, Poggio S, Osorio A, Dreyfus G, Camarena L. 2010. The flagellar protein FliL is essential for swimming in Rhodobacter sphaeroides. J Bacteriol 192:6230–6239. doi: 10.1128/JB.00655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motaleb MA, Pitzer JE, Sultan SZ, Liu J. 2011. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol 193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cusick K, Lee YY, Youchak B, Belas R. 2012. Perturbation of FliL interferes with Proteus mirabilis swarmer cell gene expression and differentiation. J Bacteriol 194:437–447. doi: 10.1128/JB.05998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belas R, Suvanasuthi R. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J Bacteriol 187:6789–6803. doi: 10.1128/JB.187.19.6789-6803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aldridge P, Jenal U. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol Microbiol 32:379–391. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]
- 28.Christen M, Christen B, Allan MG, Folcher M, Jenö P, Grzesiek S, Jenal U. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci U S A 104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minamino T, Imada K, Kinoshita M, Nakamura S, Morimoto YV, Namba K. 2011. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol 9:e1000616. doi: 10.1371/journal.pbio.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braun TF, Al-Mawsawi LQ, Kojima S, Blair DF. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35–45. doi: 10.1021/bi035406d. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Sourjik V. 2011. Assembly and stability of flagellar motor in Escherichia coli. Mol Microbiol 80:886–899. doi: 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- 34.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 35.Morimoto YV, Che YS, Minamino T, Namba K. 2010. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett 584:1268–1272. doi: 10.1016/j.febslet.2010.02.051. [DOI] [PubMed] [Google Scholar]
- 36.Ryu WS, Berry RM, Berg HC. 2000. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403:444–447. doi: 10.1038/35000233. [DOI] [PubMed] [Google Scholar]
- 37.Rajagopala SV, Titz B, Goll J, Parrish JR, Wohlbold K, McKevitt MT, Palzkill T, Mori H, Finley RL, Uetz P. 2007. The protein network of bacterial motility. Mol Syst Biol 3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. 2013. Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551-13. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lele PP, Hosu BG, Berg HC. 2013. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci U S A 110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fahrner KA, Ryu WS, Berg HC. 2003. Biomechanics: bacterial flagellar switching under load. Nature 423:938. doi: 10.1038/423938a. [DOI] [PubMed] [Google Scholar]
- 41.Yuan J, Fahrner KA, Berg HC. 2009. Switching of the bacterial flagellar motor near zero load. J Mol Biol 390:394–400. doi: 10.1016/j.jmb.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okino H, Isomura M, Yamaguchi S, Magariyama Y, Kudo S, Aizawa SI. 1989. Release of flagellar filament-hook-rod complex by a Salmonella Typhimurium mutant defective in the M ring of the basal body. J Bacteriol 171:2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanbe M, Shibata S, Umino Y, Jenal U, Aizawa SI. 2005. Protease susceptibility of the Caulobacter crescentus flagellar hook-basal body: a possible mechanism of flagellar ejection during cell differentiation. Microbiology 151:433–438. doi: 10.1099/mic.0.27386-0. [DOI] [PubMed] [Google Scholar]
- 45.McCarter L, Hilmen M, Silverman M. 1988. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 46.McCarter LL. 2004. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol 7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 47.Pearson MM, Rasko DA, Smith SN, Mobley HL. 2010. Transcriptome of swarming Proteus mirabilis. Infect Immun 78:2834–2845. doi: 10.1128/IAI.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawagishi I, Imagawa M, Imae Y, McCarter L, Homma M. 1996. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol Microbiol 20:693–699. doi: 10.1111/j.1365-2958.1996.tb02509.x. [DOI] [PubMed] [Google Scholar]
- 49.Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol 83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cairns LS, Marlow VL, Bissett E, Ostrowski A, Stanley-Wall NR. 2013. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol Microbiol 90:6–21. doi: 10.1111/mmi.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan JM, Guttenplan SB, Kearns DB. 2014. Defects in the flagellar motor increase synthesis of poly-gamma-glutamate in Bacillus subtilis. J Bacteriol 196:740–753. doi: 10.1128/JB.01217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 53.Bray D, Levin MD, Lipkow K. 2007. The chemotactic behavior of computer-based surrogate bacteria. Curr Biol 17:12–19. doi: 10.1016/j.cub.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 54.Tu Y. 2013. Quantitative modeling of bacterial chemotaxis: signal amplification and accurate adaptation. Annu Rev Biophys 42:337–359. doi: 10.1146/annurev-biophys-083012-130358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terasawa S, Fukuoka H, Inoue Y, Sagawa T, Takahashi H, Ishijima A. 2011. Coordinated reversal of flagellar motors on a single Escherichia coli cell. Biophys J 100:2193–2200. doi: 10.1016/j.bpj.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scharf BE, Fahrner KA, Turner L, Berg HC. 1998. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci U S A 95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mariconda S, Wang Q, Harshey RM. 2006. A mechanical role for the chemotaxis system in swarming motility. Mol Microbiol 60:1590–1602. doi: 10.1111/j.1365-2958.2006.05208.x. [DOI] [PubMed] [Google Scholar]
- 58.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 59.Tang H, Braun TF, Blair DF. 1996. Motility protein complexes in the bacterial flagellar motor. J Mol Biol 261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 60.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kojima S, Blair DF. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 62.Che YS, Nakamura S, Kojima S, Kami-ike N, Namba K, Minamino T. 2008. Suppressor analysis of the MotB(D33E) mutation to probe bacterial flagellar motor dynamics coupled with proton translocation. J Bacteriol 190:6660–6667. doi: 10.1128/JB.00503-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. 2008. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci U S A 105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morimoto YV, Nakamura S, Kami-ike N, Namba K, Minamino T. 2010. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol Microbiol 78:1117–1129. doi: 10.1111/j.1365-2958.2010.07391.x. [DOI] [PubMed] [Google Scholar]
- 65.Taly JF, Magis C, Bussotti G, Chang JM, Di Tommaso P, Erb I, Espinosa-Carrasco J, Kemena C, Notredame C. 2011. Using the T-Coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. Nat Protoc 6:1669–1682. doi: 10.1038/nprot.2011.393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The flagellar apparatus of Salmonella and E. coli. The entire structure from the cytoplasmic C ring to the external filament rotates (curved arrows). Stationary elements are MotAB complexes, which conduct protons and power the rotor, and the L and P rings, which act as bushings for the rod. MotAB insert and drift in the membrane until they encounter the basal MS ring structure, upon which the plugged proton channel opens. See the text for other descriptions. OM, outer membrane; PG, peptidoglycan; IM, inner membrane. Download
The effects of fliL on motility and rod fracture are similar in both Salmonella and E. coli. (A and B) Salmonella wild type (14028) and ΔfliL (UA74) and E. coli wild type (RP437) and ΔfliL (JP633a) were monitored for swimming and swarming motility in LB media by inoculating soft agar (0.3%) and swarm agar (0.6% supplemented with 0.5% glucose), respectively. Plates were incubated at 30°C (E. coli) or 37°C (Salmonella) for 6 h (swimming) or 8 h (swarming). (C) Cells were picked from swarm plates shown in the panel above and immune-stained for flagella as described in Text S1. Download
Salmonella displays more robust swimming motility than E. coli. Two different wild-type E. coli (top) and Salmonella (bottom) strains (see Table S1) were monitored for swimming motility by inoculating overnight cultures at the center of soft agar plates (LB media with 0.3% agar) and incubating them at 30°C for 6 h. This assay monitors both swimming speed and chemotactic proficiency. Download
Western blots to monitor the recovery of functional MotA-His under various conditions in Salmonella. Protein loading in whole cells and BBs was controlled for with antibodies to the periplasmic protein MBP and the BB protein FliM, respectively. Protein bands were quantified as described in the supplemental material. All strains were ΔmotA, with MotA-His provided from pJP45. BBs recovered under swimming conditions (broth-grown cells) showed 4% less MotA in the ΔfliL mutant (pJP530) and 10% more in the FliL-overexpressing strain (pJP133) than in the wild type (QW178); swim diameters of these strains followed a similar trend (see Fig. S5). BBs recovered under swarming conditions had 10% less MotA in the ΔfliL mutant and 18% more in the FliL overexpressing strain than in the wild type. The swarm diameter of the overexpression strain was also larger than that of the wild type (see Fig. S5). Blots are representative of 5 independent experiments. Download
Effect of absence or overexpression of FliL on swimming and swarming motility in Salmonella. Wild-type Salmonella (14028), an isogenic ΔfliL mutant (UA74), and a wild-type strain with pFliL (pJP133) were inoculated on swim (0.3% agar) and swarm (0.6%) plates as described in Text S1 and incubated for 6 h at 37°C. Download
MotB plug mutations increase speed of Salmonella wild-type motors. Single motors of ΔmotB strains (QW180) containing plasmids expressing the indicated MotB variants were monitored by bead assay as described for Fig. 1. Motors with wild-type MotB rotated at 58 Hz (26 ± 3 rvpm), MotBL56A averaged 63 Hz ± 4 (24 ± 4 rvpm), and MotBL60A averaged 61 Hz ± 4 (28 ± 5 rvpm). Download
(A) Strains and plasmids used in this study. (B) Extent of gene deletions in this study.
Two-hybrid analysis of basal body proteins.
Effect of FliL on growth defects associated with mutations in the MotB plug region.
Supplemental methods. Download