ABSTRACT
A relatively small number of species in the large genus Streptomyces are pathogenic; the best characterized of these is Streptomyces scabies. The pathogenicity of S. scabies strains is dependent on the production of the nitrated diketopiperazine thaxtomin A, which is a potent plant cellulose synthesis inhibitor. Much is known about the genetic loci associated with plant virulence; however, the molecular mechanisms by which S. scabies triggers expression of thaxtomin biosynthetic genes, beyond the pathway-specific activator TxtR, are not well understood. In this study, we demonstrate that binding sites for the cellulose utilization repressor CebR occur and function within the thaxtomin biosynthetic cluster. This was an unexpected result, as CebR is devoted to primary metabolism and nutritive functions in nonpathogenic streptomycetes. In S. scabies, cellobiose and cellotriose inhibit the DNA-binding ability of CebR, leading to an increased expression of the thaxtomin biosynthetic and regulatory genes txtA, txtB, and txtR. Deletion of cebR results in constitutive thaxtomin A production and hypervirulence of S. scabies. The pathogenicity of S. scabies is thus under dual direct positive and negative transcriptional control where CebR is the cellobiose-sensing key that locks the expression of txtR, the key necessary to unlock the production of the phytotoxin. Interestingly, CebR-binding sites also lie upstream of and within the thaxtomin biosynthetic clusters in Streptomyces turgidiscabies and Streptomyces acidiscabies, suggesting that CebR is most likely an important regulator of virulence in these plant-pathogenic species as well.
IMPORTANCE
What makes a microorganism pathogenic is not limited to the genes acquired for virulence. Using the main causative agent of scab lesions on root and tuber crops as an example, our work identified the subtle but essential genetic changes that generate the cis-acting elements necessary for proper timing of the expression of the cluster of genes responsible for the biosynthesis of thaxtomin A, the primary virulence factor in plant-pathogenic streptomycetes. These data illustrate a situation in which a regulator associated with primary metabolism in nonpathogens, CebR, has been coopted as a master regulator of virulence in pathogenic species. Furthermore, the manipulation of CebR-mediated control of thaxtomin production will facilitate overproduction of this natural and biodegradable herbicide for commercial purposes. Our work thus provides a concrete example of how a strictly theoretical and computational work was able to elucidate a regulatory mechanism associated with the virulence of a plant pathogen and to generate solutions to purely agro-industrial concerns.
INTRODUCTION
Streptomyces is a very large genus of Gram-positive, high-G+C-content bacteria that are largely nonpathogens and well known for the production of pharmaceutically and agriculturally important secondary metabolites (1). Although several hundred species are known to date, only a few are phytopathogenic (2). The best-studied pathogens are Streptomyces scabies, Streptomyces acidiscabies, Streptomyces turgidiscabies, and Streptomyces ipomoeae, which cause raised or pitted scab lesions on economically important root and tuber crops, like potato, radish, beet, peanut, and sweet potato. The primary virulence determinant of S. scabies, S. acidiscabies, and S. turgidiscabies is the phytotoxin thaxtomin A (3). It is a member of a family of nitrated diketopiperazines formed by nonribosomal peptide synthases out of the main components 4-nitro tryptophan and phenylalanine (4). The biosynthetic pathway is unusual in that 4-nitro tryptophan is derived via a nitric oxide synthase from arginine (3, 5).
Plant-pathogenic streptomycetes are neither host nor tissue specific (3). In keeping with this broad host range, thaxtomin A has a highly conserved target—the plant cell wall. Thaxtomin affects the structure of the cell wall in dividing and expanding plant cells through an alteration of expression of cell wall biosynthesis-related genes and depletion of cellulose synthase complexes from the plasma membrane. This causes extensive cell wall remodeling characterized by reduced incorporation of crystalline cellulose into the plant cell wall, which is compensated for by an increased amount of pectins and hemicelluloses (6, 7).
The thaxtomin biosynthetic genes are highly conserved across plant-pathogenic streptomycetes and lie on a pathogenicity island that is mobilized in some species (3). However, acquisition of genes required for virulence is only the first step on the way to a pathogenic lifestyle. Subtle genetic changes are necessary to adapt the expression of newly acquired genes to the environment and the life cycle of the recipient microorganism. A limited number of mutations in regulatory regions and especially the distribution of cis-acting elements are key to developing a strain-specific transcriptional response. These DNA motifs are targeted by transcription factors that sense environmental signals through direct interaction with membrane sensors or indirect association with elicitor transporters. The production of thaxtomin A itself is under strict transcriptional control, including at least five global regulators belonging to the bld gene family, involved in morphological differentiation and/or the secondary metabolism of Streptomyces (8) in addition to the thaxtomin biosynthesis pathway-specific transcriptional activator TxtR (9). The multiplicity of global and specific regulators associated with thaxtomin production suggests that S. scabies may respond to multiple triggers that originate from plant material, such as xylan degradation products (10), suberin (11), and, more importantly, cellobiose, a product of cellulose degradation and a well-known elicitor of thaxtomin biosynthesis (4, 10) (it directly targets TxtR [9]). In this work, we describe an additional level of control of S. scabies pathogenicity. By recruiting CebR, the cellulose utilization regulator (12–14), we illustrate how the plant pathogen causes a trans-acting factor to digress from its primary function in order to use it to lock the expression of thaxtomin biosynthesis genes, thaxtomin production, and, therefore, its virulent behavior.
RESULTS AND DISCUSSION
Identification of CebR-binding sites associated with thaxtomin biosynthetic genes.
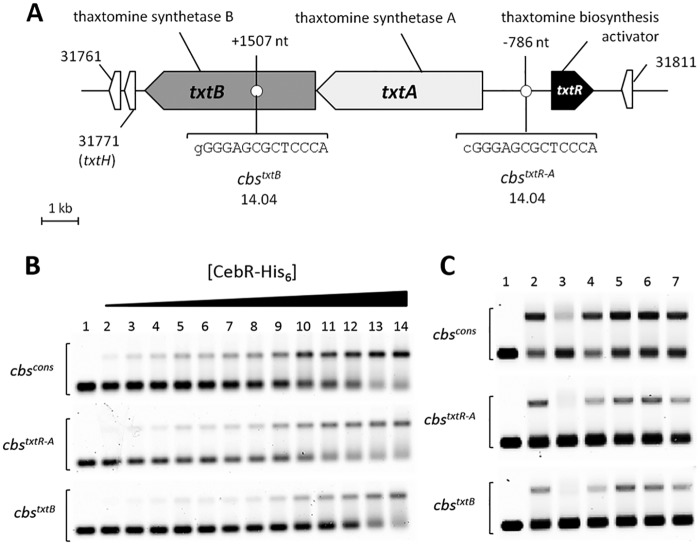
The transcription factors involved in the control of thaxtomin A production were identified by scrutinizing the genome of S. scabies for the occurrence of putative cis-acting elements of well-characterized DNA-binding proteins. Position weight matrices were generated from the compilation of DNA motifs bound by a series of regulators that were selected due to their notorious direct or indirect (implied) role in secondary-metabolite biosynthesis and/or morphological differentiation in streptomycetes. The PREDetector software (15) identified two 14-bp almost-perfect palindromic sequences highly similar to CebR boxes (henceforth called cbs for CebR-binding sites; see Fig. S1 in the supplemental material and reference 14) which are associated with the thaxtomin biosynthetic genes txtA and txtB and the pathway-specific regulatory gene txtR (Fig. 1A). The CGGGAGCGCTCCCA sequence (cbstxtR-A) lies in the intergenic region between txtR and txtA at nucleotide (nt) positions −786 and −900 upstream of txtR and txtA, respectively, whereas the GGGGAGCGCTCCCA sequence (cbstxtB) lies at nt position +1507 within the coding sequence of txtB (Fig. 1A). Both sequences display only a single mismatch with the previously reported 14-bp cbs palindromic consensus sequence TGGGAGCGCTCCCA (cbscons) (14). CebR has been identified as the repressor of cellulose/cellooligosaccharides/cellobiose utilization in streptomycetes (12–14), and in addition, this repressor has been shown to trigger morphogenesis in Streptomyces griseus (14). Binding of cellopentaose or cellobiose to CebR of S. reticuli or S. griseus, respectively, relieves binding of CebR to cbs and allows transcription of the downstream genes (12, 14). Interestingly, cellobiose and cellotriose are also known as the best elicitors of thaxtomin A production in S. scabies, S. acidiscabies, and S. turgidiscabies (9, 10), which strengthens the possible role of CebR in controlling thaxtomin biosynthesis.
FIG 1 .
CebR binding to cbs associated with thaxtomin biosynthetic genes is relieved by cellobiose. (A) Sequences and positions of CebR-binding sites identified in the chromosome region of the thaxtomin biosynthetic genes in S. scabies. Numbers associated with genes/ORFs are SCAB numbers from the annotated genome of S. scabies 87-22. The lowercase letters indicate nucleotides that do not match with the cbs consensus sequence. (B) EMSA demonstrating CebR binding to DNA motifs identified in the txtRA intergenic region (cbstxtR-A) and in txtB (cbstxtB). Numbers 1 to 14 refer to increasing concentrations of pure CebR-His6, i.e., 0 (free probe, 30 nM), 80, 160, 240, 320, 400, 480, 560, 640, 720, 960, 1,200, 1,600, and 3,200 nM, respectively. (C) EMSAs demonstrating cellobiose as the best allosteric effector of CebR. Numbers 1 and 2 refer to EMSAs with free probes (6 nM) and with probes incubated with CebR-His6, respectively. Numbers 3 to 7 refer to EMSAs with CebR-His6 preincubated with oligosaccharides, i.e., cellobiose (lane 3), cellotriose (lane 4), cellotetraose (lane 5), cellopentaose (lane 6), or cellohexaose (lane 7). Quantification of the shifted bands revealed that cellobiose (lane 3), cellotriose (lane 4), and cellohexaose (lane 7) were able to impair the CebR DNA-binding ability by approximately 56, 20, and 12%, respectively.
Electromobility gel shift assays (EMSAs) performed with double-stranded Cy5-labeled probes centered on the putative cbs sequence showed that cbstxtR-A and cbstxtB are strongly bound by CebR (Fig. 1B; Fig. S2). Under the same conditions, we also observed strong CebR binding to cbscons, which was used as a positive control because it is known to be specifically bound by CebR (12, 14) (Fig. 1B). EMSAs performed with CebR preincubated with cellobiose or cellooligosaccharides [(glucose)3 to (glucose)6] revealed that cellobiose was the best allosteric effector, disabling CebR from binding to its DNA targets (Fig. 1C). Our in silico prediction of cis-acting elements and the subsequent experimental in vitro validations allowed us to propose the following working model: once inside the cell, cellobiose and, to a lesser extent, cellotriose prevent the binding of the CebR repressor to cbstxtR-A and cbstxtB, which allows expression of txtA, txtB, and txtR.
Deletion of cebR results in constitutive production of thaxtomin A.
In order to uncover the role of CebR in controlling the expression levels of the thaxtomin biosynthetic genes txtA and txtB, we generated a cebR null mutant in S. scabies (ΔcebR mutant). Quantitative real-time reverse transcription-PCR (qPCR) on RNA extracted from wild-type S. scabies (87-22) and its cebR null mutant grown on International Streptomyces Project medium 4 (ISP-4) showed that the deletion of cebR results in overexpression of txtA, txtB, and txtR, respectively (Fig. 2). CebR thus acts as a transcriptional repressor of the thaxtomin biosynthetic genes in S. scabies as it represses the expression of the cellulolytic system in S. griseus. This result confirms that, despite the unusual positions of cbstxtR-A and cbstxtB further upstream of the target txtA and txtR genes and within the coding region of txtB (Fig. 1A), respectively, the identified DNA motifs are truly functional cis-acting elements.
FIG 2 .
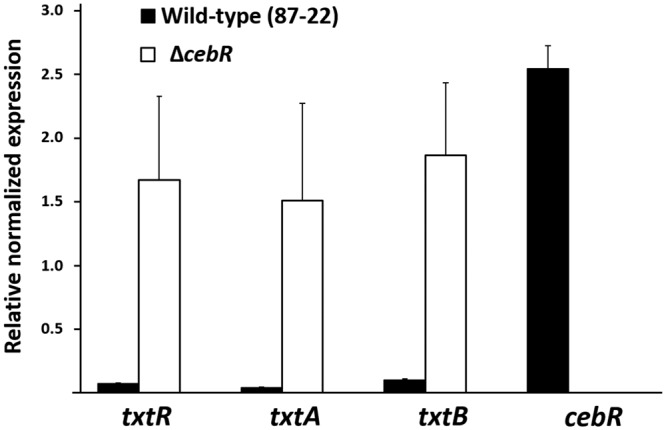
Effect of cebR deletion in S. scabies on the transcription levels of the thaxtomin biosynthetic and regulatory genes. qPCR analysis of gene expression levels in S. scabies 87-22 and in the ΔcebR strain. Data were normalized using the gyrA, murX, and hrdB genes as internal controls. Mean normalized expression levels (± standard deviations) from three biological replicates analyzed in triplicate are shown.
Moreover, the search for similar sequences in other thaxtomin A-producing Streptomyces pathogens confirmed the occurrence of cbs in their thaxtomin biosynthetic regions. In S. acidiscabies, three cbs signatures were identified within the thaxtomin biosynthetic cluster, i.e., one upstream of txtA (nt −787), one within txtA (nt +32), and one within txtB (nt +1559). In S. turgidiscabies, we found one cbs signature within txtA (nt +32) and one within txtB (nt +1508). A third cbs is found at nt position −1444 upstream of txtR. No cbs associated with thaxtomin production could be identified in S. ipomoeae, which produces mainly thaxtomin C, a phytotoxic member of the thaxtomin family; thaxtomin C production is not responsive to cellobiose or other oligosaccharides (16). Therefore, the occurrence of cbs upstream of or within the txt genes correlates with the observed cellobiose-dependent production of thaxtomin A in characterized pathogenic Streptomyces species.
We also assessed the thaxtomin production levels of both the S. scabies wild type and the cebR null mutant on oat bran agar (OBA), an undefined complex medium known to induce thaxtomin production (17), and ISP-4. The cebR mutant overproduced thaxtomin, which is bright yellow, compared to the level of its production by the wild-type strain 87-22 on OBA (Fig. 3A). More strikingly, overproduction was also observed when the ΔcebR strain was streaked onto plates containing routinely used Streptomyces ISP-4, which contains soluble starch as a carbon source (Fig. 3A). Extraction of thaxtomin from the plates and analysis by high-performance liquid chromatography (HPLC) confirmed the production of the toxin by the cebR mutant on ISP-4 (Fig. 3B). Since the wild-type strain produced only trace amounts of thaxtomin on ISP-4, this result demonstrates that we generated a mutant able to produce thaxtomin A independently of the presence of the elicitor cellobiose. Importantly, the ΔcebR mutant complemented with plasmid pRLIF8 carrying the S. scabies cebR gene and its upstream region restored the cellobiose-dependent induction of thaxtomin (see Fig. S3 in the supplemental material), demonstrating that the phenotype of the mutant is caused only by the chromosomal deletion of the S. scabies cebR gene (SCAB_57761).
FIG 3 .
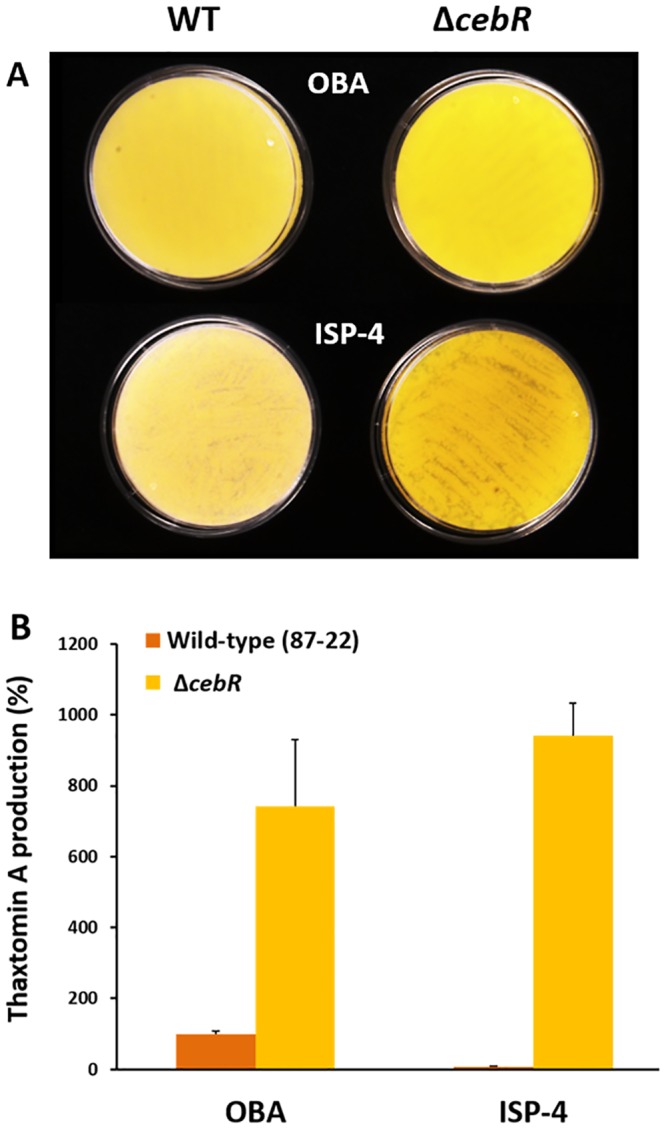
Effect of the cebR deletion on thaxtomin production in S. scabies. (A) Deletion of cebR in S. scabies resulted in higher thaxtomin production (yellow pigmentation) than in the wild type on OBA and even in its production under conditions which do not trigger thaxtomin production in the wild-type strain 87-22 (on ISP-4). (B) HPLC analysis of thaxtomin A extracted from plates shown in panel A.
Deletion of cebR leads to hypervirulence in S. scabies.
Since thaxtomin is the primary virulence determinant of the pathogenic streptomycetes, we evaluated the effect of cebR deletion on the virulence phenotype of S. scabies. Arabidopsis thaliana (ecotype Col-0) seeds grown on Murashige-Skoog (MS) agar were inoculated with spores of S. scabies 87-22 (wild-type) and its ΔcebR mutant (Fig. 4A). After 8 days of growth, seedlings inoculated with the cebR mutant presented stronger root and shoot stunting than those inoculated with the wild-type strain (Fig. 4B). In addition, we observed an increased capacity of S. scabies ΔcebR to induce pitting and necrosis of potato tuber slices compared to that of the wild-type strain 87-22 (Fig. 4C). Overall, the deletion of cebR resulted in a hypervirulent phenotype of S. scabies, confirming that CebR is the master repressor of plant pathogenicity through its impact on thaxtomin production.
FIG 4 .
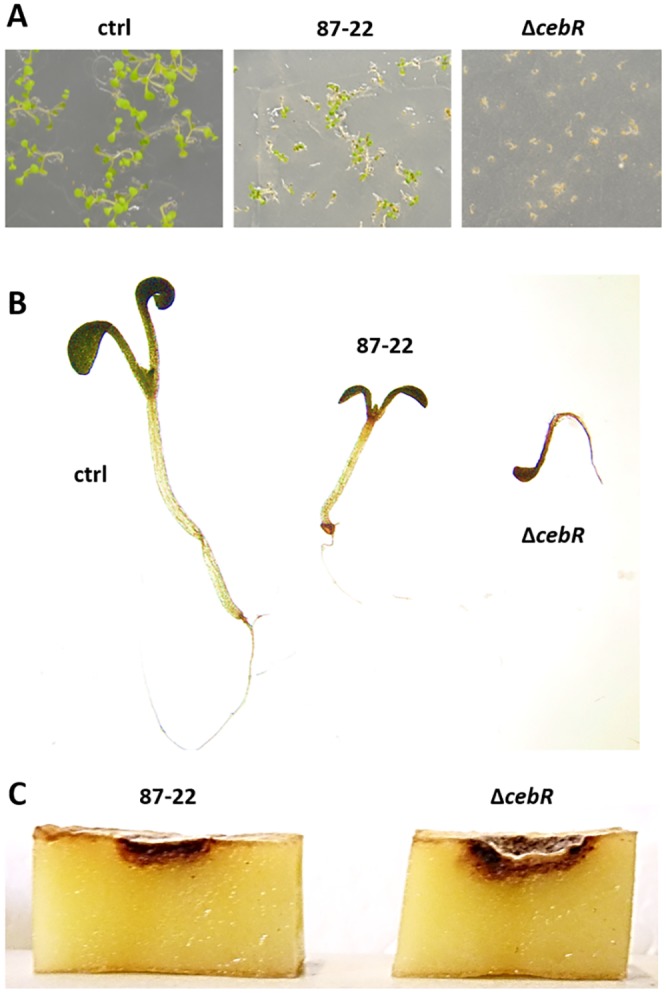
Effect of the deletion of cebR on the virulence of S. scabies. (A) Phenotype of A. thaliana grown for 8 days in the presence of S. scabies 87-22 (wild type) and its cebR null mutant; (B) closeup of representative plants grown in the MS plates shown in panel A; (C) potato tuber slice assay.
New pieces of the puzzle for thaxtomin regulation.
Regulation of virulence genes is critical for microbial pathogens and therefore often under strict control involving several layers of regulation. For thaxtomin production, one of the regulators involved is the pathway-specific TxtR protein, which acts as a transcriptional activator, inducing the expression of the thaxtomin biosynthetic genes (9). TxtR is an AraC family transcriptional regulator (AFTR) that binds cellobiose and was thought to be the primary regulator of thaxtomin production and virulence. However, enhanced transcription of txtR and thaxtomin biosynthetic genes in the cebR deletion mutant, in the absence of cellobiose, suggests a new paradigm for the molecular mechanism by which S. scabies and other thaxtomin A-producing streptomycetes utilize cellobiose as a signal. Our data show that the ability of cellobiose to induce thaxtomin production is implemented primarily through relief of CebR repression upon the transcription of the thaxtomin biosynthetic genes and that specific interaction of cellobiose with TxtR is not required for it to function as a transcriptional activator (9). Though some AFTRs require ligand binding to activate the transcription of virulence genes, this is not always the case (18). It may be that the specific interaction of cellobiose with TxtR provides a mechanism to fine-tune the regulation of thaxtomin production. However, results presented here reinforce the significance of cellobiose as a signal for thaxtomin production and, therefore, virulence and demonstrate how minor intergenic and intragenic changes in the region coding for thaxtomin biosynthesis allow CebR to act as the “gatekeeper” of pathogenicity in S. scabies. Likewise, Streptomyces coelicolor recruited DasR, the N-acetylglucosamine utilization regulator, to control the production of the antibiotic actinorhodin and the prodiginines (19), as well as siderophores (20, 21), probably as a response to sensing its own cell wall degradation during programmed cell death (22). Though noteworthy, the CebR and DasR examples in S. scabies and S. coelicolor, respectively, are not sufficient for us to state that enrolling transcription factors normally devoted to primary carbon metabolism is a common feature of streptomycetes in regulating the onset of their secondary metabolism. Finding chemical elicitors and culturing conditions that trigger the production of the so-called cryptic secondary metabolite is presented as a challenging, expensive, and time-consuming task (23–25). By deleting cebR, we have been able to create a strain that produces much higher levels of thaxtomin than the natural strains produce, which will assist in getting thaxtomin onto the market. Our studies of S. coelicolor and S. scabies thus suggest that at least some actinomycete species, by having selected by-products of the most abundant carbon and nitrogen sources on earth (cellulose and chitin or peptidoglycan) as signaling molecules, have evolved pathways simple to engineer as a means of awakening the sleeping antibiotics (26, 27).
MATERIALS AND METHODS
Bacterial strains, media, chemicals, and culture conditions.
All strains and plasmids used in this study are described in Table S1 in the supplemental material. Escherichia coli strains were cultured in Luria-Bertani (LB) medium at 37°C. Streptomyces strains were routinely grown at 28°C in tryptic soy broth (TSB; BD Biosciences) or on International Streptomyces Project medium 4 (ISP-4), 1 liter of which consists of soluble starch (10 g), K2HPO4 (1 g), MgSO4⋅7H2O (1 g), NaCl (1 g), (NH4)2SO4 (2 g), CaCO3 (2 g), FeSO4⋅7H2O (0.001 g), MnCl2⋅7H2O (0.001 g), ZnSO4⋅7H2O (0.001 g), agar (20 g) (BD Biosciences), and soy flour mannitol (SFM) agar (28). Where required, the medium was supplemented with the antibiotics apramycin (100 µg/ml), kanamycin (50 µg/ml), chloramphenicol (25 µg/ml), thiostrepton (25 µg/ml), and/or nalidixic acid (50 µg/ml). Cellobiose and cellooligosaccharides were purchased at Megazyme (Ireland).
CebR regulon prediction in streptomycetes.
The position weight matrix (PWM) was created and the CebR regulon in S. scabies was computationally predicted with the PREDetector software (15) as described previously (20). We used DNA motifs known to be bound by CebR in S. griseus (14) as a training set to generate the PWM (PWM-CebRgri) (Fig. S1) and scan the genomes of pathogenic Streptomyces species for similar DNA motifs using different types of threshold scores (29).
His-tagged CebR production in E. coli and protein purification.
The open reading frame (ORF) encoding SCAB57761 was amplified by PCR using the primers SCAB_57761+3_NdeI and SCAB_57761+1056_EcoRI (see Table S2 in the supplemental material). The corresponding PCR product was subsequently cloned into the pJET1.2/blunt cloning vector, yielding pSAJ001. After DNA sequencing, an NdeI-EcoRI DNA fragment was excised from pSAJ001 and cloned into pET-22b digested with the same restriction enzymes. E. coli Rosetta(DE3) competent cells were transformed with the resulting construct, pSAJ002. E. coli cells carrying pSAJ002 were grown at 37°C in 250 ml LB medium containing 100 µg/ml of ampicillin until the culture reached an optical density at 600 nm (OD600) of 0.6. Production of His6-tagged CebR (CebR-His6) was induced overnight (~20 h) at 16°C by addition of 1 mM isopropyl-β-d-thiogalactopyranoside. Cells were collected by centrifugation and ruptured by sonication in lysis buffer (50 mM Tris-HCl buffer; pH 7.5; supplemented with the EDTA-free cOmplete protease inhibitor cocktail (Roche). Soluble proteins were loaded onto a preequilibrated Ni2+-nitrilotriacetic acid (NTA)-agarose column (5-ml bed volume), and CebR-His6 eluted at around 150 mM imidazole. Fractions containing the pure protein were pooled (Fig. S2) and desalted using a HiTrap desalting column (GE Healthcare) with 20 mM Tris-HCl buffer.
EMSAs.
EMSAs with 34-bp double-stranded probes (Table S2) were performed using Cy5-labeled probes mainly as described previously (30). cbs probes (6 nM or 30 nM final concentration) and CebR-His6 at a final concentration of 0.08 to 3.2 µM were mixed in a total reaction volume of 50 µl. All reactions were carried out in EMSA buffer (10 mM Tris-HCl, pH 7.5, 1 mM dithiothreitol, 0.25 mM CaCl2, 0.5 mM MgCl2, 50 mM KCl, and 2% glycerol) containing an excess of nonspecific salmon sperm DNA. An EMSA with a Cy5-labeled probe containing the DasR-responsive element upstream of dasA (31) and CebR-His6 was performed as a negative control (data not shown).
Generation of the cebR gene deletion in S. scabies 87-22.
Deletion mutations in S. scabies 87-22 were created using the Redirect PCR targeting method (32) by replacing the selected gene with an antibiotic deletion cassette. These cassettes, consisting of an oriT and an antibiotic resistance gene [aac(3)IV for apramycin resistance (Table S1)] and flanked by FLP recombinase recognition target (FRT) sites were generated by PCR using primers with gene-specific homology extensions (Table S2) and pIJ773 (Table S1) as the template. The gel-purified deletion cassettes were electroporated into the E. coli BW25113 strain harboring the arabinose-inducible λ red expression plasmid pIJ790 and cosmid 833, containing the gene of interest. Transformants were recovered on apramycin-selective medium, and correct gene replacement in the cosmid was confirmed by PCR and sequencing. The resulting mutated cosmid was then transferred to S. scabies 87-22 via intergeneric conjugation after passage through the E. coli ET12567 strain harboring pUZ8002 (Table S1). Exconjugants were selected for resistance to apramycin and sensitivity to kanamycin. Genomic DNA was extracted from Streptomyces cultures grown in TSB medium using the MasterPure Gram-positive DNA purification kit (Epicentre Biotechnologies) according to the manufacturer’s instructions, and verification of the mutant isolates was performed by PCR.
qPCR.
RNA was prepared from 72-h-old mycelia grown on ISP-4 plates at 28°C using the RNeasy minikit (Qiagen) according to the manufacturer’s instructions. PCRs using the purified RNA were performed to verify the absence of genomic DNA. cDNA synthesis was performed with 1 µg of DNase-treated (Turbo DNA-free kit; Ambion) RNA using the iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time reverse transcription-PCR (qPCR) was carried out in 10 µl containing 4 µl of SsoAdvanced Sybr green supermix (Bio-Rad), 4 µl of 1/10-diluted cDNA, and 0.5 pmol of each gene-specific primer (Table S2), and the mixture was subjected to the following PCR protocol: 3 min at 95°C and 40 cycles of 30 s at 95°C, followed by 45 s at 60°C. A melting curve analysis (samples were heated from 60°C to 95°C) was performed after each qPCR run to verify specific amplification. The murX, hrdB, and gyrA genes (11) were used to normalize the amount of RNA in the samples. Each measurement was performed in triplicate with three different cebR mutant isolates.
Analysis of thaxtomin A production.
Mycelial suspensions of S. scabies strains were prepared from 48- to 72-h-old TSB-grown cultures by pelleting the mycelia, washing them twice with sterile water, and resuspending them in 1 ml of sterile water to obtain an OD600 of 1.0. Samples of 50 µl were plated out on small petri dishes (5-cm diameter) containing 12.5 ml of OBA or ISP-4 medium. After incubation for 7 days at 28°C, the medium was chopped into small cubes and soaked in 8 ml of methanol for 10 min. The supernatant was filtered through a 0.2-µm polytetrafluoroethylene (PTFE) filter and analyzed via HPLC as described previously (16). All experiments were repeated using different biological replicates of the Streptomyces strains, with three technical replicates per strain.
Virulence bioassays.
Arabidopsis thaliana (ecotype Col-0) seeds were surface sterilized with sodium hypochlorite and washed three times in deionized H2O. Seeds were suspended in 0.1% agarose and placed on Murashige-Skoog (MS) (33) agar medium supplemented with 1% sucrose. After an incubation period of one night at 4°C, seeds were inoculated with the wild-type or ΔcebR spore suspension (107 spores/ml). Plants were grown at 21 ± 2°C, with a 12-h photoperiod, for 5 to 6 days. Potato tuber slices were prepared as described previously (34) and inoculated with 5 mg of Streptomyces mycelium. Potato slices were then incubated at 23 ± 2°C in a dark and moist incubator for 4 days.
SUPPLEMENTAL MATERIAL
CebR position weight matrix (PWM). The PWM was constructed using the PREDetector software (15) with CebR-binding sites experimentally validated in Streptomyces griseus (14). Only different sequences were selected for the training set. The a priori (pi) was fixed at 71.5, as deduced from the GC content of the S. scabies 87-22 genome and using different types of threshold scores as suggested previously (29). Download
SDS-PAGE showing the level of purity of His6-tagged CebR used for electromobility gel shift assays. Lane 2, the predicted migration size of the band was around 41 kDa, which corresponds well to the calculated 40.206-kDa size of His6-tagged CebR. Lane 1, molecular weight markers (Fermentas). The protein identification was performed by liquid chromatography-electrospray ionization (LQ-ESI)-tandem mass spectrometry. The band was excised from the SDS-PAGE gel stained with Coomassie blue and reduced, alkylated, and digested within the gel slice using trypsin. The protein digest was independently analyzed on a liquid chromatograph (nano-Ultimate 3000; Dionex)-ESI ion trap (amaZon speed electron transfer dissociation; Bruker Daltonics) in positive-ion mode. Spectra were interpreted using data analysis version 4.0 (Bruker). Database searches were performed using the Mascot server version 2/2/04 and protein Scape version 3.0 (Bruker) with the NCBI database (restricted to bacterial taxonomies). Download
Genetic complementation of the cebR mutant scored by thaxtomin production on ISP-4 plates. (A) A visual inspection of ISP-4 plates indicates thaxtomin production by its typical yellow pigmentation. (B) HPLC analysis of thaxtomin production on ISP-4 plates. The cebR mutant (and the cebR mutant transformed with the empty pAU3-45 plasmid as a negative control for the complementation) produce thaxtomin under conditions that do not induce toxin production in the wild-type strain. Thaxtomin production reverts back to wild-type levels when the mutation is genetically complemented (cebR mutant transformed with pRLIF8 containing the cebR gene and its promoter region). Download
Bacterial strains, plasmids, and cosmids used in this study.
List of oligonucleotides used in this study.
ACKNOWLEDGMENTS
S.J.’s work is supported by an Aspirant grant from the FNRS. S.R. is an FRS-FNRS research associate. This work is supported in part by the Belgian program of Interuniversity Attraction Poles initiated by the Federal Office for Scientific Technical and Cultural Affairs (PAI no. P7/44) and by the FNRS (research project RFNRS.3342-T.0006.14-PDR [FRFC]). I.M.F. was supported by the Agriculture and Food Research Initiative Competitive Grants Program (grant 2010-65110-20416 from the U.S. Department of Agriculture’s National Institute of Food and Agriculture to R. Loria).
Footnotes
Citation Francis IM, Jourdan S, Fanara S, Loria R, Rigali S. 2015. The cellobiose sensor CebR is the gatekeeper of Streptomyces scabies pathogenicity. mBio 6(2):e02018-14. doi:10.1128/mBio.02018-14.
REFERENCES
- 1.Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. Oxford University Press, New York, NY. [Google Scholar]
- 2.Loria R, Kers J, Joshi M. 2006. Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol 44:469–487. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PubMed] [Google Scholar]
- 3.Loria R, Bignell DR, Moll S, Huguet-Tapia JC, Joshi MV, Johnson EG, Seipke RF, Gibson DM. 2008. Thaxtomin biosynthesis: the path to plant pathogenicity in the genus Streptomyces. Antonie Van Leeuwenhoek 94:3–10. doi: 10.1007/s10482-008-9240-4. [DOI] [PubMed] [Google Scholar]
- 4.Johnson EG, Krasnoff SB, Bignell DR, Chung WC, Tao T, Parry RJ, Loria R, Gibson DM. 2009. 4-Nitrotryptophan is a substrate for the nonribosomal peptide synthetase TxtB in the thaxtomin A biosynthetic pathway. Mol Microbiol 73:409–418. doi: 10.1111/j.1365-2958.2009.06780.x. [DOI] [PubMed] [Google Scholar]
- 5.Barry SM, Kers JA, Johnson EG, Song L, Aston PR, Patel B, Krasnoff SB, Crane BR, Gibson DM, Loria R, Challis GL. 2012. Cytochrome P450-catalyzed l-tryptophan nitration in thaxtomin phytotoxin biosynthesis. Nat Chem Biol 8:814–816. doi: 10.1038/nchembio.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheible WR, Fry B, Kochevenko A, Schindelasch D, Zimmerli L, Somerville S, Loria R, Somerville CR. 2003. An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species. Plant Cell 15:1781–1794. doi: 10.1105/tpc.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bischoff V, Cookson SJ, Wu S, Scheible WR. 2009. Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings. J Exp Bot 60:955–965. doi: 10.1093/jxb/ern344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bignell DR, Francis I, Fvans J, Loria R. 2014. Thaxtomin A production and virulence are controlled by several bld gene global regulators in Streptomyces scabies. Mol Plant Microbe Interact 27:875–885. doi: 10.1094/MPMI-02-14-0037-R. [DOI] [PubMed] [Google Scholar]
- 9.Joshi MV, Bignell DR, Johnson EG, Sparks JP, Gibson DM, Loria R. 2007. The AraC/XylS regulator TxtR modulates thaxtomin biosynthesis and virulence in Streptomyces scabies. Mol Microbiol 66:633–642. doi: 10.1111/j.1365-2958.2007.05942.x. [DOI] [PubMed] [Google Scholar]
- 10.Wach MJ, Krasnoff SB, Loria R, Gibson DM. 2007. Effect of carbohydrates on the production of thaxtomin A by Streptomyces acidiscabies. Arch Microbiol 188:81–88. doi: 10.1007/s00203-007-0225-x. [DOI] [PubMed] [Google Scholar]
- 11.Lauzier A, Simao-Beaunoir AM, Bourassa S, Poirier GG, Talbot B, Beaulieu C. 2008. Effect of potato suberin on Streptomyces scabies proteome. Mol Plant Pathol 9:753–762. doi: 10.1111/j.1364-3703.2008.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlösser A, Aldekamp T, Schrempf H. 2000. Binding characteristics of CebR, the regulator of the ceb operon required for cellobiose/cellotriose uptake in Streptomyces reticuli. FEMS Microbiol Lett 190:127–132. doi: 10.1111/j.1574-6968.2000.tb09274.x. [DOI] [PubMed] [Google Scholar]
- 13.Schlösser A, Jantos J, Hackmann K, Schrempf H. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl Environ Microbiol 65:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marushima K, Ohnishi Y, Horinouchi S. 2009. CebR as a master regulator for cellulose/cellooligosaccharide catabolism affects morphological development in Streptomyces griseus. J Bacteriol 191:5930–5940. doi: 10.1128/JB.00703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiard S, Marée R, Colson S, Hoskisson PA, Titgemeyer F, van Wezel GP, Joris B, Wehenkel L, Rigali S. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem Biophys Res Commun 357:861–864. doi: 10.1016/j.bbrc.2007.03.180. [DOI] [PubMed] [Google Scholar]
- 16.Guan D, Grau BL, Clark CA, Taylor CM, Loria R, Pettis GS. 2012. Evidence that thaxtomin C is a pathogenicity determinant of Streptomyces ipomoeae, the causative agent of streptomyces soil rot disease of sweet potato. Mol Plant Microbe Interact 25:393–401. doi: 10.1094/MPMI-03-11-0073. [DOI] [PubMed] [Google Scholar]
- 17.Johnson EG, Joshi MV, Gibson DM, Loria R. 2007. Cello-oligosaccharides released from host plants induce pathogenicity in scab-causing Streptomyces species. Physiol Mol Plant Pathol 71:18–25. doi: 10.1016/j.pmpp.2007.09.003. [DOI] [Google Scholar]
- 18.Yang J, Tauschek M, Robins-Browne RM. 2011. Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol 19:128–135. doi: 10.1016/j.tim.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep 9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craig M, Lambert S, Jourdan S, Tenconi E, Colson S, Maciejewska M, Ongena M, Martin JF, van Wezel G, Rigali S. 2012. Unsuspected control of siderophore production by N-acetylglucosamine in streptomycetes. Environ Microbiol Rep 4:512–521. doi: 10.1111/j.1758-2229.2012.00354.x. [DOI] [PubMed] [Google Scholar]
- 21.Lambert S, Traxler MF, Craig M, Maciejewska M, Ongena M, van Wezel GP, Kolter R, Rigali S. 2014. Altered desferrioxamine-mediated iron utilization is a common trait of bald mutants of Streptomyces coelicolor. Metallomics 6:1390–1399. doi: 10.1039/c4mt00068d. [DOI] [PubMed] [Google Scholar]
- 22.Chi WJ, Lee SY, Lee J. 2011. Functional analysis of SGR4635-induced enhancement of pigmented antibiotic production in Streptomyces lividans. J Microbiol 49:828–833. doi: 10.1007/s12275-011-1100-7. [DOI] [PubMed] [Google Scholar]
- 23.Aigle B, Corre C. 2012. Waking up Streptomyces secondary metabolism by constitutive expression of activators or genetic disruption of repressors. Methods Enzymol 517:343–366. doi: 10.1016/B978-0-12-404634-4.00017-6. [DOI] [PubMed] [Google Scholar]
- 24.Zhu H, Sandiford SK, van Wezel GP. 2014. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 41:371–386. doi: 10.1007/s10295-013-1309-z. [DOI] [PubMed] [Google Scholar]
- 25.Yoon V, Nodwell JR. 2014. Activating secondary metabolism with stress and chemicals. J Ind Microbiol Biotechnol 41:415–424. doi: 10.1007/s10295-013-1387-y. [DOI] [PubMed] [Google Scholar]
- 26.Świątek MA, Urem M, Tenconi E, Rigali S, van Wezel GP. 2012. Engineering of N-acetylglucosamine metabolism for improved antibiotic production in Streptomyces coelicolor A3(2) and an unsuspected role of NagA in glucosamine metabolism. Bioengineered 3:280–285. doi: 10.4161/bioe.21371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martín JF, Liras P. 2010. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol 13:263–273. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Fondation, Norwich, United Kingdom. [Google Scholar]
- 29.Rigali S, Nivelle R, Tocquin P. 6 November 2014. On the necessity and biological significance of threshold-free regulon prediction outputs. Mol Biosyst doi: 10.1039/C4MB00485J. [DOI] [PubMed] [Google Scholar]
- 30.Świątek MA, Tenconi E, Rigali S, van Wezel GP. 2012. Functional analysis of the N-acetylglucosamine metabolic genes of Streptomyces coelicolor and role in control of development and antibiotic production. J Bacteriol 194:1136–1144. doi: 10.1128/JB.06370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colson S, van Wezel GP, Craig M, Noens EE, Nothaft H, Mommaas AM, Titgemeyer F, Joris B, Rigali S. 2008. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154:373–382. doi: 10.1099/mic.0.2007/011940-0. [DOI] [PubMed] [Google Scholar]
- 32.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with Tobacco tissue cultures. Physiol Plant 15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 34.Loria R, Bukhalid RA, Creath RA, Leiner RH, Olivier M, Steffens JC. 1995. Differential production of thaxtomins by pathogenic Streptomyces species in vitro. Phytopathology 85:537–541. doi: 10.1094/Phyto-85-537. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CebR position weight matrix (PWM). The PWM was constructed using the PREDetector software (15) with CebR-binding sites experimentally validated in Streptomyces griseus (14). Only different sequences were selected for the training set. The a priori (pi) was fixed at 71.5, as deduced from the GC content of the S. scabies 87-22 genome and using different types of threshold scores as suggested previously (29). Download
SDS-PAGE showing the level of purity of His6-tagged CebR used for electromobility gel shift assays. Lane 2, the predicted migration size of the band was around 41 kDa, which corresponds well to the calculated 40.206-kDa size of His6-tagged CebR. Lane 1, molecular weight markers (Fermentas). The protein identification was performed by liquid chromatography-electrospray ionization (LQ-ESI)-tandem mass spectrometry. The band was excised from the SDS-PAGE gel stained with Coomassie blue and reduced, alkylated, and digested within the gel slice using trypsin. The protein digest was independently analyzed on a liquid chromatograph (nano-Ultimate 3000; Dionex)-ESI ion trap (amaZon speed electron transfer dissociation; Bruker Daltonics) in positive-ion mode. Spectra were interpreted using data analysis version 4.0 (Bruker). Database searches were performed using the Mascot server version 2/2/04 and protein Scape version 3.0 (Bruker) with the NCBI database (restricted to bacterial taxonomies). Download
Genetic complementation of the cebR mutant scored by thaxtomin production on ISP-4 plates. (A) A visual inspection of ISP-4 plates indicates thaxtomin production by its typical yellow pigmentation. (B) HPLC analysis of thaxtomin production on ISP-4 plates. The cebR mutant (and the cebR mutant transformed with the empty pAU3-45 plasmid as a negative control for the complementation) produce thaxtomin under conditions that do not induce toxin production in the wild-type strain. Thaxtomin production reverts back to wild-type levels when the mutation is genetically complemented (cebR mutant transformed with pRLIF8 containing the cebR gene and its promoter region). Download
Bacterial strains, plasmids, and cosmids used in this study.
List of oligonucleotides used in this study.