Background: R9C mutation of phospholamban triggers cardiomyopathy.
Results: Acute expression of R9C-phospholamban in cardiomyocytes was positively inotropic/lusitropic.
Conclusion: Loss of phospholamban inhibitory function acutely increases SERCA function but impairs SERCA regulation, resulting in a blunted stress response, leading to cardiomyopathy.
Significance: The results reconcile the pathological character of R9C with biochemical studies that showed loss of inhibition of SERCA.
Keywords: Calcium-ATPase, Cardiomyopathy, Fluorescence Resonance Energy Transfer (FRET), Heart Failure, Oxidative Stress, PLN, R9C, SERCA, SERCA2a, Phospholamban
Abstract
A naturally occurring R9C mutation of phospholamban (PLB) triggers cardiomyopathy and premature death by altering regulation of sarco/endoplasmic reticulum calcium-ATPase (SERCA). The goal of this study was to investigate the acute physiological consequences of the R9C-PLB mutation on cardiomyocyte calcium kinetics and contractility. We measured the physiological consequences of R9C-PLB mutation on calcium transients and sarcomere shortening in adult cardiomyocytes. In contrast to studies of chronic R9C-PLB expression in transgenic mice, we found that acute expression of R9C-PLB exerts a positively inotropic and lusitropic effect in cardiomyocytes. Importantly, R9C-PLB exhibited blunted sensitivity to frequency potentiation and β-adrenergic stimulation, two major physiological mechanisms for the regulation of cardiac performance. To identify the molecular mechanism of R9C pathology, we quantified the effect of R9C on PLB oligomerization and PLB-SERCA binding. FRET measurements in live cells revealed that R9C-PLB exhibited an increased propensity for oligomerization, and this was further increased by oxidative stress. The R9C also decreased PLB binding to SERCA and altered the structure of the PLB-SERCA regulatory complex. The structural change after oxidative modification of R9C-PLB was similar to that observed after PLB phosphorylation. We conclude that R9C mutation of PLB decreases SERCA inhibition by decreasing the amount of the regulatory complex and altering its conformation. This has an acute inotropic/lusitropic effect but yields negative consequences of impaired frequency potentiation and blunted β-adrenergic responsiveness. We envision a self-reinforcing mechanism beginning with phosphomimetic R9C-PLB oxidation and loss of SERCA inhibition, leading to impaired calcium regulation and heart failure.
Introduction
The sarco(endo)plasmic reticulum calcium-ATPase (SERCA)2 is an ion-motive transporter that establishes intracellular calcium (Ca2+) stores needed for cell signaling (1) and normal cardiac myocyte function (2, 3). During the cardiac cycle, sequestration of Ca2+ by SERCA2a during diastole is the fundamental mechanism for initiation of cardiac muscle relaxation (2–4). Moreover, the rate of SERCA Ca2+ uptake is one of the main determinants of the size of the Ca2+ store, so SERCA is also critical for regulating the strength of cardiac contraction during systole (2–4). Because of its key role in determining cardiac inotropy (contractility) and lusitropy (relaxation), SERCA2a activity is closely governed by an inhibitory interaction with its regulatory partner phospholamban (PLB), a 52-residue single span transmembrane peptide (3–5). PLB inhibition of SERCA is relieved by phosphorylation of PLB through protein kinase A (PKA)-dependent adrenergic signaling (3–7), increasing SERCA activity to meet increased physiological demand. A complementary mechanism for relief of inhibition is PLB phosphorylation by Ca2+/calmodulin-dependent protein kinase II (CaMKII) (6, 7). This pathway is activated by the elevation of cytosolic Ca2+ that accompanies increased pacing frequency during exercise. Thus, SERCA activity and PLB regulation of that activity allow the heart to responsively compensate for rest and stress conditions.
Disordered Ca2+ transport or regulation may cause and result from cardiac diseases such as heart failure. In particular, mutations (8–12) or deletions (13–16) of PLB give rise to human disease, underscoring the importance of this peptide and providing some insight into the molecular mechanisms of SERCA regulation by PLB. Of particular interest is the human heart failure mutant R9C-PLB that causes dilated cardiomyopathy (DCM) (8). DCM is a leading cause of cardiovascular morbidity and mortality worldwide (17–19), so there is great interest in understanding how a discrete point mutation in PLB could induce pathological dysfunction. Transgenic expression of R9C-PLB in mouse hearts recapitulates many aspects of human DCM (8, 9), but the fundamental molecular mechanism underlying the role of R9C-PLB in SERCA regulation is still unclear. Proposed mechanisms include trapping of PKA (8), disruption of PLB phosphorylation (8, 9, 20, 21), and loss of PLB inhibitory function (8, 9, 20–22). Other studies have suggested that the R9C mutation mimics PLB phosphorylation by partial unfolding of the cytoplasmic helix resulting in decreased helical conformation (23) or detachment of PLB cytoplasmic domain from the membrane surface (24, 25). Moreover, we have previously proposed that the R9C mutation induces oxidation-dependent cross-linking of adjacent R9C-PLB protomers (20). This could also mimic the effect of PLB phosphorylation in increasing its oligomerization (5, 26, 27), depleting the actively inhibitory monomeric species and relieving SERCA inhibition.
In this study, we investigated the acute physiological consequences of R9C-PLB mutation on Ca2+ kinetics and contractility using adenoviral delivery of R9C-PLB to adult rabbit cardiomyocytes. Rabbit myocytes are particularly good models of human cardiac Ca2+ cycling (28–30). We reasoned that acute expression might reveal new mechanistic information about R9C pathophysiology, complementing previous transgenesis studies that focused on the long term effects of R9C mutation. To specifically test the consequence of R9C mutation for PLB oligomerization, we modulated PLB oligomerization affinity with mutations of three transmembrane Cys residues to Ser (SSS). This set of mutations has been proposed to abolish PLB oligomerization (21) and could isolate the effect of R9C from other determinants of PLB oligomerization.
EXPERIMENTAL PROCEDURES
Molecular Biology
mCerulean (Cer) was fused to the N terminus of canine PLB or canine SERCA2a as described previously (31, 32). In addition, cyan fluorescent protein (CFP) or enhanced yellow fluorescent protein (YFP) was fused to the N terminus of canine PLB (33, 34). The canine PLB and SERCA sequences are 98% identical to human (35). Fig. 1A shows the amino acid sequence of all the mutants of PLB used in this study. All the PLB mutants were generated using the QuikChange IIXL site-directed mutagenesis kit (Stratagene, La Jolla, CA) and custom oligonucleotide primers (Eurofins MWG Operon). The nucleotide sequences were verified by DNA sequencing (ACGT, Inc.). Adenoviral vectors of canine CFP-PLB or YFP-PLB were produced using the AdEasy Adenoviral Vector System (Stratagene, La Jolla, CA).
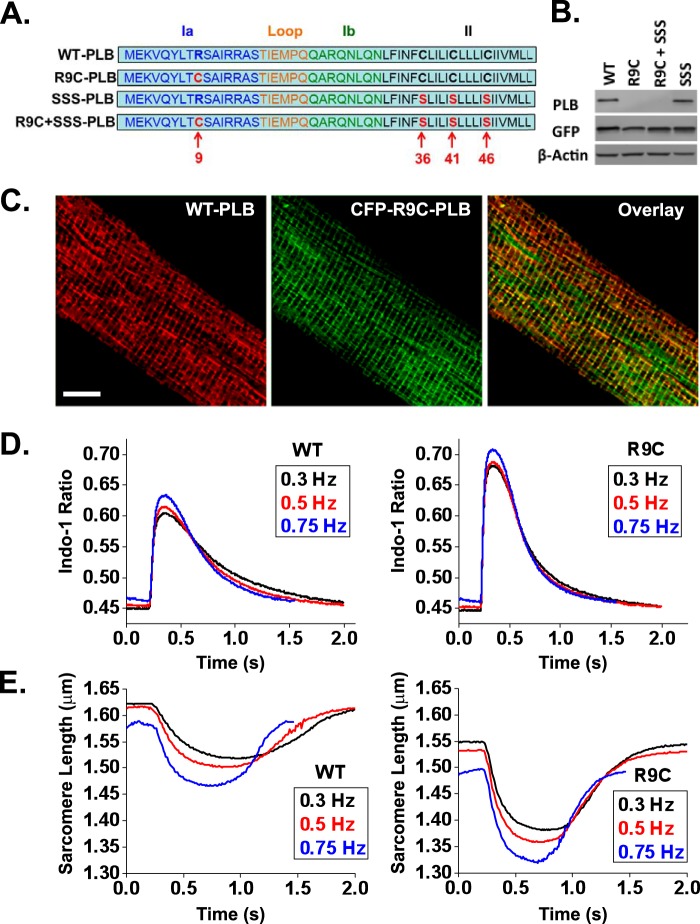
FIGURE 1.
R9C-PLB expression in cardiomyocytes induces a positively inotropic and positively lusitropic effect. A, schematic representation of primary sequence of WT-PLB and the mutant PLB constructs. The amino acid sequence of the PLB protomer shows the N-terminal cytosolic domain Ia (residues 1–16), flexible linker (residues 17–22), domain Ib (residues 23–30), and the C-terminal transmembrane domain II (residues 31–52). Cer or YFP was fused to the N terminus. The mutated residues are highlighted in red, and the residue numbers are labeled at the bottom of the sequence. B, both Cer-R9C-PLB and Cer-R9C + SSS-PLB were recognized by GFP but not by PLB 2D12 antibody. C, confocal images of an adult rabbit left ventricular myocyte expressing CFP-R9C-PLB (green) co-localized with endogenous WT-PLB (red). Scale bar, 10 μm. D and E, isolated cardiomyocytes expressing YFP-WT-PLB or YFP-R9C-PLB were used to record averaged Ca2+ transients obtained from 8 to 10 events per cell; WT (n = 17), R9C (n = 16) (D) and averaged sarcomere shortening traces obtained from 8 to 10 events per cell; WT (n = 9), R9C (n = 7) (E). Myocytes were electrically stimulated at increasing pacing frequencies of 0.3, 0.5, or 0.75 Hz.
Cell Culture
Left ventricular cardiomyocytes were enzymatically isolated from adult New Zealand White rabbits (36). All the animal protocols including cardiomyocyte isolation was approved by the Loyola University Institutional Animal Care and Use Committee. The cardiomyocytes were washed with fresh PC-1 medium (Lonza, Basel, Switzerland) and plated onto laminin-coated glass coverslips that fit into 35-mm culture dishes. Cardiomyocytes were incubated at 37 °C for 1 h, and CFP-PLB and/or YFP-PLB adenoviruses were added at a multiplicity of infection of 1000 as described previously (37). Cardiomyocytes were then paced for 48 h in culture using a C-Pace EP Pacer (IonOptix, Milton, MA) set to 10 V with a frequency of 0.1 Hz, with 5-ms pulse duration (32, 37).
AAV-293 cells were cultured in 60-mm tissue culture dishes in complete DMEM growth medium with 10% fetal bovine serum, 1% l-glutamine and incubated at 37 °C under 5% CO2. Transient transfection of cultured AAV-293 cells was performed by the CaPO4 precipitation method using the MBS mammalian transfection kit (Stratagene, La Jolla, CA) (32–34, 38). Cells were co-transfected with plasmids encoding Cer-PLB and YFP-PLB or Cer-SERCA and YFP-PLB with a molar ratio of 1:5 or 1:20, respectively (32, 34, 38). The cells were subjected to mild trypsinization and plated on glass bottom dishes coated with poly-d-lysine and allowed to adhere for 2 h before imaging (32, 38).
Fluorescence Resonance Energy Transfer (FRET) Quantification
PLB oligomerization was quantified in live cells using wide-field fluorescence microscopy (32, 38). MetaMorph software was employed for acquisition of a montage of 48 images using a motorized stage (Prior, Rockland, MA). Focus was automatically maintained by an optical feedback system (Perfect Focus System, Nikon), and image acquisition was done using ×40 objective with a numerical aperture of 0.75. The exposure time was 150 ms for each channel as follows: Cer, YFP, and FRET (Cer excitation/YFP emission). The multiple wavelength cell scoring application module in MetaMorph was used for automated quantification of fluorescence intensity. The cells were automatically selected by the software based on the criteria, including minimum fluorescent area of 50 μm2, diameter between 40 and 100 μm, and an average intensity of 100 counts above background. The average intensities of each channel were then transferred to a spreadsheet for quantifying FRET efficiency. FRET quantification was done using acceptor sensitization (E-FRET) (39), as described previously (31, 32, 38). After background subtraction, FRET efficiency was calculated according to the following formula: E = (IDA − a(IAA) − d(IDD))/(IDA − a(IAA) + (G − d) (IDD)); where IDD is the intensity of fluorescence emission from the donor channel (472/30 nm) with excitation of 427/10 nm; IAA is the intensity of fluorescence emission from the acceptor channel (542/27 nm) with excitation of 504/12 nm; and IDA is the intensity of fluorescence emission detected in the FRET channel (542/27 nm) with excitation of 427/10 nm. The constants a and d are cross-talk coefficients determined from acceptor-only or donor-only control samples, respectively, a = IDA/IAA and d = IDA/IDD. G represents the ratio of the sensitized emission to the corresponding amount of donor recovery. We obtained values of 0.082, 0.82, and 3.2 for a, d, and G ratios, respectively, as described previously (32). For time course experiments, the cells were imaged at 30-s time intervals for Cer, YFP, and FRET channels. After 5 min of image acquisition, 100 μm hydrogen peroxide (H2O2) was applied, and the cells were imaged every 30 s for an additional 20 min. The FRET efficiency for individual cells at each time point was quantified using MetaMorph, and the data from three independent experiments were averaged. The FRET ratio was calculated by dividing the intensity of the FRET channel by the intensity of Cer channel. The calculated FRET ratios for all the cells at each time point were averaged after normalizing to the first time point. FRET images were acquired by dividing the image of FRET channel by Cer fluorescence using MetaMorph.
“In-cell” Binding Assay
An in-cell binding assay was performed to estimate the parameters related to structure and binding affinity (20, 26, 31, 32, 34, 38). Briefly, the FRET efficiency of individual cells co-expressing Cer-PLB/YFP-PLB or Cer-SERCA/YFP-PLB was plotted against relative protein concentration, which was quantified from the respective YFP fluorescence intensities. The concentration dependence of FRET was fit to a hyperbolic curve of the form y = (FRETmax)x/(KD + x), with all parameters independently fit, where y is the observed FRET efficiency, and x is the protein concentration in the cell in arbitrary units. FRETmax is the intrinsic FRET of the protein complex and a measure of average distances between binding partners, providing structural information. KD is the protein concentration that yields half-FRETmax and represents the dissociation constant of the protein complex, providing an estimate of the apparent binding affinity. KD1 is the apparent dissociation constant of the PLB oligomer, and KD2 is the apparent dissociation constant of the SERCA-PLB regulatory complex. The data are pooled from 3 to 4 independent experiments for each sample. Each binding curve was developed by using an average of ∼2300 cells. Probe separation distance (R) for the SERCA-PLB regulatory complex was calculated using the Förster equation (40), r = (RO) ((1/IE) − 1)1/6), where RO is the Förster radius, and E is the measured FRETmax. Intrapentameric probe separation distance was calculated from FRETmax using a MatLab application (34), assuming a ring-shaped oligomer (41–43), with a subunit number of 5 (pentamer). The acceptor molar fraction was 0.89 for the WT-PLB pentamer and 0.92 for the R9C-PLB pentamer. For estimation of probe separation distances for both the pentamer and regulatory complex, the Förster radius of 49.8 Å was used for the Cer-YFP pair (44), and 4% nonspecific FRET was subtracted from the measured FRETmax values. Previously, we estimated nonspecific FRET to be 4%, as determined from competition with unlabeled PLB or with a fluorescently tagged PLB that is unable to participate in FRET (20, 34). FRET efficiency values and calculated distances are summarized in Table 1.
TABLE 1.
Summary of quantitative FRET data
Effect of R9C, SSS, and R9C + SSS mutations on PLB intrapentameric FRET efficiency and SERCA-PLB FRET efficiency. FRETmax is maximal FRET efficiency; KD1 is oligomer dissociation constant; KD2 is dissociation constant for the SERCA-PLB complex; AU is arbitrary units; R is distance between donor and acceptor fluorophores; ND is not determined. Data are mean ± S.E. of four independent experiments for PLB-PLB FRET and three independent experiments for SERCA-PLB FRET.
| PLB mutant |
||||
|---|---|---|---|---|
| WT | R9C | SSS | R9C + SSS | |
| PLB-PLB FRET | ||||
| Average FRET (%) | 48.3 ± 0.4 | 54.0 ± 1.1a | 45.8 ± 1.2 | 45. 9 ± 0.3 |
| FRETmax (%) | 54.4 ± 0.9 | 58.4 ± 0.7a | 61.3 ± 1.1 | 55.7 ± 0.7b |
| KD1 (AU) | 0.27 ± 0.04 | 0.12 ± 0.01a | 0.69 ± 0.05 | 0.49 ± 0.03b |
| R (Å) | 56.8 ± 0.1 | 55.9 ± 0.2a | ND | ND |
| SERCA-PLB FRET | ||||
| Average FRET (%) | 14.2 ± 0.7 | 11.6 ± 0.5a | 20.8 ± 0.3 | 17.5 ± 0.5b |
| 27.2 ± 0.7 | 25.2 ± 0.8a | 45.1 ± 1.5 | 45.5 ± 1.5 | |
| KD2 (AU) | 11.4 ± 0.7 | 14.7 ± 0.9a | 17.0 ± 1.2 | 22.6 ± 1.4b |
| R (Å) | 60.8 ± 0.4 | 62.0 ± 0.1a | ND | ND |
a p < 0.05 versus WT.
b p < 0.05 versus SSS.
IonOptix Data Acquisition and Analysis
Adult rabbit left ventricular cardiomyocytes expressing YFP-WT-PLB or YFP-R9C-PLB were loaded with 10 μm Indo-1 AM Ca2+ dye (Invitrogen) for 20 min at room temperature and washed with fresh Tyrode's solution (135 mm NaCl, 4 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm d-Glc, 10 mm HEPES, pH 7.4). Following this, cardiomyocytes were electrically stimulated with a 20 V, 6-ms pulse duration, at increasing pacing frequencies of 0.3, 0.5, and 0.75 Hz. These values were found to produce reliable entrainment without induction of Ca2+ waves in the cultured myocytes. Ca2+ transients and sarcomere shortening were recorded (IonOptix, Milton, MA) before and after 10 min of incubation with 100 nm isoproterenol (iso). Ca2+ transient recordings were obtained by measuring fluorescence intensity at excitation and emission wavelengths of 340 and 405/485 nm, respectively, and analyzed using IonOptix software. 8–10 transients were averaged for display of data.
SDS-PAGE and Western Blot Analysis
Total cell lysates were obtained by washing AAV-293 cells expressing Cer-tagged PLB-WT and mutant constructs with phosphate-buffered saline (PBS; pH 7.4), and treating with Hunter's buffer on ice (25 mm HEPES, pH 7.4, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 10% glycerol, and complete protease inhibitor mixture (Santa Cruz Biotechnology)). The lysates were sonicated and centrifuged at 14,000 rpm at 4 °C for 30 min, and the supernatants were boiled in Laemmli buffer containing β-mercaptoethanol prior to SDS-PAGE and Western blotting. The proteins were transferred onto the PVDF membranes, which were blocked for 1 h with 5% milk in TBS, 0.05% Tween 20 (TBST). The membrane was washed with TBST and incubated with anti-PLB mouse monoclonal primary antibody 2D12 (Abcam) at a dilution of 1:2000 at 4 °C overnight. After washing with TBST, the membrane was incubated with fluorescent secondary antibody, Alexa Fluor 532 goat anti-mouse IgG (Invitrogen) at a dilution of 1:10,000 at room temperature for 1 h. The membrane was then scanned using Typhoon Trio with the following settings of acquisition mode: fluorescence; emission filter, 555 BP 20 R6G, HEX, AF532; laser, Green-532; photomultiplier tube, 425; pixel size, 100 μm. The same samples were subjected to Western blot analysis using anti-GFP rabbit polyclonal antibody (Abcam) to detect Cer-PLB and anti-β-actin rabbit polyclonal antibody (Abcam) to act as a loading control.
Immunofluorescence Microscopy
After a 48-h period of adenoviral infection, cardiomyocytes in culture expressing CFP-R9C-PLB were fixed in 4% paraformaldehyde solution for 15 min, washed with PBS, and permeabilized using 0.2% Triton X-100 for 10 min at room temperature. The cells were blocked with 1% bovine serum albumin for 30 min and then incubated with the anti-PLB mouse monoclonal 2D12 antibody at 1:500 dilution overnight. After rinsing with PBS, cells were incubated with Alexa Fluor 532 goat anti-mouse antibody at 1:1000 dilution for 1 h at room temperature. The coverslips were washed with PBS and mounted on the microscopic slides using mounting media (Vector Laboratories). After immunofluorescent staining, cells were subjected to confocal imaging using an inverted Leica TCS SP5 confocal microscope with ×63 water immersion objective. CFP and Alexa Fluor 532 were sequentially excited at 458 and 543 nm to detect the localization of exogenous CFP-R9C-PLB and endogenous WT-PLB, respectively.
Statistical Analysis
Errors are reported as mean ± S.E., and statistical significance was evaluated using Student's t test, where p < 0.05 was considered significant.
RESULTS
R9C-PLB Exerts a Positive Inotropic and Positive Lusitropic Effect in Cardiomyocytes
Adenoviral delivery of R9C-PLB tagged with CFP or YFP to enzymatically isolated adult rabbit cardiac myocytes yielded fluorescence detectable by confocal or wide-field fluorescence microscopy after 48 h in culture. We observed PLB localization in the perinuclear region and in longitudinal streaks and cross-striations (Fig. 1C) as observed previously for fluorescently labeled wild-type (WT)-PLB (32). The fluorescence pattern is consistent with localization in the sarcoplasmic reticulum and is similar to that previously observed for fluorescently labeled SERCA2a (32, 37). R9C mutation is within the epitope region for the PLB-2D12 antibody (between amino acid residues 9 and 17 of canine PLB), and we observed that this mutation abolishes 2D12 reactivity (Fig. 1B). This permitted comparison of localization of exogenous R9C-PLB with endogenous WT-PLB by immunofluorescence microscopy. We observed that the localization of endogenous WT-PLB labeled with Alexa Fluor 532 secondary antibody was similar to CFP-R9C-PLB (Fig. 1C). Moreover, immunofluorescence data indicate that exogenous PLB does not fully replace endogenous PLB. Thus, the present experimental system may be considered a model of mixed expression, R9C-PLB against a WT-PLB background. R9C-PLB and WT-PLB signals were not completely co-localized, as evidenced from subcellular regions with relatively more CFP or Alexa Fluor 532 signal (Fig. 1C). This is likely due to nonuniform decoration of the myocytes by the primary or secondary antibodies used to visualize endogenous WT-PLB.
Chronic R9C-PLB expression in transgenic mouse models results in depressed Ca2+ handling and decreased myocyte contractility (8, 9). To determine the physiological effect of acute R9C-PLB expression after adenoviral delivery, we compared Indo-1 Ca2+ transients and sarcomere shortening kinetics of myocytes expressing YFP-R9C-PLB or YFP-WT-PLB at increasing pacing frequencies of 0.3, 0.5, and 0.75 Hz (Fig. 1, D and E). R9C-PLB-expressing myocytes showed markedly accelerated Ca2+ handling as evidenced from an elevated peak Ca2+ and decreased Ca2+ transient duration compared with WT-PLB-expressing myocytes (Fig. 1D). There was a corresponding increase in contractility for R9C-PLB-expressing myocytes as evidenced from an increased peak amplitude and decreased peak duration compared with WT-PLB-expressing myocytes (Fig. 1E). The baseline for Ca2+ transients and sarcomere length was not significantly different between WT-PLB- and R9C-PLB-expressing cells. We noted that the resting sarcomere length of the cultured myocytes was shorter than the 1.7–1.8-μm value observed for freshly isolated cardiac myocytes as a consequence of 2 days of maintenance in culture. Ca2+ transient parameters and sarcomere shortening data are summarized in Fig. 2 and Tables 2 and 3. R9C-PLB-expressing cells were significantly hyperdynamic compared with those expressing WT-PLB, with a 19% decrease in the Ca2+ transient time to 50% baseline (Fig. 2A) and a 20% decrease in the peak duration of the Ca2+ transient (Fig. 2C). The corresponding lusitropic effect was evidenced by faster sarcomere relengthening for R9C-PLB-expressing myocytes, with a 23% decrease in the sarcomere shortening time to 50% relaxation (Fig. 2B) and a 19% decrease in the peak duration of the sarcomere shortening transient compared with WT-PLB-expressing myocytes (Fig. 2D).
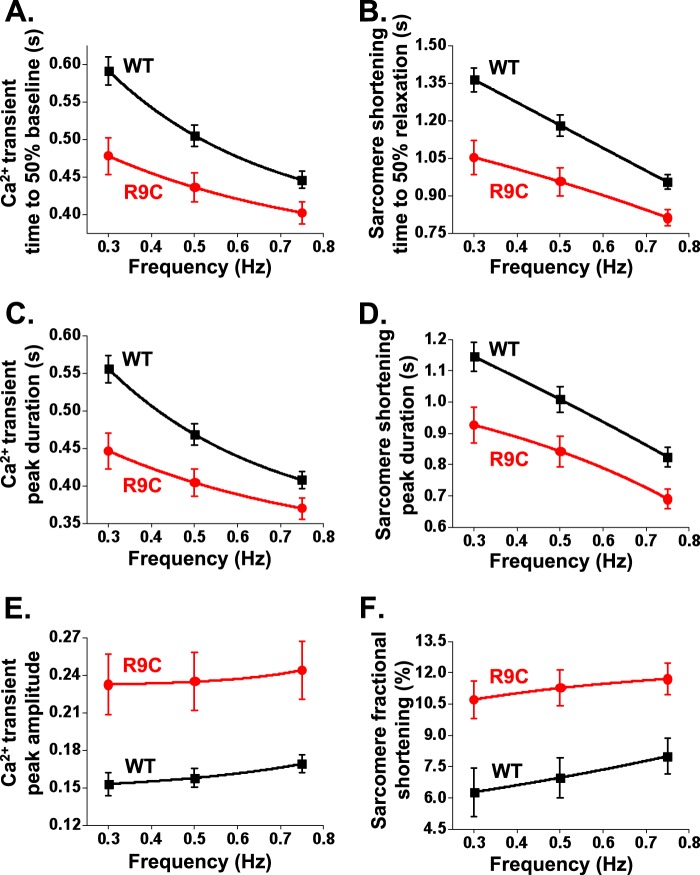
FIGURE 2.
Quantification of inotropic and lusitropic effects of R9C-PLB. A–F, kinetics of intracellular Ca2+ transients (A, C, and E) and sarcomere shortening (B, D, and F) of isolated cardiomyocytes expressing YFP-WT-PLB or YFP-R9C-PLB. A–D, WT-expressing cells responded strongly to increased pacing frequency, whereas R9C-expressing cells exhibited a blunted sensitivity to frequency potentiation. We observed a significant difference between R9C and WT for all parameters shown in this figure at each pacing frequency, p < 0.05 (Tables 2 and 3). Values are mean ± S.E.
TABLE 2.
Summary of quantitative Ca2+ transients data
Effect of frequency potentiation on the Ca2+ handling parameters in R9C-PLB-expressing cardiomyocytes compared with WT-PLB-expressing cardiomyocytes. Data are mean ± S.E.; WT (n = 17), R9C (n = 16). AU = arbitrary units.
| Ca2+ transients parameters | 0.3 Hz |
0.5 Hz |
0.75 Hz |
||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | R9C | p value | WT | R9C | p value | WT | R9C | p value | |
| Baseline (AU) | 0.452 ± 0.007 | 0.45 ± 0.006 | 0.795 | 0.458 ± 0.007 | 0.456 ± 0.006 | 0.842 | 0.465 ± 0.008 | 0.465 ± 0.007 | 0.986 |
| Departure velocity (AU/s) | 6.336 ± 0.444 | 9.682 ± 1.011 | 0.004 | 6.745 ± 0.541 | 10.39 ± 1.07 | 0.004 | 6.106 ± 0.566 | 10.06 ± 1.261 | 0.006 |
| Departure velocity time (s) | 0.024 ± 0.001 | 0.021 ± 9E-04 | 0.037 | 0.024 ± 0.002 | 0.02 ± 8E-04 | 0.021 | 0.024 ± 0.001 | 0.02 ± 7E-04 | 0.014 |
| Peak (AU) | 0.606 ± 0.015 | 0.683 ± 0.026 | 0.014 | 0.616 ± 0.014 | 0.691 ± 0.026 | 0.014 | 0.634 ± 0.014 | 0.709 ± 0.027 | 0.017 |
| Peak amplitude (AU) | 0.153 ± 0.009 | 0.233 ± 0.024 | 0.003 | 0.158 ± 0.007 | 0.235 ± 0.023 | 0.003 | 0.169 ± 0.007 | 0.244 ± 0.023 | 0.003 |
| Fractional change (%) | 33.62 ± 1.616 | 51.62 ± 5.089 | 0.002 | 34.3 ± 1.21 | 51.27 ± 4.763 | 0.001 | 36.21 ± 1.138 | 52.19 ± 4.55 | 0.001 |
| Time to peak (s) | 0.152 ± 0.009 | 0.136 ± 0.006 | 0.144 | 0.149 ± 0.008 | 0.133 ± 0.007 | 0.134 | 0.145 ± 0.006 | 0.131 ± 0.007 | 0.115 |
| Return velocity (AU/s) | −0.251 ± 0.015 | −0.57 ± 0.101 | 0.003 | −0.29 ± 0.017 | −0.63 ± 0.101 | 0.001 | −0.38 ± 0.017 | −0.72 ± 0.108 | 0.003 |
| Return velocity time (s) | 0.332 ± 0.033 | 0.325 ± 0.025 | 0.882 | 0.355 ± 0.019 | 0.331 ± 0.021 | 0.382 | 0.333 ± 0.013 | 0.315 ± 0.017 | 0.398 |
| Time to 10.0% peak (s) | 0.115 ± 0.02 | 0.113 ± 0.014 | 0.925 | 0.119 ± 0.016 | 0.117 ± 0.014 | 0.947 | 0.099 ± 0.016 | 0.094 ± 0.015 | 0.821 |
| Time to 50.0% peak (s) | 0.035 ± 0.002 | 0.031 ± 0.001 | 0.05 | 0.037 ± 0.002 | 0.032 ± 0.001 | 0.025 | 0.038 ± 0.002 | 0.032 ± 0.001 | 0.014 |
| Time to 90.0% peak (s) | 0.072 ± 0.004 | 0.064 ± 0.003 | 0.13 | 0.074 ± 0.003 | 0.067 ± 0.003 | 0.139 | 0.076 ± 0.003 | 0.066 ± 0.003 | 0.045 |
| Time to 10.0% baseline (s) | 0.273 ± 0.012 | 0.247 ± 0.013 | 0.135 | 0.253 ± 0.011 | 0.24 ± 0.012 | 0.439 | 0.246 ± 0.009 | 0.233 ± 0.01 | 0.338 |
| Time to 50.0% baseline (s) | 0.591 ± 0.018 | 0.478 ± 0.024 | 0.001 | 0.505 ± 0.015 | 0.436 ± 0.019 | 0.007 | 0.446 ± 0.011 | 0.402 ± 0.015 | 0.025 |
| Time to 90.0% baseline (s) | 1.388 ± 0.045 | 1.141 ± 0.099 | 0.028 | 1.093 ± 0.037 | 0.936 ± 0.057 | 0.026 | 0.861 ± 0.018 | 0.765 ± 0.031 | 0.013 |
| Transient decay time constant (s) | 0.613 ± 0.032 | 0.443 ± 0.037 | 0.001 | 0.492 ± 0.023 | 0.386 ± 0.028 | 0.006 | 0.434 ± 0.02 | 0.348 ± 0.022 | 0.006 |
| Peak duration (s) | 0.556 ± 0.018 | 0.447 ± 0.024 | 0.001 | 0.468 ± 0.014 | 0.404 ± 0.018 | 0.01 | 0.408 ± 0.011 | 0.37 ± 0.014 | 0.041 |
TABLE 3.
Summary of quantitative sarcomere shortening data
Effect of frequency potentiation on the sarcomere shortening parameters in R9C-PLB-expressing cardiomyocytes compared with WT-PLB-expressing cardiomyocytes. Data are mean ± S.E.; WT (n = 9), R9C (n = 7).
| Sarcomere shortening parameters | 0.3 Hz |
0.5 Hz |
0.75 Hz |
||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | R9C | p value | WT | R9C | p value | WT | R9C | p value | |
| Baseline (μm) | 1.62 ± 0.03 | 1.546 ± 0.03 | 0.11 | 1.615 ± 0.031 | 1.53 ± 0.028 | 0.068 | 1.594 ± 0.031 | 1.496 ± 0.026 | 0.033 |
| Contraction velocity (μm/s) | −0.37 ± 0.1 | −1.12 ± 0.205 | 0.003 | −0.52 ± 0.095 | −1.22 ± 0.178 | 0.002 | −0.77 ± 0.129 | −1.12 ± 0.13 | 0.084 |
| Contraction velocity time (s) | 0.153 ± 0.026 | 0.093 ± 0.012 | 0.085 | 0.121 ± 0.009 | 0.075 ± 0.006 | 0.002 | 0.089 ± 0.006 | 0.077 ± 0.007 | 0.232 |
| Peak (μm) | 1.517 ± 0.028 | 1.381 ± 0.033 | 0.007 | 1.501 ± 0.026 | 1.358 ± 0.031 | 0.003 | 1.465 ± 0.021 | 1.321 ± 0.03 | 0.001 |
| Peak amplitude (μm) | 0.103 ± 0.021 | 0.165 ± 0.013 | 0.032 | 0.114 ± 0.017 | 0.172 ± 0.012 | 0.02 | 0.129 ± 0.016 | 0.174 ± 0.01 | 0.041 |
| Fractional shortening (%) | 6.282 ± 1.174 | 10.72 ± 0.905 | 0.013 | 6.974 ± 0.95 | 11.27 ± 0.855 | 0.006 | 8.003 ± 0.867 | 11.71 ± 0.755 | 0.008 |
| Time to peak (s) | 0.787 ± 0.04 | 0.589 ± 0.043 | 0.005 | 0.703 ± 0.042 | 0.53 ± 0.031 | 0.007 | 0.559 ± 0.028 | 0.481 ± 0.021 | 0.054 |
| Relaxation velocity (μm/s) | 0.199 ± 0.045 | 0.449 ± 0.077 | 0.011 | 0.257 ± 0.048 | 0.491 ± 0.078 | 0.017 | 0.456 ± 0.122 | 0.667 ± 0.092 | 0.212 |
| Relaxation velocity time (s) | 1.329 ± 0.055 | 1.022 ± 0.065 | 0.003 | 1.149 ± 0.038 | 0.922 ± 0.047 | 0.002 | 1.015 ± 0.054 | 0.801 ± 0.032 | 0.007 |
| Time to 10.0% contraction (s) | 0.077 ± 0.007 | 0.237 ± 0.123 | 0.16 | 0.146 ± 0.079 | 0.179 ± 0.088 | 0.784 | 0.054 ± 0.006 | 0.043 ± 0.004 | 0.167 |
| Time to 50.0% contraction (s) | 0.219 ± 0.017 | 0.127 ± 0.013 | 0.001 | 0.173 ± 0.008 | 0.113 ± 0.009 | 0.0002 | 0.13 ± 0.008 | 0.106 ± 0.004 | 0.037 |
| Time to 90.0% contraction (s) | 0.488 ± 0.026 | 0.305 ± 0.03 | 0.0004 | 0.397 ± 0.021 | 0.277 ± 0.018 | 0.001 | 0.32 ± 0.014 | 0.272 ± 0.017 | 0.046 |
| Time to 10.0% relaxation (s) | 1.085 ± 0.057 | 0.847 ± 0.045 | 0.007 | 0.929 ± 0.047 | 0.769 ± 0.038 | 0.023 | 0.769 ± 0.045 | 0.642 ± 0.028 | 0.044 |
| Time to 50.0% relaxation (s) | 1.364 ± 0.048 | 1.053 ± 0.067 | 0.002 | 1.181 ± 0.042 | 0.956 ± 0.055 | 0.005 | 0.954 ± 0.028 | 0.813 ± 0.032 | 0.005 |
| Time to 90.0% relaxation (s) | 1.635 ± 0.054 | 1.355 ± 0.117 | 0.086 | 1.497 ± 0.06 | 1.217 ± 0.086 | 0.016 | 1.16 ± 0.028 | 1.016 ± 0.051 | 0.02 |
| Relaxation time constant (s) | 0.78 ± 0.075 | 0.778 ± 0.079 | 0.991 | 0.881 ± 0.058 | 0.642 ± 0.059 | 0.013 | 0.619 ± 0.064 | 0.55 ± 0.067 | 0.474 |
| Peak duration (s) | 1.145 ± 0.046 | 0.926 ± 0.057 | 0.009 | 1.008 ± 0.041 | 0.843 ± 0.049 | 0.021 | 0.825 ± 0.032 | 0.691 ± 0.031 | 0.013 |
R9C-PLB Exhibits Blunted Sensitivity to Frequency Potentiation and β-Adrenergic Stimulation
Compared with WT-PLB-expressing cells, R9C-PLB-expressing cells did not show as large an increase in Ca2+ reuptake or contractility as pacing frequency increased from 0.3 to 0.75 Hz (Fig. 1, D and E, Fig. 2). The blunted frequency response is apparent in Fig. 2, A and D, as a decrease in the slope of the R9C-PLB frequency response compared with WT-PLB. Residual sensitivity to frequency potentiation may be due to the mixed expression of R9C and endogenous WT-PLB. The data suggest that R9C mutation of PLB stimulates Ca2+ uptake and cell relaxation, and additional stimulation by increased pacing frequency provides only a marginal additive effect. We also observed a 52% increase in the amplitude of the peak of the Ca2+ transient in R9C-PLB-expressing cells compared with WT-PLB (Fig. 2E), suggesting an increase in myocyte sarcoplasmic reticulum Ca2+ load. This increase in Ca2+ release resulted in positive inotropy, with a 71% increase in myocyte fractional shortening (Fig. 2F). Most of the Ca2+ handling and sarcomere shortening parameters quantified here and in Tables 2 and 3 showed the same pattern of a blunted frequency response, suggesting that R9C-PLB-expressing cells were already maximally stimulated, with little additional capacity for frequency-dependent lusitropy or inotropy.
Similarly, we observed decreased responsiveness of R9C-PLB-expressing cells to β-adrenergic stimulation with iso. Although WT-PLB-expressing cells showed a robust increase in peak Ca2+ and a faster Ca2+ transient decay in response to iso (Fig. 3A), R9C-PLB-expressing cells were already hyperdynamic, and iso caused no additional increase in Ca2+ handling kinetics (Fig. 3B). Instead, we observed a modest decrease in peak Ca2+, possibly as a result of troponin I phosphorylation or increased Na+-K+-ATPase activity. Baseline Ca2+ levels were not significantly different for WT or R9C after iso treatment. Fig. 3, C and D, shows the corresponding effects of iso stimulation on sarcomere shortening for WT-PLB- and R9C-PLB-expressing cells, respectively. Overall, the acute physiological effect of the R9C mutation of PLB is positively inotropic and lusitropic, consistent with a model of disinhibition of SERCA as a result of a loss of inhibitory function for R9C-PLB (8, 9, 20–22).
FIGURE 3.
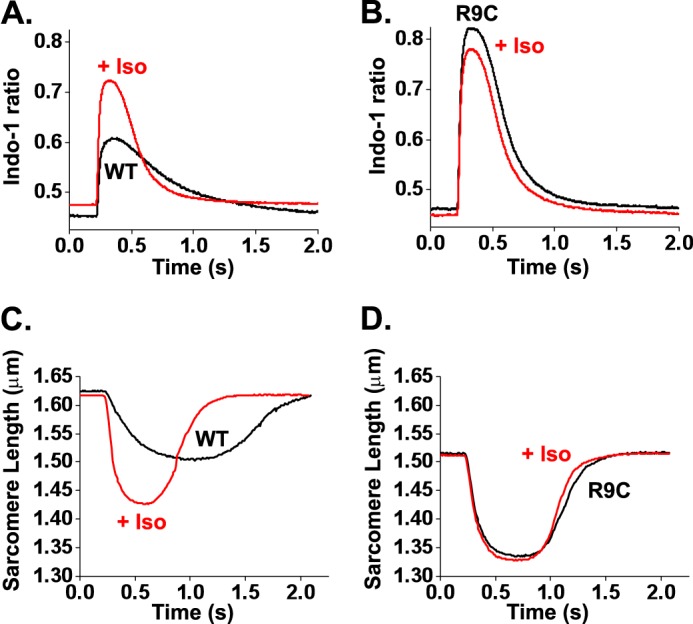
R9C-PLB exhibits lack of responsiveness to β-adrenergic stimulation. A and B, averaged Ca2+ transients (obtained from 8 to 10 events per cell) recorded from isolated cardiomyocytes expressing YFP-WT-PLB (A) or YFP-R9C-PLB (B) in the presence (WT (n = 13), R9C (n = 7)) and absence (WT (n = 5), R9C (n = 5)) of 100 nm iso. C and D, averaged sarcomere shortening traces (obtained from 8 to 10 events per cell) recorded from isolated cardiomyocytes expressing YFP-WT-PLB (C) or YFP-R9C-PLB (D) in the presence (WT (n = 9), R9C (n = 4)) and absence (WT (n = 5), R9C (n = 4)) of 100 nm iso.
R9C-PLB Increased Oligomerization and Decreased SERCA Binding Despite Pentamer Destabilization by SSS Mutation
We have previously attributed the loss-of-function character of R9C to increased PLB oligomerization secondary to oxidative cross-linking of the introduced cysteine in adjacent protomers of PLB pentamers (20). To investigate the change in PLB oligomerization energetics in more detail, we compared the relative effects of R9C mutation with substitution of transmembrane Cys residues 36, 41, and 46 with Ser (SSS) (Fig. 1A). The SSS mutation is known to destabilize the PLB pentamer (45), and this mutant runs as a monomer in PAGE (21). We performed E-FRET (39) measurements of large populations of AAV-293 cells (500–1500 cells per experiment) as described previously (32, 38). We observed a 12% increase in the average intrapentameric FRET for R9C-PLB compared with WT (Table 1), consistent with previous observations (20). SSS-PLB also exhibited FRET, suggesting that despite running as a monomer on SDS-PAGE (21), the SSS mutant can form pentamers in the membrane environment. However, SSS average FRET was 5% less than WT, suggesting that oligomerization was weakened by the mutations of the transmembrane domain (Table 1). To quantify PLB-PLB binding affinity, we used an in-cell binding assay described previously (20, 26, 31, 32, 34, 38). The heterogeneous protein expression level of the transiently transfected population of cells provided insight into the dependence of FRET on protein concentration. FRET increased with protein expression up to a maximal level (Fig. 4A) that reflected the intrinsic FRET of the pentamer (FRETmax). The concentration of protein that yielded half-maximal FRET is a measure of the apparent dissociation constant (KD1) of the PLB pentamer. Thus, the binding curve reveals the relative contributions of oligomerization and protein structural changes to the observed increase in intrapentameric FRET with mutations. As observed previously, R9C increased FRETmax, suggesting that the R9C-PLB pentamer had a more compact conformation (Fig. 4A and Table 1). This observation is consistent with a model in which disulfide cross-linking of introduced Cys residues on adjacent protomers brings the cytoplasmic domains and Cer/YFP fusion tags into closer proximity. The R9C mutation also increased the affinity of PLB oligomerization, as shown by a left shift of the binding curve of R9C (Fig. 4A), accounting for 55% decrease in KD1 (Fig. 4B) compared with WT. In contrast, destabilization of the PLB pentamer by SSS right-shifted the binding curve compared with WT (Fig. 4A). Notably, addition of the R9C mutation increased the stability of the SSS-PLB pentamers, as shown by a left shift of the R9C + SSS-PLB binding curve (Fig. 4A), and a 29% decrease in KD1 (Fig. 4B) relative to SSS. The data are summarized in Fig. 4B and Table 1. We conclude that R9C enhances PLB oligomerization both in WT and SSS background, indicating that R9C potentiates PLB oligomerization even for weakly oligomeric variants.
FIGURE 4.
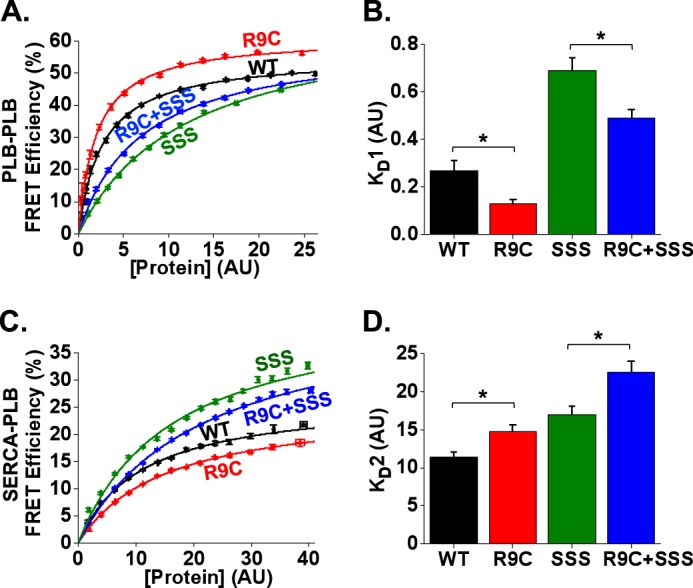
R9C-PLB causes increased oligomerization and decreased SERCA binding both in WT and SSS backgrounds. A, in-cell intrapentameric FRET efficiency measurements for WT and mutant constructs of PLB. B, R9C exhibited a decrease in oligomer dissociation constant (KD1) compared with WT, and R9C + SSS showed a decrease in KD1 compared with SSS. Data are mean ± S.E. of four independent experiments; *, p < 0.05, AU, arbitrary units. C, in-cell SERCA:PLB FRET efficiency measurements for WT and mutant constructs of PLB. D, R9C exhibited an increase in the dissociation constant of the SERCA-PLB complex (KD2) compared with WT, and R9C + SSS exhibited an increase in KD2 compared with SSS. Data are mean ± S.E. of three independent experiments; *, p < 0.05, AU, arbitrary units.
Increased oligomerization of R9C-PLB had the predicted consequence for binding of R9C-PLB to SERCA. Because SERCA is regulated by monomeric PLB (3, 4), depletion of the monomer pool by increased oligomerization was expected to decrease PLB-SERCA binding. Indeed, we observed a right shift of the R9C-PLB-SERCA binding curve (Fig. 4C) and a 29% increase in the apparent dissociation constant KD2 (Fig. 4D) compared with WT. The data are summarized in Fig. 4D and Table 1. The effect of the SSS mutation was less clear. We expected increased binding of SSS to SERCA versus WT, but because the binding curve did not saturate, we could not accurately quantify FRETmax and KD2 for this mutant. The failure to saturate may be due to increased nonspecific FRET for the more monomeric PLB species (46). Although the absolute value of KD2 is not certain, we did observe the expected relative change with the addition of R9C. Specifically, the combined mutant R9C-SSS showed a right-shifted binding curve relative to SSS (Fig. 4C), with an increase in KD2 relative to SSS (Fig. 4D). We also observed a decrease in FRETmax for R9C relative to WT (Fig. 4C and Table 1). This parameter represents the intrinsic FRET efficiency of the bound PLB-SERCA regulatory complex. A decrease in FRETmax suggests a change in the conformation of the regulatory complex that moves the FRET acceptor (YFP) farther from the donor (Cer). The functional significance of this structural change is not clear, but it is reminiscent of the decrease in PLB-SERCA FRETmax observed with phosphomimetic mutations of PLB (26). Overall, we conclude that increased PLB oligomerization reduced SERCA regulation, accounting for the observed hyperdynamic Ca2+ handling of the R9C-PLB-expressing cardiac myocytes.
R9C-PLB Exhibits Increased Sensitivity to Oxidative Stress
To determine the role of Cys oxidation in the observed effect of R9C substitution on PLB oligomerization, cardiac myocytes were treated with 100 μm H2O2 (47) during observation by fluorescence imaging. Wide-field fluorescence microscopy did not reveal a change in the localization of PLB (data not shown), and we did not observe any aggregation of protein after treatment with H2O2. However, oxidation significantly increased in the relative emission of YFP/CFP (with CFP excitation). The observed 14% increase in FRET ratio is evident in Fig. 5A (+ H2O2) and supplemental video 1 as a transition to warmer colors, and it is quantified in Fig. 5B. There was no change in the FRET ratio for cardiac myocytes expressing WT CFP/YFP-PLB (Fig. 5B), consistent with previous observations in AAV-293 cells (20). To differentiate the effect of oxidation of cytoplasmic domain Cys-9 from the transmembrane cysteines, we measured PLB-PLB FRET after mutating the three transmembrane cysteines to serine residues (SSS) (Fig. 1A). A quantitative comparison of average FRET efficiency of these mutants in AAV-293 cells showed that the R9C-PLB is already increasingly oligomeric before the addition of H2O2 (29% FRET versus 26% for WT), and oxidation further increased FRET to a maximum of 35% (Fig. 5C, red). WT-PLB intrapentameric FRET does not increase with oxidation (Fig. 5C, black). SSS (Fig. 5C, green) is likewise unresponsive to H2O2, and average FRET is reduced compared with WT consistent with destabilization of SSS pentamers. Interestingly, the combination of R9C + SSS (Fig. 5C, blue) yields an intermediate level of FRET that is markedly increased by H2O2 oxidation, up to the same maximal 35% FRET efficiency observed for R9C-PLB. The data, summarized in Fig. 5D, suggest that oxidation of R9C on the native transmembrane domain or SSS background results in maximal oligomerization of PLB regardless of the initial level of oligomerization. We conclude that the oxidation of Cys-9 thiol (not the transmembrane cysteines) is the primary cause of increase in R9C-PLB oligomerization.
FIGURE 5.
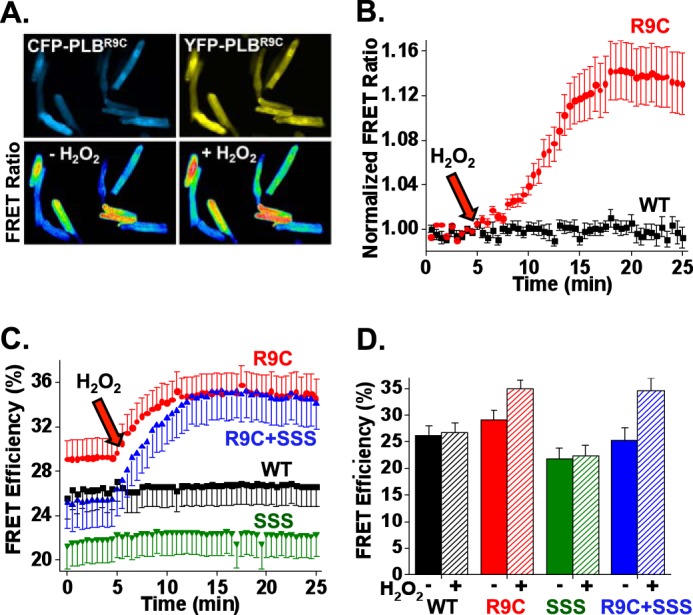
R9C-PLB exhibits increased sensitivity to oxidative stress. A, fluorescence microscopy images of live adult cardiomyocytes co-expressing CFP-R9C-PLB and YFP-R9C-PLB. Application of a 100 μm H2O2 increased FRET, as reflected by an increase in measured FRET ratio. B, quantification of A. R9C-PLB expressed in cardiac myocytes showed a time-dependent increase in intrapentameric FRET after application of 100 μm H2O2, whereas the FRET ratio for WT was unaffected; WT (n = 8), R9C (n = 8). C, quantitative FRET for PLB expressed in HEK cells. 100 μm H2O2 increased FRET efficiency for both R9C and R9C + SSS, indicating that Cys-9 is the primary cause of oxidation-dependent R9C-PLB oligomerization. Arrows indicate the time of addition of 100 μm H2O2. Data are mean ± S.E. of three independent experiments; WT (n = 17), R9C (n = 15), R9C + SSS (n = 11), SSS (n = 12). D, summary of C, the FRET efficiency before (solid) and after (striped) addition of H2O2.
DISCUSSION
Chronic Versus Acute Effects of R9C Mutation of PLB
Previous studies of R9C-PLB showed that R9C-PLB is loss-of-function with respect to SERCA inhibition (8, 9, 20–22). Such in vitro observations lead to the expectation of enhanced SERCA activity and positive inotropy/lusitropy in vivo, but this is not observed in transgenic models or in human patients. Specifically, R9C-PLB transgenic mouse myocytes show slower Ca2+ handling kinetics (8, 9) compared with WT. We reasoned that chronic exposure to R9C-PLB may elicit compensatory changes, and the long term disease evolution may not reveal the fundamental mechanistic defect. To investigate the acute effect of R9C mutation of PLB, we introduced the mutated protein to rabbit cardiac myocytes using an adenoviral vector delivery. On a time scale of hours to days, the physiological effect of R9C-PLB is as predicted from its loss-of-function character, with YFP-labeled R9C-PLB-expressing cardiac myocytes showing positive inotropy and lusitropy compared with YFP-WT-PLB-expressing myocytes (Figs. 1 and 2). Thus, the impaired hemodynamics of the mutant mouse may be long term consequences of secondary changes such as decreased SERCA2a mRNA and protein expression, progressive intracellular stress responses, cardiac remodeling, apoptotic signaling (48), or other changes in gene expression or cell/organ structure that evolve over weeks or months. We hypothesize that the fundamental pathological triggers of this disease progression is blunted sensitivity to frequency potentiation and β-adrenergic stimulation, as observed in the present acute physiological measurements (Fig. 3). These two mechanisms are the main physiological means of regulation of cardiac performance, and the lack of a functional response to stress is expected to induce cardiac remodeling and related decompensation processes. We conclude that transgenesis provides insight into the progression of R9C-PLB disease (and heart failure more generally), while acute delivery of mutant PLB provides insight into the fundamental Ca2+-handling defects that initiate this disease process.
Disordered Structure/Function Mechanisms of R9C-PLB
The molecular basis of R9C-PLB pathology is of great interest, and several possible mechanisms have been proposed. There is an emerging consensus that R9C-PLB is a loss-of-function with respect to inhibition of SERCA (8, 9, 20–22), resulting in dysregulation of calcium cycling (8, 9, 48). There is also evidence for impaired phosphorylation of R9C-PLB by PKA (8, 9, 20, 21). However, a cytoplasmic domain fragment of R9C-PLB was phosphorylated by PKA with normal kinetics (20), suggesting that there is not an intrinsic defect of PKA recognition of the R9C-PLB substrate. Schmitt et al. (8) observed increased co-immunoprecipitation of R9C-PLB with PKA, suggesting the mutated PLB physically traps and inactivates the kinase, leading to impaired signaling and heart failure. Although other studies have failed to detect PKA cross-linking to R9C-PLB (20), it is possible that PKA precipitation occurs secondary to other aggregation events, as discussed below. Other possible mechanisms for R9C-PLB pathology include decreased open probability of a putative PLB channel (49) or disruption of hydrophobic balance of the PLB cytoplasmic domain leading to loss of inhibitory function (22). In addition, Gramolini et al. (48) reported that the impaired Ca2+ handling by the R9C mouse may be a long term consequence of secondary changes such as decreased SERCA2a mRNA and protein expression or activation of the endoplasmic reticulum stress response, cytoskeletal remodeling, and apoptosis. In addition to these diverse pathological mechanisms, we have previously proposed that the introduced Cys residue may induce pathological intersubunit cross-linking, stabilizing the R9C-PLB pentamer (20). Evidence for this mechanism includes an increase in intrapentameric FRET for the R9C-PLB mutant (Figs. 4A and 5), which is further enhanced by oxidative stress (Fig. 5). In this regard, R9C-PLB cross-linking may mimic one of the functional effects of PLB phosphorylation, which also enhances PLB oligomerization (5, 26, 27). The expected functional consequence of increased PLB oligomerization is to reduce the availability of the inhibitory monomeric species, decreasing SERCA inhibition. The acute physiological result of decreased SERCA inhibition is apparent in the present experiments as a positive inotropic and lusitropic effect (Figs. 1 and 2). The data are consistent with our previous observations in AAV-293 cells (20), and in this study, we extend that analysis using pentamer-destabilizing mutations of the transmembrane domain (45) to more clearly isolate the effect of R9C in stabilizing PLB pentamers. SSS-PLB has been presumed to be a fully monomeric mutant based on SDS-PAGE mobility (21), but we demonstrate here that SSS-PLB can still form low affinity pentamers (Fig. 4A). Combining SSS and R9C mutations yielded an oxidation-sensitive mutant with a basal level of oligomerization that was intermediate between WT-PLB and SSS-PLB (Fig. 5C). After oxidation with H2O2, SSS-R9C-PLB FRET increased dramatically, achieving the same final value as R9C-PLB with a wild-type transmembrane domain (Fig. 5C). The data suggest that the degree of oligomerization is dominated by cross-linking of the cytoplasmic Cys-9 residue of the R9C-PLB pentamer.
Another aspect of R9C mutation that is reminiscent of PLB phosphorylation is the small decrease in FRETmax for the R9C-PLB-SERCA regulatory complex (Fig. 4C and Table 1), a difference that did not achieve statistical significance in a previous study (20). The change in FRETmax suggests a conformational change that increases the distance between the donor and acceptor fluorescent probes. This is similar to the structural change that results from phosphomimetic mutations of the PKA site (Ser-16) and CaMKII site (Thr-17) in the PLB cytoplasmic domain (26). The R9C-dependent structural transition and the phosphorylation-dependent conformational change are similar in direction and magnitude (+1.2 and +4 Å, respectively). A regulatory complex structural change is the primary mechanism for relief of inhibition of SERCA by PLB phosphorylation (50, 51). How the R9C mutation may imitate phosphorylation of PLB is not clear, but we speculate that there may be modification of the Cys at position 9 to a negatively charged species that resembles phosphorylated residues of neighboring PKA/CaMKII sites on the PLB. Likely modifications include Cys deprotonation to a negatively charged thiolate or hyperoxidation to sulfenic, sulfinic, or sulfonic acid. By this mechanism, the functional consequence of oxidative cross-linking of PLB protomers in the pentamer would be reinforced by loss-of-function oxidative modifications of the remaining monomeric PLB. The data suggest that the fundamental molecular mechanism underlying the R9C pathology is increased sensitivity to oxidative challenge, which occurs periodically in the healthy heart under conditions of physiological stress. Because these oxidative changes are poorly reversible (20), damage from transient oxidative episodes may accumulate over time, leading to chronically impaired SERCA regulation, disordered Ca2+ handling, and eventual heart failure.
Summary
The structural and functional consequences of the R9C-PLB mutation are summarized in Fig. 6. This scheme parallels our previous mechanistic model for the relief of SERCA inhibition by phosphorylation (26). We propose that R9C mutation enhances transitions away from the inhibited SERCA-PLB complex (Fig. 6, high FRET), leaving disinhibited SERCA (Fig. 6, low FRET). R9C mutation altered the PLB pentamer conformation (Fig. 6A), increased oligomerization affinity (Fig. 6B), and altered the structure of regulatory complex (Fig. 6D). We also observed a decrease in binding of PLB to SERCA (Fig. 6C); however, this may be an indirect effect of decreased availability of monomeric PLB, rather than a change in the intrinsic affinity of PLB for SERCA. The functional consequences of these structural changes are also summarized in Fig. 6. Impaired regulation of SERCA results in blunted sensitivity to local regulation (frequency potentiation) and humoral (β-adrenergic) stimulation. This fundamental lack of responsiveness to physiological stress leads to pathological remodeling and heart failure caused by this mutant. The failing heart suffers from prevailing oxidative stress (52), exacerbating the oxidative modification of R9C-PLB that initiated the pathological pathway.
FIGURE 6.
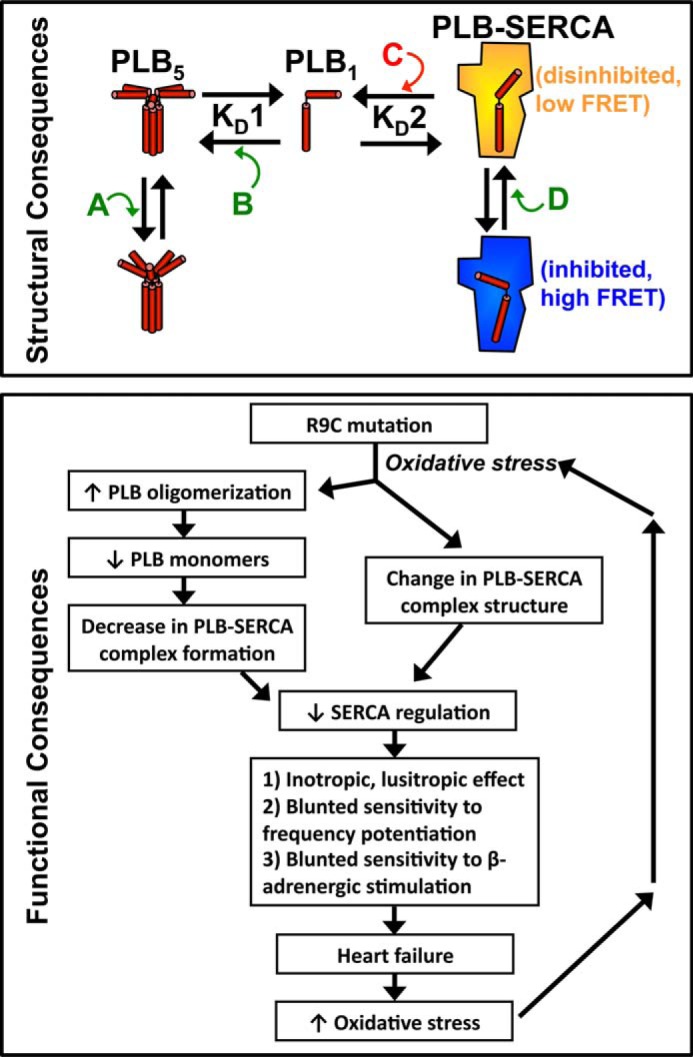
Model of the structural and functional consequences of R9C-PLB. For structural consequences, the R9C mutation alters the PLB pentamer structure (A), oligomerization affinity (B), PLB-SERCA binding (an indirect effect) (C), and the PLB-SERCA regulatory complex structure (D). For functional consequences, we propose that R9C mutation leads to increased PLB oligomerization, decreased PLB-SERCA binding, and alteration of the structure of the regulatory complex. These effects result in decreased SERCA regulation, consistent with the observed increase in myocyte contractility observed in the present acute experiments. The long term inability of R9C-PLB to regulate SERCA exerts a pathological effect by blunting sensitivity to frequency potentiation and β-adrenergic stimulation, with consequent impaired cardiac performance and eventual heart failure. Heart failure exacerbates oxidative stress conditions, further enhancing R9C-PLB oligomerization, thus reinforcing the pathological effects of the R9C mutation.
Acknowledgments
We are grateful for technical assistance from Zhanjia Hou, Daniel Blackwell, Stefan Mazurek, Elisa Bovo, and Younss Ait Mou. We also thank Jody Martin for production of adenoviral constructs and Aleksey Zima for providing the rabbit adult ventricular cardiomyocytes.
This work was supported, in whole or in part, by National Institutes of Health Grants HL-092321 (to S. L. R.) and HL-62426 and HL-75494 (to P. dT.).

This article contains Supplemental Video 1.
- SERCA
- sarco(endo)plasmic reticulum calcium ATPase
- PLB
- phospholamban
- CaMKII
- calcium/calmodulin-dependent protein kinase II
- DCM
- dilated cardiomyopathy
- SSS
- C36S/C41S/C46S
- Cer
- mCerulean
- CFP
- cyan fluorescent protein
- R
- probe separation distance
- iso
- isoproterenol.
REFERENCES
- 1. Lipskaia L., Hulot J. S., Lompré A. M. (2009) Role of sarco/endoplasmic reticulum calcium content and calcium ATPase activity in the control of cell growth and proliferation. Pflugers Arch. 457, 673–685 [DOI] [PubMed] [Google Scholar]
- 2. Periasamy M., Huke S. (2001) SERCA pump level is a critical determinant of Ca2+ homeostasis and cardiac contractility. J. Mol. Cell. Cardiol. 33, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 3. Kranias E. G., Hajjar R. J. (2012) Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ. Res. 110, 1646–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacLennan D. H., Kranias E. G. (2003) Phospholamban: a crucial regulator of cardiac contractility. Nat. Rev. Mol. Cell Biol. 4, 566–577 [DOI] [PubMed] [Google Scholar]
- 5. Simmerman H. K., Jones L. R. (1998) Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiol. Rev. 78, 921–947 [DOI] [PubMed] [Google Scholar]
- 6. Hagemann D., Xiao R. P. (2002) Dual site phospholamban phosphorylation and its physiological relevance in the heart. Trends Cardiovasc. Med. 12, 51–56 [DOI] [PubMed] [Google Scholar]
- 7. Wegener A. D., Simmerman H. K., Lindemann J. P., Jones L. R. (1989) Phospholamban phosphorylation in intact ventricles. Phosphorylation of serine 16 and threonine 17 in response to β-adrenergic stimulation. J. Biol. Chem. 264, 11468–11474 [PubMed] [Google Scholar]
- 8. Schmitt J. P., Kamisago M., Asahi M., Li G. H., Ahmad F., Mende U., Kranias E. G., MacLennan D. H., Seidman J. G., Seidman C. E. (2003) Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 299, 1410–1413 [DOI] [PubMed] [Google Scholar]
- 9. Schmitt J. P., Ahmad F., Lorenz K., Hein L., Schulz S., Asahi M., Maclennan D. H., Seidman C. E., Seidman J. G., Lohse M. J. (2009) Alterations of phospholamban function can exhibit cardiotoxic effects independent of excessive sarcoplasmic reticulum Ca2+-ATPase inhibition. Circulation 119, 436–444 [DOI] [PubMed] [Google Scholar]
- 10. Haghighi K., Kolokathis F., Pater L., Lynch R. A., Asahi M., Gramolini A. O., Fan G. C., Tsiapras D., Hahn H. S., Adamopoulos S., Liggett S. B., Dorn G. W., 2nd, MacLennan D. H., Kremastinos D. T., Kranias E. G. (2003) Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J. Clin. Invest. 111, 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Medeiros A., Biagi D. G., Sobreira T. J., de Oliveira P. S., Negrão C. E., Mansur A. J., Krieger J. E., Brum P. C., Pereira A. C. (2011) Mutations in the human phospholamban gene in patients with heart failure. Am. Heart J. 162, 1088–1095 [DOI] [PubMed] [Google Scholar]
- 12. Landstrom A. P., Adekola B. A., Bos J. M., Ommen S. R., Ackerman M. J. (2011) PLN-encoded phospholamban mutation in a large cohort of hypertrophic cardiomyopathy cases: summary of the literature and implications for genetic testing. Am. Heart J. 161, 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haghighi K., Kolokathis F., Gramolini A. O., Waggoner J. R., Pater L., Lynch R. A., Fan G. C., Tsiapras D., Parekh R. R., Dorn G. W., 2nd, MacLennan D. H., Kremastinos D. T., Kranias E. G. (2006) A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 103, 1388–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeWitt M. M., MacLeod H. M., Soliven B., McNally E. M. (2006) Phospholamban R14 deletion results in late-onset, mild, hereditary dilated cardiomyopathy. J. Am. Coll. Cardiol. 48, 1396–1398 [DOI] [PubMed] [Google Scholar]
- 15. Posch M. G., Perrot A., Geier C., Boldt L. H., Schmidt G., Lehmkuhl H. B., Hetzer R., Dietz R., Gutberlet M., Haverkamp W., Ozcelik C. (2009) Genetic deletion of arginine 14 in phospholamban causes dilated cardiomyopathy with attenuated electrocardiographic R amplitudes. Heart Rhythm 6, 480–486 [DOI] [PubMed] [Google Scholar]
- 16. van Rijsingen I. A., van der Zwaag P. A., Groeneweg J. A., Nannenberg E. A., Jongbloed J. D., Zwinderman A. H., Pinto Y. M., Dit Deprez R. H., Post J. G., Tan H. L., de Boer R. A., Hauer R. N., Christiaans I., van den Berg M. P., van Tintelen J. P., Wilde A. A. (2014) Outcome in phospholamban R14del carriers: results of a large multicentre cohort study. Circ. Cardiovasc. Genet. 7, 455–465 [DOI] [PubMed] [Google Scholar]
- 17. Jefferies J. L., Towbin J. A. (2010) Dilated cardiomyopathy. Lancet 375, 752–762 [DOI] [PubMed] [Google Scholar]
- 18. Dellefave L., McNally E. M. (2010) The genetics of dilated cardiomyopathy. Curr. Opin. Cardiol. 25, 198–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parvari R., Levitas A. (2012) The mutations associated with dilated cardiomyopathy. Biochem. Res. Int. 2012, 639250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ha K. N., Masterson L. R., Hou Z., Verardi R., Walsh N., Veglia G., Robia S. L. (2011) Lethal Arg9Cys phospholamban mutation hinders Ca2+-ATPase regulation and phosphorylation by protein kinase A. Proc. Natl. Acad. Sci. U.S.A. 108, 2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ceholski D. K., Trieber C. A., Holmes C. F., Young H. S. (2012) Lethal, hereditary mutants of phospholamban elude phosphorylation by protein kinase A. J. Biol. Chem. 287, 26596–26605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ceholski D. K., Trieber C. A., Young H. S. (2012) Hydrophobic imbalance in the cytoplasmic domain of phospholamban is a determinant for lethal dilated cardiomyopathy. J. Biol. Chem. 287, 16521–16529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paterlini M. G., Thomas D. D. (2005) The α-helical propensity of the cytoplasmic domain of phospholamban: a molecular dynamics simulation of the effect of phosphorylation and mutation. Biophys. J. 88, 3243–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu X., Lorigan G. A. (2013) Probing the interaction of Arg9Cys mutated phospholamban with phospholipid bilayers by solid-state NMR spectroscopy. Biochim. Biophys. Acta 1828, 2444–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu X., Lorigan G. A. (2014) Secondary structure, backbone dynamics, and structural topology of phospholamban and its phosphorylated and Arg9Cys-mutated forms in phospholipid bilayers utilizing 13C and 15N solid-state NMR spectroscopy. J. Phys. Chem. B 118, 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hou Z., Kelly E. M., Robia S. L. (2008) Phosphomimetic mutations increase phospholamban oligomerization and alter the structure of its regulatory complex. J. Biol. Chem. 283, 28996–29003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wegener A. D., Jones L. R. (1984) Phosphorylation-induced mobility shift in phospholamban in sodium dodecyl sulfate-polyacrylamide gels. Evidence for a protein structure consisting of multiple identical phosphorylatable subunits. J. Biol. Chem. 259, 1834–1841 [PubMed] [Google Scholar]
- 28. Hasenfuss G. (1998) Animal models of human cardiovascular disease, heart failure and hypertrophy. Cardiovasc. Res. 39, 60–76 [DOI] [PubMed] [Google Scholar]
- 29. Pattison J. S., Waggoner J. R., James J., Martin L., Gulick J., Osinska H., Klevitsky R., Kranias E. G., Robbins J. (2008) Phospholamban overexpression in transgenic rabbits. Transgenic Res. 17, 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bers D. M. (2002) Cardiac excitation-contraction coupling. Nature 415, 198–205 [DOI] [PubMed] [Google Scholar]
- 31. Hou Z., Robia S. L. (2010) Relative affinity of calcium pump isoforms for phospholamban quantified by fluorescence resonance energy transfer. J. Mol. Biol. 402, 210–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bidwell P., Blackwell D. J., Hou Z., Zima A. V., Robia S. L. (2011) Phospholamban binds with differential affinity to calcium pump conformers. J. Biol. Chem. 286, 35044–35050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robia S. L., Campbell K. S., Kelly E. M., Hou Z., Winters D. L., Thomas D. D. (2007) Forster transfer recovery reveals that phospholamban exchanges slowly from pentamers but rapidly from the SERCA regulatory complex. Circ. Res. 101, 1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelly E. M., Hou Z., Bossuyt J., Bers D. M., Robia S. L. (2008) Phospholamban oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase binding measured by fluorescence resonance energy transfer in living cells. J. Biol. Chem. 283, 12202–12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Autry J. M., Jones L. R. (1998) High-level coexpression of the canine cardiac calcium pump and phospholamban in Sf21 insect cells. Ann. N. Y. Acad. Sci. 853, 92–102 [DOI] [PubMed] [Google Scholar]
- 36. Domeier T. L., Blatter L. A., Zima A. V. (2009) Alteration of sarcoplasmic reticulum Ca2+ release termination by ryanodine receptor sensitization and in heart failure. J. Physiol. 587, 5197–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pallikkuth S., Blackwell D. J., Hu Z., Hou Z., Zieman D. T., Svensson B., Thomas D. D., Robia S. L. (2013) Phosphorylated phospholamban stabilizes a compact conformation of the cardiac calcium-ATPase. Biophys. J. 105, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abrol N., Smolin N., Armanious G., Ceholski D. K., Trieber C. A., Young H. S., Robia S. L. (2014) Phospholamban C-terminal residues are critical determinants of the structure and function of the calcium ATPase regulatory complex. J. Biol. Chem. 289, 25855–25866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zal T., Gascoigne N. R. (2004) Photobleaching-corrected FRET efficiency imaging of live cells. Biophys. J. 86, 3923–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Förster T. (1948) Intermolecular energy migration and fluorescence. Ann. Phys. 2, 55–75 [Google Scholar]
- 41. Li M., Reddy L. G., Bennett R., Silva N. D., Jr., Jones L. R., Thomas D. D. (1999) A fluorescence energy transfer method for analyzing protein oligomeric structure: application to phospholamban. Biophys. J. 76, 2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robia S. L., Flohr N. C., Thomas D. D. (2005) Phospholamban pentamer quaternary conformation determined by in-gel fluorescence anisotropy. Biochemistry 44, 4302–4311 [DOI] [PubMed] [Google Scholar]
- 43. Runnels L. W., Scarlata S. F. (1995) Theory and application of fluorescence homotransfer to melittin oligomerization. Biophys. J. 69, 1569–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gadella T. W. (2009) in FRET and FLIM Techniques (Pillai S., van der Vliet P. C., series eds) 1st Ed., p. 193, Elsevier, Amsterdam, Boston [Google Scholar]
- 45. Fujii J., Maruyama K., Tada M., MacLennan D. H. (1989) Expression and site-specific mutagenesis of phospholamban. Studies of residues involved in phosphorylation and pentamer formation. J. Biol. Chem. 264, 12950–12955 [PubMed] [Google Scholar]
- 46. King C., Sarabipour S., Byrne P., Leahy D. J., Hristova K. (2014) The FRET signatures of noninteracting proteins in membranes: simulations and experiments. Biophys. J. 106, 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schröder E., Eaton P. (2008) Hydrogen peroxide as an endogenous mediator and exogenous tool in cardiovascular research: issues and considerations. Curr. Opin. Pharmacol. 8, 153–159 [DOI] [PubMed] [Google Scholar]
- 48. Gramolini A. O., Kislinger T., Alikhani-Koopaei R., Fong V., Thompson N. J., Isserlin R., Sharma P., Oudit G. Y., Trivieri M. G., Fagan A., Kannan A., Higgins D. G., Huedig H., Hess G., Arab S., et al. (2008) Comparative proteomics profiling of a phospholamban mutant mouse model of dilated cardiomyopathy reveals progressive intracellular stress responses. Mol. Cell. Proteomics 7, 519–533 [DOI] [PubMed] [Google Scholar]
- 49. Smeazzetto S., Saponaro A., Young H. S., Moncelli M. R., Thiel G. (2013) Structure-function relation of phospholamban: modulation of channel activity as a potential regulator of SERCA activity. PLoS One 8, e52744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dong X., Thomas D. D. (2014) Time-resolved FRET reveals the structural mechanism of SERCA-PLB regulation. Biochem. Biophys. Res. Commun. 449, 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gustavsson M., Verardi R., Mullen D. G., Mote K. R., Traaseth N. J., Gopinath T., Veglia G. (2013) Allosteric regulation of SERCA by phosphorylation-mediated conformational shift of phospholamban. Proc. Natl. Acad. Sci. U.S.A. 110, 17338–17343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choudhary G., Dudley S. C., Jr. (2002) Heart failure, oxidative stress, and ion channel modulation. Congest. Heart Fail. 8, 148–155 [DOI] [PubMed] [Google Scholar]