Background: Whether papain-like protease (PLpro) from avian infectious bronchitis virus (IBV) that infects poultry, causing huge losses, can perform deubiquitination is unknown.
Results: IBV PLpro cleaves Lys48- and Lys63-linked polyubiquitin. The crystal structure reveals a markedly different set of residues for binding substrates.
Conclusion: IBV PLpro performs deubiquitination with catalytic efficiencies that are different from other PLpros.
Significance: Results provide a framework for developing antivirals.
Keywords: Crystal Structure, Enzyme Mechanism, Infectious Disease, Protease, Virus
Abstract
Papain-like protease (PLpro) of coronaviruses (CoVs) carries out proteolytic maturation of non-structural proteins that play a role in replication of the virus and performs deubiquitination of host cell factors to scuttle antiviral responses. Avian infectious bronchitis virus (IBV), the causative agent of bronchitis in chicken that results in huge economic losses every year in the poultry industry globally, encodes a PLpro. The substrate specificities of this PLpro are not clearly understood. Here, we show that IBV PLpro can degrade Lys48- and Lys63-linked polyubiquitin chains to monoubiquitin but not linear polyubiquitin. To explain the substrate specificities, we have solved the crystal structure of PLpro from IBV at 2.15-Å resolution. The overall structure is reminiscent of the structure of severe acute respiratory syndrome CoV PLpro. However, unlike the severe acute respiratory syndrome CoV PLpro that lacks blocking loop (BL) 1 of deubiquitinating enzymes, the IBV PLpro has a short BL1-like loop. Access to a conserved catalytic triad consisting of Cys101, His264, and Asp275 is regulated by the flexible BL2. A model of ubiquitin-bound IBV CoV PLpro brings out key differences in substrate binding sites of PLpros. In particular, P3 and P4 subsites as well as residues interacting with the β-barrel of ubiquitin are different, suggesting different catalytic efficiencies and substrate specificities. We show that IBV PLpro cleaves peptide substrates KKAG-7-amino-4-methylcoumarin and LRGG-7-amino-4-methylcoumarin with different catalytic efficiencies. These results demonstrate that substrate specificities of IBV PLpro are different from other PLpros and that IBV PLpro might target different ubiquitinated host factors to aid the propagation of the virus.
Introduction
Avian infectious bronchitis virus (IBV)3 is an enveloped, single-stranded RNA virus belonging to the Coronavirus genus (1, 2). The virus infects the upper respiratory tract of chickens, spreading progressively to the digestive and urinary tracts during the later stages of the infection (3). IBV is highly contagious, and each year outbreaks of IBV infections result in huge economic losses to the poultry industry globally. Currently available vaccines do not provide adequate protection against IBV because the strains show variability (4). There are no IBV-specific therapeutics available on the market. Therefore, it is important to study proteins of the virus that aid its replication and assist in circumventing the host immune surveillance machinery. Selective abrogation of function of such proteins could help prevent the spread of the virus. The papain-like protease (PLpro) of IBV is one such protein that could be targeted for development of effective antivirals.
IBV replicates in the cytoplasm of the cell using endogenous replicase proteins translated from the genomic RNA (5, 6). Initially, two large viral polypeptides are translated that are proteolytically processed into 15 non-structural proteins (nsp1–15) that play a role in the replication of the viral genome (7). Two viral proteases, a PLpro and a main protease, are known to perform the proteolytic maturation of the nsps of IBV (8–10). In particular, PLpro is known to cleave at sites between nsp2 and nsp3 as well as between nsp3 and nsp4 (10–12). So far, genomic RNA of IBV has been shown to encode only one functional PLpro that shows similarity to PL2pro from other viruses (13, 14). In addition to proteolytic maturation of nsps, PLpro of coronaviruses is known to perform deubiquitination and removal of interferon stimulated gene 15 (ISG15) from of host proteins (15–19). For instance, the PLpro of SARS CoV is known to deubiquitinate interferon regulatory transcription factor 3, which stalls its migration to the nucleus, preventing the initiation of host antiviral responses (15, 20, 21). Similarly, the PLpro from MERS CoV has been shown to degrade polyubiquitin chains associated with MDA-5 and RIG-I, preventing activation of host antiviral signaling pathways (14, 22). Consequently, inhibition of PLpro by small molecule inhibitors is considered a viable strategy to stall the life cycle of the virus (23–25). Whether the PLpro from IBV can perform similar dual functions (proteolytic maturation of viral nsps and deubiquitination of host cell factors) is currently not known.
Here, we describe the crystal structure of PLpro from IBV refined to 2.15-Å resolution. Comparison of the structure with PLpros from other viruses reveals conservation of catalytic residues. However, differences are observed in amino acids of the substrate binding site, suggesting differences in substrate specificities. We show that, unlike the PLpro from SARS CoV, the PLpro from IBV prefers Lys63-linked polyubiquitin over Lys48 for degradation. However, both PLpros do not cleave linear polyubiquitin chains. Furthermore, the PLpros differ in kinetics of degradation of peptide substrates. The reasons underlying these differences are discussed.
EXPERIMENTAL PROCEDURES
Expression and Purification of IBV Papain-like Protease
IBV PLpro (polyprotein residues 1174–1483) cloned into pGEX-6p-1 vector was expressed in Escherichia coli BL21(DE3) cells (Novagen). Cells were grown at 37 °C to an A600 nm of ∼0.5. The culture was cooled, and recombinant protein production was initiated by inducing the culture with 1 mm isopropyl d-thiogalactoside for 4 h at 28 °C under shaking conditions. At the end of the incubation time, cells were pelleted by centrifugation and resuspended in 40 ml of ice-cold lysis buffer (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 5 mm DTT). Cells were lysed using an ultra-high pressure cell disrupter (JNBIO, Guangzhou, China). Following centrifugation to remove cell debris and unbroken cells, recombinant protein was purified from the supernatant by affinity chromatography using GST resin. After thorough washing to remove contaminating protein, recombinant protein bound to the resin was eluted using a buffer containing 50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 5 mm DTT, 15 mm reduced glutathione. Recombinant GST-human rhinovirus 3c protease fusion protein was added to the eluted protein, and the mixture was dialyzed overnight against 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm DTT buffer. The dialyzed solution was passed over GST resin to remove the GST-human rhinovirus 3c and free GST polypeptide. Recombinant IBV PLpro was then further purified using an ion exchange chromatography step. Recombinant protein bound to the Mono Q column was eluted using a linear gradient of NaCl. Fractions containing the protein were pooled, concentrated, and loaded onto a Superdex 75 gel filtration column (GE Healthcare). IBV PLpro eluted in a single peak. Fractions containing the protein were pooled, concentrated to 10–15 mg/ml, and stored at −20 °C until further use. Selenomethionine-labeled IBV PLpro(1174–1483) was produced following a procedure published previously (26) and purified as described above. The PLpro catalytic domains from SARS CoV and MERS CoV were expressed and purified separately according to methods published previously (14). All purification procedures were carried out at 4 °C.
Crystallization
Crystallization of IBV PLpro was performed at 18 °C using the hanging drop vapor diffusion technique. Crystals were obtained by mixing 1 μl of protein solution (10 mg/ml) with an equal volume of reservoir solution. The crystallization drops were equilibrated against 200 μl of reservoir solution. Crystals of native IBV PLpro were obtained in a solution containing 100 mm Bis-Tris, pH 6.5, 25–30% polyethylene glycol monomethyl ether 2000. Crystals grew to maximum dimensions of 100 × 50 × 150 μm in 3 days. Selenomethionine-derivatized IBV PLpro crystallized under similar conditions.
Data Collection, Processing, and Structure Determination
Crystals were transferred into a harvesting solution containing the respective precipitant solutions plus 5% (v/v) glycerol before being flash frozen in liquid nitrogen. Data were collected under cryogenic conditions at 100 K. The selenomethionine single wavelength anomalous dispersion data set of IBV PLpro at the selenium peak wavelength was collected at Shanghai Synchrotron Radiation Facility (China) beamline BL17U1 to 1.5-Å resolution. All data sets were processed using the HKL-3000 package (27). Crystals belonged to space group I222 with cell parameters a = 71.3 Å, b = 95.1 Å, c = 118.4 Å, α = β = γ = 90°. The only selenium atom was located and refined, and the single wavelength anomalous dispersion data phases were calculated and substantially improved by solvent flattening using the PHENIX program (28). A model was manually built into the modified experimental electron density using Coot (29) and further refined in PHENIX. Refinement statistics for the final model are summarized in Table 1. Model geometry was verified using the program MolProbity (30). Structural figures were drawn using the program PyMOL (31).
TABLE 1.
Data collection and refinement statistics
Numbers in parentheses are corresponding values for the highest resolution shell. r.m.s.d., root mean square deviation; Res., residues; Se-SAD, selenomethionine single wavelength anomalous dispersion.
| Parameters | Se-SAD peak |
|---|---|
| Data collection statistics | |
| Cell parameters | |
| a (Å) | 71.3 |
| b (Å) | 95.1 |
| c (Å) | 118.4 |
| α, β, γ (°) | 90, 90, 90 |
| Space group | I222 |
| Wavelength used (Å) | 0.97855 |
| Resolution (Å) | 50.00-2.00 (2.07-2.00) |
| No. of all reflections | 52,949 (5,254) |
| No. of unique reflections | 8,146 (809) |
| Completeness (%) | 100.0 (100.0) |
| Average I/σ(I) | 26.0 (2.8) |
| Rmergea (%) | 12.4 (73.5) |
| Refinement statistics | |
| No. of reflections used (σ(F) > 0) | 22,249 |
| Rworkb (%) | 17.5 |
| Rfreeb (%) | 21.6 |
| r.m.s.d. bond distance (Å) | 0.008 |
| r.m.s.d. bond angle (°) | 1.027 |
| Average B-value (Å2) | 37.1 |
| No. of protein atoms | 2,504 |
| No. of ligand atoms | 1 |
| No. of solvent atoms | 128 |
| Ramachandran plot | |
| Res. in favored regions (%) | 97.6 |
| Res. in allowed regions (%) | 2.4 |
| Res. in outlier regions (%) | 0 |
a Rmerge = ΣhΣl|Iih − Ih|/ΣhΣIIh where Ih is the mean of the observations Iih of reflection h.
b Rwork = Σ(‖Fp(obs)| − |Fp(calc)‖)/Σ|Fp(obs)|. Rfree is an R factor for a preselected subset (5%) of reflections that was not included in refinement.
Steady-state Kinetic Studies
The rates of IBV PLpro-catalyzed reactions were determined using a modified version of methods described previously (18, 22, 32). Briefly, the rate of hydrolysis of peptide substrates carboxybenzyl-Leu-Arg-Gly-Gly-7-amino-4-methylcoumarin (Z-LRGG-AMC), carboxybenzyl-Lys-Ala-Gly-Gly-7-amino-4-methylcoumarin (Z-KAGG-AMC), and carboxybenzyl-Lys-Lys-Ala-Gly-7-amino-4-methylcoumarin (Z-KKAG-AMC) (Life Sensors, Inc.) was determined by measuring the increase in fluorescence of the AMC group released (excitation λ = 340 nm; emission λ = 460 nm) at different time points. The reaction mixtures consisted of various concentrations of peptide substrate (10–160 μm) suspended in 20 mm Tris-HCl, pH 7.5, 0.1 mg/ml BSA, 150 mm NaCl, 2 mm DTT. Assays were initiated by the addition of 0.2 μm pure IBV PLpro, and the incubation was carried out at 25 °C. The intensity of the fluorescence observed was converted to the amount of substrate hydrolyzed by comparing it with the fluorescence of known standards. The ability of SARS CoV PLpro and MERS CoV PLpro to hydrolyze Z-LRGG-AMC was tested using the same assay with the addition of 0.2 μm SARS CoV PLpro or 1.6 μm MERS CoV PLpro instead of IBV PLpro. Active sites of all three enzymes could not be saturated using the peptide substrates. Therefore, initial rates of the reactions for different concentration of substrate were fit to the Michaelis-Menten equation using linear regression with SigmaPlot (Systat Software, Inc.) to obtain the apparent kcat/Km values.
Ubiquitin Chain Cleavage Experiments
Proteolytic cleavage of linear Ub4, Lys48-Ub4 and Lys63-Ub4 (all from UBPBio) was carried out as follows. 50 nm purified protein (IBV PLpro) was incubated with 40 μg of linear Ub4, Lys48-Ub4, or Lys63-Ub4 at 25 °C in a 50-μl volume containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm DTT. A control reaction was incubated under identical conditions with the exclusion of enzyme. At various time points, 8 μl were removed, and the reaction was quenched with the addition of SDS-PAGE sample loading buffer. The samples were analyzed by electrophoresis on a 4–15% SDS-polyacrylamide gel and stained with Coomassie dye.
RESULTS
Overall Structure
IBV PLpro (polyprotein residues 1174–1483) could be overexpressed as soluble protein in E. coli. The protein was purified to homogeneity using GST affinity, ion exchange, and size exclusion chromatography steps. Pure IBV PLpro crystallized in a number of crystallization conditions. The structure was solved by the selenomethionine single wavelength anomalous dispersion method by collecting a data set of selenomethionine-labeled IBV PLpro crystal (I222 space group) at the selenium peak wavelength. Clear electron density permitted unambiguous modeling of residues. The final model consisting of residues 4–304 was refined to 2.15-Å resolution (Rwork = 17.5% and Rfree = 21.6%). Statistics pertaining to data collection and refinement are listed in Table 1.
The overall structure of the IBV PLpro consists of four distinct domains (Fig. 1, A and B). The first 56 residues fold into a ubiquitin-like (Ubl) domain. The Ubl domain is well separated from the other three domains that contact each other and form a compact globular structure. This Ubl domain adopts a β-grasp fold similar to ubiquitin and is followed by three other domains representing the thumb, fingers, and palm of an extended right hand, an arrangement previously observed for deubiquitinating enzymes. The thumb domain is predominantly α-helical. It contains six α-helices and two short β-strands that are arranged in a β-hairpin. The thumb domain makes contact with the palm domain using this β-hairpin. Amino acids 173–310 fold into the finger and palm domains. This region of the protein consists of mostly β-strands. Although the finger domain is made up of three long (β6, β7, and β10) and two short β-strands, the palm domain is made up of seven β-strands. One Zn2+ ion coordinated by four cysteine residues originating from two β-hairpins is located between strands β6 and β10 of the finger domain. The conformation of the side chains of cysteines and the arrangement of the β-hairpins are similar to those observed for members belonging to the zinc ribbon fold group. The three long β-strands of the finger domain protrude out of the protein. This facilitates formation of a hydrophobic cavity at the junction of thumb and finger domains, which has implications for the function of the protein.
FIGURE 1.
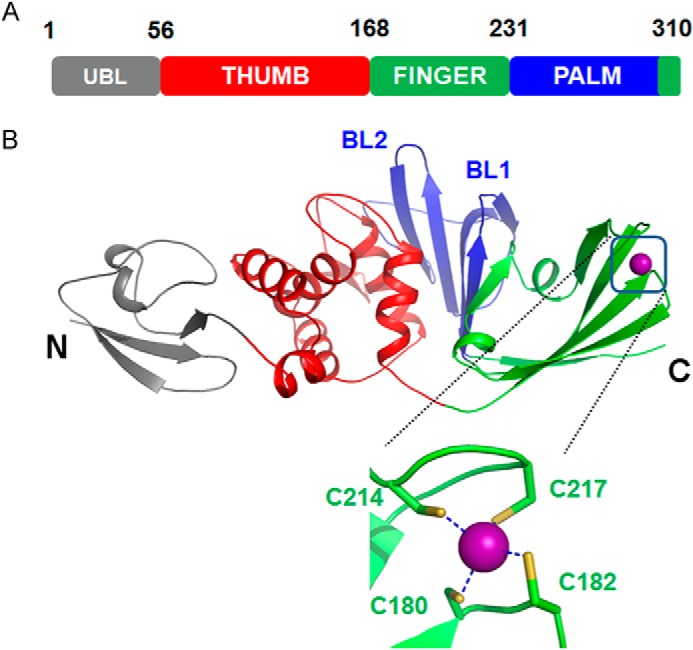
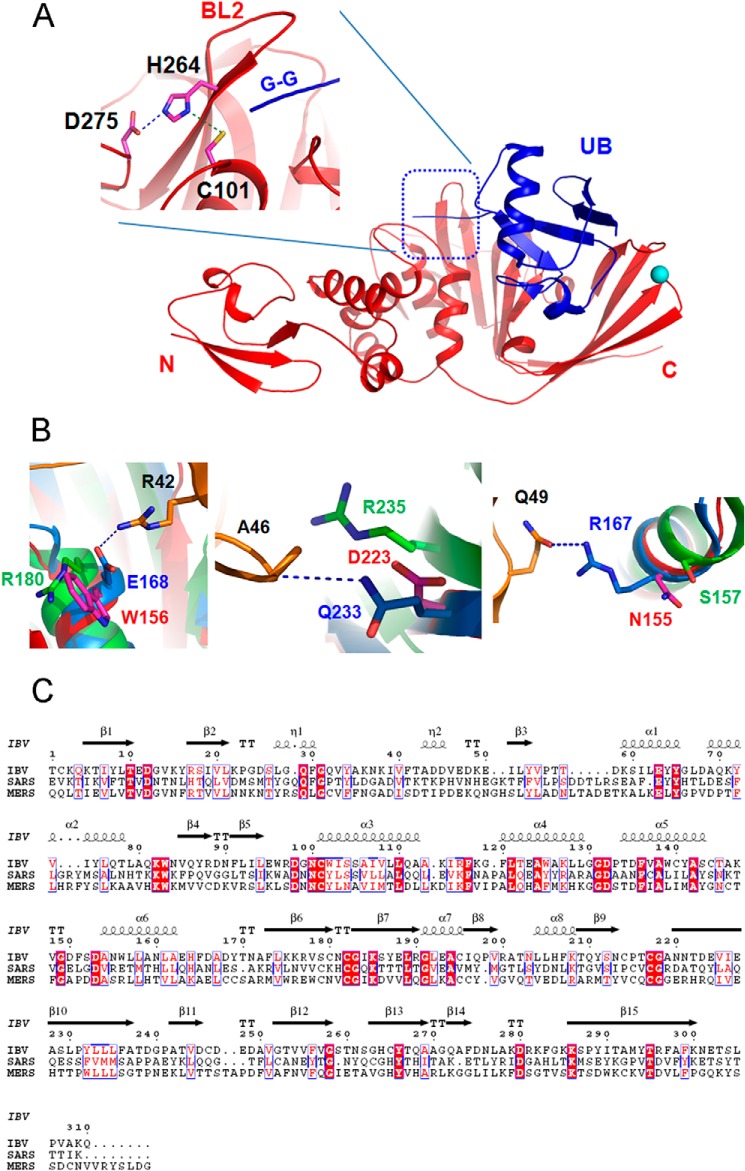
Overall structure of PLpro from IBV. A, diagrammatic representation of the organization of domains of IBV PLpro inferred from the crystal structure. B, schematic representation of the crystal structure of IBV PLpro. The locations of BL1 and BL2 as well as the termini of the protein are marked. Zn2+ ion bound by the protein is shown as a sphere. The Zn2+ ion is coordinated by four cysteine residues (inset) shown as sticks.
Comparison of the Overall Fold
Despite sharing only 12–15% sequence identity with ubiquitin-specific proteases (USPs) for which crystal structures are available, the overall architecture and domain organization of the IBV PLpro is similar to USPs. Unlike USPs, PLpros are dual functional enzymes and therefore their substrate binding sites are designed to accommodate two different substrates. For instance, in addition to cleaving ubiquitin, the IBV PLpro cleaves the nascent viral polypeptide between nsp2 and nsp3 as well as between nsp3 and nsp4. Comparison of the structures of USPs with IBV PLpro reveals differences in the region connecting the finger domain with the palm domain that permits promiscuity in catalysis by IBV PLpro. In particular, “blocking loop” (BL) 1, connecting β8 with β9, which is known to regulate the de-ubiquitinating activity of USP14, is made up of 22 residues and is part of the substrate-binding surface. The corresponding loop in IBV PLpro is much shorter (Asp245–Val251) and connects β11 with β12. As a consequence, the substrate binding site is larger in IBV PLpro than in USPs. This probably enables the enzyme to not only bind ubiquitin but also viral polyprotein and operate as a dually functional enzyme.
We searched the Protein Data Bank for structural homologues of IBV PLpro using the DALI server. The search retrieved the structure of PLpro from SARS CoV as the top match (Z score = 28.6) with a root mean square deviation of 2.4 Å for 292 overlapping Cα atoms. Although the overall structure is similar, superimposition studies revealed four regions with significant differences between the two coronaviral PLpros (Fig. 2, A and B). The first structural difference is observed in the Ubl domain. Strands β1 and β2 move inward in IBV PLpro. As a consequence, the loop connecting β2 with β3 is displaced and does not align well with a similar region of SARS CoV PLpro. The second structural difference involves the α1 helix of the thumb domain. This helix is much shorter than the corresponding helix in the SARS CoV PLpro. Because of this, the loop connecting the Ubl domain with the thumb domain is much longer in IBV PLpro. The third structural difference is observed in the palm domain. Although PLpro from IBV exhibits a BL1-like loop connecting strands β11 with β12, the PLpro from SARS CoV shows a complete lack of a BL1-like loop. Lastly, strand β16 of the palm domain of SARS CoV PLpro is missing in IBV PLpro. In addition, the strand following this region (β16) in IBV PLpro is much shorter (Fig. 2B).
FIGURE 2.
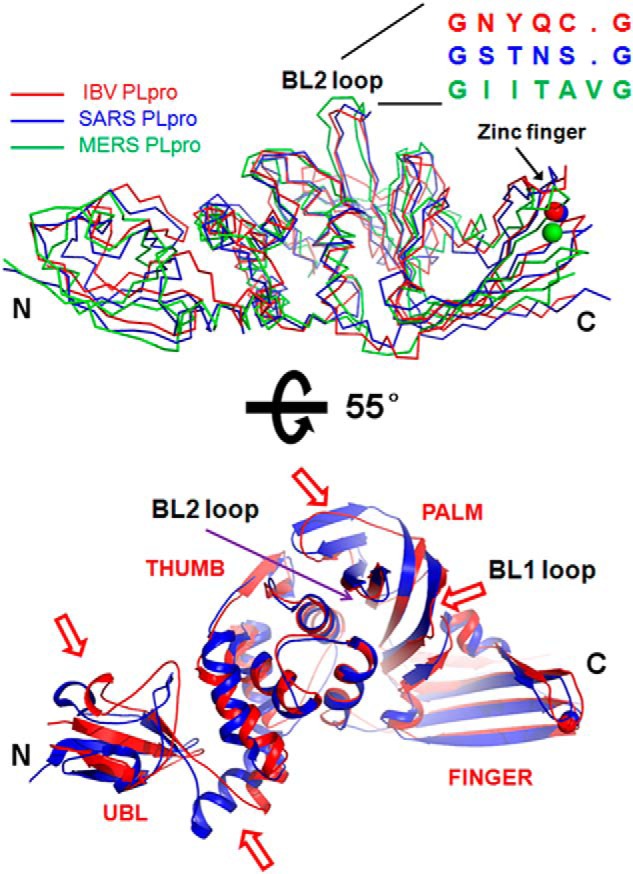
Comparison of the overall structures of PLpros from different CoVs. A, crystal structures of PLpros from SARS CoV (blue ribbon; Protein Data Bank code 4M0W) (19) and MERS (green ribbon; Protein Data Bank code 4RF1) CoVs (36) were superimposed over the structure of PLpro from IBV (red ribbon). The location of BL2 and the primary sequences of the PLpros corresponding to this region are shown. Zn2+ ion is shown as a sphere. B, superimposition of the structures of PLpro from SARS CoV (blue schematic) over IBV PLpro (red schematic). The view is the same as in A but rotated by 55° around the x axis. Regions that show significant deviation are marked by a red arrow. The N and C termini of the protein are marked as N and C, respectively.
Comparison of the structure of IBV PLpro with that of MERS CoV PLpro revealed differences in the thumb domain. MERS CoV PLpro has an extra β-hairpin in this region. An additional difference involves the zinc finger motif. Specifically, the Zn2+ ion of PLpro from MERS CoV has moved inward from its location in PLpros from IBV and SARS CoV (Fig. 2A). Interestingly, a feature shared between IBV and MERS CoV PLpros but missing in SARS CoV PLpro is the BL1-like loop. MERS CoV PLpro has a BL1-like loop at a location similar to that observed in IBV PLpro. Thus, the PLpro from IBV, SARS, and MERS CoVs shows some differences that probably impart different substrate specificities and possibly govern the ability of the virus to target different signaling pathways of host cells to assist propagation of the virus.
Ubiquitin Binding Site of IBV PLpro
Similar to PLpro from SARS and MERS CoVs, IBV PLpro also probably targets ubiquitinated host factors to aid viral replication. To gain information about the mode of binding of substrate, in particular ubiquitin, to IBV PLpro, we superimposed the structure of SARS CoV PLpro bound with ubiquitin over the structure of IBV PLpro. Because the overall structures of the two PLpros are highly similar, it is not surprising to note that in this model ubiquitin binds at a similar location, the junction of thumb, palm, and finger domains, in IBV PLpro (Fig. 3A). The β-barrel of the ubiquitin is primarily engaged by the finger domain, whereas the C-terminal tail containing the diglycine motif extends into a cavity formed at the junction of thumb and palm domains. More importantly, the diglycine is in proximity of BL2 and key catalytic residues that are conserved across PLpros (Fig. 3A). Although the overall mode of engaging the ubiquitin is similar in PLpros from IBV, SARS, and MERS CoVs, the nature of amino acids of PLpro from these CoVs interacting with ubiquitin is different. These differences are illustrated in the following examples involving one salt bridge and several other hydrogen bonding interactions. The side-chain guanidinium group of Arg42 of ubiquitin forms a salt bridge with the side-chain carboxylate group of Glu168 of SARS CoV PLpro. A tryptophan (Trp156) occupies the position of Glu168 in IBV PLpro, whereas an arginine is located at a similar position in MERS CoV PLpro 2 (Fig. 3, B and C). Three hydrogen bonds are formed between ubiquitin and the PLpro from SARS CoV. 1) The main-chain amide nitrogen atom of Ala46 of ubiquitin hydrogen bonds with the side chain of Gln233 from SARS CoV PLpro. 2) The main-chain carbonyl oxygen atom of Gly47 hydrogen bonds with the main-chain amide of Met209. 3) The side chain of Gln49 forms a hydrogen bond with the side chain of Arg167. The equivalent residues of IBV PLpro interacting with ubiquitin, Asp223 (arginine in MERS CoV PLpro), Val199, and Asn155 (serine in MERS CoV PLpro), respectively, are all different (Fig. 3, B and C). In addition, substitution of His172, Tyr208, and Tyr269 of SARS CoV PLpro with Asn160 (threonine in MERS CoV PLpro), Val199 (valine in MERS CoV PLpro), and Thr260 (threonine in MERS CoV PLpro), respectively, further illustrates that PLpros use different amino acids to tether ubiquitin and possibly peptide substrates. Alternatively, differences in substrate binding residues are likely to help occlude non-substrate ligands, increase affinity for canonical peptide ligands, and impact catalytic efficiencies.
FIGURE 3.
Substrate binding site of IBV PLpro. A, ubiquitin (UB; blue schematic) from the structure of PLpro of SARS CoV bound with ubiquitin (Protein Data Bank code 4M0W) was superimposed over the structure of PLpro from IBV (red schematic). The location of the diglycine motif (G-G) with respect to the catalytic triad (shown as sticks) and BL2 is shown (inset). Interaction between the catalytic residues is shown by dashed lines. B, comparison of amino acids of different PLpros interacting with ubiquitin (orange color). Interactions of amino acids of ubiquitin such Arg42, Ala46, and Gln49 (orange sticks) with corresponding amino acids of the PLpro from IBV (magenta sticks), SARS (blue sticks), and MERS (green sticks) CoVs are shown to illustrate the differences in residues of the ligand binding site of the PLpros from different sources. C, alignment of primary sequences of PLpro from IBV, SARS, and MERS CoVs. Secondary structural elements of PLpro from IBV are shown at the top. Absolutely conserved residues are highlighted with a red background. Conserved residues are shown in red.
PLpros from IBV and SARS CoV Exhibit Different Catalytic Efficiencies
Because superimposition studies revealed that the amino acids of IBV PLpro interacting with ubiquitin are different from those of SARS CoV PLpro interacting with ubiquitin, suggesting different substrate specificities, we tested the ability of IBV PLpro to degrade linear, Lys48- and Lys63-linked polyubiquitin chains. Similar to SARS CoV PLpro, IBV PLpro could degrade Lys48- and Lys63-linked polyubiquitin chains but not linear ubiquitin chains (Fig. 4). Remarkably, but consistent with the inference gained from the structural studies that the substrate specificity of IBV PLpro could be different from that of SARS CoV PLpro, the protease from IBV showed a preference for Lys63-linked polyubiquitin chains over Lys48-linked polyubiquitin chains. Specifically, Lys63-linked tetraubiquitin could be degraded almost completely within 5 min of the incubation time of the assay mixture to lower order oligomers such as triubiquitin, diubiquitin, and monoubiquitin. Within 30 min of the reaction time, most of the oligomers were degraded to monoubiquitin. In contrast, the degradation of Lys48-linked tetraubiquitin under identical conditions was relatively slower. At the end of 5 min of incubation time, a significant amount of substrate remained uncleaved. In fact, even after 240 min of incubation, the substrate could not be completely degraded to monoubiquitin. In addition to the substrate, tri- and diubiquitin products persisted throughout the incubation period. Thus, lower order oligomers of Lys48-linked ubiquitin seem to impose product inhibition on IBV PLpro. No cleavage of linear tetraubiquitin chains was observed even after prolonged incubation (4 h) under identical conditions of the assay. Previously, SARS CoV PLpro has been shown to degrade Lys48-linked polyubiquitin chains more rapidly than Lys63-linked polyubiquitin chains. Thus, results of enzymatic assays are consistent with inferences gained from structural studies and confirm that PLpros from IBV and SARS CoVs have different substrate specificities.
FIGURE 4.
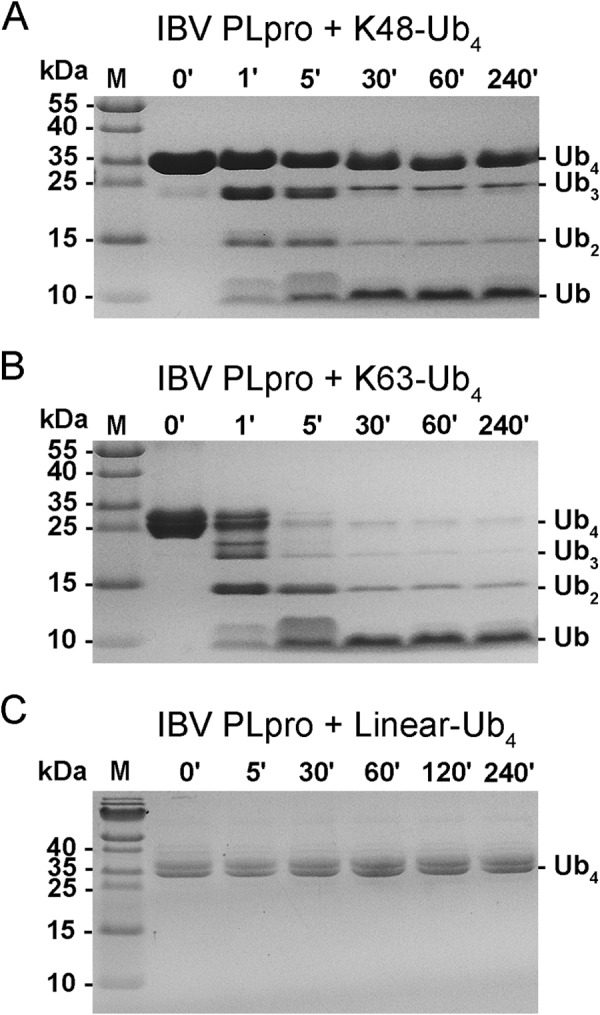
Cleavage of polyubiquitin chains by PLpro from IBV. A, picture of the gel from SDS-PAGE analysis of reaction products of Lys48-linked tetraubiquitin chains treated with IBV PLpro at different time intervals. B, analysis similar to that in A except the substrate used was Lys63-linked tetraubiquitin. C, SDS-PAGE gel picture of reaction products of linear ubiquitin chains treated with PLpro from IBV. The protease could not cleave linear ubiquitin under reaction conditions similar to those used for cleaving Lys48- and Lys63-linked tetraubiquitin. M, molecular mass markers; ′, minutes.
PLpro from IBV has been shown to cleave peptides at KAGG motifs (11, 12), whereas SARS CoV has been shown to cleave at LXGG (X represents any amino acid) sites (32, 33). We tested the catalytic ability of IBV PLpro to cleave at these motifs using peptide substrates KAGG-AMC, KKAG-AMC, and LRGG-AMC. Intriguingly, IBV PLpro could not cleave KAGG-AMC, suggesting that this peptide substrate is not a close mimic of the physiological substrate. Either a larger peptide substrate is essential for correctly orienting the site of cleavage to release AMC, or at least a larger side chain like lysine or arginine is required at the P3 position. Previous studies have shown that IBV PLpro can cleave polypeptide with an alanine at the P2 position instead of a glycine (11). In agreement with this observation, we could monitor activity of IBV PLpro using KKAG-AMC as the peptide substrate. The PLpro from IBV could cleave KKAG-AMC and LRGG-AMC. However, the catalytic efficiency of the PLpro toward KKAG-AMC was 5 times higher than that for LRGG-AMC (Table 2). We next compared the catalytic efficiencies of PLpros from IBV and SARS CoVs against their preferred peptide substrates. IBV PLpro exhibited a catalytic efficiency of 3.23 ± 0.72 s−1 mm−1 against KKAG-AMC, which was comparable with that of the PLpro from SARS CoV that showed a catalytic efficiency of 4.41 ± 0.50 s−1 mm−1 against LRGG-AMC when assayed under identical conditions (Table 2). Thus, the catalytic efficiencies of the PLpros from IBV and SARS CoVs are similar against their preferred peptide substrates. Because the peptide substrates preferred by both the PLpros are markedly different, the results further support the fact that PLpros from different sources differ in substrate specificities.
TABLE 2.
Comparison of the kinetic parameters of PLpros from different CoVs
| PLpro | Peptide | Cleavage | kcat/Kma |
|---|---|---|---|
| s−1 mm−1 | |||
| IBV | Lys48-Ub4 | Yes | |
| Lys63-Ub4 | Yes | ||
| Linear Ub4 | No | ||
| LRGG-AMC | Yes | 0.67 ± 0.11 | |
| KKAG-AMC | Yes | 3.23 ± 0.72 | |
| KAGG-AMC | No | NDb | |
| SARS CoV | LRGG-AMC | Yes | 4.41 ± 0.50 |
| MERS CoV | LRGG-AMC | Yes | 0.05 ± 0.01 |
a kcat/Km values shown here are apparent kcat/Km values obtained by fitting data to the Michaelis-Menten equation using linear regression. The active sites of the enzyme could not be saturated with substrate.
b Not detected.
DISCUSSION
In this study, we show that the PLpro from IBV can degrade Lys48- and Lys63-linked polyubiquitin chains completely to monoubiquitin. The PLpro could not cleave ubiquitin molecules from linear polyubiquitin chains even after prolonged incubation. Furthermore, the PLpro from IBV could cleave two different peptide substrates, KKAG-AMC and LRGG-AMC. The nature of the P3 and P4 amino acids of the peptide substrates KKAG-AMC and LRGG-AMC cleaved by IBV PLpro is significantly different; in particular, the amino acid occupying the P4 position of the substrate that is buried in the protease during catalysis is very different in both peptide substrates. Despite this, the PLpro from IBV can cleave both the peptides, conceivably with different efficiencies. Thus, the active site pocket offers some promiscuity but at the cost of catalytic efficiency. The peptide substrate LRGG-AMC was degraded less efficiently when compared with KKAG-AMC. Examination of the structure of IBV PLpro with one molecule of ubiquitin from the structure of SARS CoV PLpro modeled in the active site by superimposition reveals that substrate selection is probably governed by three regions: 1) the region around BL2 that positions and orients the diglycine region (P1-P2 residues) of substrates for proteolysis, 2) the region between the finger and palm domain of the PLpro that binds the β-barrel of ubiquitin or the viral peptide substrate, and 3) the region behind BL2 that holds the part of the peptide downstream of the cleavage site. The diglycine binding region of PLpros from IBV and SARS CoVs is quite similar, suggesting a shared mechanism for optimally orienting the peptide bond and its cleavage. In SARS CoV PLpro, P1 (Gly76 of ubiquitin) forms a hydrogen bonding interaction with Gly272. In additional, the C-terminal carboxylate group of Gly76 interacts with Trp107, Cys112, and Tyr113 (16, 19). In IBV PLpro, except for Tyr113 that is replaced by a Trp, other spatially equivalent residues, Gly263, Trp96, and Cys101, respectively, are all conserved. Phe151 (Leu163 in SARS CoV PLpro), Asp153 (conserved), Gly263 (conserved), and Cys265 (Tyr266 in SARS CoV PLpro) are involved in shaping the equally restricted S2 subsite, which again is specific for glycine.
Compared with the S1 and S2 subsites that are mostly buried, the residue of the peptide substrate occupying the S3 subsite is partially solvent-exposed, and the side chain at this position makes hardly any contact with the PLpro. Examination of the region around the P3 of ubiquitin-bound SARS CoV PLpro shows the main-chain amide of Arg at this position forming two hydrogen bonds, one each with Gly272 and the other with the side-chain hydroxyl of Tyr265. Gly272 is conserved in PLpro from SARS CoV as well as IBV, but Tyr265 is replaced by Phe256 in IBV PLpro. More importantly, the side chain of Arg74 of ubiquitin (P3 site) is involved in only one intermolecular interaction with the backbone carbonyl oxygen of Leu163 (Fig. 5). A smaller side chain at the P3 position of the substrate, for instance replacement of arginine with alanine as observed in the KAGG motif cleaved by IBV PLpro, could prevent steric clashes with Leu163 and facilitate easy access to catalytic residues during binding of substrate.
FIGURE 5.
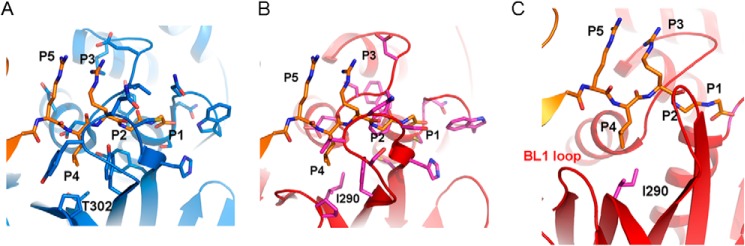
Putative mode of ligand binding. The mode of binding of ligands by SARS CoV PLpro (A) and IBV PLpro (B and C) is different. A, P1–P4 amino acids (orange sticks) of ubiquitin tethered by SARS CoV PLpro. Amino acids of PLpro within 4 Å of the ligand are shown as sticks. B and C, mode of binding of P1–P4 residues of ubiquitin (orange sticks) by IBV PLpro inferred by superimposing the ubiquitin from the structure of SARS CoV PLpro bound with ubiquitin (Protein Data Bank code 4M0W) on the structure of IBV PLpro (red schematic). Amino acids within 4 Å of the ligand are shown (magenta sticks). The location of the BL1-like loop and its proximity to the P4 residue are shown in C.
In contrast to the S3 site, the S4 site is buried, and the corresponding P4 residues occupying this site during proteolysis of viral peptide substrates are very different in IBV and SARS CoV PLpros. Therefore, the P4 residue probably has a profound effect on the catalytic efficiencies. Although SARS CoV PLpro has a Leu at this position, the IBV PLpro has a Lys at this position. Accordingly, the amino acids forming the S4 subsite are different in both PLpros to accommodate these two very different side chains. Amino acids such as Asp165, Pro249, Tyr265, Tyr269, Tyr274, and Thr302 of SARS CoV PLpro are observed to form a pocket for entrenching Leu73 of ubiquitin (Fig. 5A). Residues of IBV PLpro occupying equivalent positions include Asp153, Pro241, Phe256, Thr260, Cys265, and Ileu290, respectively (Fig. 5B). Notably, the substitution of Thr302 with Ile is likely to obstruct the placement of the side chain of a lysine residue of the KAGG site cleaved by IBV PLpro in the pocket. To offset this steric clash, the PLpro from IBV has BL1 located immediately adjacent to Ile290, creating a larger pocket possibly to accommodate the side chain of a P3 lysine (Fig. 5C). The equivalent region in SARS CoV PLpro is rigid and made up of long β-strands. Thus, differences are observed in the active site pocket, especially in the S3-S4 subsite region between the two PLpros. These differences coupled with differences in amino acids involved in tethering the region of the substrate adjacent to the P4–P1 residues, i.e. the β-barrel of ubiquitin during deubiquitination or the viral polypeptide adjacent to the proteolytic site during proteolytic maturation of nsps, probably give rise to differences in substrate specificities and catalytic efficiencies observed for the PLpros.
The differences in substrate specificities observed probably confer different abilities on the PLpros and consequently the virus to scuttle different aspects of the host antiviral responses. For instance, SARS CoV PLpro shows a preference for Lys48-linked polyubiquitin over Lys63-linked ubiquitin. This PLpro cleaves the Lys48-linked polyubiquitin chains of interferon regulatory transcription factor 3 and degrades the Lys48-linked polyubiquitin chains tagged on IκBα that prevent its proteasomal degradation (20, 21). Both these actions block the activation of host antiviral signaling pathways. In contrast, the MERS CoV PLpro has been shown to process Lys48- and Lys63-linked polyubiquitin chains with similar rates, producing monoubiquitin species (22). This PLpro has been shown to degrade Lys63-linked polyubiquitin chains associated with RIG-I and MDA-5, preventing activation of antiviral responses mediated by both these proteins. Thus, differences in substrate specificities of PLpros probably seem to help the viruses target different aspects of host immune responses. Further studies, for example comparison of the abilities of different PLpros in targeting the same as well as different proteins for deubiquitination and evaluation of their effect on immune responses, are likely to shed more light on the roles of different PLpros. Because PLpros modulate immune responses, they also probably impact the virulence of viruses.
The structure of IBV PLpro presented here provides a clear view of the protease at the molecular level and brings out several interesting features of this PLpro that need to be investigated further. In particular, it would be interesting to know the choice of ubiquitinated host factors targeted by this PLpro and the outcome of degradation of these ubiquitinated substrates. Proteomics studies have identified thousands of proteins involved in diverse signaling pathways that are modified by ubiquitin including Lys48- and Lys63-linked polyubiquitin chains that have been shown to be degraded in vitro by IBV PLpro in this study (34). In particular, activation of nuclear factor κB transcription factor by Toll-like receptors, nucleotide oligomerization domain-like receptors, RIG-I, and stimulator of interferon genes among others is dependent on modification of proteins participating in these antiviral signaling pathways by polyubiquitin chains (35). Perhaps IBV PLpro derails activation of host antiviral signaling pathways by degrading Lys48- and Lys63-linked polyubiquitin chains associated with these proteins. In addition, ubiquitination is not limited to immune responses. Therefore, PLpros are probably capable of modulating other aspects of the host cell regulated by ubiquitination, for example the cell cycle, to assist survival of the virus that await discovery.
Acknowledgment
We gratefully acknowledge the staff of Shanghai Synchrotron Radiation Facility (China) for assistance with x-ray diffraction data collection.
This work is supported by National Major Project (973 Program) Grants 2014CB542800 and 2013CB911100, National Natural Science Foundation of China Grants 81330036 and 81322023, and Tsinghua University Initiative Scientific Research Program Grant 20141080146.
The atomic coordinates and structure factors (code 4X2Z) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- IBV
- infectious bronchitis virus
- PLpro
- papain-like protease
- CoV
- coronavirus
- SARS
- severe acute respiratory syndrome
- AMC
- 7-amino-4-methylcoumarin
- BL
- blocking loop
- nsp
- non-structural protein
- MERS
- Middle East respiratory syndrome
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Z
- carboxybenzyl
- Ub
- ubiquitin
- Ubl
- ubiquitin-like
- USP
- ubiquitin-specific protease
- RIG-I
- retinoic acid inducible gene I.
REFERENCES
- 1. Stern D. F., Sefton B. (1984) Organization of the IBV genome. Adv. Exp. Med. Biol. 173, 11–23 [DOI] [PubMed] [Google Scholar]
- 2. Stern D. F., Sefton B. M. (1984) Coronavirus multiplication: locations of genes for virion proteins on the avian infectious bronchitis virus genome. J. Virol. 50, 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cavanagh D. (2007) Coronavirus avian infectious bronchitis virus. Vet. Res. 38, 281–297 [DOI] [PubMed] [Google Scholar]
- 4. Cavanagh D., Casais R., Armesto M., Hodgson T., Izadkhasti S., Davies M., Lin F., Tarpey I., Britton P. (2007) Manipulation of the infectious bronchitis coronavirus genome for vaccine development and analysis of the accessory proteins. Vaccine 25, 5558–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hodgson T., Britton P., Cavanagh D. (2006) Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 80, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casais R., Davies M., Cavanagh D., Britton P. (2005) Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 79, 8065–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Ginkel F. W., van Santen V. L., Gulley S. L., Toro H. (2008) Infectious bronchitis virus in the chicken Harderian gland and lachrymal fluid: viral load, infectivity, immune cell responses, and effects of viral immunodeficiency. Avian Dis. 52, 608–617 [DOI] [PubMed] [Google Scholar]
- 8. Ng L. F., Liu D. X. (2002) Membrane association and dimerization of a cysteine-rich, 16-kilodalton polypeptide released from the C-terminal region of the coronavirus infectious bronchitis virus 1a polyprotein. J. Virol. 76, 6257–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lim K. P., Liu D. X. (1998) Characterisation of a papain-like proteinase domain encoded by ORF1a of the coronavirus IBV and determination of the C-terminal cleavage site of an 87 kDa protein. Adv. Exp. Med. Biol. 440, 173–184 [DOI] [PubMed] [Google Scholar]
- 10. Lim K. P., Liu D. X. (1998) Characterization of the two overlapping papain-like proteinase domains encoded in gene 1 of the coronavirus infectious bronchitis virus and determination of the C-terminal cleavage site of an 87-kDa protein. Virology 245, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lim K. P., Ng L. F., Liu D. X. (2000) Identification of a novel cleavage activity of the first papain-like proteinase domain encoded by open reading frame 1a of the coronavirus avian infectious bronchitis virus and characterization of the cleavage products. J. Virol. 74, 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ziebuhr J., Thiel V., Gorbalenya A. E. (2001) The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J. Biol. Chem. 276, 33220–33232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han Y. S., Chang G. G., Juo C. G., Lee H. J., Yeh S. H., Hsu J. T., Chen X. (2005) Papain-like protease 2 (PLP2) from severe acute respiratory syndrome coronavirus (SARS-CoV): expression, purification, characterization, and inhibition. Biochemistry 44, 10349–10359 [DOI] [PubMed] [Google Scholar]
- 14. Mielech A. M., Kilianski A., Baez-Santos Y. M., Mesecar A. D., Baker S. C. (2014) MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 450–451, 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chaudhuri R., Tang S., Zhao G., Lu H., Case D. A., Johnson M. E. (2011) Comparison of SARS and NL63 papain-like protease binding sites and binding site dynamics: inhibitor design implications. J. Mol. Biol. 414, 272–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ratia K., Kilianski A., Baez-Santos Y. M., Baker S. C., Mesecar A. (2014) Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating activity of SARS-CoV papain-like protease. PLoS Pathog. 10, e1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B. S., Johnson M. E., Baker S. C., Ghosh A. K., Mesecar A. D. (2008) A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U.S.A. 105, 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lei J., Mesters J. R., Drosten C., Anemüller S., Ma Q., Hilgenfeld R. (2014) Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features. Antiviral Res. 109, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chou C. Y., Lai H. Y., Chen H. Y., Cheng S. C., Cheng K. W., Chou Y. W. (2014) Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease. Acta Crystallogr. D Biol. Crystallogr. 70, 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z. (2014) SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 5, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clementz M. A., Chen Z., Banach B. S., Wang Y., Sun L., Ratia K., Baez-Santos Y. M., Wang J., Takayama J., Ghosh A. K., Li K., Mesecar A. D., Baker S. C. (2010) Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84, 4619–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Báez-Santos Y. M., Mielech A. M., Deng X., Baker S., Mesecar A. D. (2014) Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J. Virol. 88, 12511–12527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee H., Cao S., Hevener K. E., Truong L., Gatuz J. L., Patel K., Ghosh A. K., Johnson M. E. (2013) Synergistic inhibitor binding to the papain-like protease of human SARS coronavirus: mechanistic and inhibitor design implications. ChemMedChem 8, 1361–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghosh A. K., Takayama J., Rao K. V., Ratia K., Chaudhuri R., Mulhearn D. C., Lee H., Nichols D. B., Baliji S., Baker S. C., Johnson M. E., Mesecar A. D. (2010) Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand x-ray structure and biological evaluation. J. Med. Chem. 53, 4968–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghosh A. K., Takayama J., Aubin Y., Ratia K., Chaudhuri R., Baez Y., Sleeman K., Coughlin M., Nichols D. B., Mulhearn D. C., Prabhakar B. S., Baker S. C., Johnson M. E., Mesecar A. D. (2009) Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome-coronavirus papain-like protease. J. Med. Chem. 52, 5228–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 27. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 28. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 32. Ratia K., Saikatendu K. S., Santarsiero B. D., Barretto N., Baker S. C., Stevens R. C., Mesecar A. D. (2006) Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U.S.A. 103, 5717–5722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A. D., Baker S. C. (2005) The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79, 15189–15198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shaw N., Ouyang S., Liu Z. J. (2013) Binding of bacterial secondary messenger molecule c di-GMP is a STING operation. Protein Cell 4, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bailey-Elkin B. A., Knaap R. C., Johnson G. G., Dalebout T. J., Ninaber D. K., van Kasteren P. B., Bredenbeek P. J., Snijder E. J., Kikkert M., Mark B. L. (2014) Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 289, 34667–34682 [DOI] [PMC free article] [PubMed] [Google Scholar]