Abstract
Background:
Type 2 diabetes mellitus (T2DM) is a worldwide problem that threatens the public health and economies of all countries. A multifactorial etiology and interaction between environmental factors and genetic components are responsible for triggering and progression of T2DM. Recently, rs7754840 single nucleotide polymorphism (SNP) in the CDKAL1 gene was reported to be associated with T2DM in various populations. However, due to inconsistent results in various populations about the association of rs7754840 with T2DM, and lack of information in the Iranian population, we have evaluated its association with T2DM in a subset of the Iranian population from Isfahan province, central part of Iran.
Materials and Methods:
The study included 140 patients and 140 controls selected based on the World Health Organization guidelines. Genomic DNA was extracted from blood samples and the rs7754840 SNP was genotyped using a polymerase chain reaction-restriction fragment length polymorphism assay with specific primers and restriction enzyme (Ac1I).
Results:
The frequency of the C allele in the cases was higher than that in the controls (72.9% vs. 65%; P = 0.045). Using logistic regression analysis, we found a significant risk association of CC genotype with T2DM susceptibility (OR = 2.319, 95% CI = 1.436-3.744, P = 0.001). Furthermore, compared with the CC genotype, individuals with the GC genotype had a lower risk (protective association) of developing T2DM (OR = 0.332, 95% CI = 0.202-0.547, P < 0.001).
Conclusions:
We confirmed that there is a significant risk association between rs7754840 polymorphism and development of T2DM in a subset of the Iranian population from Isfahan province.
Keywords: CDKAL1 gene, single nucleotide polymorphism, type 2 diabetes mellitus
INTRODUCTION
Type 2 diabetes mellitus (T2DM) is a global public health crisis that threatens the economies of all nations, and its prevalence continues to increase in many countries, including Iran.[1,2] Although the exact underlying molecular mechanisms of T2DM development have not been fully revealed, a mixture of various genetic and environmental elements is deliberated to be involved in pathogenesis of the disease.[3,4] In addition to the important contribution of environmental factors, including dietary patterns and lifestyle, genetic determinants also play a major role in T2DM susceptibility.[3,5] In recent years, many genes associated with T2DM were recognized by genome-wide association studies (GWAS).[6,7,8,9,10,11] From these, cyclin-dependent kinase 5 regulatory subunit-associated protein 1 - like 1 (CDKAL1) gene was newly known as a predisposition gene for T2DM through GWAS in white European and Asian populations.[11,12,13,14,15,16] This gene encodes CDKAL1 protein that might influence the action of the CDK5 protein, which promotes insulin production, and may impact on other functions in the insulin-producing β-cells of the pancreas gland.[17,18] Moreover, it is supposed that elevated activity of CDK5 in the absence of CDKAL1 has a role in the pathophysiology of β-cell defect.[17,19,20] However, precise molecular mechanisms of susceptibility to T2DM by interactions between these proteins are not yet clear.[20,21] Replication studies reproducibly evaluated the association between T2DM and rs7754840 SNP within the CDKAL1 gene in multiple populations of Europe and Asian.[12,13,15,16,22,23,24,25,26,27,28] Most of the studies confirmed this significant association.[12,13,15,16,22,25,26,29] However, some studies failed to detect a significant association, suggesting variability in the contribution of this SNP to the risk of T2DM in different ethnic tribes.[27,28] In spite of the regular associations among Europeans and Asians, the contribution of this genetic variant in some ethnic groups such as Iran is not determined. Therefore, we conducted a replication study of candidate rs7754840 SNP in CDKAL1 gene in a subset of the Iranian population from the Isfahan province in relation to the T2DM.
MATERIALS AND METHODS
Study population
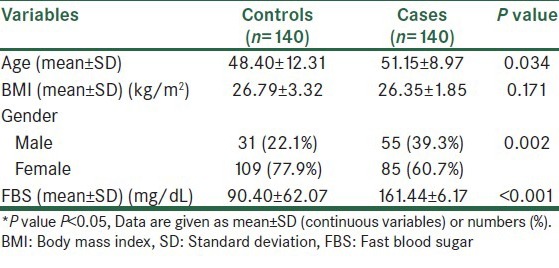
A total of 280 individuals, including 140 T2DM patients and 140 normal controls, were involved in the current study. Diabetes was defined according to the World Health Organization criteria (fasting plasma glucose 7.0 mmol/L and/or 2-h plasma glucose ≥ 11.1 mmol/L) and only confirmed cases were included in the study. We also randomly selected and examined 140 control subjects from the general population who were enrolled from an annual health check directed at the same center (31 men and 109 women; age 48.40 ± 12.31 years; fasting plasma glucose 90.40 ± 6.17 mg/dL; BMI 26.3 ± 1.8 kg/m2; Table 1). For control individuals, lack of history of diabetes in the subjects and among their first-degree relatives was carefully monitored. Informed consent was obtained from all participants by a standard questionnaire and the Ethics Committee of the Isfahan University of Medical Sciences approved this study.
Table 1.
Demographic parameters in the cases and controls
SNP genotyping
Peripheral blood samples of participants were collected and then genomic DNA was isolated from whole blood samples by the DNG-plus DNA extraction kit (Cinnagen, Iran) according to the manufacturer's instructions. Genotyping was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). PCR was performed using the following primers: Forward 5’-GAGTTTCAAATTGTCCAG-3’ and reverse 5’-TCTGTTATTGACTGAGGTAT-3’. The PCR cycling conditions were adjusted as follows: Denaturation at 95°C for 6 min followed by 30 cycles at 94°C for 50 s, 57°C for 50 s and 72°C for 50 s, followed by a final extension at 72°C for 5 min. The amplified products were separated by electrophoresis through 2% agarose gel stained with ethidium bromide. For RFLP of rs7754840 polymorphism, the PCR products were digested with the AclI restriction endonuclease (Fermentas, Lithuania) for 3 h at 37°C. Following digestion, the appearance of two bands of 112 bp and 80 bp was indicative of the CC genotype, whereas 192 bp digestion products were indicative of the GG genotype. Also, when heterozygous genotypes exist, three bands were observed: 192 bp, 112 bp and 80 bp [Figure 1]. For confirmation of the determined genotype by PCR-RFLP, we performed direct sequencing on 10% of the randomly selected samples.
Figure 1.
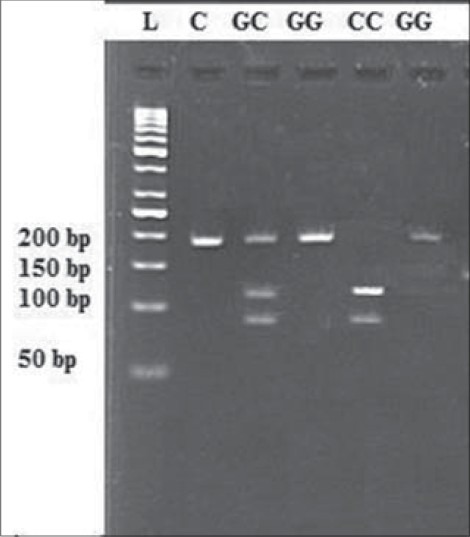
Lane 1 (L): DNA ladder, lanes 2; (C=Control) PCR product (undigested); lane 3; GC Heterozygous, lanes 4 and 6; GG genotypes, lane 5; CC Homozygous
Statistical analyses
Statistical analyses were achieved using SPSS for Windows software (version 18.0; SPSS, Chicago, IL, USA). The allele and genotype frequencies were tested for Hardy-Weinberg equilibrium using the x2test. Logistic regression analysis was accomplished to calculate distributions and risk allele/genotype-specific odds ratios (ORs), 95% confidential intervals (CIs) and analogous P values after adjustment for gender, age and body mass index (BMI) as covariates between cases. All continuous variables were expressed as the mean ± standard deviation (SD). Student's t test was used to compare the continuous variables between the T2DM and non-diabetic control groups. Pearson's v2 test was used to evaluate the difference in the prevalence of T2DM among the genotypes.
RESULTS
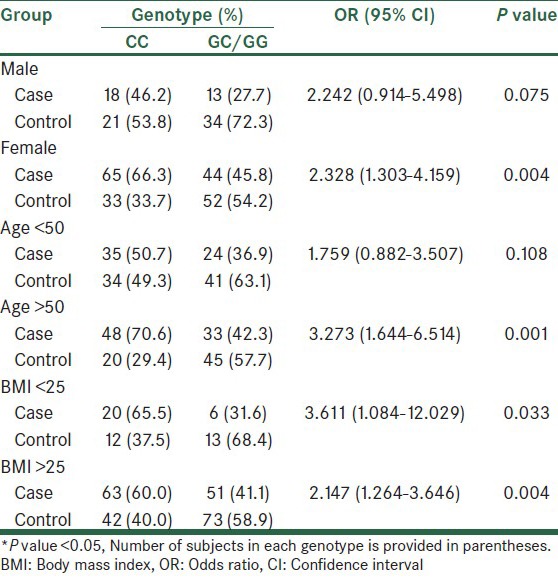
Information of clinical and biochemical characteristics of the participants are shown in Table 1. The mean age, BMI and fasting plasma glucose (FPG) level were 51.15 ± 8.97 years, 27 ± 2.4 kg/m2 and 161.44 ± 62.07 mg/dL for the diabetic patients, respectively, and 48.40 ± 12.31 years, 26.3 ± 1.8 kg/m2 and 90.40 ± 6.17 mg/dL for the controls, respectively. A higher proportion of females than males were observed in the case (60.7% vs. 39.3%) and control groups (77.9% vs. 22.1%), which may be as a result of a contribution bias in our population. The association results of genotype and allele frequencies of the T2DM-susceptibility rs7754840 variant in patients and control individuals is shown in Table 2. According to our results, frequencies of the CC, CG and GG genotypes of rs7754840 were 38.6, 52.9 and 8.6% in controls and 59.3, 27.1 and 13.6% in cases, respectively. Among these three genotypes of the rs7754840 SNP in the CDKAL1 gene, CC was found to be meaningfully associated with an increased risk [OR = 2.921 (95% CI 1.789-4.771), P = <0.001] of T2DM in our population. Also, the frequency of the C allele in cases (72.9%) was more than that in the healthy control group (27.1%), acting as a risk allele [OR = 2.319 (95% CI 1.436-3.744); P = 0.001]. The risk allele (C allele) was the same direction as the previous studies, but this risk allele was the minor allele in Asian groups. Beside this, we found that compared with the CC genotype (as risk genotype), individuals with the GC heterozygote genotype had a lower risk and protective association to develop T2DM (as protective genotype; OR = 0.332, 95% CI = 0.202–0.547, P < 0.001). We then tested the association between genotypes and T2DM-related traits such as gender, age (under and over 50 years) and BMI (under and over 25 kg/m2) to examine whether these genotypes conferred a risk of T2DM through their effects on any of these traits [Table 3]. We found consistent results for all variables except for individuals with age under 50 years who showed no significant association (OR = 1.759, 95% CI = 0.882-3.507, P = 0.108) and males who had a marginal association.
Table 2.
Allele and genotype distribution of rs7754840 SNP in cases and controls and its association with T2DM
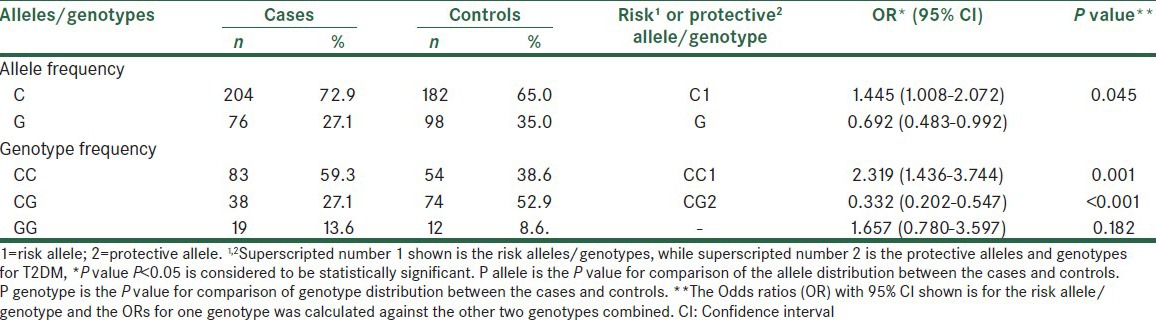
Table 3.
Stratification analysis of rs7754840 SNP genotype frequency in cases and controls based on T2DM-related traits
DISCUSSION
Recent GWAS studies have recognized rs7754840 SNP in the CDKAL1 gene associated with T2DM in white Europeans and Asians.[10,11,27,30,31] The association between CDKAL1 rs7754840 SNP and T2DM has been broadly studied in different populations.[12,13,15,16,22,24,27,29,32] Most replication studies proved a significant relationship,[13,16,22,24,26,29] nevertheless, some studies failed to detect a noteworthy association,[27,28] proposing variability in the impact of this polymorphism to conferring the risk of T2DM in various populations. To explore this issue, we performed the present replication study to evaluate and replicate the association of rs7754840 SNP within CDKAL1 gene in a subset of Iranians from Isfahan province. Our results showed that the carriers of C allele and CC homozygous genotypes of the rs7754840 SNP had an increased risk and were more likely to develop T2DM compared with those with GG homozygous and GC heterozygous genotypes (adjusted OR = 2.921 [95% CI 1.789-4.771], P = <0.001). Consistent with our study, a significant association between C allele and CC genotypes of this SNP and T2DM was seen in most ethnic groups in the European[12,16] and Asian populations.[13,15] Among these, the CC genotype was significantly associated with T2DM in the Finnish,[18] Ashkenazi,[33] Han Chinese people from Hong Kong,[34] Korean[25] and Japanese populations,[24,35] although a marginal relationship was found (P = 0.05 and P = 0.076) in the two Japanese studies.[35,36] Also, in spite of the significant results from our work and the aforementioned studies, some studies failed to find any association; for instance, no statistically significant association of the rs7754840 variant with T2DM was reported in two studies from Europe,[9,33] Indian Sikhs[37] and Pima Indians.[28] These inconsistencies findings propose that rs7754840 SNP might has an even more important interaction in diabetes susceptibility basis on different genetic background of diverse ethnic groups, lifestyles and environmental variables in different populations. To the best of our knowledge, the present work is the first replication study to evaluate the association of CDKAL1 and T2DM-susceptibility in the Iranian population. In addition, for this SNP, the C risk allele was a major allele in our population, consistent with former reports in European populations,[15] while the C allele was shown as a minor allele in the Asian groups.[12] Therefore, these variances might reveal the differences in the outcomes for the association of rs7754840 SNP in the CDKAL1 gene with T2DM. Ethnic differences in risk allele frequencies for rs7754840 SNP may lead to differences in the attributable risk allele and thus alter its effect on different populations. Interestingly, we also detected a novel significant association finding in our study: Compared with the CC genotype (as risk genotype), individuals with the GC heterozygote genotype had a lower risk and protective association to develop T2DM (as protective genotype), suggesting that each genotype has a distinctive effect related to T2DM; however, more replication studies and meta-analyses are required for the evaluation of this genotype-specific effect in different populations. Furthermore, when we performed stratification analysis on the basis of T2DM-related traits such as gender, age (under and over 50 years) and BMI status (under and over 25 kg/m2) regarding the CC risk genotype, another association was detected. The CC genotype only in subgroups of over 50 years is a risk genotype, and, in the subgroup of under 50 years, was not related to T2DM susceptibility. This suggests that genetic susceptibility caused by this genetic variant in our population is often associated with a late age of onset (age specific) and may play a more important role in the development of T2DM among older patients in the Iranian population. This finding is also consistent with this fact that multifactorial diseases such as diabetes to be revealed with elevated age due to the interaction between genetic background and environmental factors. In conclusion, our finding confirms the association between rs7754840 polymorphism in the CDKAL1 gene and sporadic T2DM risk and also the GC heterozygote genotype of the polymorphism is associated with reduced risk (protective effect) of T2DM development compared with the CC genotype. Furthermore, our study suggests an association between the CC genotype of the rs7754840 polymorphism and people with late-onset T2DM. This genetic susceptibility may be a valuable marker for differentiating people with an elevated risk of T2DM, who could then be subjected to a further careful or earlier routine screening for T2DM. Also, to further confirm this association, additional studies are needed to elucidate the exact functional mechanism of this variant.
ACKNOWLEDGMENT
We thank the financial support of Vice-Chancellor for Research, the Isfahan University of Medical Sciences. We are also grateful to the head and stuff of Isfahan Endocrinology and Metabolism Research Center for their help in sample collection.
Footnotes
Source of Support: We thank the financial support of Vice-Chancellor for Research, Isfahan University of Medical Sciences, Isfahan, Iran. We are also grateful to the head and stuff of Isfahan Endocrinology and Metabolism Research Center for their help in sample collection
Conflict of Interest: None declared.
REFERENCES
- 1.Esteghamati A, Meysamie A, Khalilzadeh O, Rashidi A, Haghazali M, Asgari F, et al. Third national Surveillance of Risk Factors of Non-Communicable Diseases (SuRFNCD-2007) in Iran: Methods and results on prevalence of diabetes, hypertension, obesity, central obesity, and dyslipidemia. BMC Public Health. 2009;9:167. doi: 10.1186/1471-2458-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–53. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 3.Hu FB. Globalization of diabetes: The role of diet, lifestyle, and genes. Diabetes Care. 2011;34:1249–57. doi: 10.2337/dc11-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant RW, Moore AF, Florez JC. Genetic architecture of type 2 diabetes: Recent progress and clinical implications. Diabetes Care. 2009;32:1107–14. doi: 10.2337/dc08-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363:2339–50. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 6.Wheeler E, Barroso I. Genome-wide association studies and type 2 diabetes. Brief Funct Genomics. 2011;10:52–60. doi: 10.1093/bfgp/elr008. [DOI] [PubMed] [Google Scholar]
- 7.Tabara Y, Osawa H, Kawamoto R, Onuma H, Shimizu I, Miki T, et al. Replication study of candidate genes associated with type 2 diabetes based on genome-wide screening. Diabetes. 2009;58:493–8. doi: 10.2337/db07-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–62. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 9.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 10.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–5. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 12.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Li H, Loos RJ, Yu Z, Ye X, Chen L, et al. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57:2834–42. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan JT, Ng DP, Nurbaya S, Ye S, Lim XL, Leong H, et al. Polymorphisms identified through genome-wide association studies and their associations with type 2 diabetes in Chinese, Malays, and Asian-Indians in Singapore. J Clin Endocrinol Metab. 2010;95:390–7. doi: 10.1210/jc.2009-0688. [DOI] [PubMed] [Google Scholar]
- 15.Dehwah M, Wang M, Huang QY. CDKAL1 and type 2 diabetes: A global meta-analysis. Genet Mol Res. 2010;9:1109–20. doi: 10.4238/vol9-2gmr802. [DOI] [PubMed] [Google Scholar]
- 16.Grarup N, Rose CS, Andersson EA, Andersen G, Nielsen AL, Albrechtsen A, et al. Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: Validation and extension of genome-wide association studies. Diabetes. 2007;56:3105–11. doi: 10.2337/db07-0856. [DOI] [PubMed] [Google Scholar]
- 17.Miyaki K, Oo T, Song Y, Lwin H, Tomita Y, Hoshino H, et al. Association of a cyclin-dependent kinase 5 regulatory subunit-associated protein 1-like 1 (CDKAL1) polymorphism with elevated hemoglobin A1c levels and the prevalence of metabolic syndrome in Japanese men: Interaction with dietary energy intake. Am J Epidemiol. 2010;172:985–91. doi: 10.1093/aje/kwq281. [DOI] [PubMed] [Google Scholar]
- 18.Stancáková A, Pihlajamäki J, Kuusisto J, Stefan N, Fritsche A, Häring H, et al. EUGENE2 Consortium. Single-nucleotide polymorphism rs7754840 of CDKAL1 is associated with impaired insulin secretion in nondiabetic offspring of type 2 diabetic subjects and in a large sample of men with normal glucose tolerance. J Clin Endocrinol Metab. 2008;93:1924–30. doi: 10.1210/jc.2007-2218. [DOI] [PubMed] [Google Scholar]
- 19.Ohara-Imaizumi M, Yoshida M, Aoyagi K, Saito T, Okamura T, Takenaka H, et al. Deletion of CDKAL1 affects mitochondrial ATP generation and first-phase insulin exocytosis. PLoS One. 2010;5:e15553. doi: 10.1371/journal.pone.0015553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Qiao W, Zhao X, Tao M. Quantitative assessment of the influence of hematopoietically expressed homeobox variant (rs1111875) on type 2 diabetes risk. Mol Genet Metab. 2011;102:194–9. doi: 10.1016/j.ymgme.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Pascoe L, Tura A, Patel SK, Ibrahim IM, Ferrannini E, Zeggini E, et al. RISC Consortium: U.K. Type 2DiabetesGenetics Consortium. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes. 2007;56:3101–4. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- 22.Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008;57:2226–33. doi: 10.2337/db07-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han X, Luo Y, Ren Q, Zhang X, Wang F, Sun X, et al. Implication of genetic variants near SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, FTO, TCF2, KCNQ1, and WFS1 in type 2 diabetes in a Chinese population. BMC Med Genet. 2010;11:81. doi: 10.1186/1471-2350-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omori S, Tanaka Y, Takahashi A, Hirose H, Kashiwagi A, Kaku K, et al. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008;57:791–5. doi: 10.2337/db07-0979. [DOI] [PubMed] [Google Scholar]
- 25.Lee YH, Kang ES, Kim SH, Han SJ, Kim CH, Kim HJ, et al. Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. J Hum Genet. 2008;53:991–8. doi: 10.1007/s10038-008-0341-8. [DOI] [PubMed] [Google Scholar]
- 26.Jiang JY, Qiu H, Zhao GM, Zhou Y, Mo M, Shu L, et al. Association of CDKAL1 genetic polymorphism with glycosylated hemoglobin A1clevel among non-diabetic Chinese adults. J Diabetes Metab. 2011;2:166. [Google Scholar]
- 27.Hertel JK, Johansson S, Raeder H, Midthjell K, Lyssenko V, Groop L, et al. Genetic analysis of recently identified type 2 diabetes loci in 1,638 unselected patients with type 2 diabetes and 1,858 control participants from a Norwegian population-based cohort (the HUNT study) Diabetologia. 2008;51:971–7. doi: 10.1007/s00125-008-0982-3. [DOI] [PubMed] [Google Scholar]
- 28.Rong R, Hanson RL, Ortiz D, Wiedrich C, Kobes S, Knowler WC, et al. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes. 2009;58:478–88. doi: 10.2337/db08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng MC, Tam CH, So WY, Ho JS, Chan AW, Lee HM, et al. Implication of genetic variants Near NEGR1, SEC16B, TMEM18, ETV5/DGKG, GNPDA2, LIN7C/BDNF, MTCH2, BCDIN3D/FAIM2, SH2B1, FTO, MC4R, and KCTD15 with obesity and type 2 diabetes in 7705 Chinese. J Clin Endocrinol Metab. 2010;95:2418–25. doi: 10.1210/jc.2009-2077. [DOI] [PubMed] [Google Scholar]
- 30.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Wellcome Trust Case Control Consortium (WTCCC).Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeuchi F, Serizawa M, Yamamoto K, Fujisawa T, Nakashima E, Ohnaka K, et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58:1690–9. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Kilpeläinen TO, Liu C, Zhu J, Liu Y, Hu C, et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55:981–95. doi: 10.1007/s00125-011-2370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cauchi S, Meyre D, Durand E, Proença C, Marre M, Hadjadj S, et al. Post genome-wide association studies of novel genes associated with type 2 diabetes show gene-gene interaction and high predictive value. PloS One. 2008;3:e2031. doi: 10.1371/journal.pone.0002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y, Li P, Cai L, Zhang B, Tang X, Zhang X, et al. Association study of genetic variants in eight genes/loci with type 2 diabetes in a Han Chinese population. BMC Med Genet. 2010;11:97. doi: 10.1186/1471-2350-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horikoshi M, Hara K, Ito C, Shojima N, Nagai R, Ueki K, et al. Variations in the HHEX gene are associated with increased risk of type 2 diabetes in the Japanese population. Diabetologia. 2007;50:2461–6. doi: 10.1007/s00125-007-0827-5. [DOI] [PubMed] [Google Scholar]
- 36.Horikawa Y, Miyake K, Yasuda K, Enya M, Hirota Y, Yamagata K, et al. Replication of genome-wide association studies of type 2 diabetes susceptibility in Japan. J Clin Endocrinol Metab. 2008;93:3136–41. doi: 10.1210/jc.2008-0452. [DOI] [PubMed] [Google Scholar]
- 37.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, et al. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]


