Background: Dysbindin and Disrupted-in-schizophrenia 1 (DISC1) are major schizophrenia susceptibility factors.
Results: DISC1 enhances stability of dysbindin, which is critical for neurite outgrowth.
Conclusion: Dysbindin and DISC1 form a physiologically functional complex that is essential for normal neurite outgrowth.
Significance: Our findings indicate the existence of a protein complex composed of multiple schizophrenia susceptibility factors functioning in a pathway for neurite outgrowth.
Keywords: Neurite Outgrowth, Protein Complex, Protein Stability, Schizophrenia, Ubiquitylation (Ubiquitination), DISC1, Dysbindin
Abstract
Dysbindin and DISC1 are schizophrenia susceptibility factors playing roles in neuronal development. Here we show that the physical interaction between dysbindin and DISC1 is critical for the stability of dysbindin and for the process of neurite outgrowth. We found that DISC1 forms a complex with dysbindin and increases its stability in association with a reduction in ubiquitylation. Furthermore, knockdown of DISC1 or expression of a deletion mutant, DISC1 lacking amino acid residues 403–504 of DISC1 (DISC1Δ403–504), effectively decreased levels of endogenous dysbindin. Finally, the neurite outgrowth defect induced by knockdown of DISC1 was partially reversed by coexpression of dysbindin. Taken together, these results indicate that dysbindin and DISC1 form a physiologically functional complex that is essential for normal neurite outgrowth.
Introduction
Neurite outgrowth is a critical process in neuronal development. Abnormalities in this process have been linked to the biological basis of various mental illnesses (1–3). Especially, defects in neurite outgrowth have been proposed to underlie the reduced neuropil hypothesis stating that schizophrenia is involved in a decrease in axons and dendrites (4–8). In support of this hypothesis, the multiple schizophrenia susceptibility factors, including Disrupted-in-schizophrenia 1 (DISC1) and dystrobrevin-binding protein 1 (dysbindin, DTNBP1) (9–11), are involved in neurite outgrowth (12, 13).
Dysbindin was initially identified as an interactor of dystrobrevin in muscle and brain (14). Subsequent genetic studies have associated dysbindin with schizophrenia (10, 15–18). Consistent with these findings, the sandy (sdy) mouse strain, which lacks functional dysbindin, displays significant schizophrenia-like behaviors, including deficits in social interaction and working memory (19–22). Dysbindin mRNA and protein are reduced significantly in schizophrenic patients (23, 24). Notably, dysbindin is also involved in several neuronal developmental processes, including neurite outgrowth. Knockdown of dysbindin results in disorganization of the actin cytoskeleton and shortening of neurites, and sandy mice show defective neurite outgrowth and abnormal growth cone morphology (12, 13, 25). Moreover, a recent report shows that dendritic spine morphogenesis depends on the interaction of dysbindin with WAVE2 and Abi-1 (26), underscoring the essential function of dysbindin.
DISC1 has emerged as a susceptibility factor in mental disorders on the basis of the cosegregation of a balanced chromosomal translocation (t1;11) that disrupts the DISC1 gene in multiple psychiatric conditions, including schizophrenia (10). Subsequently, mice that are defective in DISC1 display neuroanatomical and behavioral abnormalities relevant to schizophrenia (27, 28). Furthermore, DISC1 plays multiple roles during neurodevelopment, including neuronal proliferation (29), neurite outgrowth (3, 30–32), neuronal migration (29, 33, 34), and synapse formation (35), all implicated in the pathophysiology of schizophrenia.
A considerable number of studies have characterized interacting partners of dysbindin and DISC1. Interestingly, several reports show that the protein interaction networks associated with dysbindin and DISC1 share common components, implying a common physiological pathway for these two major schizophrenia susceptibility factors (36, 37). Consistent with this, dysbindin and DISC1 physically associate in vitro, although the biological relevance of this has not been investigated (38). In this study, we demonstrate the functional collaboration of dysbindin and DISC1 in the neurite outgrowth process which involves fine-tuning of dysbindin degradation by DISC1, and provide evidence of a convergent pathway where dysbindin and DISC1, two schizophrenia-associated proteins, cooperate.
EXPERIMENTAL PROCEDURES
Plasmids
For yeast two-hybrid assays, pPC97-DISC1 and pPC86-NDEL1 were prepared as described previously (39). The human dysbindin coding sequence was amplified by PCR from a human dysbindin cDNA clone (clone ID 4139934, Open Biosystems, GenBank accession number BC011912) and inserted into the pPC86 vector (Invitrogen) using the SalI and NotI sites. Full-length human and mouse DISC1 constructs were prepared as described previously (39). For constructs of DISC1 fragments, corresponding regions of human DISC1 (hDISC1) were amplified by PCR and inserted into the pEGFP-C3 (Clontech) and pFLAG-CMV2 (Sigma). For mouse dysbindin constructs, the full-length dysbindin coding sequence was amplified using reverse transcription products generated from CAD2 mouse neuroblastoma cells, subcloned into pEGFP-N1 (Clontech), pUB-GFP (Addgene), pFLAG-CMV2 (Sigma), and pCDNA3.1 Myc-His (Invitrogen). To construct the deletion mutant of dysbindin, a region of human dysbindin corresponding to the designated codon was amplified by PCR using EGFP-dysbindin as a template and inserted into pEGFP-N1. Sequences were verified by DNA sequencing. For shRNA constructs, mouse DISC1 shRNA was prepared following a previous description (39). The oligonucleotide sequences used for the constructs were GTGATAAGTCAAGA GAAGCTTCAAGAGAGCTTCTCTTGACTTACACTTTTTT and TCGAGAAAAAAGTGATAA GTCAAGAGAAGCTCTCTTGAAGCTTCTCTTGACTTAT CACA for mouse dysbindin. These oligonucleotides were annealed and ligated into the pLentiLox3.7 vector using the HpaI and XhoI sites. To generate the shRNA-resistant GFP-mouse DISC1 and mouse dysbindin constructs by inserting silent mutations in the shRNA target sequence in pUB-GFP (Addgene), site-directed mutagenesis of DISC1 and dysbindin was performed using PCR-based methods. The PCR primer sequences used were as follows: 5′-AAG ACT TTA AGT GAC AAA TCA AGG GAG GCA AAAGTGAAA-3′ for dysbindin and 5′-CGCACAATGAAGGCCAATACCGTCAAATGTATGGAAGTGTTG-3′ for DISC1.
Cells and Transfections
HEK293T, HEK293, COS7, and CAD cells were grown in DMEM supplemented with 10% fetal bovine serum and antibiotics under 5% CO2 at 37 °C. Cells were transfected using Lipofectamine 2000 (Invitrogen) or VivaMagic (Vivagen) according to the instructions of the manufacturer. For neuronal cultures, developing cortices were dissected from embryonic day 15 Institute for Cancer Research (CrljBgi:CD-1) mouse embryos in Hanks' balanced salt solution (Invitrogen). Dissected tissue was dissociated by treatment with DNase I (0.1%) and Trypsin (0.25%) for 6 min at 37 °C. The dissociated cells were diluted in plating medium, Neurobasal medium (Invitrogen) containing 10 mm HEPES (pH 7.4) and 10% horse serum (Gibco), and then plated onto cover slips coated with poly-d-lysine. Plating media was replaced with culture media (Neurobasal media supplemented with 2% B27 (Invitrogen), 2 mm glutamine, and antibiotics).
Antibodies and Immunoblot Analyses
Rabbit polyclonal anti-dysbindin was used as described previously (26). Anti-mDISC1 antibodies were a gift from Dr. Kaibuchi (University of Nagoya). A rabbit polyclonal anti-GFP protein (Molecular Probes), anti-FLAG (ABR), and mouse monoclonal anti-FLAG M2 (Sigma) were used. Anti-α tubulin (catalog no. DM1A), anti-GAPDH (catalog no. 6C5), anti-GFP (catalog no. B-2), anti-DISC1 (catalog no. N-16), and anti-Myc (catalog no. 9E10) antibodies were purchased from Santa Cruz Biotechnology. For immunoblotting, cells were lysed in Nonidet P-40 lysis buffer (50 mm Tris (pH 8.0), 150 mm NaCl, 1% Nonidet P-40, 5 mm EDTA, 5 mm glycerol-2-phosphate, 2 mm sodium pyrophosphate, 5 mm NaF, 2 mm Na3VO4, 1 mm DTT, and EDTA-free protease inhibitor mixture (Roche)) and precleared by centrifugation for 10 min at 12,000 × g. Supernatants were denatured in SDS sample buffer by boiling for 5 min and subjected to SDS-PAGE followed by immunoblotting.
Immunoprecipitation and in Vitro Binding Assay
Whole brain tissues from an Institute for Cancer Research mouse were homogenized in Nonidet P-40 lysis buffer and precleared by centrifugation for 15 min at 12,000 × g. Supernatants were incubated with antibody (0.5–1 μg) on a rocking platform for 2 h or overnight at 4 °C, and then 100 μl of 10% protein A-agarose (GE Healthcare) resuspended in the same lysis buffer was added and incubated with gentle shaking for an additional 90 min at 4 °C. The precipitate was washed three times with lysis buffer and resuspended in 2× SDS sample loading buffer. For the in vitro binding assay, transfected HEK293T cell lysates were incubated with 1 μg of anti-FLAG M2 (Sigma) antibody for 2 h at 4 °C, and then protein A-agarose beads (GE Healthcare) were added and incubated for an additional 90 min at 4 °C. Precipitates were washed three times with lysis buffer and incubated with 5 μg of GST-dysbindin for 2 h at 4 °C. Beads were collected by centrifugation at 2000 × g and rinsed four times with lysis buffer. Precipitates were subjected to anti-GST and anti-FLAG immunoblotting.
Immunocytochemistry
Cells were fixed for 10 min in 4% paraformaldehyde in PBS and incubated for 30 min in blocking solution (2% goat serum and 1% Triton X-100 in PBS). Cells were incubated with anti-Myc (1:200, Santa Cruz Biotechnology) and rabbit anti-GFP antibodies (1:2000, Molecular Probes) for 1.5 h at room temperature, followed by incubation with Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 568-conjugated goat anti-mouse IgG secondary antibodies (Molecular Probes) for 1 h at room temperature. Hoechst dye was used for nuclear staining, and cells on coverslips were rinsed three times with PBS and mounted in antifade medium (Molecular Probes). Pictures were taken with a confocal microscope (Olympus, FluoView-1000).
Ubiquitylation Analysis
HEK293 cells were transfected with HA-ubiquitin (a gift from Dr. Chin Ha Chung, Seoul National University, Seoul, Korea) along with Myc-dysbindin and EGFP-DISC1 constructs. 48 h after transfection, cells were treated with 25 μm MG132 (Enzo) for 8 h prior to lysis. The cell lysates were prepared in Nonidet P-40 lysis buffer with 10 mm N-ethylmaleimide and incubated with anti-Myc antibodies for 4 h. Protein A-agarose beads were washed three times with lysis buffer, and immunoprecipitated proteins were eluted in 2× SDS/PAGE sampling buffer and analyzed by immunoblotting with anti-Myc, anti-GFP, and anti-HA antibodies.
Neurite Outgrowth Assay
CAD cells were cultured in DMEM containing 10% FBS and antibiotics for 1 day and then transfected with the indicated constructs using VivaMagic (Vivagen). After 48 h of transfection, cells were detached using trypsin, diluted in DMEM with antibiotics, seeded again onto 12-well plates, and cultured for 72 h. Neurites on 150–250 fluorescent cells from three independent experiments were selected randomly, and neurites longer than the diameter of the soma were measured by ImageJ software (National Institute of Health). The length of the longest neurite and total neurite length were used for analyses. For cultured primary neurons, control red fluorescent protein (RFP) vector or RFP-expressing shRNAs were transfected into cells to visualize the neuronal morphology using Lipofectamine 2000 at days in vitro 0 and cultured for 3 days. Neurons were fixed, and neurite length was measured in randomly selected microscopic fields using a ×20 objective. Neurites on 190–249 transfected neurons from three independent experiments were measured by ImageJ software and analyzed blindly.
Statistical Analysis
Data were analyzed using GraphPad Prism 5 software and are presented as mean ± S.E. Statistical significance was determined by ANOVA or Student's t test.
RESULTS
Dysbindin Interacts with DISC1
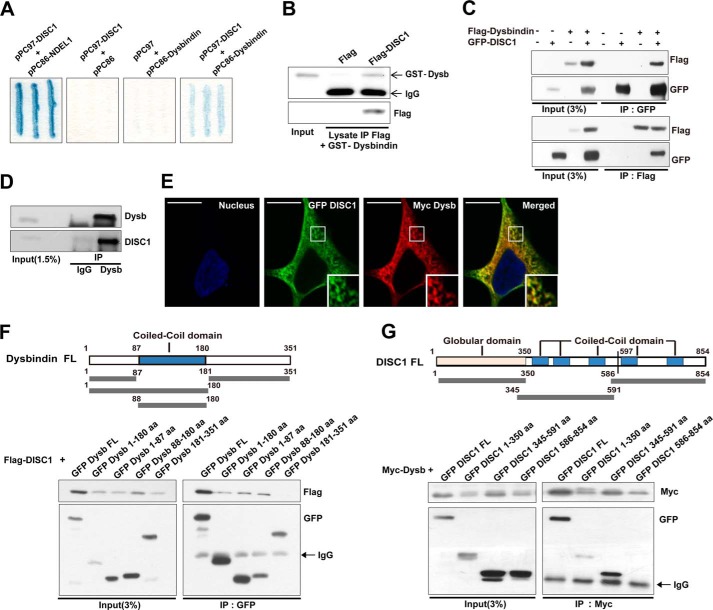
To examine whether dysbindin interacts with DISC1, we employed a yeast-two-hybrid assay. We found that cotransformants containing dysbindin and DISC1 showed a positive signal, indicating interaction-dependent β-galactosidase activity (Fig. 1A). DISC1 and NDEL1 (nudE nuclear distribution gene E homolog (A. nidulans)-like 1) were used as positive controls in this assay. In addition, GST-dysbindin also interacted directly with FLAG-DISC1 purified by immunoprecipitation (Fig. 1B). Moreover, reciprocal coimmunoprecipitation of dysbindin and DISC1 was detected consistently in transfected HEK293T cell lysates (Fig. 1C), indicating potential protein complex formation in the cellular context. Consistently, DISC1 and dysbindin were coimmunoprecipitated from mouse brain lysates (Fig. 1D). We also examined the subcellular localization of these two proteins using immunocytochemistry in HEK293 cells. Confocal microscopy images showed significant colocalization between Myc-dysbindin and EGFP-DISC1 in the cytoplasm (Fig. 1E). Taken together, these results suggest that dysbindin and DISC1 form a physiologically functional complex.
FIGURE 1.
Dysbindin interacts with DISC1. A, yeast two-hybrid assay using a β-galactosidase reporter. Cotransformation with pPC97-DISC1 and pPC86-NDEL1 was used as a positive control. B, interaction of dysbindin (Dysb) and DISC1 in an in vitro binding assay. IP, immunoprecipitation. C, reciprocal coimmunoprecipitation of dysbindin with DISC1. Lysates from HEK293T cells transfected with dysbindin and DISC1 were immunoprecipitated with anti-GFP (top panel) or anti-Flag (bottom panel) antibodies. Immunoprecipitates were analyzed by immunoblotting with anti-FLAG and anti-GFP antibodies. D, coimmunoprecipitation of endogenous dysbindin and DISC1 from mouse brain lysates. Anti-dysbindin immunoprecipitates were analyzed by immunoblotting with anti-dysbindin and anti-DISC1 antibodies. E, colocalization of dysbindin and DISC1 in HEK293 cells shown by immunocytochemistry. Myc-dysbindin and EGFP-DISC1 were stained with anti-Myc (red) and anti-GFP (green) antibodies. Nuclei were stained with Hoechst dye (blue). Scale bars = 10 μm. F, schematic of dysbindin fusion proteins used for coimmunoprecipitation analyses. aa, amino acids; FL, full-length. G, coimmunoprecipitation assays with DISC1 fragments and Myc-dysbindin. A schematic of DISC1 fusion proteins is also shown.
We next attempted to identify the protein subregions mediating the interaction between dysbindin and DISC1. A series of deletion mutants of dysbindin were generated on the basis of domain predictions, and their interactions with DISC1 were assayed by coimmunoprecipitation in HEK293T cells. As shown in Fig. 1F, the unstructured N-terminal and coiled-coil domains of dysbindin (residues 1–87 (dysbindin1–87) and 88–180 (dysbindin88–180), respectively) were coimmunoprecipitated with full-length DISC1, whereas the dysbindin C terminus (residues 181–351, dysbindin181–351) was not. Conversely, we also analyzed the interaction of full-length dysbindin with EGFP-tagged fragments of DISC1 (residues 1–350, 345–591, and 586–854). DISC11–350 and DISC1345–591 were both coimmunoprecipitated with full-length dysbindin, whereas DISC1586–854 was not (Fig. 1G). Therefore, the interaction between dysbindin and DISC1 involves a rather broad interface encompassing residues 1–180 of dysbindin and residues 1–591 of DISC1.
DISC1 Protects Dysbindin from the Ubiquitin-Proteasome System
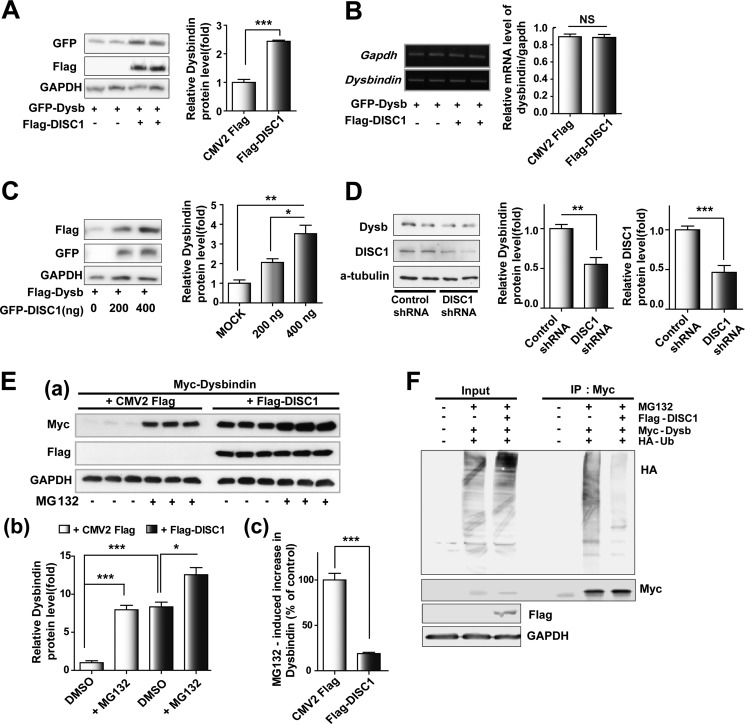
The reduction of dysbindin protein in schizophrenic patients has been shown previously (23, 24), implying that the abundance of dysbindin can be associated with the disease. It has been reported recently that TRIM32, a ubiquitin E3 ligase, ubiquitylates dysbindin and facilitates its degradation (40). Moreover, some reports hint at the possibility that DISC1 affects the ubiquitylation and stability of multiple DISC1-interacting proteins (39, 41). In this line, we examined whether DISC1 regulates stability of dysbindin protein by monitoring dysbindin level transiently transfected in the presence or absence of DISC1 coexpression. The results showed a remarkable increase in dysbindin following transfection with DISC1 (Fig. 2A) without alteration of the dysbindin mRNA level (Fig. 2B). In addition, the degree of increase in dysbindin showed a strong correlation with the DISC1 expression level (Fig. 2C). To ascertain whether DISC1 is essential for maintaining the endogenous dysbindin protein level, we introduced shRNA constructs targeting DISC1 into Cath.a-differentiated (CAD) mouse neuroblastoma cells. The level of dysbindin protein was reduced significantly following knockdown of DISC1 (Fig. 2D).
FIGURE 2.
DISC1 up-regulates dysbindin. A, up-regulation of dysbindin (Dysb) protein by DISC1 coexpression. 30 h after transfection with the indicated plasmids. Cells were subjected to immunoblotting analyses. Error bars show mean ± S.E. ***, p <0.001 by Student's t test (n = 3). B, no changes in the level of dysbindin mRNA by DISC1 coexpression. Dysbindin mRNA levels were measured by RT-PCR and normalized to GAPDH mRNA. NS, not significant by Student's t test (n = 3). C, dose-dependent increases in dysbindin protein by DISC1. Dysbindin protein levels were normalized to GAPDH. Error bars show mean ± S.E. *, p < 0.05; **, p < 0.01 by Student's t test (n = 3). D, down-regulation of dysbindin protein by knockdown of DISC1 in differentiated CAD cells. Error bars show mean ± S.E. **, p < 0.01; ***, p < 0.001 by Student's t test (n = 6). E, reduction of MG132 efficacy on dysbindin by coexpression of DISC1. 30 h after transfection, MG132 (10 μm) was treated for 8 h in a. Protein levels relative to control cells are shown in b, and the relative increase in dysbindin upon MG132 treatment is shown in c. Error bars show mean ± S.E. *, p < 0.05; ***, p < 0.001; Student's t test (n = 3). DMSO, dimethyl sulfoxide. F, decreased ubiquitylation of dysbindin following DISC1 coexpression. HEK293 cells were cotransfected with the indicated plasmids, treated with MG132 for 8 h, followed by immunoprecipitation (IP) and immunoblotting as indicated. Ub, ubiquitin.
Dysbindin is degraded by the ubiquitin-proteasome system (UPS) (40). Therefore, we used the proteasome inhibitor MG132 to test whether DISC1 affects dysbindin stability via the UPS. As shown in Fig. 2E, dysbindin was up-regulated dramatically following MG132 treatment (Fig. 2E, a and b). Indeed, this up-regulation was decreased under coexpression of DISC1 (Fig. 2E, c). Moreover, a significant decrease in dysbindin ubiquitylation was consistently detected upon coexpression of DISC1 (Fig. 2F). In sum, these results indicate that DISC1 enhances the stability of dysbindin, at least in part, by protecting dysbindin from ubiquitylation-dependent proteasomal degradation.
Amino Acid Residues 403–504 of DISC1 Are Involved in the Regulation of Dysbindin Stability
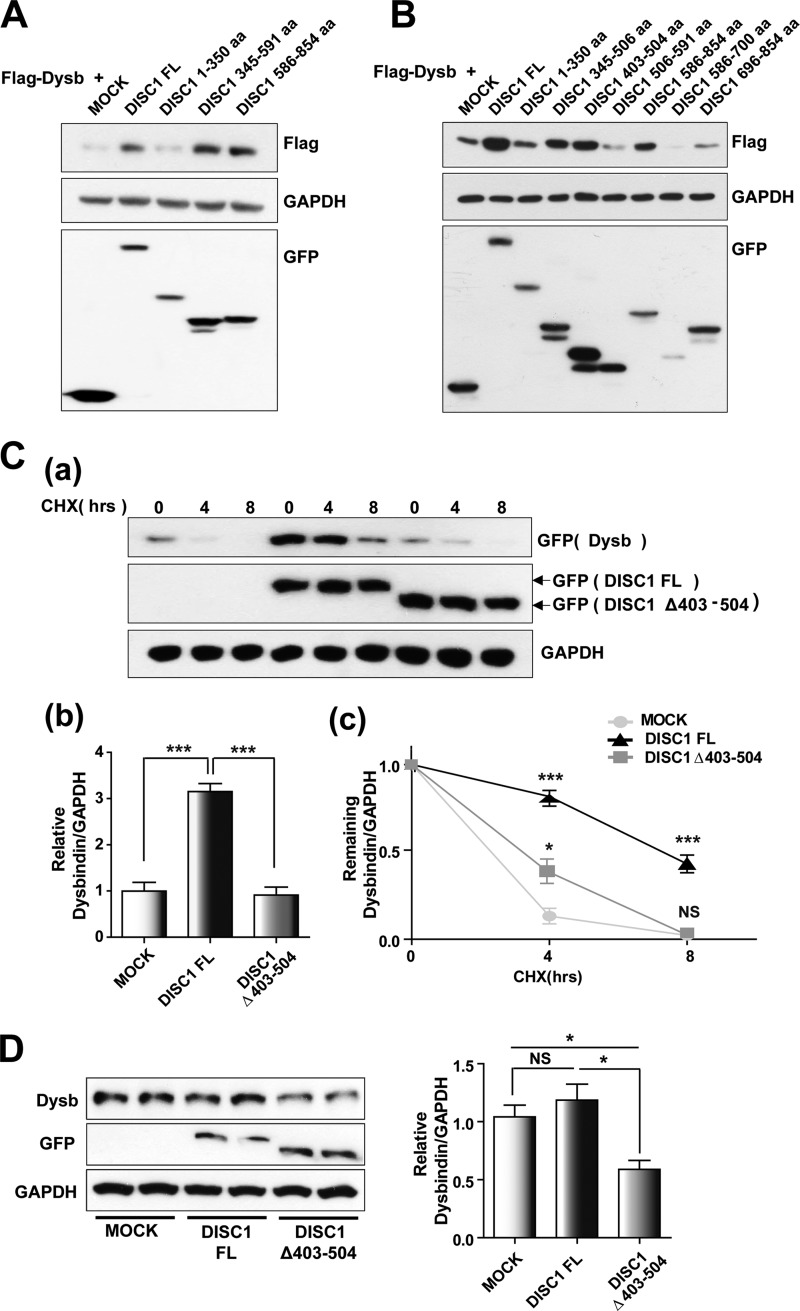
We next investigated which regions of DISC1 were important for regulating dysbindin. Interestingly, the level of dysbindin protein was up-regulated upon coexpression of DISC1345–591 and DISC1586–854, comparable with the effect of full-length DISC1 coexpression (Fig. 3A), indicating that the effect of DISC1 on dysbindin stability was dependent on amino acid residues 345–854. Although the C-terminal part of DISC1 is not responsible for interaction with dysbindin, amino acid residues 345–591 are involved in the physical interaction and have the capacity to protect dysbindin from degradation. Therefore, we attempted to further narrow down the region of DISC1 responsible for regulating dysbindin stability. We separated the region into five fragments, amino acid residues 345–506, 403–504, 506–591, 586–700, and 696–854 (DISC1345–506, DISC1403–504, DISC1506–591, DISC1586–700, and DISC1696–854, respectively) without disrupting the predicted coiled-coil structures and tested the impact on dysbindin stability upon coexpression. Although both fragments of C-terminal DISC1 (DISC1586–700 and DISC1696–854) did not result in a noticeable effect on dysbindin, DISC1345–506 and DISC1403–504 caused an up-regulation of dysbindin levels (Fig. 3B). This result may reflect that DISC1403–504 can mediate the enhancement of dysbindin stability by direct interaction, which is consistent with a recent report showing that incubation of purified dysbindin with a recombinant DISC1 fragment containing amino acid residues 316–854 with a deletion of amino acid residues 403–504 did not result in an additional higher molecular peak in size exclusion chromatography (supplemental data in Ref. 38). On the basis of this information, we introduced DISC1 lacking this region, DISC1Δ403–504, into the analysis. We observed a consistent decrease in dysbindin protein upon coexpression of DISC1Δ403–504 compared with full-length DISC1, suggesting that amino acid residues 403–504 of DISC1 are critical for preventing dysbindin from degradation (Fig. 3C, a and b). To further confirm this, we compared the stability of dysbindin upon coexpression of either full-length DISC1 or DISC1Δ403–504 in cycloheximide chase experiments. In these experiments, the dysbindin level declined more rapidly upon coexpression of DISC1Δ403–504 compared with full-length DISC1 (Fig. 3C, c). Furthermore, expression of DISC1Δ403–504 caused a reduction in endogenous dysbindin (Fig. 3D), further supporting the notion that amino acid residues 403–504 of DISC1 are important for protecting dysbindin from proteasome-mediated degradation.
FIGURE 3.
DISC1 enhances dysbindin stability. A, up-regulation of dysbindin is mediated by DISC1 amino acid residues 345–854. FL, full-length; aa, amino acids. B, increased expression of dysbindin (Dysb) upon coexpression with DISC1403–504. 30 h after transfection with the indicated plasmids, cells were harvested for immunoblotting analyses. C, effect of DISC1Δ403–504 coexpression on dysbindin stability. HEK293T cells were treated with 100 μg/ml cycloheximide (CHX) 30 h after cotransfection of EGFP-dysbindin constructs with either full-length DISC1 or DISC1Δ403–504. Dysbindin levels were quantified by immunoblotting. Representative data are shown in a, and the relative dysbindin protein level at time 0 is shown in b. Error bars show mean ± S.E. ***, p < 0.001 by one-way ANOVA with Bonferroni's multiple comparison test (n = 4); NS, not significant. c, the dynamics indicating the remaining protein levels of dysbindin were measured as a ratio of the value at time 0 in the same group. Error bars show mean ± S.E. *, p < 0.05; ***, p < 0.001 versus EGFP-dysbindin with empty vector (MOCK) by one-way ANOVA with Bonferroni's multiple comparison test (n = 4). D, down-regulation of dysbindin protein by overexpression of DISC1Δ403–504. COS7 cells were transfected with either EGFP-DISC1 or EGFP-DISC1Δ403–504 and subjected to immunoblotting. Error bars show mean ± S.E. *, p < 0.05 by one-way ANOVA with Bonferroni's multiple comparison test (n = 3).
DISC1 Regulates Dysbindin-mediated Neurite Outgrowth
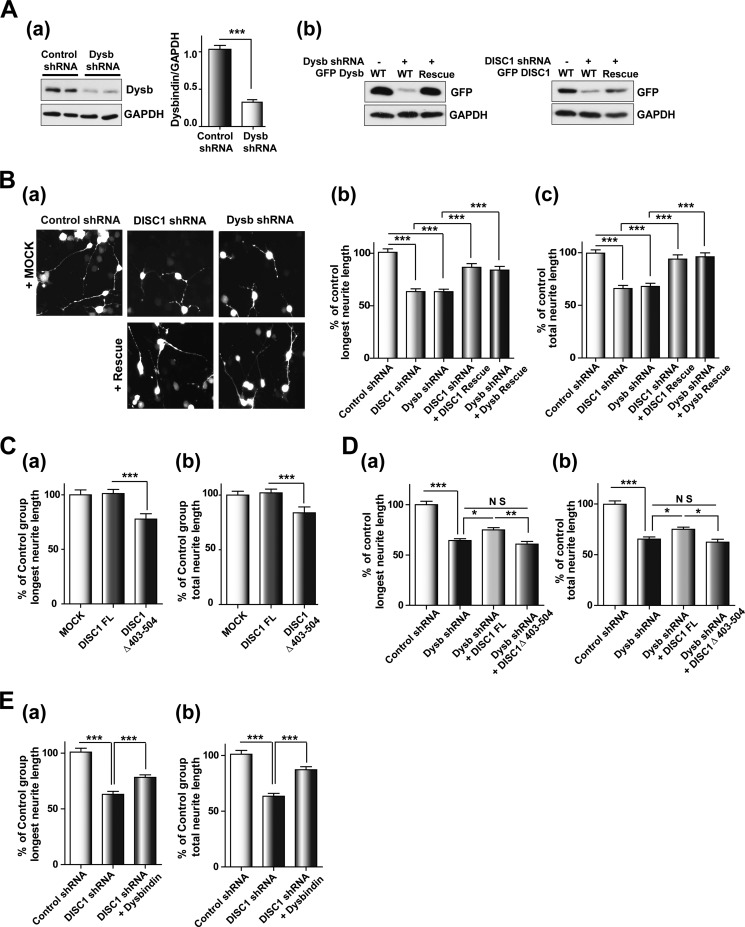
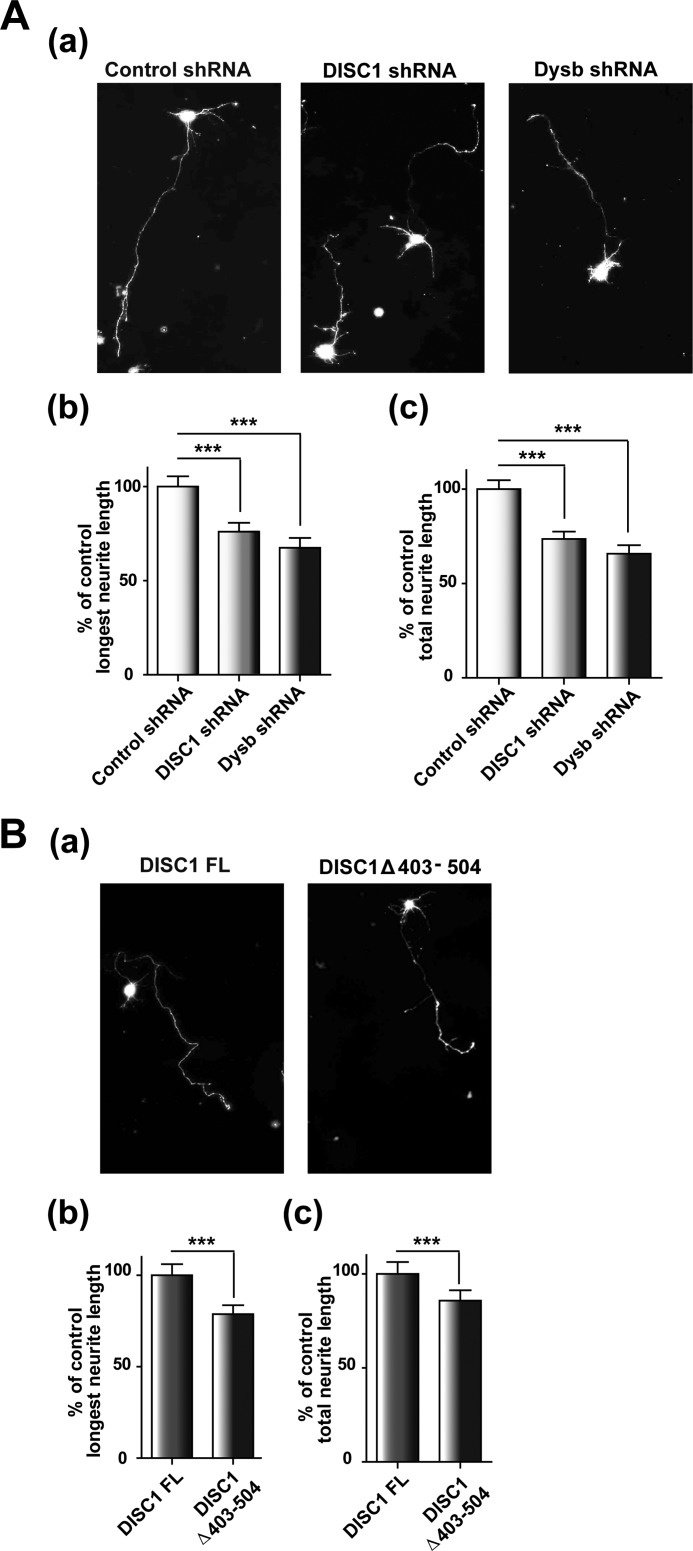
Dysbindin is involved in neurite outgrowth in differentiated neuroblastoma cells and primary neurons (12, 13, 25). Comparable neurite outgrowth defects are caused by deficiencies in DISC1 function (3, 30–32). Therefore, we attempted to test whether DISC1 and dysbindin are involved in neurite outgrowth through a common pathway. For this, shRNA against the mouse form of dysbindin (Fig. 4A, a), shRNA-resistant mouse dysbindin, and DISC1 constructs (Fig. 4A, b) were generated and utilized for neurite outgrowth assays in CAD cells. Consistent with previous observations, the longest neurite and total neurite length were both reduced significantly by shRNA-mediated knockdown of either dysbindin or DISC1. A remarkable restoration of neurite outgrowth by coexpression with shRNA-resistant DISC1 and dysbindin in knockdown cells was also observed, verifying the target specificity of knockdown constructs (Fig. 4B). Notably, although the full-length DISC1 did not influence neurite length, small but significant reductions of the longest neurite and total neurite length were observed upon overexpression of DISC1Δ403–504 (Fig. 4C). Although the stability of dysbindin in HEK293 cells was increased upon coexpression of full-length DISC1, the neurite outgrowth in CAD cells was not affected, implying that the endogenous level of DISC1 in CAD cells appears sufficient to ensure maximal neurite outgrowth. Moreover, we tested whether DISC1 can affect neurite outgrowth in CAD cells by stabilization of dysbindin protein held at low levels in dysbindin knockdown cells. Indeed, the defects in neurite outgrowth in dysbindin knockdown cells were partially rescued by coexpression of full-length DISC1 but not by DISC1Δ403–504, supporting the hypothesis that DISC1 affects the availability of dysbindin protein in the cell (Fig. 4D). To characterize the cooperation of DISC1 and dysbindin, we examined whether the DISC1 knockdown phenotype can be restored by dysbindin coexpression. Indeed, the neurite outgrowth defects caused by DISC1 knockdown were significantly restored by dysbindin coexpression (Fig. 4E). These results collectively indicate that DISC1 and dysbindin collaborate in the regulation of neurite outgrowth.
FIGURE 4.
DISC1 affects the dysbindin-mediated neurite outgrowth process. A, characterization of shRNA constructs. a, knockdown of endogenous dysbindin (Dysb) by dysbindin shRNA in differentiated CAD cells (n = 3). Error bars show mean ± S.E. ***, p < 0.001 by Student's t test. b, expression of shRNA-resistant DISC1 and dysbindin. B, neurite outgrowth defects in DISC1 and dysbindin knockdown CAD cells. Shown are representative images of differentiated CAD cells transfected with shRNA constructs alone or in combination with rescue constructs of DISC1 and dysbindin (a). The longest neurite length (b) and total neurite length (c) were measured using ImageJ software and normalized to the control group (n = 243 for control shRNA, n = 237 for DISC1 shRNA, n = 245 for dysbindin shRNA, n = 215 for DISC1 shRNA + DISC1 rescue, and n = 217 for dysbindin shRNA + dysbindin rescue). Error bars show mean ± S.E. ***, p < 0.001 by one-way ANOVA with Bonferroni's multiple comparison test. C, inhibition of neurite outgrowth by coexpression of DISC1Δ403–504. The longest neurite length (a) and total neurite length (b) were analyzed in CAD cells differentiated 3 days (n = 193 for MOCK, n = 237 for DISC1 FL, and n = 223 for DISC1Δ403–504). Error bars show mean ± S.E. ***, p < 0.001 by one-way ANOVA with Bonferroni's multiple comparison test. D, partial rescue of the dysbindin knockdown phenotype by coexpression of DISC1. The longest neurite length (a) and total neurite length (b) are shown (n = 211 for control shRNA, n = 210 for dysbindin shRNA, n = 193 for dysbindin shRNA + DISC1 FL, and n = 106 for dysbindin shRNA + DISC1Δ403–504). Error bars show mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not significant by one-way ANOVA with Bonferroni's multiple comparison test. E, reversal of the neurite outgrowth defects by dysbindin coexpression in DISC1 knockdown cells. The longest neurite length (a) and total neurite length (b) are shown (n = 192 for control shRNA, n = 219 for DISC1 shRNA, and n = 219 for DISC1 shRNA + dysbindin). Error bars show mean ± S.E. ***, p < 0.001 by one-way ANOVA with Bonferroni's multiple comparison test.
We also examined neurite outgrowth in primary cultured mouse cortical neurons. The longest neurite and total neurite length were both reduced significantly upon knockdown of either dysbindin or DISC1 (Fig. 5A). Moreover, significantly shorter neurite lengths in the longest neurite and total neurite length were observed upon overexpression of DISC1Δ403–504 compared with the overexpression of full-length DISC1 (Fig. 5B). These results are largely consistent with the CAD cell results and support the notion that DISC1 and dysbindin not only play roles but also cooperate in the regulation of neurite outgrowth.
FIGURE 5.
Compromised neurite outgrowth by deficiencies in DISC1 and dysbindin in primary cortical neurons. A, neurite outgrowth defects in DISC1- and dysbindin (Dysb) shRNA-transfected cortical neurons. Shown are representative images of cortical neurons transfected with indicated shRNA constructs (a). The longest neurite length (b) and total neurite length (c) were analyzed. Neurites on ∼249 transfected neurons from three independent experiments were measured and normalized to control cells (n = 214 for control shRNA, n = 249 for DISC1 shRNA, and n = 192 for dysbindin shRNA). Error bars show mean ± S.E. ***, p < 0.001 by Student's t test. B, inhibition of neurite outgrowth by DISC1Δ403–504 coexpression. Shown are representative images of cortical neurons transfected with full-length DISC1 or DISC1Δ403–504 (a). The longest neurite length (b) and total neurite length (c) were analyzed (n = 193 for DISC1 FL and n = 223 for DISC1Δ403–504). Neurons were transfected with constructs for GFP-DISC1 FL or GFP-DISC1Δ403–504 in combination with the red fluorescent protein (RFP) construct (ratio 3:1) at days in vitro 0 to visualize neuronal morphology. Error bars show mean ± S.E. ***, p < 0.001 by Student's t test.
DISCUSSION
In this study, we describe the physical and functional interaction between DISC1 and dysbindin, two major schizophrenia susceptibility factors. We found that DISC1 physically interacts with dysbindin and stabilizes the protein by decreasing its ubiquitylation. Although the interaction was mediated by a broad region of DISC1, its capacity to up-regulate dysbindin appeared to be restricted to a relatively specific region (amino acid residues 403–504). Finally, we demonstrated that DISC1 and dysbindin cooperate in the neurite outgrowth process. The potential functional interaction between DISC1 and dysbindin has been suggested on the basis of their close proximity in the protein-protein interaction network of schizophrenia susceptibility factors (36, 37). Moreover, a recent report showed a colocalization of DISC1 and dysbindin in cellular aggresomes and the association of purified recombinant dysbindin and DISC1 (38). However, it was unclear whether DISC1 and dysbindin interact under more physiological conditions and what its functional relevance might be. In this line, our results provide additional mechanistic insights, supporting the idea that multiple schizophrenia susceptibility factors form a common biological pathway associated with the etiology of schizophrenia and related mental illnesses.
We provided evidence indicating that DISC1 affects dysbindin functions by enhancing its stability. Studies have revealed extended roles for the UPS in neuronal growth and development, synaptic function and plasticity, and pathway-specific modulators (42–44). However, only few UPS-associated players have been studied in neurons so far, and many components and functions of the UPS in the neuronal system have yet to be identified. Interestingly, in addition to the DISC1 region responsible for the interaction with dysbindin, the C-terminal region of DISC1 (amino acid residues 586–854) appears to be involved in the dysbindin level in an interaction-independent manner. This is consistent with previous reports showing that the C-terminal region of DISC1 participates in controlling the stability of the counterpart-interacting proteins by affecting their ubiquitylation levels (39, 41). These results raise the possibility that there might be a common ubiquitin-associated mechanism involving the C-terminal region of DISC1. The ubiquitylation state of a protein is determined by the antagonistic activities of the ubiquitin-conjugating enzyme (E2)-ubiquitin ligase (E3) complex and deubiquitylating enzymes (45, 46). Therefore, it is intriguing to speculate that DISC1 might affect the function of either the ubiquitin-conjugating enzyme complex or deubiquitylating enzymes to alter the stability of its binding partners.
Although dysbindin was initially identified as a component of the dystrophin-dystroglycan complex, subsequent studies showed that dysbindin in the brain exists as a stable component of biogenesis of lysosome-related organelles complex 1 (BLOC-1), a multiprotein complex containing at least eight components (dysbindin (dystrobrevin-binding protein 1, DTNBP1), muted (MUTED), pallidin (PLDN), cappuccino (CNO), snapin (SNARE-associated protein, SNAPAP), and biogenesis of lysosome-related organelles complex 1, subunit 1–3 (BLOC1S1–3)) with functions in organelle biogenesis and intracellular membrane trafficking (12, 47–49). It is notable that other BLOC-1 subunits, BLOC1S2, BLOCIS3, and MUTED, have been reported to be potential susceptibility factors of schizophrenia (2, 50, 51) and that a deficiency in BLOC-1 induces neurite outgrowth defects in rodent hippocampal neurons (12). Interestingly, destabilization of dysbindin protein has been observed consistently in muted (mu) (51, 52) and pallid (pa) mice (12, 52), implying the potential importance of decreased dysbindin stability as a common cellular correlative associated with schizophrenia. Taken together, the findings in this study suggest that DISC1 may regulate the integrity of BLOC-1 and that dysbindin mediates this interplay among multiple schizophrenia susceptibility factors.
This work was supported by Grants (NRF-2012R1A2A2A01012923, NRF-2012R1A4A1028200, and NRF-2013R1A1A2074251 from the ministry of science, ICT, and future planning (MSIP) and by the Framework of International Cooperation Program managed by national research foundation (NRF) of Korea (Grant NRF-2012K2A1A2033117). This work was also supported by the POSTECH basic science research institute (BSRI) Fund.
- CAD
- Cath.a-differentiated
- EGFP
- enhanced GFP
- ANOVA
- analysis of variance
- UPS
- ubiquitin-proteasome system
- FL
- full-length.
REFERENCES
- 1. Freedman R. (2003) Schizophrenia. N. Engl. J. Med. 349, 1738–1749 [DOI] [PubMed] [Google Scholar]
- 2. Ross C. A., Margolis R. L., Reading S. A., Pletnikov M., Coyle J. T. (2006) Neurobiology of schizophrenia. Neuron 52, 139–153 [DOI] [PubMed] [Google Scholar]
- 3. Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., Sawamura N., Park U., Kudo C., Okawa M., Ross C. A., Hatten M. E., Nakajima K., Sawa A. (2005) A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 7, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 4. Selemon L. D. (2004) Increased cortical neuronal density in schizophrenia. Am. J. Psychiatry 161, 1564. [DOI] [PubMed] [Google Scholar]
- 5. Selemon L. D., Goldman-Rakic P. S. (1999) The reduced neuropil hypothesis: a circuit based model of schizophrenia. Biol. Psychiatry 45, 17–25 [DOI] [PubMed] [Google Scholar]
- 6. Buxhoeveden D., Roy E., Switala A., Casanova M. F. (2000) Reduced interneuronal space in schizophrenia. Biol. Psychiatry 47, 681–683 [DOI] [PubMed] [Google Scholar]
- 7. Arnold S. E., Lee V. M., Gur R. E., Trojanowski J. Q. (1991) Abnormal expression of two microtubule-associated proteins (MAP2 and MAP5) in specific subfields of the hippocampal formation in schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 88, 10850–10854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalus P., Müller T. J., Zuschratter W., Senitz D. (2000) The dendritic architecture of prefrontal pyramidal neurons in schizophrenic patients. Neuroreport 11, 3621–3625 [DOI] [PubMed] [Google Scholar]
- 9. Millar J. K., Wilson-Annan J. C., Anderson S., Christie S., Taylor M. S., Semple C. A., Devon R. S., St Clair D. M., Muir W. J., Blackwood D. H., Porteous D. J. (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423 [DOI] [PubMed] [Google Scholar]
- 10. Straub R. E., Jiang Y., MacLean C. J., Ma Y., Webb B. T., Myakishev M. V., Harris-Kerr C., Wormley B., Sadek H., Kadambi B., Cesare A. J., Gibberman A., Wang X., O'Neill F. A., Walsh D., Kendler K. S. (2002) Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am. J. Hum. Genet. 71, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Donovan M. C., Williams N. M., Owen M. J. (2003) Recent advances in the genetics of schizophrenia. Hum. Mol. Genet. 12, 125–133 [DOI] [PubMed] [Google Scholar]
- 12. Ghiani C. A., Starcevic M., Rodriguez-Fernandez I. A., Nazarian R., Cheli V. T., Chan L. N., Malvar J. S., de Vellis J., Sabatti C., Dell'Angelica E. C. (2010) The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol. Psychiatry 15, 204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma X., Fei E., Fu C., Ren H., Wang G. (2011) Dysbindin-1, a schizophrenia-related protein, facilitates neurite outgrowth by promoting the transcriptional activity of p53. Mol. Psychiatry 16, 1105–1116 [DOI] [PubMed] [Google Scholar]
- 14. Benson M. A., Newey S. E., Martin-Rendon E., Hawkes R., Blake D. J. (2001) Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J. Biol. Chem. 276, 24232–24241 [DOI] [PubMed] [Google Scholar]
- 15. Schwab S. G., Knapp M., Mondabon S., Hallmayer J., Borrmann-Hassenbach M., Albus M., Lerer B., Rietschel M., Trixler M., Maier W., Wildenauer D. B. (2003) Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. Am. J. Hum. Genet. 72, 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Den Bogaert A., Schumacher J., Schulze T. G., Otte A. C., Ohlraun S., Kovalenko S., Becker T., Freudenberg J., Jönsson E. G., Mattila-Evenden M., Sedvall G. C., Czerski P. M., Kapelski P., Hauser J., Maier W., Rietschel M., Propping P., Nöthen M. M., Cichon S. (2003) The DTNBP1 (dysbindin) gene contributes to schizophrenia, depending on family history of the disease. Am. J. Hum. Genet. 73, 1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang J. X., Zhou J., Fan J. B., Li X. W., Shi Y. Y., Gu N. F., Feng G. Y., Xing Y. L., Shi J. G., He L. (2003) Family-based association study of DTNBP1 in 6p22.3 and schizophrenia. Mol. Psychiatry 8, 717–718 [DOI] [PubMed] [Google Scholar]
- 18. Funke B., Finn C. T., Plocik A. M., Lake S., DeRosse P., Kane J. M., Kucherlapati R., Malhotra A. K. (2004) Association of the DTNBP1 locus with schizophrenia in a U. S. population. Am. J. Hum. Genet. 75, 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng Y. Q., Zhou Z. Y., He X., Wang H., Guo X. L., Hao C. J., Guo Y., Zhen X. C., Li W. (2008) Dysbindin deficiency in sandy mice causes reduction of snapin and displays behaviors related to schizophrenia. Schizophr. Res. 106, 218–228 [DOI] [PubMed] [Google Scholar]
- 20. Papaleo F., Yang F., Garcia S., Chen J., Lu B., Crawley J. N., Weinberger D. R. (2012) Dysbindin-1 modulates prefrontal cortical activity and schizophrenia-like behaviors via dopamine/D2 pathways. Mol. Psychiatry 17, 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carr G. V., Jenkins K. A., Weinberger D. R., Papaleo F. (2013) Loss of dysbindin-1 in mice impairs reward-based operant learning by increasing impulsive and compulsive behavior. Behav. Brain Res. 241, 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hattori S., Murotani T., Matsuzaki S., Ishizuka T., Kumamoto N., Takeda M., Tohyama M., Yamatodani A., Kunugi H., Hashimoto R. (2008) Behavioral abnormalities and dopamine reductions in sdy mutant mice with a deletion in Dtnbp1, a susceptibility gene for schizophrenia. Biochem. Biophys. Res. Commun. 373, 298–302 [DOI] [PubMed] [Google Scholar]
- 23. Weickert C. S., Rothmond D. A., Hyde T. M., Kleinman J. E., Straub R. E. (2008) Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophr. Res. 98, 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Talbot K., Eidem W. L., Tinsley C. L., Benson M. A., Thompson E. W., Smith R. J., Hahn C. G., Siegel S. J., Trojanowski J. Q., Gur R. E., Blake D. J., Arnold S. E. (2004) Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J. Clin. Invest. 113, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kubota K., Kumamoto N., Matsuzaki S., Hashimoto R., Hattori T., Okuda H., Takamura H., Takeda M., Katayama T., Tohyama M. (2009) Dysbindin engages in c-Jun N-terminal kinase activity and cytoskeletal organization. Biochem. Biophys. Res. Commun. 379, 191–195 [DOI] [PubMed] [Google Scholar]
- 26. Ito H., Morishita R., Shinoda T., Iwamoto I., Sudo K., Okamoto K., Nagata K. (2010) Dysbindin-1, WAVE2 and Abi-1 form a complex that regulates dendritic spine formation. Mol. Psychiatry 15, 976–986 [DOI] [PubMed] [Google Scholar]
- 27. Jaaro-Peled H. (2009) Gene models of schizophrenia: DISC1 mouse models. Prog. Brain Res. 179, 75–86 [DOI] [PubMed] [Google Scholar]
- 28. Kuroda K., Yamada S., Tanaka M., Iizuka M., Yano H., Mori D., Tsuboi D., Nishioka T., Namba T., Iizuka Y., Kubota S., Nagai T., Ibi D., Wang R., Enomoto A., Isotani-Sakakibara M., Asai N., Kimura K., Kiyonari H., Abe T., Mizoguchi A., Sokabe M., Takahashi M., Yamada K., Kaibuchi K. (2011) Behavioral alterations associated with targeted disruption of exons 2 and 3 of the Disc1 gene in the mouse. Hum. Mol. Genet. 20, 4666–4683 [DOI] [PubMed] [Google Scholar]
- 29. Ishizuka K., Kamiya A., Oh E. C., Kanki H., Seshadri S., Robinson J. F., Murdoch H., Dunlop A. J., Kubo K., Furukori K., Huang B., Zeledon M., Hayashi-Takagi A., Okano H., Nakajima K., Houslay M. D., Katsanis N., Sawa A. (2011) DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature 473, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hattori T., Baba K., Matsuzaki S., Honda A., Miyoshi K., Inoue K., Taniguchi M., Hashimoto H., Shintani N., Baba A., Shimizu S., Yukioka F., Kumamoto N., Yamaguchi A., Tohyama M., Katayama T. (2007) A novel DISC1-interacting partner DISC1-binding zinc-finger protein: implication in the modulation of DISC1-dependent neurite outgrowth. Mol. Psychiatry 12, 398–407 [DOI] [PubMed] [Google Scholar]
- 31. Hattori T., Shimizu S., Koyama Y., Yamada K., Kuwahara R., Kumamoto N., Matsuzaki S., Ito A., Katayama T., Tohyama M. (2010) DISC1 regulates cell-cell adhesion, cell-matrix adhesion and neurite outgrowth. Mol. Psychiatry 15, 778, 798–809 [DOI] [PubMed] [Google Scholar]
- 32. Kamiya A., Tomoda T., Chang J., Takaki M., Zhan C., Morita M., Cascio M. B., Elashvili S., Koizumi H., Takanezawa Y., Dickerson F., Yolken R., Arai H., Sawa A. (2006) DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum. Mol. Genet. 15, 3313–3323 [DOI] [PubMed] [Google Scholar]
- 33. Meyer K. D., Morris J. A. (2009) Disc1 regulates granule cell migration in the developing hippocampus. Hum. Mol. Genet. 18, 3286–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fukuda T., Sugita S., Inatome R., Yanagi S. (2010) CAMDI, a novel disrupted in schizophrenia 1 (DISC1)-binding protein, is required for radial migration. J. Biol. Chem. 285, 40554–40561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hayashi-Takagi A., Takaki M., Graziane N., Seshadri S., Murdoch H., Dunlop A. J., Makino Y., Seshadri A. J., Ishizuka K., Srivastava D. P., Xie Z., Baraban J. M., Houslay M. D., Tomoda T., Brandon N. J., Kamiya A., Yan Z., Penzes P., Sawa A. (2010) Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat. Neurosci. 13, 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Camargo L. M., Collura V., Rain J. C., Mizuguchi K., Hermjakob H., Kerrien S., Bonnert T. P., Whiting P. J., Brandon N. J. (2007) Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry 12, 74–86 [DOI] [PubMed] [Google Scholar]
- 37. Mead C. L., Kuzyk M. A., Moradian A., Wilson G. M., Holt R. A., Morin G. B. (2010) Cytosolic protein interactions of the schizophrenia susceptibility gene dysbindin. J. Neurochem. 113, 1491–1503 [DOI] [PubMed] [Google Scholar]
- 38. Ottis P., Bader V., Trossbach S. V., Kretzschmar H., Michel M., Leliveld S. R., Korth C. (2011) Convergence of two independent mental disease genes on the protein level: recruitment of dysbindin to cell-invasive disrupted-in-schizophrenia 1 aggresomes. Biol. Psychiatry 70, 604–610 [DOI] [PubMed] [Google Scholar]
- 39. Park Y. U., Jeong J., Lee H., Mun J. Y., Kim J. H., Lee J. S., Nguyen M. D., Han S. S., Suh P. G., Park S. K. (2010) Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc. Natl. Acad. Sci. U.S.A. 107, 17785–17790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Locke M., Tinsley C. L., Benson M. A., Blake D. J. (2009) TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum Mol. Genet 18, 2344–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma T. M., Abazyan S., Abazyan B., Nomura J., Yang C., Seshadri S., Sawa A., Snyder S. H., Pletnikov M. V. (2013) Pathogenic disruption of DISC1-serine racemase binding elicits schizophrenia-like behavior via d-serine depletion. Mol. Psychiatry 18, 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W. (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219 [DOI] [PubMed] [Google Scholar]
- 43. Tursun B., Schlüter A., Peters M. A., Viehweger B., Ostendorff H. P., Soosairajah J., Drung A., Bossenz M., Johnsen S. A., Schweizer M., Bernard O., Bach I. (2005) The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 19, 2307–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pak D. T., Sheng M. (2003) Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science 302, 1368–1373 [DOI] [PubMed] [Google Scholar]
- 45. Pickart C. M., Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 46. van Wijk S. J., Timmers H. T. (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24, 981–993 [DOI] [PubMed] [Google Scholar]
- 47. Di Pietro S. M., Falcón-Pérez J. M., Tenza D., Setty S. R., Marks M. S., Raposo G., Dell'Angelica E. C. (2006) BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell 17, 4027–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nazarian R., Starcevic M., Spencer M. J., Dell'Angelica E. C. (2006) Reinvestigation of the dysbindin subunit of BLOC-1 (biogenesis of lysosome-related organelles complex-1) as a dystrobrevin-binding protein. Biochem. J. 395, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guo A. Y., Sun J., Riley B. P., Thiselton D. L., Kendler K. S., Zhao Z. (2009) The dystrobrevin-binding protein 1 gene: features and networks. Mol. Psychiatry 14, 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morris D. W., Murphy K., Kenny N., Purcell S. M., McGhee K. A., Schwaiger S., Nangle J. M., Donohoe G., Clarke S., Scully P., Quinn J., Meagher D., Baldwin P., Crumlish N., O'Callaghan E., Waddington J. L., Gill M., Corvin A. P. (2008) Dysbindin (DTNBP1) and the biogenesis of lysosome-related organelles complex 1 (BLOC-1): main and epistatic gene effects are potential contributors to schizophrenia susceptibility. Biol. Psychiatry 63, 24–31 [DOI] [PubMed] [Google Scholar]
- 51. Iizuka Y., Sei Y., Weinberger D. R., Straub R. E. (2007) Evidence that the BLOC-1 protein dysbindin modulates dopamine D2 receptor internalization and signaling but not D1 internalization. J. Neurosci. 27, 12390–12395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li W., Zhang Q., Oiso N., Novak E. K., Gautam R., O'Brien E. P., Tinsley C. L., Blake D. J., Spritz R. A., Copeland N. G., Jenkins N. A., Amato D., Roe B. A., Starcevic M., Dell'Angelica E. C., Elliott R. W., Mishra V., Kingsmore S. F., Paylor R. E., Swank R. T. (2003) Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 35, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]