ABSTRACT
Entry into cells is critical for virulence of the human bacterial pathogens Shigella spp. Shigella spp. induce membrane ruffle formation and macropinocytic uptake, but the events instigating this process are incompletely understood. The host small GTPase ADP-ribosylation factor 6 (ARF6) functions in membrane trafficking at the plasma membrane and activates membrane ruffle formation. We demonstrate that ARF6 is required for efficient Shigella flexneri entry, is activated by S. flexneri dependent on the phosphatase activity of the type III secreted effector IpgD, and depends on cytohesin guanine nucleotide exchange factors (GEFs) for recruitment to entry sites. The cytohesin GEF ARF nucleotide binding site opener (ARNO) is recruited to these sites, also dependent on IpgD phosphatase activity. ARNO recruitment is independent of ARF6, indicating that, in addition to the described recruitment of ARNO by ARF6, ARNO is recruited upstream of ARF6. Our data provide evidence that ARF6, IpgD, phosphoinositide species, and ARNO constitute a previously undescribed positive feedback loop that amplifies ARF6 activation at bacterial entry sites, thereby promoting efficient S. flexneri uptake.
IMPORTANCE
Shigella spp. cause diarrhea and dysentery by infection of epithelial cells in the human colon. Critical to disease is the ability of Shigella to enter into cells, yet the mechanisms involved in entry are incompletely understood. We demonstrate that the small GTPase ADP-ribosylation factor 6 (ARF6) is required for efficient cellular entry of Shigella flexneri and that activation of ARF6 depends on the phosphatase activity of the Shigella protein IpgD, which is introduced into cells via the bacterial type III secretion system. We further show that IpgD phosphatase activity is required for recruitment of the ARF6 guanine nucleotide exchange factor (GEF) ARF nucleotide binding site opener (ARNO) to bacterial entry sites and that ARNO lies upstream of ARF6 activation. These relationships define a positive feedback loop that contributes to activation of ARF6 at S. flexneri entry sites and leads to local amplification of signals that promote bacterial entry.
INTRODUCTION
Shigella spp. are Gram-negative bacterial pathogens that cause diarrheal disease by invading and spreading through the colonic epithelium. Shigella entry into intestinal epithelial cells is critical to the disease process (1, 2) and requires a type III secretion system (T3SS) and secreted effectors (2, 3). Upon contact with nonphagocytic cells, delivery of T3SS effector proteins causes rearrangements of the actin cytoskeleton, which induce plasma membrane ruffling that leads to macropinocytic uptake of the pathogen (4).
Several bacterial and host proteins have been shown to be involved in Shigella entry. Disruption of the activity of individual proteins leads to defects but not to complete abrogation of entry, indicating that entry pathways are partially redundant. Shigella IpaA promotes entry by binding vinculin, but an ipaA mutant still invades at low frequency (5). Actin rearrangements promoted by the small Rho GTPases Cdc42 and Rac1 contribute to entry, yet expression of dominant negative forms of either decreases entry by only ~70% (6). Activation of the Rac1 guanine exchange factor (GEF) DOCK180/ELMO by Shigella IpgB1 contributes to entry, yet an ipgB1 mutant invades at 35% of the efficiency of the wild type (WT) (7, 8).
GTPases are molecular switches that transduce signals to regulate functions as diverse as gene expression, vesicular trafficking, and cytoskeletal rearrangement (9) and are manipulated by several bacterial pathogens (10). GTPases of the ADP-ribosylation factor (ARF) family act at the plasma membrane and at endosomes to regulate membrane trafficking, phosphoinositide and lipid metabolism, and actin remodeling (11, 12). Distinct from other ARF GTPases in amino acid sequence and intracellular localization, GTP-bound ARF6 is localized to the plasma membrane and on endosomes, where it activates phosphatidylinositol 4-phosphate 5-kinase (PIP5K), generating phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (13–15). In motile cells, ARF6 also promotes actin remodeling via activation of Rac1, dependent on the GEFs ARF nucleotide binding site opener (ARNO) and DOCK180/ELMO complex (16–19).
The localization of ARF6 at the plasma membrane and its role in membrane trafficking and actin remodeling during endocytosis place it in an ideal position to participate in pathogen uptake and endosome remodeling (18–22). The mechanism of ARF6 recruitment and activation has not been described for any pathogen.
We report a role for ARF6 in a previously undescribed positive feedback loop that amplifies ARF6 activation to promote Shigella flexneri entry. We find that ARF6 is activated during S. flexneri entry and is required for efficient entry. We show that the ARF GEF ARNO lies upstream of ARF6 in this pathway and that the activity of the T3SS effector IpgD, an inositol 4-phosphatase (23–25), is required for recruitment of both ARF6 and ARNO to bacterial entry sites. These findings provide new insights into the mechanism of GTPase recruitment and activation during bacterial entry.
RESULTS
ARF6 is required for efficient entry of S. flexneri.
To test whether ARF6 contributes to S. flexneri infection, we quantified the efficiency of infection in HeLa cells transfected with wild-type (WT) hemagglutinin (HA)-tagged ARF6 or ARF6 N122I, a dominant negative mutant unable to bind guanine nucleotides (21). At 2 h of infection, the number of intracellular bacteria in cells expressing ARF6 N122I was 30% less than in cells expressing WT ARF6 (7.2 ± 0.1 versus 5.1 ± 0.2 intracellular bacteria per transfected cell, P = 0.001) (Fig. 1A), demonstrating that entry and/or early intracellular replication of S. flexneri is less efficient in the presence of dominant negative ARF6. To test whether ARF6 per se is required for efficient infection, we examined infection in mouse embryonic fibroblasts (MEFs) that harbor a stable lentiviral short hairpin RNA (shRNA) construct targeting Arf6 mRNA (ARF6K/D) (26), such that levels of ARF6 are reduced by >90% compared to control cells (ARF6+) that harbor a nontargeting lentiviral insertion (see Fig. S1A in the supplemental material). Viable intracellular S. flexneri levels in ARF6K/D MEFs were 2-fold decreased compared to ARF6+ MEFs at 1 and 2 h of infection (at 1 h, 1,327 ± 311 CFU versus 651 ± 147 CFU, P = 0.03; at 2 h, 11,027 ± 1,430 CFU versus 5,188 ± 193 CFU, P = 0.03) (Fig. 1B). Thus, in both HeLa cells and MEFs, in the absence of ARF6, entry and/or early intracellular replication of S. flexneri is less efficient.
FIG 1 .
ARF6 is required for efficient entry of S. flexneri. (A) Efficiency of infection of HeLa cells transfected with ARF6-HA or dominant negative ARF6 N122I-HA. Number of intracellular bacteria per cell at 2 h of infection, by differential staining. (B) S. flexneri recovered after infection (1 or 2 h) of ARF6K/D or ARF6+ MEFs, by gentamicin protection assay. (C and D, left graph) Early infection (40 min) of ARF6K/D or ARF6+ MEFs, with differential staining of extracellular versus intracellular bacteria. Extracellular bacteria stained with antibody to LPS (green) and all bacteria identified by DAPI (DNA, blue). Phalloidin staining of polymerized actin (red). Images are representative. Arrows, intracellular bacteria. Arrowheads, extracellular bacteria. Bar, 10 µm. (D, right graph) Rescue of S. flexneri entry by transient transfection of ARF6K/D MEFs with ARF6-HA. Efficiency of entry into ARF6-HA-transfected cells was for the subset of cells that expressed ARF6-HA. Mean ± standard error of the mean of at least 3 independent experiments.
S. flexneri entry into cells occurs as early as 15 min after bacterial contact with the host cell (27, 28). To test whether ARF6 is required for S. flexneri entry per se, we quantified the percentage of intracellular bacteria 40 min after initial contact with cells, before significant replication of intracellular bacteria would have occurred, differentiating extracellular bacteria from those that were intracellular by differential labeling. Extracellular bacteria were identified by labeling with S. flexneri lipopolysaccharide (LPS) prior to cell permeabilization, whereas all bacteria were identified by staining DNA with 4′,6-diamidino-2-phenylindole (DAPI). At 40 min of infection, the number of cells infected by S. flexneri was decreased by 75% in ARF6K/D MEFs compared to ARF6+ MEFs (32% ± 8.6% versus 8% ± 0.6% of cells were infected, P = 0.03) (Fig. 1C and D, left graph). When transiently transfected with an HA-tagged WT ARF6 construct, ARF6K/D MEFs exhibited partial restoration of ARF6 protein levels (see Fig. S1A in the supplemental material). The relatively minor rescue in levels was likely due to the combination of only a portion of the cells having been transfected and the susceptibility of the ARF6-HA RNA to the shRNA expressed in the ARF6K/D cells. Nevertheless, transient transfection of this construct resulted in rescue of bacterial entry to that in ARF6+ MEFs (Fig. 1D, right graph), suggesting that relatively low levels of ARF6 are sufficient to promote entry and demonstrating that efficient S. flexneri entry is dependent on ARF6.
Virulent S. flexneri activates ARF6 during early infection.
Like most small GTPases, in resting cells, ARF6 is predominantly in the GDP-bound inactive form. The requirement for ARF6 for efficient S. flexneri entry suggested that ARF6 might be activated during this process. To quantify ARF6 activation, GTP-bound ARF6 was precipitated from lysates of infected cells using beads conjugated to the ARF binding domain of Golgi complex-associated, gamma adaptin ear-containing ARF binding protein 3 (GGA3), which specifically binds GTP-bound ARF6 (29). WT S. flexneri induced activation of ARF6 as early as 20 min after contact with HeLa cells, whereas a strain that is noninvasive by virtue of lacking the virulence plasmid did not activate ARF6 at either time assessed (Fig. 2A), indicating that S. flexneri activates ARF6 soon after cell contact and in a manner that depends on virulence plasmid-encoded factors.
FIG 2 .
Virulent S. flexneri activates ARF6 early during infection. (A) ARF6 activation determined by pulldown of GTP-bound ARF6 from HeLa cells 20 or 40 min after initial contact of WT or noninvasive S. flexneri (top panel) with cells, versus total ARF6 in each lysate (bottom panel). GTPγS or GDP added to lysates of uninfected cells and uninfected cells alone (uninfect) are positive and negative controls. Ratio of ARF6-GTP to total ARF6 for blots shown, by densitometry. Blots are representative. (B and C) ARF6-HA recruitment to AFA-I-expressing WT or noninvasive S. flexneri in HeLa cells infected (40 min), fixed, and stained with antibody to HA (green), DAPI (blue), and phalloidin (red). ND, not detectable. Mean ± standard error of the mean for 3 independent experiments. Images are representative. Arrows, ARF6 recruitment to entering bacteria. Arrowheads, bacteria without ARF6 recruitment. Bar, 10 µm.
Exchange of bound GDP for GTP activates ARF6 and recruits it from a cytosolic pool to the plasma membrane (30–32), suggesting that ARF6 activation during S. flexneri entry might be associated with its recruitment to entry sites. WT S. flexneri recruited HA-tagged ARF6 to bacteria at the plasma membrane early in infection (40 min), but the noninvasive strain did not (85% versus <1% of cells displayed ARF6 recruitment to bacteria, P = 0.03) (Fig. 2B and C). In this experiment, to ensure that bacterial contact with the plasma membrane occurred independently of type III secretion, we used strains that adhered tightly as a result of heterologous production of the uropathogenic Escherichia coli afimbrial adhesin (AFA-I); as a result, every cell displayed adherent bacteria. S. flexneri similarly recruited ARF6-HA in MEFs (18% versus <1% of cells with bacteria associated showed ARF6 recruitment to bacteria, P = 0.001) (see Fig. S2 in the supplemental material); here, only a subset of cells displayed adherent bacteria, as AFA-I was not used because its receptor is absent in murine cells. These results show that ARF6 activation by S. flexneri is associated with its recruitment to bacterial entry sites and is dependent not simply on contact with the plasma membrane but rather on virulence plasmid-encoded factors.
The phosphoinositide phosphatase activity of Shigella T3SS effector IpgD is required for efficient ARF6 activation at entry sites.
We tested whether any of several effector proteins secreted into cells by the S. flexneri virulence plasmid-encoded type III secretion system (T3SS) were required for activation of ARF6. We postulated that the involved effector was likely to either possess GTPase exchange factor (GEF) activity or recruit an ARF GEF to entry sites. We therefore focused on the effectors IpgB1, IpgB2, IpgD, and IpaJ. IpgB1 and IpgB2 possess GEF activity; IpgB1 principally activates the host GTPases Rac1 and Cdc42, and IpgB2 principally activates RhoA, yet the two effectors are homologous with some overlap in function (8, 33, 34). IpgD is an inositol 4-phosphatase with no known homology to effectors with GEF activity but, like IpgB1, is translocated early upon bacterial contact with host cells and participates in actin remodeling at entry sites (7, 23, 24). Moreover, the substrate of IpgD, PI(4,5)P2, is concentrated at the plasma membrane (35). IpaJ demyristoylates ARF1 (36), an ARF GTPase that is 66% identical to ARF6 (11).
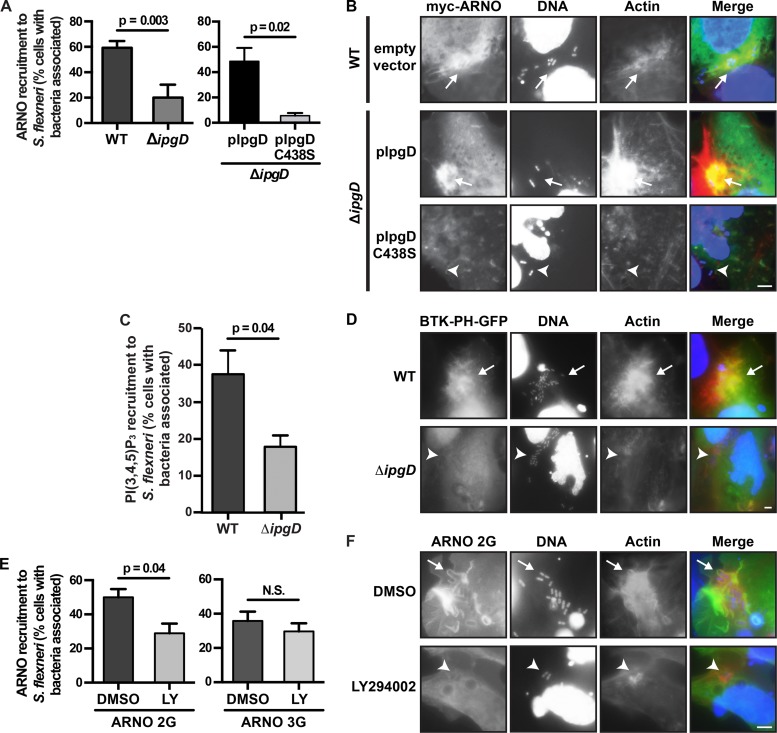
To test whether any of these effectors is required for S. flexneri-induced ARF6 activation, we examined the ability of strains lacking each individually to activate ARF6 during entry. Whereas the other mutant strains examined activated ARF6 at levels comparable to the WT strain, the ΔipgD mutant was defective in ARF6 activation (Fig. 3A). We then tested whether IpgD was required for ARF6 recruitment to bacterial entry sites by specifically assessing the efficiency of ARF6 recruitment to sites where bacteria were adherent to cells and/or within a membrane ruffle, reasoning that these constituted the bulk of the bacterial population potentially undergoing cellular entry. Compared to WT, the ΔipgD mutant was defective in ARF6 recruitment (57% versus 38% of cells with bacteria associated showed ARF6 recruitment, P = 0.04) (Fig. 3B, left graph). The ARF6 recruitment defect was rescued by ipgD expressed in trans, but not by expression of ipgD C438S, a catalytically dead mutant with no phosphatase activity (24) (54% and 7% of cells with bacteria associated showed ARF6 recruitment, respectively, P = 0.003) (Fig. 3B, right graph, and C). These results demonstrate that IpgD phosphatase activity is critical for ARF6 activation and for ARF6 recruitment to bacterial entry sites.
FIG 3 .
The phosphoinositide phosphatase activity of Shigella T3SS effector IpgD is required for efficient ARF6 activation at entry sites. (A) ARF6 activation determined by pulldown of GTP-bound ARF6 from HeLa cells infected with indicated S. flexneri strains for 40 min after initial bacterial contact with cells (top panel) versus total ARF6 in each lysate (bottom panel). Ratio of ARF6-GTP to total ARF6 for blots shown, by densitometry. Uninfect, uninfected; Non-invas, noninvasive strain; ΔipgB1 ΔB2, ΔipgB1 ΔipgB2 mutant. Blots are representative. (B to E) Infection of ARF6-HA-transfected HeLa cells (40 min) with WT, ΔipgD, ΔipgD pIpgD, or ΔipgD pIpgD C438S S. flexneri, fixed and labeled with antibody to HA (green), DAPI (blue), and phalloidin (red). (B) Percentage of cells with bacteria associated that show ARF6 recruitment to bacteria. (C) Representative images. Arrows, bacteria with ARF6 recruitment. Arrowheads, bacteria without ARF6 recruitment. Bar, 10 µM. (D and E) Percentage of cells infected or with ruffles. Mean ± standard error of the mean of at least 3 independent experiments.
The ΔipgD mutant was defective in entry (17% of cells infected by ΔipgD mutant versus 31% of cells infected by WT, P = 0.002) (Fig. 3D, left graph), and the entry defect was rescued by WT IpgD but not by IpgD C438S (68% versus 21% of cells infected, respectively; P = 0.0006) (Fig. 3D, right graph). Thus, IpgD phosphatase activity is required both for activation of ARF6 and for efficient entry. The ΔipgD mutant was defective in the formation of plasma membrane ruffles at entry sites (25% versus 12% of cells displayed bacteria in a ruffle; P = 0.001) (Fig. 3E, left graph). IpgD but not IpgD C438S rescued the ruffling defect (46% and 4% of cells displayed bacteria in a ruffle, respectively; P = 0.0016) (Fig. 3E, right graph). Both the ΔipgD mutant complemented with WT IpgD and that complemented with IpgD C438S produced IpgD at higher levels than the WT strain (see Fig. S3 in the supplemental material), likely due to plasmid copy number. In the presence of overexpression of WT IpgD, entry and ruffle formation were more efficient, whereas overexpression of IpgD C438S was associated with an apparent dominant negative effect on entry and ruffle formation, further confirming a role for IpgD and IpgD phosphatase activity in S. flexneri entry.
Efficient ARF6 recruitment to entering S. flexneri requires cytohesin GEFs.
GEFs activate small GTPases such as ARF6 by stimulating GDP dissociation, which allows GTP binding. In the absence of homology that might suggest intrinsic GEF activity, a mechanism by which IpgD might promote ARF6 activation is by recruiting and/or activating an ARF6 GEF at S. flexneri entry sites. Known GEFs of ARF6 include BRAG1, GEP100/BRAG2, EFA6, and the cytohesin GEFs, cytohesin-1, ARNO (cytohesin-2), and GRP1 (cytohesin-3) (37–41). We tested whether cytohesin GEFs are required for S. flexneri entry and for ARF6 recruitment to entering bacteria by treating cells with SecinH3, a small-molecule cytohesin family-specific inhibitor (42) that inhibits all cytohesin family GEFs. Cytohesin-1 is highly expressed in natural killer T lymphocytes (43), whereas ARNO and GRP1 have roles in epithelial cells (17, 44). SecinH3 inhibited S. flexneri entry (30% versus 11% of cells were infected; P = 0.04) (Fig. 4A) and ruffle formation (29% versus 10% of cells displayed bacteria in a ruffle; P = 0.02) (Fig. 4B), indicating that one or more cytohesin GEFs contribute to entry. SecinH3 inhibited S. flexneri entry not only in the presence of ARF6 (29% versus 14% of ARF6+ MEFs were infected, P = 0.03) but also in its absence (16% versus 8% of ARF6K/D MEFs were infected, P = 0.02) (data not shown); since ARNO can activate ARF1 downstream of ARF6 (18, 19), we speculate that this SecinH3-induced decrease in infection of ARF6K/D MEFs may be due to inhibition of ARNO-dependent activation of ARF1.
FIG 4 .
Efficient ARF6 recruitment to entering S. flexneri requires cytohesin GEFs. Recruitment of ARF6-HA to HeLa cells infected with S. flexneri for 40 min in the presence of SecinH3 or dimethyl sulfoxide (DMSO). (A) Percentage of cells infected. (B) Percentage of cells with ruffles. (C) Percentage of cells with bacteria associated showing ARF6 recruitment to bacteria. Mean ± standard error of the mean of at least 3 independent experiments. (D) Representative images with labeling with antibody to HA (green), DAPI (blue), and phalloidin (red). Arrows, bacteria with ARF6 recruitment. Arrowheads, bacteria without ARF6 recruitment. Bar, 10 µm.
To determine whether cytohesin GEFs were functioning upstream of ARF6 in S. flexneri entry, we tested whether ARF6 recruitment depended on cytohesin activity. In the presence of SecinH3, ARF6 recruitment to potential entry sites was decreased (44% versus 15% of cells with bacteria associated showed ARF6 recruitment to bacteria; P = 0.001) (Fig. 4C and D), demonstrating that SecinH3 blocked efficient ARF6 activation by S. flexneri. Thus, cytohesin GEF activity is critical to efficient ARF6 activation by entering S. flexneri and to efficient bacterial entry, consistent with the inhibitory effect of SecinH3 on entry being due at least in part to inhibition of ARF6 activation.
Interaction of IpgB1 with ELMO, a component of the Rac1 GEF DOCK180/ELMO complex, promotes bacterial uptake (7). ARNO is required for activation of DOCK180/ELMO (17). Whether ARNO activation of DOCK180/ELMO depends on ARF6 is unclear. These observations, in conjunction with the observed effects of SecinH3 on S. flexneri activation of ARF6 and entry, led us to postulate that ARNO might participate in activation of ARF6. Myc-tagged ARNO was recruited to S. flexneri entry sites. Experimental optimization suggested that membrane ruffling and ARF6 recruitment both peak within 30 to 40 min after initial bacterial contact with HeLa cells, whereas ARNO recruitment peaks approximately 10 min earlier (data not shown). ARNO recruitment in HeLa cells depended on the presence of the virulence plasmid, even for bacteria that expressed the AFA-I adhesin (55% of cells for WT versus <1% of cells for noninvasive strain displayed ARNO recruitment to bacteria; P = 0.001) (Fig. 5A and B).
FIG 5 .
ARNO is recruited to entering S. flexneri independent of ARF6. (A and B) Recruitment of Myc-ARNO 3G to AFA-I-expressing WT or noninvasive S. flexneri in HeLa cells, infected for 30 min. Representative images labeled with antibody to Myc (green), DAPI (blue), and phalloidin (red). (C and D) Recruitment of Myc-ARNO 2G or Myc-ARNO 3G to S. flexneri entering ARF6K/D or ARF6+ MEFs, infected for 25 min. Labeling as in panel B. Arrows, entering bacteria with ARNO recruitment. Arrowheads, bacteria without ARNO recruitment. ND, not detectable. NS, not significant. Mean ± standard error of the mean of at least 3 independent experiments. Bars, 10 µm.
Since ARNO recruitment apparently preceded ARF6 recruitment, we tested whether ARNO recruitment might be independent of ARF6 by examining its recruitment in ARF6+ and ARF6K/D MEFs. ARNO exists as two splice variants—ARNO 2G and ARNO 3G—that differ by the addition of a single glycine residue in the phosphoinositide-binding pocket of the ARNO pleckstrin homology (PH) domain (45). Because the two variants display distinct binding preferences for phosphoinositide species at the membrane (45, 46), we separately examined the recruitment of each. Each ARNO variant was independent of ARF6 for recruitment to entering bacteria (42% each of cells with bacteria associated showed ARNO 2G recruitment to bacteria in ARF6+ and ARF6K/D MEFs, respectively; P = 1.0; for ARNO 3G, 42% versus 35%, respectively; P = 0.3) (Fig. 5C and D), indicating that ARNO is recruited upstream of ARF6, which positions it to potentially contribute to activation of ARF6 at sites of S. flexneri entry. These findings are notably different from what has been observed during Salmonella infection, where ARNO recruitment to entry sites is defective in the absence of ARF6 (19); the reasons for these organism-based differences are at present unclear.
IpgD and IpgD phosphatase activity are required for ARNO recruitment to entry sites and S. flexneri entry.
IpgD-induced PI(5)P accumulation activates PI 3-kinase (PI3K), leading to generation of phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] (47). The PH domains of ARF GEFs bind phosphoinositides with various affinities; these interactions recruit GEFs to the plasma membrane (37, 48–50). We considered that recruitment of ARNO to S. flexneri entry sites might be enhanced by IpgD-mediated generation of phosphoinositide species that bind ARNO with high affinity. IpgD was required for recruitment of ARNO to entry sites (cells with bacteria associated showing ARNO recruitment to bacteria, 59% for WT versus 20% for ΔipgD mutant, P = 0.001) (Fig. 6A, left graph). IpgD but not IpgD C438S rescued ARNO recruitment by the ΔipgD mutant (44% and 5% of cells with bacteria associated showed ARNO recruitment to bacteria, respectively) (Fig. 6A, right graph, and B). In the absence of IpgD, ARNO recruitment was more dramatically reduced than ARF6 recruitment (Fig. 6A, left graph, versus 3B), likely due to a contribution of other SecinH3-sensitive ARF6 GEFs to ARF6 recruitment, since knockdown of ARNO alone did not lead to significant decreases in ARF6 recruitment (see Fig. S4 in the supplemental material). As above (Fig. 3D and E), the phosphatase activity of IpgD was required for both efficient entry and membrane ruffling at entry sites (see Fig. S5 in the supplemental material). Moreover, PI(3,4,5)P3 was generated at S. flexneri entry sites, as assessed by recruitment of a green fluorescent protein (GFP)-tagged pleckstrin homology domain from Bruton’s tyrosine kinase (BTK-PH-GFP), which specifically binds PI(3,4,5)P3 (51), and efficient generation of PI(3,4,5)P3 was dependent on IpgD (ΔipgD mutant, 18%, versus WT, 37%, for cells displaying BTK-PH-GFP recruitment to entry sites, P = 0.04) (Fig. 6C and D). Thus, the phosphatase activity of IpgD is critical for both generation of PI(3,4,5)P3 at entry sites and ARNO recruitment, and IpgD and SecinH3-sensitive GEFs are each required for ARF6-mediated S. flexneri entry.
FIG 6 .
IpgD phosphatase activity is required for ARNO recruitment to entry sites, and PI 3-kinase contributes to ARNO 2G but not ARNO 3G recruitment. (A and B) WT, ΔipgD, ΔipgD pIpgD, and ΔipgD pIpgD C438S S. flexneri infection for 30 min of HeLa cells transfected with Myc-ARNO 3G. (A) Percentage of cells with ARNO recruitment to entering bacteria. (B) Representative images, labeled with antibody to Myc (green), DAPI (blue), and phalloidin (red). (C and D) Recruitment of PI(3,4,5)P3 to bacterial entry sites upon WT or ΔipgD infection for 40 min of HeLa cells transfected with BTK-PH-GFP, which specifically binds PI(3,4,5)P3. (C) Percentage of cells with BTK-PH-GFP recruitment to entering bacteria. (D) Representative images, labeled with DAPI (blue), and phalloidin (red), and with GFP signal. (E and F) Inhibition of recruitment of ARNO 2G, but not ARNO 3G, upon treatment with LY294002 upon WT infection of ARF6K/D MEFs for 25 min. (E) Percentage of cells with ARNO recruitment to entering bacteria. (F) Representative images, labeled as in panel B. Arrows, bacteria with ARNO recruitment. Arrowheads, bacteria without ARNO recruitment. Bars, 10 µm. NS, not significant. Mean ± standard error of the mean of at least 3 independent experiments. DMSO, dimethyl sulfoxide; LY, LY294002.
Recruitment to entry sites of ARNO 2G but not ARNO 3G is defective upon inhibition of PI3K.
These data raised the possibility that IpgD-dependent changes in phosphoinositide composition, specifically PI3K-dependent generation of PI(3,4,5)P3, contribute to recruitment of ARNO to bacterial entry sites. We tested whether inhibition of PI3K by addition of the chemical inhibitor LY294002 to cells during bacterial entry altered recruitment of ARNO. Recruitment of ARNO 2G, which preferentially binds PI(3,4,5)P3, was defective in the presence of LY294002 (29% versus 50% of cells with bacteria attached displaying ARNO recruitment, P = 0.04) (Fig. 6E, left graph, and F), whereas recruitment of ARNO 3G, which binds PI(4,5)P2 with high affinity (45, 46), was unaffected (30% versus 36%, P = 0.4) (Fig. 6E, right graph). Treatment of cells with LY294002 was associated with defective phosphorylation of Akt (see Fig. S6 in the supplemental material), confirming that the conditions used inhibited PI 3-kinase activity. Thus, IpgD-dependent generation of PI(3,4,5)P3 during entry of S. flexneri enhances recruitment of the 2G variant of the ARF6 GEF ARNO.
DISCUSSION
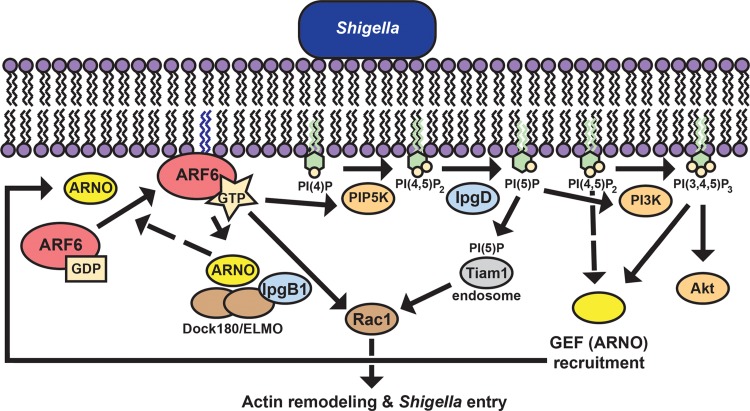
Our findings, combined with published data, support the presence of a positive feedback loop that promotes ARF6 activation during S. flexneri entry into cells. We demonstrate that activation of ARF6 is required for efficient entry of S. flexneri into both epithelial cells and MEFs, that ARF6 activation depends on phosphatase activity of the T3SS effector IpgD, and that the cytohesin ARF6 GEF ARNO is recruited upstream of ARF6 where it contributes to ARF6 activation (Fig. 7). ARF6 activation is known to drive Rac1-dependent actin remodeling, which promotes membrane ruffle formation and bacterial entry (16, 17). Thus, the amplification of signals immediately beneath sites of bacterial contact with the plasma membrane that result from this positive feedback loop would be expected to promote rapid local changes that facilitate bacterial entry.
FIG 7 .
Model of IpgD-dependent positive feedback loop that amplifies ARF6 activation during entry of S. flexneri. Extracellular S. flexneri recruits ARNO to the plasma membrane in part via IpgD-induced local accumulation of PI(3,4,5)P3. ARNO-dependent activation of ARF6 induces PIP5K-mediated changes in phosphoinositide composition at the membrane that promote additional ARNO recruitment, mediating further activation of ARF6. Solid arrows, relationships supported by data presented here or previously published. Dashed arrows, postulated relationships.
In a positive feedback loop, amplification allows the output signal to reach a critical threshold at which the system switches from one state to another, traditionally basal to active (52). Positive feedback loops have been reported for Rab (53, 54) and ARF (55, 56) GTPase signaling. ARNO recruitment to the plasma membrane is by two distinct mechanisms: binding to phosphoinositides and phosphatidylserine and binding to ARF6 (57, 58). In an ARF6 feedback loop, GTP-bound ARF6 stimulates ARNO, and ARNO activates ARF1 and is then stimulated by GTP-bound ARF1 (55). Cohen et al. found that GTP-bound ARF6 binds the ARNO PH domain, leading to ARNO recruitment to the plasma membrane and activation of ARF1 (57). Cascades of GEFs and GTPases can thus constitute positive feedback loops that feed back onto themselves.
The positive feedback loop that our data support is distinct from described GEF cascades in that IpgD phosphatase activity generates signals that contribute to GEF recruitment. These signals include the generation of membrane-anchored phosphoinositide species that bind ARNO. Since PI(4,5)P2, the substrate for IpgD phosphatase activity, is generated by ARF6-mediated activation of PIP5K, by our model, IpgD activity is fueled by its ability to generate phosphoinositides that enhance ARNO recruitment. Moreover, because inactive ARF6 does not bind the ARNO PH domain (57), IpgD-generated phosphoinositides may function as the seed for ARNO recruitment and subsequent ARF6 recruitment and activation. This model, supported by the data presented here, predicts that IpgD activity expands the existing GEF-ARF cascade in a manner that further amplifies ARF6 activation.
Our findings establish a substantial expansion of the model of ARF GTPase participation in entry, previously described for Salmonella enterica serovar Typhimurium by Humphreys et al. (18, 19). Data from the prior studies support a model in which ARF6 recruits ARNO, ARNO in turn recruits ARF1, and ARF1 promotes actin polymerization via the WAVE regulatory complex (18, 19). In our work here, in addition to showing that entry by S. flexneri parallels that of S. Typhimurium with respect to participation of ARF GTPases, we characterize for the first time a mechanism of activation of ARF6 in this process and demonstrate the presence of the previously undescribed positive feedback loop that likely amplifies ARF6 activation. Moreover, our results identify an important difference from the entry of S. Typhimurium, wherein ARNO recruitment is dependent on ARF6 (19), in that during entry of S. flexneri, ARNO recruitment is independent of and occurs upstream of ARF6. Although the reasons are currently unclear, these findings may highlight the existence of important functional divergence between Shigella and Salmonella.
An important insight that emerges from our data together with published data (18, 19) is that during bacterial entry, ARNO participates in the activation of both ARF6 and ARF1, which lies downstream of ARF6. Thus, ARF GEFs such as ARNO (but perhaps not restricted to ARNO) may participate promiscuously at multiple steps during bacterial infection. ARNO recruitment was absolutely dependent on IpgD. The effector IpgB1 recruits the Rac1 GEF DOCK180/ELMO to entry sites (7). DOCK180/ELMO is activated by ARF6 via ARF1 and dependent on ARNO GEF activity (17, 19).
PI(4,5)P2 is relatively abundant in both resting and stimulated cells, whereas PI(3,4,5)P3 is detectable only following stimulation (59). IpgD phosphatase activity leads to the recruitment of PI(3,4,5)P3 (Fig. 6C and D). PI(4,5)P2 and PI(3,4,5)P3 bind PH domains of ARF GEFs. The high-affinity interaction of membrane-anchored PI(3,4,5)P3 with the ARNO 2G variant is sufficient to recruit it to the plasma membrane, whereas the low-affinity interaction of PI(3,4,5)P3 with the ARNO 3G variant is not sufficient (45, 60, 61). In brain tissue, the 3G variant of ARNO is most abundant (46); the relative abundance of the two variants in other tissues is unknown.
Recruitment of ARNO to S. flexneri entry sites is dependent on IpgD phosphatase activity (Fig. 6A to C) and independent of ARF6 (Fig. 5). The reduction in recruitment of the 2G variant of ARNO upon chemical inhibition of PI 3-kinase (Fig. 6E and F) is consistent with a model in which transient PI(3,4,5)P3 accumulation, induced by IpgD-dependent activation of PI 3-kinase, leads to efficient recruitment of ARNO 2G via its high-affinity interaction with PI(3,4,5)P3. However, recruitment of ARNO 3G is independent of PI(3,4,5)P3 accumulation (Fig. 6D) and thus must be mediated by other factors that accumulate at entry sites in response to IpgD phosphatase activity.
Our data demonstrate for the first time that IpgD is required for efficient S. flexneri entry. Of note, although statistically significant, the defect in entry of the ipgD mutant is relatively minor (Fig. 3D). That the relatively small defect is meaningful is supported by the enhanced entry observed upon overexpression of IpgD (Fig. 3D). The presence of only a minor defect is also consistent with several lines of evidence that point to Shigella entry resulting from multiple partially redundant mechanisms mediated by various T3SS effectors (5, 7, 8). The relatively small decrease observed, along with differences in strain backgrounds, likely explains why the role of IpgD in entry has been unappreciated in other experimental systems (62, 63).
IpgD phosphatase activity and the phosphoinositides that it generates have previously been implicated in S. flexneri activation of Akt (47), regulation of vesicular trafficking (64), recruitment of Rab-11-containing vesicles to bacterial uptake vacuoles (63), inhibition of chemokine-induced migration of T cells (65), inhibition of ATP release through connexin hemichannels (66), and activation of the GEF Tiam1 in Rac1-mediated actin rearrangements (67). Our data now add to the list a role in entry via activation of ARF6. The multiple effects of IpgD during S. flexneri infection highlight the central role that phosphoinositides play in intracellular signaling and the extent to which S. flexneri has evolved mechanisms to manipulate these signaling pathways.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids are listed in Table 1. All S. flexneri strains are isogenic to serotype 2a wild-type strain 2457T (68). S. flexneri bacteria were grown at 37°C in tryptic soy broth from individual Congo red-positive colonies. Antibiotics were ampicillin (Amp), 100 µg/ml; chloramphenicol (Cm), 25 µg/ml; and kanamycin (Km), 50 µg/ml. To generate pIpgD and pIpgD C438S, the coding sequences of ipgD, its promoter, and its chaperone ipgE were cloned into pACYC184 with site-directed mutagenesis of pIpgD (QuikChange mutagenesis kit; Stratagene).
TABLE 1 .
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Bacterial strains | ||
| Wild type | Serotype 2a S. flexneri strain 2457t | 68 |
| Noninvasive | BS103 (2457T cured of its virulence plasmid) | 71 |
| Wild type with adhesin | 2457T pIL22, Ampr | Lab stock |
| Noninvasive with adhesin | BS103 pIL22, Ampr | Lab stock |
| ΔipgD mutant | 2457T ipgD::FRT-Kmr-FRTa | Gift of C. Lesser |
| ΔipgD pIpgD mutant | 2457T ipgD::FRT-Kmr-FRT pACYC184 ipgD-ipgE | This study |
| ΔipgD pIpgD C438S mutant | 2457T ipgD::FRT-Kmr-FRT pACYC184 ipgD C438S-ipgE | This study |
| ΔipgB1 mutant | 2457T ipgB1::FRT | Lab stock |
| ΔipgB1 ΔipgB2 mutant | 2457T ipgB1::FRT ipgB2::FRT | Lab stock |
| ΔipaJ mutant | 2457T ipaJ::FRT | Lab stock |
| Plasmids | ||
| pACYC184 | Cmr | Lab stock |
| pIpgD | pACYC184 ipgD-ipgE, Cmr | This study |
| pIpgD C438S | pACYC184 ipgD C438S-ipgE, Cmr | This study |
| pIL22 | pBR322 encoding Escherichia coli afimbrial adhesin AFA-I, Ampr | 72 |
| ARF6-HA | pcDNA HA-tagged ARF6, Ampr | 21 |
| ARF6 N122I-HA | pcDNA HA-tagged ARF6 defective for nucleotide binding, Ampr | 21 |
| BTK-PH-GFP | pEGFP-N1-BTK-PH-GFP, Kmr | 51 |
| Myc-ARNO 3G | pcDNA3 Myc-tagged ARNO, 3G variant, Ampr | 29 |
| Myc-ARNO 2G | pcDNA3 Myc-tagged ARNO, 2G variant, Ampr | Gift of L. Santy |
FRT, FLP recombination target.
Cell culture.
HeLa cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco), and ARF6K/D and ARF6+ mouse embryonic fibroblasts (MEFs) (gift of Wim Annaert [26]) were cultured in DMEM–nutrient mixture F-12, each with 10% fetal bovine serum. Monolayers were transfected 24 h prior to infection with 1.5 µg DNA and Fugene 6 transfection reagent (Promega). Knockdown of ARNO was performed using the ARNO-specific small interfering RNAs (siRNAs) Hs_PSCD2_2 SI00061292, Hs_PSCD2_3 SI00061299, Hs_PSCD2_6 SI03050894, and Hs_PSCD2_7 SI03072629 or AllStars negative-control siRNA SI03650318 (Qiagen) and HiPerFect transfection reagent (Qiagen).
Infection of cells.
Bacteria (optical density at 600 nm [OD600], 0.3 to 0.4) were added to cell monolayers at a multiplicity of infection (MOI) of 200 to 300:1 (bacteria to cell) or, for strains containing pIL22, at an MOI of 10:1. Cells were centrifuged at 700 × g for 10 min and then incubated at 37°C for a specified additional period of time. SecinH3 (2.5 μM; Calbiochem) or LY294002 (50 µM; Cell Signaling; catalog no. 9901S) was added immediately prior to addition of bacteria. Infected monolayers were washed, fixed in 3.7% paraformaldehyde for 15 to 20 min, and permeabilized with 1% Triton X-100. Labeling was performed using antibodies to S. flexneri 2a lipopolysaccharide (LPS; Gibco) or HA (Covance MMS-101P) or with Myc (Clontech 631206), phalloidin (Invitrogen), or DAPI (100 μM; Invitrogen).
Gentamicin protection assay.
Cells seeded at 3 × 105 per well of a 6-well dish were infected at an MOI of 0.02:1 (69); incubated at 37°C for 5 or 50 min; washed three times; incubated for an additional 45 or 60 min, respectively, in DMEM containing 50 µg/ml gentamicin to kill extracellular bacteria; and lysed with 0.5 to 1% Triton X-100. Bacteria in cell lysates and in the initial inoculum were determined by plating.
Protein production and secretion.
Synthesis and secretion of IpgD were assayed essentially as described previously (70). Proteins were resolved by SDS-PAGE and probed with IpgD (gift of A. Phalipon [24]) or DnaK (StressGen) antibody.
Pulldown assay.
GTP-bound ARF6 in infected monolayers was measured per the manufacturer’s instructions (Cytoskeleton, Inc.). Briefly, cells were infected at an MOI of 300:1 (as described above), followed by incubation for an additional 10 or 30 min, lysis on ice, and harvesting and snap-freezing of the lysate. Two hundred fifty micrograms of each lysate was incubated with beads covalently conjugated to the ARF6 binding domain of GGA3 for 1 h at 4°C. Beads were washed and recovered. Bound ARF6 was resolved by SDS-PAGE and analyzed using ARF6 antibody (gift of J. Donaldson). Other antibodies used were phosphor-Akt S473 and pan-Akt (Cell Signaling) and ARNO (Abcam ab56510).
Microscopy and data analysis.
Microscopy was performed on a Nikon Eclipse TE300 or TE2000 microscope with Chroma Technology filters. For all sets of experiments, three or more independent experiments were performed. Samples were blinded. At least 10 infected cells were analyzed for each condition. A bacterium was counted as entering the cell if it was found associated with a membrane ruffle, determined by actin staining and phase microscopy, and as potentially undergoing bacterial entry if it was adherent and/or within a membrane ruffle. A cell was counted as infected if at least one bacterium was entering or present within the cell. The significance of differences between two-way comparisons was determined by paired t test. The significance of differences among multiple sets of data was determined by one-way analysis of variance (ANOVA) followed by a post hoc Tukey comparison.
SUPPLEMENTAL MATERIAL
Arf6 knockdown. Whole-cell lysates of ARF6K/D and ARF6+ MEFs, transfected with ARF6-HA where indicated. Immunoblotting with ARF6 or β-actin antibodies. Download
S. flexneri recruits ARF6 to entry sites in MEFs. (A) Recruitment of ARF6-HA to WT or noninvasive S. flexneri in ARF6+ MEFs infected for 25 min. ND, not detectable. Mean ± standard error of the mean of 3 independent experiments. (B) Representative images, labeled with antibody to HA (green), DAPI (blue), and phalloidin (red). Arrows, entering bacteria with ARF6 recruitment. Arrowheads, bacteria without ARF6 recruitment. Bar, 10 µm. Download
IpgD production in complemented ΔipgD S. flexneri. Congo red-induced secretion of S. flexneri strains. Pellets and precipitated supernatants immunoblotted with antibodies to IpgD or DnaK. Download
Recruitment of ARF6-HA to entering bacteria following siRNA depletion of ARNO in HeLa cells. (A) Depletion of ARNO by siRNA. Immunoblotting with ARNO or β-actin antibodies. (B) Percentage of cells with bacteria associated showing ARF6-HA recruitment to bacteria. Mean ± standard error of the mean of at least 3 independent experiments. NS, not significant. Download
In cells transfected with ARNO constructs, IpgD phosphatase activity is required for efficient S. flexneri entry and ruffle formation. (A) Percentage of cells infected. (B) Percentage of cells with ruffles. Mean ± standard error of the mean of at least 3 independent experiments. Download
LY294002 treatment that inhibits recruitment of ARNO 2G is associated with inhibition of Akt activation. Levels of phospho-Akt S473 and total Akt under conditions used for data presented in Fig. 6. Western blotting with β-actin as loading control. Download
ACKNOWLEDGMENTS
We acknowledge C. R. Yi and S. Y. Lee for bacterial strain construction and B. B. Herrera for technical support. We thank W. Annaert, J. G. Donaldson, L. C. Santy, R. R. Isberg, A. Phalipon, H. Ploegh, and C. F. Lesser for providing reagents; V. W. Hsu for helpful advice; and J. R. Mayers for critical reading of the manuscript.
This work was supported by Public Health Service grant R01AI081724 (to M.B.G.), trainee support on T32GM007753 (to A.C.G.-M.), and T32AI007061 (to K.A.M. and B.C.R.) from the National Institutes of Health.
Footnotes
Citation Garza-Mayers AC, Miller KA, Russo BC, Nagda DV, Goldberg MB. 2015. Shigella flexneri regulation of ARF6 activation during bacterial entry via an IpgD-mediated positive feedback loop. mBio 6(2):e2584-14. doi:10.1128/mBio.02584-14.
REFERENCES
- 1.Hale TL. 1991. Genetic basis of virulence in Shigella species. Microbiol Rev 55:206–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sansonetti PJ, Kopecko DJ, Formal SB. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun 35:852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasakawa C, Kamata K, Sakai T, Makino S, Yamada M, Okada N, Yoshikawa M. 1988. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J Bacteriol 170:2480–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroeder GN, Hilbi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 21:134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran Van Nhieu G, Ben-Ze’ev A, Sansonetti PJ. 1997. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J 16:2717–2729. doi: 10.1093/emboj/16.10.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mounier J, Laurent V, Hall A, Fort P, Carlier MF, Sansonetti PJ, Egile C. 1999. Rho family GTPases control entry of Shigella flexneri into epithelial cells but not intracellular motility. J Cell Sci 112:2069–2080. [DOI] [PubMed] [Google Scholar]
- 7.Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, Fukui Y, Sasakawa C.. 2007. Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat Cell Biol 9:121–128. doi: 10.1038/ncb1526. [DOI] [PubMed] [Google Scholar]
- 8.Hachani A, Biskri L, Rossi G, Marty A, Ménard R, Sansonetti P, Parsot C, Van Nhieu GT, Bernardini ML, Allaoui A. 2008. IpgB1 and IpgB2, two homologous effectors secreted via the Mxi-Spa type III secretion apparatus, cooperate to mediate polarized cell invasion and inflammatory potential of Shigella flexneri. Microbes Infect 10:260–268. doi: 10.1016/j.micinf.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Takai Y, Sasaki T, Matozaki T. 2001. Small GTP-binding proteins. Physiol Rev 81:153–208. doi: 10.1016/S0074-7696(08)61861-6. [DOI] [PubMed] [Google Scholar]
- 10.Baxt LA, Garza-Mayers AC, Goldberg MB. 2013. Bacterial subversion of host innate immune pathways. Science 340:697–701. doi: 10.1126/science.1235771. [DOI] [PubMed] [Google Scholar]
- 11.D’Souza-Schorey C, Chavrier P. 2006. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson JG, Jackson CL. 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y. 1999. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99:521–532. doi: 10.1016/S0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 14.D’Souza-Schorey C, Li G, Colombo MI, Stahl PD. 1995. A regulatory role for ARF6 in receptor-mediated endocytosis. Science 267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- 15.Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, Donaldson JG, Klausner RD. 1995. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol 128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. 1999. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J Cell Sci 112:855–866. [DOI] [PubMed] [Google Scholar]
- 17.Santy LC, Ravichandran KS, Casanova JE. 2005. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol 15:1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 18.Humphreys D, Davidson A, Hume PJ, Koronakis V. 2012. Salmonella virulence effector SopE and host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe 11:129–139. doi: 10.1016/j.chom.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V. 2013. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc Natl Acad Sci U S A 110:16880–16885. doi: 10.1073/pnas.1311680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balañá ME, Niedergang F, Subtil A, Alcover A, Chavrier P, Dautry-Varsat A. 2005. ARF6 GTPase controls bacterial invasion by actin remodelling. J Cell Sci 118:2201–2210. doi: 10.1242/jcs.02351. [DOI] [PubMed] [Google Scholar]
- 21.Wong KW, Isberg RR. 2003. Arf6 and phosphoinositol-4-phosphate-5-kinase activities permit bypass of the Rac1 requirement for beta1 integrin-mediated bacterial uptake. J Exp Med 198:603–614. doi: 10.1084/jem.20021363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, Tomchick DR, Alto NM. 2011. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature 469:107–111. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Gall T, Mavris M, Martino MC, Bernardini ML, Denamur E, Parsot C. 2005. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 151:951–962. doi: 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- 24.Niebuhr K, Giuriato S, Pedron T, Philpott DJ, Gaits F, Sable J, Sheetz MP, Parsot C, Sansonetti PJ, Payrastre B. 2002. Conversion of PtdIns(4,5)P(2) into PtdIns(5)P by the S. flexneri effector IpgD reorganizes host cell morphology. EMBO J 21:5069–5078. doi: 10.1093/emboj/cdf522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niebuhr K, Jouihri N, Allaoui A, Gounon P, Sansonetti PJ, Parsot C. 2000. IpgD, a protein secreted by the type III secretion machinery of Shigella flexneri, is chaperoned by IpgE and implicated in entry focus formation. Mol Microbiol 38:8–19. doi: 10.1046/j.1365-2958.2000.02041.x. [DOI] [PubMed] [Google Scholar]
- 26.Sannerud R, Declerck I, Peric A, Raemaekers T, Menendez G, Zhou L, Veerle B, Coen K, Munck S, De Strooper B, Schiavo G, Annaert W. 2011. ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc Natl Acad Sci U S A 108:E559–E568. doi: 10.1073/pnas.1100745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sansonetti PJ, Ryter A, Clerc P, Maurelli AT, Mounier J. 1986. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect Immun 51:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray K, Bobard A, Danckaert A, Paz-Haftel I, Clair C, Ehsani S, Tang C, Sansonetti P, Tran GV, Enninga J. 2010. Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell Microbiol 12:545–556. doi: 10.1111/j.1462-5822.2010.01428.x. [DOI] [PubMed] [Google Scholar]
- 29.Santy LC, Casanova JE. 2001. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol 154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonny B, Beraud-Dufour S, Chardin P, Chabre M. 1997. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- 31.Gaschet J, Hsu VW. 1999. Distribution of ARF6 between membrane and cytosol is regulated by its GTPase cycle. J Biol Chem 274:20040–20045. doi: 10.1074/jbc.274.28.20040. [DOI] [PubMed] [Google Scholar]
- 32.Yang CZ, Heimberg H, D’Souza-Schorey C, Mueckler MM, Stahl PD. 1998. Subcellular distribution and differential expression of endogenous ADP-ribosylation factor 6 in mammalian cells. J Biol Chem 273:4006–4011. doi: 10.1074/jbc.273.7.4006. [DOI] [PubMed] [Google Scholar]
- 33.Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, Chai J, Alto NM. 2009. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol Biol 16:853–860. doi: 10.1038/nsmb.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, Dixon JE. 2006. Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 35.Lu R, Goldberg MB. 2010. Bacterial exploitation of host cell signaling. Sci Transl Med 2:51ps48. doi: 10.1126/scitranslmed.3001612. [DOI] [PubMed] [Google Scholar]
- 36.Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, Selyunin AS, Way SS, Patrie SM, Alto NM. 2013. Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature 496:106–109. doi: 10.1038/nature12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langille SE, Patki V, Klarlund JK, Buxton JM, Holik JJ, Chawla A, Corvera S, Czech MP. 1999. ADP-ribosylation factor 6 as a target of guanine nucleotide exchange factor GRP1. J Biol Chem 274:27099–27104. doi: 10.1074/jbc.274.38.27099. [DOI] [PubMed] [Google Scholar]
- 38.Frank S, Upender S, Hansen SH, Casanova JE. 1998. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem 273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- 39.Myers KR, Wang G, Sheng Y, Conger KK, Casanova JE, Zhu JJ. 2012. Arf6-GEF BRAG1 regulates JNK-mediated synaptic removal of GluA1-containing AMPA receptors: a new mechanism for nonsyndromic X-linked mental disorder. J Neurosci 32:11716–11726. doi: 10.1523/JNEUROSCI.1942-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikenouchi J, Umeda M. 2010. FRMD4A regulates epithelial polarity by connecting Arf6 activation with the PAR complex. Proc Natl Acad Sci U S A 107:748–753. doi: 10.1073/pnas.0908423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabe H, Hashimoto S, Morishige M, Ogawa E, Hashimoto A, Nam JM, Miura K, Yano H, Onodera Y. 2009. The EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer invasion and metastasis. Traffic 10:982–993. doi: 10.1111/j.1600-0854.2009.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hafner M, Schmitz A, Grüne I, Srivatsan SG, Paul B, Kolanus W, Quast T, Kremmer E, Bauer I, Famulok M. 2006. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature 444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 43.Liu L, Pohajdak B. 1992. Cloning and sequencing of a human cDNA from cytolytic NK/T cells with homology to yeast SEC7. Biochim Biophys Acta 1132:75–78. doi: 10.1016/0167-4781(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Malaby AW, Famulok M, Sabe H, Lambright DG, Hsu VW. 2012. Grp1 plays a key role in linking insulin signaling to glut4 recycling. Dev Cell 22:1286–1298. doi: 10.1016/j.devcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klarlund JK, Tsiaras W, Holik JJ, Chawla A, Czech MP. 2000. Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J Biol Chem 275:32816–32821. doi: 10.1074/jbc.M002435200. [DOI] [PubMed] [Google Scholar]
- 46.Ogasawara M, Kim SC, Adamik R, Togawa A, Ferrans VJ, Takeda K, Kirby M, Moss J, Vaughan M. 2000. Similarities in function and gene structure of cytohesin-4 and cytohesin-1, guanine nucleotide-exchange proteins for ADP-ribosylation factors. J Biol Chem 275:3221–3230. doi: 10.1074/jbc.275.5.3221. [DOI] [PubMed] [Google Scholar]
- 47.Pendaries C, Tronchère H, Arbibe L, Mounier J, Gozani O, Cantley L, Fry MJ, Gaits-Iacovoni F, Sansonetti PJ, Payrastre B. 2006. PtdIns5P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. EMBO J. 25:1024–1034. doi: 10.1038/sj.emboj.7601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. 1997. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 49.Venkateswarlu K, Oatey PB, Tavaré JM, Cullen PJ. 1998. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol 8:463–466. doi: 10.1016/S0960-9822(98)70181-2. [DOI] [PubMed] [Google Scholar]
- 50.Nagel W, Schilcher P, Zeitlmann L, Kolanus W. 1998. The PH domain and the polybasic c domain of cytohesin-1 cooperate specifically in plasma membrane association and cellular function. Mol Biol Cell 9:1981–1994. doi: 10.1091/mbc.9.8.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manna D, Albanese A, Park WS, Cho W. 2007. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J Biol Chem 282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- 52.Brandman O, Meyer T. 2008. Feedback loops shape cellular signals in space and time. Science 322:390–395. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng S, Knödler A, Ren J, Zhang J, Zhang X, Hong Y, Huang S, Peränen J, Guo W. 2012. A Rab8 guanine nucleotide exchange factor-effector interaction network regulates primary ciliogenesis. J Biol Chem 287:15602–15609. doi: 10.1074/jbc.M111.333245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortiz D, Medkova M, Walch-Solimena C, Novick P. 2002. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol 157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stalder D, Barelli H, Gautier R, Macia E, Jackson CL, Antonny B. 2011. Kinetic studies of the Arf activator Arno on model membranes in the presence of Arf effectors suggest control by a positive feedback loop. J Biol Chem 286:3873–3883. doi: 10.1074/jbc.M110.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richardson BC, McDonold CM, Fromme JC. 2012. The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev Cell 22:799–810. doi: 10.1016/j.devcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. 2007. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell 18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Macia E, Paris S, Chabre M. 2000. Binding of the PH and polybasic C-terminal domains of ARNO to phosphoinositides and to acidic lipids. Biochemistry 39:5893–5901. doi: 10.1021/bi992795w. [DOI] [PubMed] [Google Scholar]
- 59.Auger KR, Cantley LC. 1991. Novel polyphosphoinositides in cell growth and activation. Cancer Cells 3:263–270. [PubMed] [Google Scholar]
- 60.DiNitto JP, Cronin TC, Lambright DG. 2003. Membrane recognition and targeting by lipid-binding domains. Sci STKE 2003:re16. doi: 10.1126/stke.2132003re16. [DOI] [PubMed] [Google Scholar]
- 61.Cronin TC, DiNitto JP, Czech MP, Lambright DG. 2004. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J 23:3711–3720. doi: 10.1038/sj.emboj.7600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allaoui A, Ménard R, Sansonetti PJ, Parsot C. 1993. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect Immun 61:1707–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mellouk N, Weiner A, Aulner N, Schmitt C, Elbaum M, Shorte SL, Danckaert A, Enninga J. 2014. Shigella subverts the host recycling compartment to rupture its vacuole. Cell Host Microbe 16:517–530. doi: 10.1016/j.chom.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 64.Ramel D, Lagarrigue F, Pons V, Mounier J, Dupuis-Coronas S, Chicanne G, Sansonetti PJ, Gaits-Iacovoni F, Tronchère H, Payrastre B. 2011. Shigella flexneri infection generates the lipid PI5P to alter endocytosis and prevent termination of EGFR signaling. Sci Signal 4:ra61. doi: 10.1126/scisignal.2001619. [DOI] [PubMed] [Google Scholar]
- 65.Konradt C, Frigimelica E, Nothelfer K, Puhar A, Salgado-Pabon W, di Bartolo V, Scott-Algara D, Rodrigues CD, Sansonetti PJ, Phalipon A. 2011. The Shigella flexneri type three secretion system effector IpgD inhibits T cell migration by manipulating host phosphoinositide metabolism. Cell Host Microbe 9:263–272. doi: 10.1016/j.chom.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 66.Puhar A, Tronchère H, Payrastre B, Nhieu GT, Sansonetti PJ. 2013. A Shigella effector dampens inflammation by regulating epithelial release of danger signal ATP through production of the lipid mediator PtdIns5P. Immunity 39:1121–1131. doi: 10.1016/j.immuni.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 67.Viaud J, Lagarrigue F, Ramel D, Allart S, Chicanne G, Ceccato L, Courilleau D, Xuereb JM, Pertz O, Payrastre B, Gaits-Iacovoni F. 2014. Phosphatidylinositol 5-phosphate regulates invasion through binding and activation of Tiam1. Nat Commun 5:4080. doi: 10.1038/ncomms5080. [DOI] [PubMed] [Google Scholar]
- 68.Labrec EH, Schneider H, Magnani TJ, Formal SB. 1964. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. J. Bacteriol. 88:1503–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Isberg RR, Falkow S. 1985. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 70.Parsot C, Ménard R, Gounon P, Sansonetti PJ. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol 16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 71.Marquart ME, Picking WL, Picking WD. 1996. Soluble invasion plasmid antigen C (IpaC) from Shigella flexneri elicits epithelial cell responses related to pathogen invasion. Infect Immun 64:4182–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Labigne-Roussel AF, Lark D, Schoolnik G, Falkow S. 1984. Cloning and expression of an afimbrial adhesin (AFA-I) responsible for P blood group-independent, mannose-resistant hemagglutination from a pyelonephritic Escherichia coli strain. Infect Immun 46:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Arf6 knockdown. Whole-cell lysates of ARF6K/D and ARF6+ MEFs, transfected with ARF6-HA where indicated. Immunoblotting with ARF6 or β-actin antibodies. Download
S. flexneri recruits ARF6 to entry sites in MEFs. (A) Recruitment of ARF6-HA to WT or noninvasive S. flexneri in ARF6+ MEFs infected for 25 min. ND, not detectable. Mean ± standard error of the mean of 3 independent experiments. (B) Representative images, labeled with antibody to HA (green), DAPI (blue), and phalloidin (red). Arrows, entering bacteria with ARF6 recruitment. Arrowheads, bacteria without ARF6 recruitment. Bar, 10 µm. Download
IpgD production in complemented ΔipgD S. flexneri. Congo red-induced secretion of S. flexneri strains. Pellets and precipitated supernatants immunoblotted with antibodies to IpgD or DnaK. Download
Recruitment of ARF6-HA to entering bacteria following siRNA depletion of ARNO in HeLa cells. (A) Depletion of ARNO by siRNA. Immunoblotting with ARNO or β-actin antibodies. (B) Percentage of cells with bacteria associated showing ARF6-HA recruitment to bacteria. Mean ± standard error of the mean of at least 3 independent experiments. NS, not significant. Download
In cells transfected with ARNO constructs, IpgD phosphatase activity is required for efficient S. flexneri entry and ruffle formation. (A) Percentage of cells infected. (B) Percentage of cells with ruffles. Mean ± standard error of the mean of at least 3 independent experiments. Download
LY294002 treatment that inhibits recruitment of ARNO 2G is associated with inhibition of Akt activation. Levels of phospho-Akt S473 and total Akt under conditions used for data presented in Fig. 6. Western blotting with β-actin as loading control. Download