Background: Enzymes of the FAH superfamily catalyze a multitude of diverse chemical reactions.
Results: Using molecular modeling followed by biochemical investigations, FAHD1 was identified as oxaloacetate decarboxylase.
Conclusion: Our findings suggest that ODx activity can be found in eukaryotic members of the FAH superfamily.
Significance: Our results identify a mammalian ODx enzyme as a so far undescribed player in mitochondrial metabolism.
Keywords: Computer Modeling, Decarboxylase, Energy Metabolism, Mitochondria, Pyruvate, FAH Domain, FAHD1, Oxaloacetate Decarboxylase
Abstract
Fumarylacetoacetate hydrolase (FAH) domain-containing proteins occur in both prokaryotes and eukaryotes, where they carry out diverse enzymatic reactions, probably related to structural differences in their respective FAH domains; however, the precise relationship between structure of the FAH domain and the associated enzyme function remains elusive. In mammals, three FAH domain-containing proteins, FAHD1, FAHD2A, and FAHD2B, are known; however, their enzymatic function, if any, remains to be demonstrated. In bacteria, oxaloacetate is subject to enzymatic decarboxylation; however, oxaloacetate decarboxylases (ODx) were so far not identified in eukaryotes. Based on molecular modeling and subsequent biochemical investigations, we identified FAHD1 as a eukaryotic ODx enzyme. The results presented here indicate that dedicated oxaloacetate decarboxylases exist in eukaryotes.
Introduction
The fumarylacetoacetate hydrolase (FAH)2 protein superfamily branches out through all organisms, from prokaryotes to humans, comprising a diverse set of enzymatic functions. Its distinguishing feature is the highly conserved catalytic center designated as the FAH fold. It is named after the founding member of the family, the well characterized mammalian enzyme FAH, which hydrolyzes fumarylacetoacetate to fumarate and acetoacetate (1), constituting the last step in the metabolic breakdown of tyrosine. In the genetic disorder hereditary tyrosinemia type 1, this reaction is compromised due to mutations in the FAH gene (2). The accumulation of toxic metabolites caused by the disruption of FAH activity leads to severe liver and renal disease (3).
Although the FAH fold is highly conserved, FAH superfamily members cover a wide range of diverse enzymatic activities. Several members possess β-diketone hydrolase activity, including FAH itself. HpcE from Escherichia coli is a bifunctional decarboxylase/isomerase, catalyzing the decarboxylation of 2-oxo-5-carboxy-hept-3-ene-1,7-dioic acid to 2-hydroxy-hepta-2,4-diene-1,7-dioic acid, followed by the conversion into 2-oxo-hept-3-ene-1,7-dioic acid (4). Other members that were characterized in both structure and enzymatic function include KdaD from Sulfolobus solfataricus (5), dehydrating 2-keto-3-deoxy-d-arabinonate to 2,5-dioxopentanoate, and MhpD from E. coli (6), which hydrates 2-hydroxypenta-2,4-dienoic acid to 4-hydroxy-2-ketopentanoic acid. Overall, the FAH superfamily includes many prokaryotic members with very distinct functions that lack homologs in eukaryotes. This can be explained by the fact that these enzymes are part of highly specialized metabolic pathways, involving chemical compounds that higher organisms are unable to convert and utilize for their metabolism (5). One recently identified prokaryotic member of the FAH superfamily found in Corynebacterium glutamicum, referred to as Cg1458, was characterized as a novel soluble oxaloacetate decarboxylase (ODx) (7, 8); however, eukaryotic ODx enzymes were not identified so far.
Besides FAH itself, two other members of the superfamily have been described in humans, FAHD1 and FAHD2, with orthologs in most eukaryotic organisms. The 25-kDa FAH domain-containing protein 1 (FAHD1) has been studied by x-ray crystallography (9), revealing a homodimeric structure and a mixed β-sandwich roll fold bearing a metal ion binding site, similar to other members of the FAH protein family. At that time, there were no further publications about FAHD1, and its enzymatic function was still unknown. Based on the structure of the active site, a hydroxylase or decarboxylase function was proposed. Also, a common function shared with other so far uncharacterized FAH proteins, E. coli YcgM and Thermus thermophilus TTHA0809, was suggested due to structural similarities (9). FAHD2 shows moderate sequence homology with FAHD1, but no further data are available for this enzyme at the moment.
Due to the broad range of enzymatic functions present within the FAH superfamily despite the structural similarities, it proved challenging to narrow down possible functions for FAHD1 from sequence and structural information. In a recent study (10), we utilized a bioinformatics approach to show for the first time that FAHD1 is closely related to the prokaryotic fumarylpyruvate hydrolase NagK (11). This finding was derived from the fact that charged residues present in the catalytic center of FAHD1 and likely involved in its enzymatic function (9) are fully conserved between NagK and FAHD1. Enzymatic assays performed with the purified protein confirmed that FAHD1 is able to hydrolyze substrates of the acylpyruvate class in vitro. In the case of acetylpyruvate, Vmax was determined as 0.14 μmol min−1 mg−1, with a Km value of 4.6 μm. The reaction rate was found to be dependent on the type of the bound divalent metal ion, with Mg2+ and Mn2+ yielding the highest rates. Immunoblot analysis indicated widespread expression of FAHD1 in various murine tissues, with highest expression in kidney and liver, as well as in a wide range of human cell lines; immunofluorescence studies showed clear mitochondrial localization (10).
As there is no existing evidence that acylpyruvates play any significant role in the metabolism of higher organisms, the in vivo relevance of the acylpyruvase activity associated with FAHD1 still awaits confirmation. In this study, we conducted a further search for possible in vivo substrates by means of bioinformatics and structural modeling, trying to clarify the relevance of FAHD1 for mammalian metabolism.
EXPERIMENTAL PROCEDURES
Structural Investigation of the FAH Domain Active Sites
The Protein Data Bank (PDB) database (12) was screened for entries associated with the Pfam accession number PF01557 to identify the FAH family. The 32 resulting entries were structurally compared in the Molecular Operating Environment (MOE) software (Chemical Computing Group, MOE release 2013.08, Montreal, Canada). The study was limited to chain A of PDB entries containing multiple structures in a unit cell. Superposition of the proteins was first performed based on all C-α atoms. Subsequently, a refined superposition on residues within 9 Å around the Mg2+ ion in 1SAW was performed. This inner sphere was defined as the active site. Structural similarity is calculated based on the root mean square deviation of C-α atom positions included within this sphere. Low values of root mean square deviation indicate very similar structures. Based on the similarity matrix between the 32 structures, a hierarchical clustering was performed resulting in a tree representation of FAH relations.
Modeling of FAHD1 Active Site
The x-ray structure of human FAHD1 (PDB code 1SAW) (9, 13) lacks 11 highly flexible residues next to the active site (Asp-29 to Leu-39). We constructed this region using the “Loop modeler” tool of MOE. To allow the loop region to extend into the active site, as expected for a closed lid conformation, four water positions were deleted in the active site: HOH314, HOH316, HOH356, and HOH359. The missing loop was defined between Val-21 and Val-43 to identify potential loop candidates from the PDB. A tolerant maximal walk step of 5 amino acids allowed varying potential anchoring points between residue 16 and residue 48. All other parameters were kept as default. 94 structured loop regions were identified as potential templates with these parameters. We used the best scored candidate from the putative FAH protein from Yersinia pestis (PDB code 3S52) as the template structure for loop construction. The parameters for refined loop modeling included an adaption of the environment by side chain repacking (default parameters).
Bacterial Recombinant Expression and Purification of FAHD1
N-terminally His- and S-tagged versions of human wild-type FAHD1 and three mutants (H30A, E33A, and D102A/R106A) were recombinantly expressed in E. coli and purified as reported previously (10).
Oxaloacetate Decarboxylase Assay Utilizing Lactate Dehydrogenase and NADH
ODx activity was measured with an Infinite M200 spectrophotometer (Tecan) in a disposable UV cuvette (1-cm path length, Brand) at 25 °C. Each reaction was performed in 1 ml of assay buffer (50 mm Tris-HCl, 100 mm KCl, 1 mm MgCl2, 200 μm NADH, pH 7.4) containing 12 μg of recombinant FAHD1 and 0.1 units of l-lactate dehydrogenase. The reaction was started by adding oxaloacetate to a final concentration of 100 μm. The decreasing absorbance of NADH was recorded between 300 and 380 nm in regular time intervals.
Direct Oxaloacetate Decarboxylase Assay
ODx activity was measured with an Infinite M200 spectrophotometer (Tecan), similar to Ran et al. (8). The assay was performed in a disposable UV cuvette (1-cm path length, Brand) at 25 °C in 1 ml of assay buffer (50 mm Tris-HCl, 100 mm KCl, 1 mm MgCl2, pH 7.4) containing varying amounts of recombinant FAHD1 (3–60 μg). The reaction was started by adding oxaloacetate to a final concentration of up to 1 mm. The decreasing absorbance at 255 nm (ϵ = 1070 m−1 cm−1 for oxaloacetate) was measured over several minutes in regular time intervals and used to calculate initial reaction rates. For the inhibitor assay, varying concentrations of oxalate were included. All reactions were performed three times. Reaction mixtures containing no substrate were used as blank. All rates were corrected for appearance of pyruvate and for auto-decarboxylation under assay conditions (14).
Analysis of Oxaloacetate Decarboxylase Reaction by HPLC
1 ml of assay buffer containing oxaloacetate (1 mm) and purified recombinant FAHD1 protein (120 μg) was incubated at room temperature for 30 min. A control lacking FAHD1 was incubated analogously. The conversion mixture and control of the FAHD1 reaction were analyzed by HPLC using an ÄKTA purifier system (GE Healthcare) equipped with a Bio-Rad Aminex HPX-87H column (300 × 7.8 mm). Detection was at 210 nm. 84 μl of sample were injected and eluted with 5 mm H2SO4 as the eluent at a flow rate of 0.5 ml min−1 at room temperature. Identification of peaks was based on the characteristic retention times of high purity standards (>99%) of oxaloacetate and pyruvate.
Mouse Work
F2 generation C57BL/6 mice heterozygous for a LoxP-flanked FAHD1 gene were established by inGenious Targeting Laboratory. FAHD1−/− mice were generated by crossing FAHD1flox/+ with Cre0/+ transgenic mice followed by outcrossing of Cre alleles and crossing of FAHD1−/+ mice. The knock-out was verified by PCR and immunoblot. The mice were housed in a temperature-controlled room at 22 °C with a 12-h light/12-h dark cycle. All mice were given unlimited access to water and standard chow (ssniff V1536-00). Individually housed male mice, aged 6–8 weeks, were divided into three groups; control mice were provided with standard chow ad libitum, and food was withdrawn for 18 h for fasted and refed mice, who were given access to standard chow for 4 h after fasting. Water was provided ad libitum. After the mice were sacrificed via cervical dislocation, desired organs were harvested, snap-frozen, and stored in liquid nitrogen.
Preparation of RNA and Quantitative Real Time PCR
Total RNA from mouse liver was isolated using TRIzol® reagent (Sigma) according to the manufacturer's protocol. For cDNA synthesis, 1 μg of total RNA was reverse transcribed with the RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific) and diluted 1:5. Amplification was carried out with specific primers for FAHD1, PC (pyruvate carboxylase), and PCK1 (phosphoenolpyruvate carboxykinase 1). Real time quantitative PCR was performed in triplicates using LightCycler® 480 SYBR Green I master mix (Roche Applied Science).
Western Blot
Frozen mouse organs were homogenized in PBS supplemented with protease inhibitors (one Complete Mini EDTA-free tablet per 10 ml, Roche Diagnostics). Lysate supernatants (30 μg of total protein) were separated by SDS-PAGE and blotted onto a PVDF membrane. Rabbit monoclonal anti-mouse FAHD1 antibody (purpose-made, 14 μg/ml), rabbit polyclonal anti-PEPCK antibody recommended for detection of PCK1 and PCK2 (Santa Cruz Biotechnology sc-32879, 1:2,000), rabbit polyclonal anti-PC antibody (Proteintech 16588-1-AP, 1:2,000), and anti-rabbit HRP-conjugated secondary antibody (Dako P0399, 1:2,500) were applied by standard Western blot protocol. α-Tubulin antibody (Sigma T5168, 1:10,000) was used for loading control with an anti-mouse HRP-conjugated secondary antibody (Dako P0447, 1:20,000). Detection was achieved by ECL Prime (GE Healthcare). For relative protein quantification, specific bands were analyzed using Fiji software (15).
Analysis of Oxaloacetate Levels in Mouse Tissue
3-month-old female mice (wild-type and FAHD1−/− littermates) were sacrificed by cervical dislocation, and the desired organs (kidney and liver) were immediately excised and shock-frozen in liquid nitrogen. Frozen organs were homogenized in ice-cold 5% perchloric acid (1 ml per 100 mg of tissue) and assayed according to the method described by Parvin et al. (16). Briefly, after hydrolysis of endogenous acetyl-CoA and subsequent neutralization, a 25-μl aliquot was included in a 200-μl reaction containing 0.6 units of citrate synthase (Sigma) and 3 pmol of [3H]acetyl-CoA (39 nCi, Moravek Biochemicals) to transform endogenous oxaloacetate into [3H]citrate. After adsorption of unreacted [3H]acetyl-CoA to activated charcoal (Sigma), samples were measured in a liquid scintillation counter (LS 6500, Beckman).
RESULTS
Identification of FAHD1 as Candidate Oxaloacetate Decarboxylase by Molecular Modeling
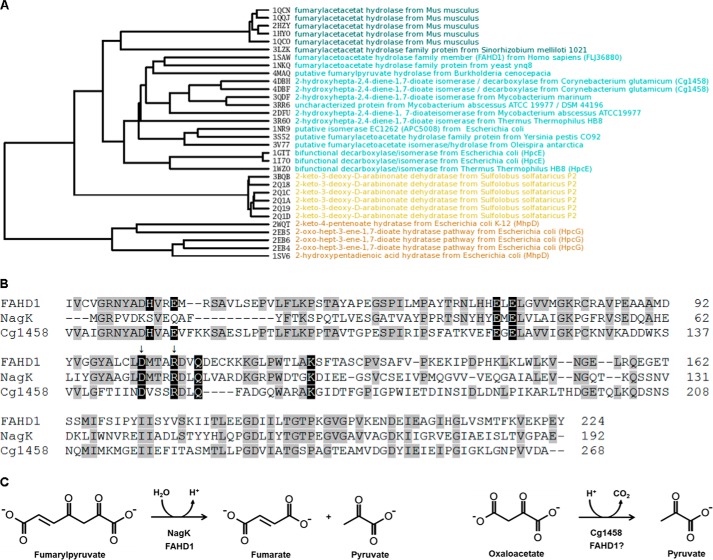
To systematically explore potential catalytic activities inherent to the FAH domain of FAHD1, we performed a detailed structural comparison of FAHD1 with 32 entries from the PDB assigned to the FAH superfamily (Fig. 1A). We focused the comparison on residues around the active site only as the structures of the FAH domains differ in various insertions extending from the FAH core. In this clustering by structural similarity, the human FAHD1 segregates in one branch together with FAH domains from lower organisms but not with FAH, the founding member of the FAH superfamily. One of the proteins within the same branch as FAHD1 was the oxaloacetate decarboxylase Cg1458 from C. glutamicum (Fig. 1A). The crystal structure of Cg1458 was described recently (8), and the importance of eight charged amino acids deemed vital for the enzymatic function was analyzed via mutants, resulting in mostly inactive variants. Sequence alignment of FAHD1 and Cg1458 reveals that all of these charged residues are conserved between the two enzymes, whereas only six of these amino acids are conserved in bacterial fumarylpyruvate hydrolase NagK (Fig. 1B). In Fig. 1C, the known catalytic activities of NagK, Cg1458, and FAHD1 are schematically depicted, as well as the hypothetical ODx activity of FAHD1. The catalytic mechanism of Cg1458 was derived from an x-ray structure with the inhibitor oxalate bound to the active site (8). Binding of the inhibitor induces the closure of a lid region over the binding site (Fig. 2A). This region contains the catalytic histidine residue stabilized by a glutamate residue essential for the activity. In the available structure of FAHD1, the corresponding region is missing, probably due to high mobility. By computational loop modeling utilizing a putative FAH family protein from Y. pestis, we propose a closed structure of FAHD1 where His-30 and Glu-33 complete the active site (Fig. 2B) in equivalence to the structure of Cg1458.
FIGURE 1.
Homology between FAHD1 and specific FAH family members. A, root mean square deviation-based active site clustering between structures of different FAH domains indicates that FAHD1 from Homo sapiens (PDB code 1SAW) is more closely related to prokaryotic FAH domains (cyan) than to FAH from higher organisms (dark cyan, e.g. PDB code 2HCY). For the structural comparison of the multidomain enzyme HpcE, the C-terminal domain was considered in the alignment. The proteins MhpD and HpcG (brown) have the highest structural difference to FAHD1. The catalytic mechanism of these hydrating enzymes does not include the cleavage of a carbon-carbon bond. B, primary sequence alignment of human FAHD1 with functionally related members of the FAH superfamily. From top to bottom: H. sapiens FAHD1 (amino acids 20–224), Ralstonia sp. strain U2 NagK fumarylpyruvate hydrolase, C. glutamicum Cg1458 oxaloacetate decarboxylase (amino acids 63–268). Conserved amino acids shared with FAHD1 are marked in gray, and charged amino acids reported to be important for ODx activity in Cg1458 are marked in black. Arrows denote amino acids altered in the FAHD1 mutant. C, chemical structures and reaction schemes outlining the hydrolysis of fumarylpyruvate and the decarboxylation of oxaloacetate.
FIGURE 2.
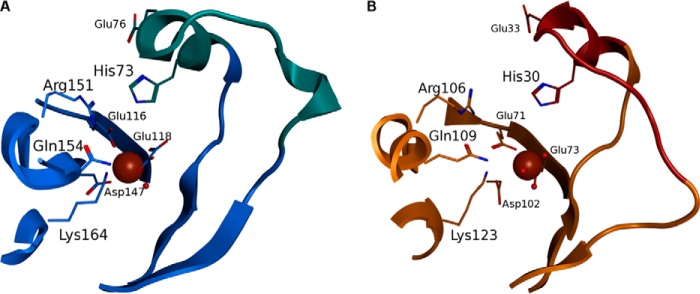
Model generation of human FAHD1 with closed lid structure. A and B, comparison of the conserved active site geometry of Cg1458 (A) and FAHD1 (B). The lid region (cyan) carrying the catalytic histidine and an activating glutamate residue is closed in Cg1458 upon binding of the inhibitor oxalate (shown as sticks). The model of the closed lid region in FAHD1 (brown) completes the active site of FAHD1 around the magnesium ion (brown sphere). Conserved residues around the active site are shown as sticks and labeled. Secondary structure elements are represented as graphic elements for Cg1458 and FAHD1 in blue and orange, respectively. Two water molecules (red spheres) are co-crystallized in FAHD1 at the binding site of the oxalate, suggesting a similar binding mechanism.
Oxaloacetate Decarboxylation by FAHD1 in Vitro
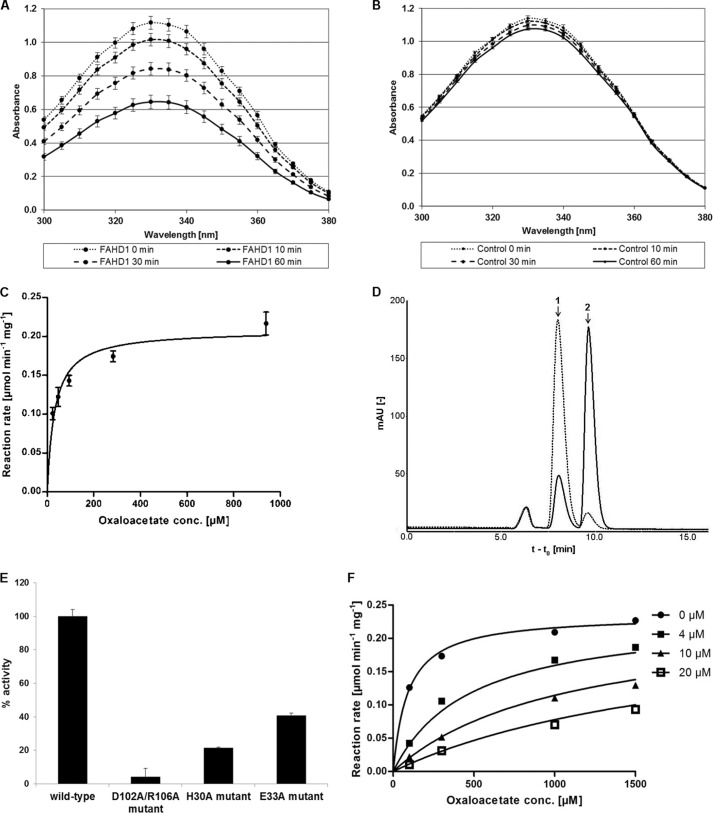
To confirm the hypothesis that FAHD1 exhibits ODx activity, we tested recombinant purified FAHD1 in a photometric assay by monitoring the follow-up reaction by lactate dehydrogenase metabolizing pyruvate (Fig. 3A). In this assay, FAHD1 showed ODx activity, which was clearly discernible from the observed rate of auto-decarboxylation (Fig. 3B). In a direct oxaloacetate decarboxylation assay, a Vmax of 0.21 μmol min−1 mg−1 and a Km value of 32 μm were determined for the wild-type enzyme (Fig. 3C). HPLC analysis, after incubation of the substrate in the presence or absence of the purified enzyme, confirmed the conversion of oxaloacetate to pyruvate by FAHD1 (Fig. 3D). A catalytically dead double mutant reported before (10) displayed only residual ODx activity, whereas mutation of loop residues His-30 and Glu-33, conserved and of known importance in Cg1458, led to a significant reduction in each case (Fig. 3E). To further characterize the ODx activity inherent to FAHD1, we used oxalate, an established inhibitor of Cg1485 (8). Indeed, oxalate caused potent competitive inhibition of oxaloacetate decarboxylation by FAHD1, with an inhibitor constant (KI) of about 1 μm (Fig. 3F).
FIGURE 3.
Characterization of in vitro ODx activity of FAHD1. A and B, degradation of 100 μm oxaloacetate at 25 °C in 1 ml of assay buffer by 12 μg of recombinant FAHD1 (A) or without enzyme (B). Actual curves show follow-up consumption of 200 μm NADH by lactate dehydrogenase metabolizing pyruvate. C, effect of substrate concentration on ODx reaction rate in the presence of purified FAHD1, determined by photometric analysis at 25 °C. Nonlinear regression analysis of Michaelis-Menten kinetics was performed with Prism 5 software (GraphPad Software). Data are represented as mean ± S.D. (n = 3). D, HPLC analysis of oxaloacetate breakdown in the presence (solid line) and absence (dashed line) of purified FAHD1. Retention times for oxaloacetate (peak 1) and pyruvate (peak 2) standards are indicated by arrows. E, ODx activity of wild-type FAHD1, and of several FAHD1 mutants. Data are represented as mean ± S.D. (n = 3). F, nonlinear regression analysis of inhibition by oxalate, performed with Prism 5 software (KI = 0.93 μm, R2 = 0.99). Oxaloacetate conc., oxaloacetate concentration.
Expression and Evidence for in Vivo Activity in Mice
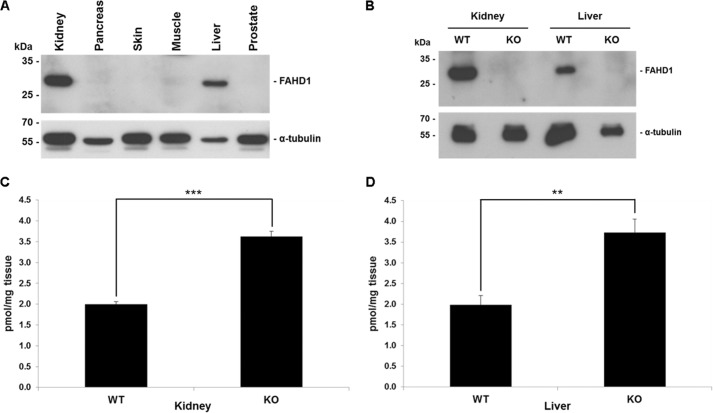
Previous studies, using a polyclonal antiserum raised against recombinant FAHD1 protein, indicated a widespread expression pattern for FAHD1 in many mouse tissues, based on two distinct bands revealed by Western blot (10). In view of the specific ODx activity for FAHD1 revealed here, the expression pattern of FAHD1 was reinvestigated by improved immunological reagents. In particular, we utilized a purpose-made monoclonal antibody to evaluate endogenous FAHD1 levels in a selection of mouse tissues by immunoblotting. FAHD1 was detected as a single band of apparent molecular mass of ∼29 kDa, and among the tested tissues, expression was predominantly observed in kidney and liver (Fig. 4A). Decarboxylation of oxaloacetate was observed previously in extracts from rat liver mitochondria (17, 18); however, the identity of the protein(s) responsible for this activity has remained unclear and has not since been further investigated. To investigate whether oxaloacetate is a relevant in vivo substrate for FAHD1, the FAHD1 gene was deleted in the mouse by homologous recombination, and FAHD1 knock-out was confirmed by Western blot of kidney and liver lysates (Fig. 4B). Metabolites were extracted from both the kidney and the liver of FAHD1−/− mice and wild-type littermates. The concentration of oxaloacetate in these extracts was determined in an enzymatic assay utilizing the reaction with 3H-labeled acetyl-CoA to form citrate (16). The concentration of oxaloacetate was significantly increased in both the kidney (Fig. 4C) and the liver (Fig. 4D) of FAHD1−/− mice, indicating that oxaloacetate is indeed a relevant in vivo substrate for FAHD1 in mice. No other overt phenotypical alterations relative to wild type were observed in these mice, at least at young age.3 This may be the consequence of compensatory mechanisms or a non-essential role of FAHD1 in mice, or it may depend on the nutritional and metabolic conditions the mice are experiencing.
FIGURE 4.
Comparison of oxaloacetate levels in organs of WT and FAHD1 KO mice. A, Western blot showing FAHD1 levels in wild-type mouse tissues. Tubulin levels are shown as loading control. B, Western blot confirming lack of FAHD1 protein expression in kidney and liver lysates of the FAHD1−/− mouse, with wild-type as control. C and D, oxaloacetate levels determined in kidney (C) and liver (D) extracts of 3-month-old female wild-type and FAHD1 knock-out mice. Data are represented as mean ± S.D. (n = 3). **, p < 0.01; ***, p < 0.001.
Regulation of FAHD1 and Metabolically Related Genes
In light of the newly ascribed ODx function of FAHD1, expression levels may well be influenced by different metabolic states. To take a first look into possible regulatory effects, we compared the expression of FAHD1, PC, and phosphoenolpyruvate carboxykinase (PEPCK) during a fed/fasted/refed cycle in the mouse liver, on both the mRNA (Fig. 5A) and the protein level (Fig. 5, B and C). Unlike PEPCK, which is strongly up-regulated during fasting conditions on both the mRNA and the protein level, FAHD1 showed only a very mild and marginally significant down-regulation of mRNA levels in fasted mice. PC mRNA was up-regulated during fasting, but this effect disappeared when looking at protein levels, as observed before (19).
FIGURE 5.
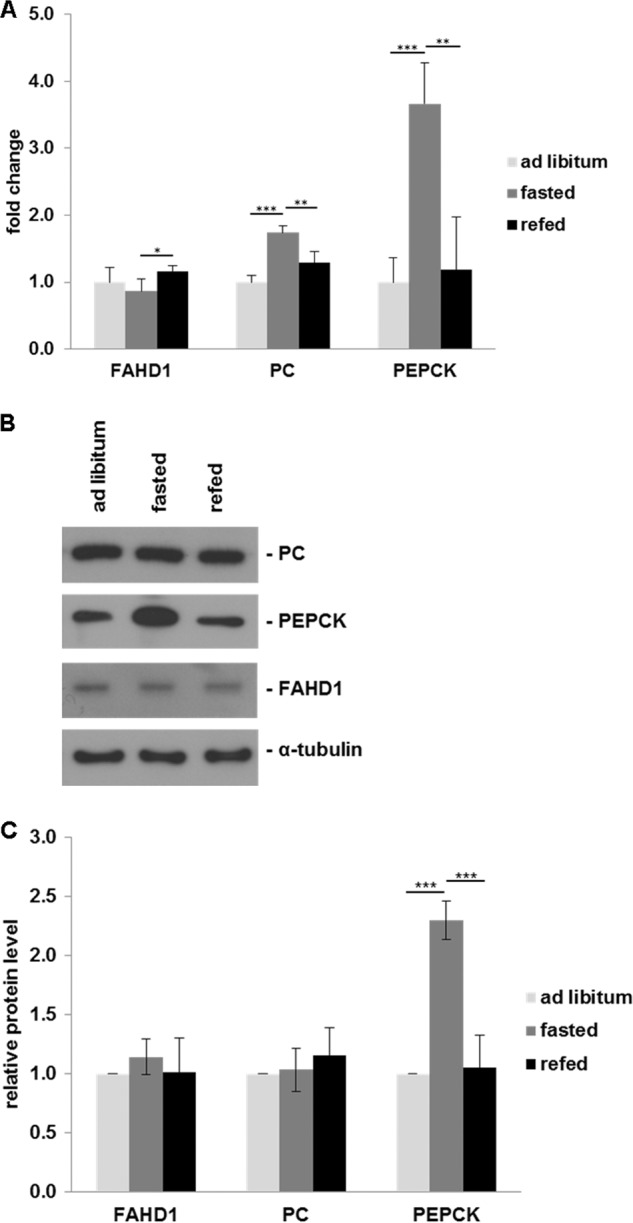
Regulation of gene expression of selected enzymes during fed/fasted cycle. A, mRNA expression of FAHD1, PC, and PEPCK was examined by real time quantitative PCR in the liver of ad libitum fed, fasted, and refed mice. For each gene, the fold induction of mRNA expression in fasted and refed mice as compared with ad libitum fed mice is indicated. Results indicate mean ± S.D. of three triplicate runs (n = 5). B, representative Western blot showing FAHD1, PC, and PEPCK protein levels in liver of ad libitum fed, fasted, and refed mice; α-tubulin levels are shown as a loading control. C, specific band intensities were quantified using Fiji software and normalized to tubulin. Data are represented as mean ± S.D. (n = 5). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
The results reported here provide new insight into structure-function relationships within the superfamily of FAH domain-containing proteins. This family contains members with a large variety of distinct catalytic activities, many of which are however currently hypothetical. In a structure-based clustering analysis, FAHD1 clustered together with prokaryotic members of the FAH superfamily, including fumarylpyruvate hydrolases and the oxaloacetate decarboxylase Cg1458 from C. glutamicum. Oxaloacetate decarboxylases from prokaryotic organisms are well studied and are either soluble and divalent cation-dependent (like Cg1458) or membrane-bound and dependent on sodium and biotin (20). Molecular modeling based on the established x-ray structure of FAHD1 (9) revealed that indeed the catalytic centers of FAHD1 and Cg1458 are very similar and support a common catalytic mechanism.
ODx activity in the mammalian organism, especially in regard to mitochondria, has been discussed in a small number of studies, but the majority of these were published many decades ago. The responsible enzyme(s) were detected and characterized in these works, typically from rat liver mitochondria, but not conclusively identified (17, 18, 21). Our own enzymatic studies were performed in vitro with recombinantly expressed and purified FAHD1. The catalytic rates we observed may seem low in relation to prokaryotic ODx enzymes like Cg1458 (7), with Vmax values that are several magnitudes higher, but they compare well with the results from earlier studies on mammalian tissue mentioned above. It seems reasonable that at least some of these earlier studies were actually based on FAHD1, although the enzyme itself was not identified.
Also, it is difficult to judge the actual in vivo activity from our in vitro results as additional parameters such as cofactors or post-translational modifications could play an important role. To address the question of whether the inclusion of protein tags or recombinant expression in a prokaryotic system might be detrimental to the enzymatic activity of FAHD1, an untagged variant was purified from E. coli and a His-tagged variant was obtained from baculoviral expression in SF9 insect cells. These preparations did not differ in their enzymatic properties when tested.3 The in vitro experiments reported here also revealed an inhibitory effect of oxalate on FAHD1 that had already been reported for Cg1458 (8), yielding further evidence for the similarity of their active sites.
PC replenishes oxaloacetate that is withdrawn from the Krebs cycle for biosynthesis in different pathways (19). The tissue-dependent expression pattern of mouse FAHD1 reported here shows correlation with the published expression pattern of PC (22). More importantly, we could observe elevated oxaloacetate levels in FAHD1 knock-out mice, giving reasonable evidence that FAHD1 directly counteracts PC activity, contributing to the balance between pyruvate and oxaloacetate levels (Fig. 6). Although both PEPCK and to a lesser extent PC expression are known to be altered in liver during a fasting cycle (19, 23), no clear transcriptional regulation could be observed for FAHD1 under these circumstances in mice.
FIGURE 6.
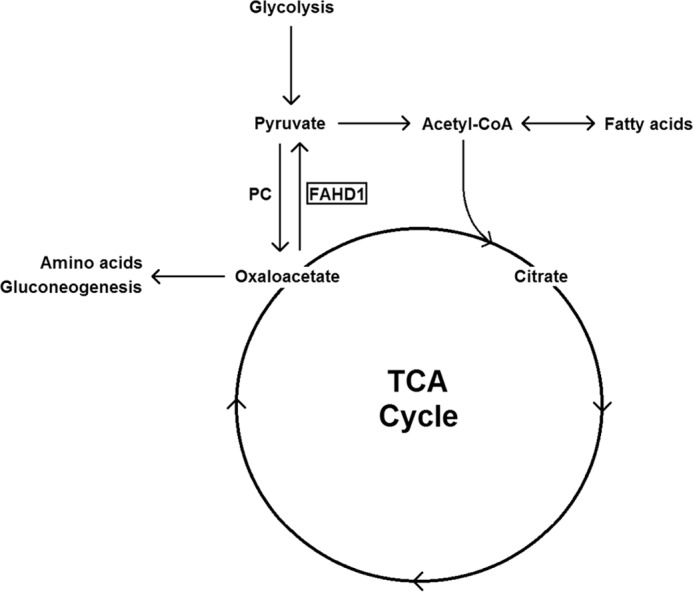
Putative role of FAHD1 in central metabolism as an antagonist of PC. TCA cycle, tricarboxylic acid cycle.
Finally, oxaloacetate is known to act as an inhibitor of several mitochondrial enzymes (18); thus ODx activity might play a vital role in preventing inactivation of metabolic pathways by keeping oxaloacetate levels in check. Additional studies are now required to further elucidate the relevance of FAHD1 for eukaryotic metabolism and to look into possible compensatory mechanisms in the case of FAHD1 depletion.
Acknowledgments
We thank Michael Neuhaus for expert technical assistance. We are grateful to Grit D. Straganz for initial contributions to the FAHD1 project.
This work was supported by a grant from the Austrian Science Funds (FWF; NFN S93) (to P. J.-D.) and by the European Union through the FP6 Integrated Project MiMAGE (to P. J.-D.).
H. Pircher, T. Diener, C. Metzger, E. Albertini, A. Taferner, and P. Jansen-Dürr, unpublished results.
- FAH
- fumarylacetoacetate hydrolase
- FAHD1
- fumarylacetoacetate hydrolase domain-containing protein 1
- ODx
- oxaloacetate decarboxylase
- PC
- pyruvate carboxylase
- PEPCK
- phosphoenolpyruvate carboxykinase.
REFERENCES
- 1. Hsiang H. H., Sim S. S., Mahuran D. J., Schmidt D. E., Jr. (1972) Purification and properties of a diketo acid hydrolase from beef liver. Biochemistry 11, 2098–2102 [DOI] [PubMed] [Google Scholar]
- 2. Phaneuf D., Labelle Y., Bérubé D., Arden K., Cavenee W., Gagné R., Tanguay R. M. (1991) Cloning and expression of the cDNA encoding human fumarylacetoacetate hydrolase, the enzyme deficient in hereditary tyrosinemia: assignment of the gene to chromosome 15. Am. J. Hum. Genet. 48, 525–535 [PMC free article] [PubMed] [Google Scholar]
- 3. Mitchell G. A., Grompe M., Lambert M., Tanguay R. M. (2001) Hypertyrosinemia. in The Metabolic and Molecular Bases of Inherited Diseases (Scriver C. R., Beaudet A. L., Sly W. S., Valle D., eds), 8th Ed., pp. 1777–1805, McGraw-Hill, New York [Google Scholar]
- 4. Tame J. R., Namba K., Dodson E. J., Roper D. I. (2002) The crystal structure of HpcE, a bifunctional decarboxylase/isomerase with a multifunctional fold. Biochemistry 41, 2982–2989 [DOI] [PubMed] [Google Scholar]
- 5. Brouns S. J., Barends T. R., Worm P., Akerboom J., Turnbull A. P., Salmon L., van der Oost J. (2008) Structural insight into substrate binding and catalysis of a novel 2-keto-3-deoxy-d-arabinonate dehydratase illustrates common mechanistic features of the FAH superfamily. J. Mol. Biol. 379, 357–371 [DOI] [PubMed] [Google Scholar]
- 6. Pollard J. R., Bugg T. D. (1998) Purification, characterisation and reaction mechanism of monofunctional 2-hydroxypentadienoic acid hydratase from Escherichia coli. Eur. J. Biochem. 251, 98–106 [DOI] [PubMed] [Google Scholar]
- 7. Klaffl S., Eikmanns B. J. (2010) Genetic and functional analysis of the soluble oxaloacetate decarboxylase from Corynebacterium glutamicum. J. Bacteriol. 192, 2604–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ran T., Gao Y., Marsh M., Zhu W., Wang M., Mao X., Xu L., Xu D., Wang W. (2013) Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for the FAH family. Biochem. J. 449, 51–60 [DOI] [PubMed] [Google Scholar]
- 9. Manjasetty B. A., Niesen F. H., Delbrück H., Götz F., Sievert V., Büssow K., Behlke J., Heinemann U. (2004) X-ray structure of fumarylacetoacetate hydrolase family member Homo sapiens FLJ36880. Biol. Chem. 385, 935–942 [DOI] [PubMed] [Google Scholar]
- 10. Pircher H., Straganz G. D., Ehehalt D., Morrow G., Tanguay R. M., Jansen-Dürr P. (2011) Identification of human fumarylacetoacetate hydrolase domain-containing protein 1 (FAHD1) as a novel mitochondrial acylpyruvase. J. Biol. Chem. 286, 36500–36508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou N. Y., Fuenmayor S. L., Williams P. A. (2001) nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183, 700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berman H., Henrick K., Nakamura H. (2003) Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10, 980. [DOI] [PubMed] [Google Scholar]
- 13. Timm D. E., Mueller H. A., Bhanumoorthy P., Harp J. M., Bunick G. J. (1999) Crystal structure and mechanism of a carbon-carbon bond hydrolase. Structure 7, 1023–1033 [DOI] [PubMed] [Google Scholar]
- 14. Wolfenden R., Lewis C. A., Jr., Yuan Y. (2011) Kinetic challenges facing oxalate, malonate, acetoacetate, and oxaloacetate decarboxylases. J. Am. Chem. Soc. 133, 5683–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parvin R., Caramancion M. N., Pande S. V. (1980) Convenient rapid determination of picomole amounts of oxaloacetate and aspartate. Anal. Biochem. 104, 296–299 [DOI] [PubMed] [Google Scholar]
- 17. Corwin L. M. (1959) Oxaloacetic decarboxylase from rat liver mitochondria. J. Biol. Chem. 234, 1338–1341 [PubMed] [Google Scholar]
- 18. Wojtczak A. B., Walajtys E. (1974) Mitochondrial oxaloacetate decarboxylase from rat liver. Biochim. Biophys. Acta 347, 168–182 [DOI] [PubMed] [Google Scholar]
- 19. Jitrapakdee S., St Maurice M., Rayment I., Cleland W. W., Wallace J. C., Attwood P. V. (2008) Structure, mechanism and regulation of pyruvate carboxylase. Biochem. J. 413, 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lietzan A. D., St Maurice M. (2014) Functionally diverse biotin-dependent enzymes with oxaloacetate decarboxylase activity. Arch. Biochem. Biophys. 544, 75–86 [DOI] [PubMed] [Google Scholar]
- 21. Dean B., Bartley W. (1973) Oxaloacetate decarboxylases of rat liver. Biochem. J. 135, 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ballard F. J., Hanson R. W., Reshef L. (1970) Immunochemical studies with soluble and mitochondrial pyruvate carboxylase activities from rat tissues. Biochem. J. 119, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanson R. W., Reshef L. (1997) Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 66, 581–611 [DOI] [PubMed] [Google Scholar]