Background: Eight glycoside hydrolases (GHs) encoded by genes belonging to one operon have been identified in the phytopathogenic bacterium Xanthomonas campestris.
Results: Five of these GHs are involved in the sequential degradation of a plant N-glycan in vitro.
Conclusion: This is the first evidence of N-glycan degradation by plant pathogen enzymes.
Significance: Our results suggest that N-glycans may be metabolized by phytopathogenic bacteria.
Keywords: Bacteria, Carbohydrate Processing, Enzyme Kinetics, Glycoside Hydrolase, N-Linked Glycosylation, Plant, Phytopathogen, Xanthomonas
Abstract
N-Glycans are widely distributed in living organisms but represent only a small fraction of the carbohydrates found in plants. This probably explains why they have not previously been considered as substrates exploited by phytopathogenic bacteria during plant infection. Xanthomonas campestris pv. campestris, the causal agent of black rot disease of Brassica plants, possesses a specific system for GlcNAc utilization expressed during host plant infection. This system encompasses a cluster of eight genes (nixE to nixL) encoding glycoside hydrolases (GHs). In this paper, we have characterized the enzymatic activities of these GHs and demonstrated their involvement in sequential degradation of a plant N-glycan using a N-glycopeptide containing two GlcNAcs, three mannoses, one fucose, and one xylose (N2M3FX) as a substrate. The removal of the α-1,3-mannose by the α-mannosidase NixK (GH92) is a prerequisite for the subsequent action of the β-xylosidase NixI (GH3), which is involved in the cleavage of the β-1,2-xylose, followed by the α-mannosidase NixJ (GH125), which removes the α-1,6-mannose. These data, combined to the subcellular localization of the enzymes, allowed us to propose a model of N-glycopeptide processing by X. campestris pv. campestris. This study constitutes the first evidence suggesting N-glycan degradation by a plant pathogen, a feature shared with human pathogenic bacteria. Plant N-glycans should therefore be included in the repertoire of molecules putatively metabolized by phytopathogenic bacteria during their life cycle.
Introduction
In the context of host-bacteria interactions, i.e. symbiosis or pathogenesis, the question of the degradation of host N-glycoproteins by microorganisms is relevant, and only a few studies demonstrating orchestrated processes involving catabolic enzymes have been reported. Glycosylation is the most widespread post-translational modification of proteins found in nature (1). It concerns not only eukaryotes but also some bacteria and archaea for which several examples of sugar attachment on proteins have been reported (2, 3). In eukaryotes, two types of glycosylation are described, N- and O-glycosylation. Whereas O-glycosylation can display a huge diversity, in terms of structure and of the sequence surrounding the amino acid carrier, N-glycosylation is more conserved (4). N-Glycosylation occurs in the secretory pathway, by adding a N-glycan to the amide group of asparagine in the Asn-Xaa-Ser/Thr context, where Xaa can be any amino acid except Pro. It starts in the endoplasmic reticulum by the co-translational transfer of a preformed lipid-linked oligosaccharide onto the nascent polypeptide. Then maturation steps involving glycosidases and glycosyltransferases take place in the Golgi apparatus through secretion of proteins to their final destination. Resulting N-glycans can be of three types, either high mannose-, hybrid-, or complex-type (4). This global mechanism is common to most eukaryotic systems, but the action of specific maturation enzymes leads to different complex N-glycan structures. For instance, in plants, the first GlcNAc of the core can be substituted with an α-1,3-fucose, instead of an α-1,6-fucose for human N-glycans. In addition, the β-mannose of the core can be decorated with a β-1,2-xylose in plants, whereas a β-1,4-GlcNAc can be found at this position in human glycans. From a functional point of view, N-glycans can modify the folding, the activity, or the stability of proteins and subsequently play key roles in a number of physiological processes, such as signal transduction, targeting, cell-cell recognition, infection, and immunity (5–7).
Investigation into the alteration of N-glycan structure, because of either aberrant N-glycosylation build-up or degradation, has focused on a better understanding of the molecular basis of these biological processes. N-Glycan degradation involves carbohydrate-active enzymes, in particular glycoside hydrolases (GHs)8 presently classified into 133 families (8). In addition, synergic processes may also occur, in particular involving glycoside phosphorylases (9, 10). To date, the most documented model of glycoconjugate breakdown concerns the gut bacterium Bacteroides thetaiotaomicron, which displays a broad repertoire of endo- and exo-glycosidases designed to hydrolyze plant polysaccharides, as well as human N-glycans (11–14). The biological role of GHs involved in N-glycan degradation is unclear, but it may consist of mannose foraging for basic metabolic inputs. In Bacteroides fragilis, the don locus, which confers a selective advantage during extraintestinal infections, is involved in transferrin deglycosylation (15). Enterococcus faecalis, a nosocomial human pathogen, produces an endo-β-N-acetylglucosaminidase able to release high mannose-type N-glycans from glycoproteins. This activity, associated with mannosidase action, may play a role in the survival and persistence of the pathogen in vivo (16). Similarly, Streptococcus oralis, responsible for infection of immunocompromised patients, was shown to deglycosylate complex-type N-glycans through a sequential mechanism. Released monosaccharides are assumed to be used by the bacteria to sustain growth (17). However, S. oralis enzymes responsible for N-glycan processing have not been identified. More recently, enzymes from Streptococcus pyogenes and Streptococcus pneumoniae classified in the GH38 (α-1,3-mannosidase) and GH125 (α-1,6-mannosidase) families, respectively, have been identified. These enzymes are active on N-glycans, highlighting the processing of N-glycans by Streptococcus bacteria (18, 19). Finally, it was shown that the human pathogen Capnocytophaga canimorsus deglycosylates surface glycoproteins from the host and supports its growth on the released glycan moiety (20). A large enzymatic complex has been identified, and a functional model of deglycosylation processing has been proposed (21).
Despite fast growing advances in understanding N-glycan processing by human pathogenic bacteria, nothing is known about the capacity of plant pathogens to achieve such mechanisms. In the plant pathogenic bacterium Xanthomonas campestris pv. campestris (Xcc) strain ATCC33913, we have recently identified an operon coding for eight GHs (NixE to NixL; N-acetylglucosamine-induced in Xcc) that belongs to the GlcNAc exploitation system. At the transcriptional level, the expression of nixE to nixL is induced in the presence of GlcNAc, and nixE to nixH are under the control of the LacI family NagR repressor, the repressive effect of which is relieved in the presence of GlcNAc (22). This operon was shown to be involved in the exploitation of GlcNAc-containing molecules derived from the plant during infection (22). Based on their predicted functions, Nix GHs were proposed to be involved in the cleavage of glycosidic bonds found in plant N-glycans, and the nixE-nixL cluster was called N-glycan cluster (22).
The objective of the present work was to experimentally demonstrate the involvement of Nix enzymes in plant N-glycan degradation. The predicted activities were first confirmed on pNP synthetic substrates, and the kinetic parameters were determined. Then Nix enzymes were shown to sequentially process an N-glycopeptide. Our data provide the first evidence of N-glycan degradation by a plant pathogenic bacterium in vitro and broaden the source of substrates putatively exploited by phytopathogens like Xanthomonas strains during infection. This study constitutes a new example of N-glycan processing by bacteria and provides new insights into the deglycosylation mechanism carried out by GHs.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The Xcc strains and plasmids used in this study are listed in Table 1. Xcc cells were grown at 28 °C in MOKA-rich medium (23) or in minimal medium for hrp gene expression (MME) (24). Escherichia coli cells were grown on LB medium at 37 °C. For solid medium, agar was added to 1.5% (w/v) final concentration. Antibiotics were used at the following concentrations for Xcc: 50 μg·ml−1 rifampicin, 50 μg·ml−1 kanamycin, and 5 μg·ml−1 tetracycline. Antibiotics were used at the following concentrations for E. coli: 50 μg·ml−1 ampicillin, 50 μg·ml−1 kanamycin, and 10 μg·ml−1 tetracycline.
TABLE 1.
Plasmids and strains used or generated in this study
| Plasmids and strains | Descriptiona | Location of mutations or Xcc cloned sequences relative to the putative start codon | Reference |
|---|---|---|---|
| X. campestris pv. campestris | |||
| WT | Wild-type strain; rifampicin-resistant strain derivative of Xcc LMG568/ATCC33913 | Ref. 54 | |
| ΔnagR | ΔXCC3412; RifR | From start to stop | Ref. 28 |
| ΔnixE-nixL | ΔXCC2888–2895; RifR | From start of XCC2888 to stop of XCC2895 | Ref. 22 |
| ΔxpsD | ΔXCC0670 | From start to stop | This study |
| ΔxcsD | ΔXCC3425 | From start to stop | This study |
| ΔxpsD-xcsD | ΔXCC0670 ΔXCC3425 | From start to stop | This study |
| E. coli | |||
| TG1 | K-12 F′ [traD36 proAB+ lacIq lacZΔM15]supE thi-1 Δ (lac-proAB) Δ (mcrB-hsdSM)5, (rK−mK−) | Lucigen® | |
| Plasmids | |||
| pK18mobsacB | Mobilizable pBR322 derivative containing a genetically modified sacB gene; KmR | Ref. 30 | |
| pCZ917 | pFAJ1700 derivative, containing 2094 bp of pSC150 with lacI, Ptac promoter and T7 terminator; TetR AmpR | Ref. 28 | |
| p0; pCZ1016 | pCZ917 derivative, containing Ptac and T7 terminator, and without LacI gene; TetR AmpR | Ref. 27 | |
| pC-nixE-nixL | pCZ1016-XCC2888-XCC2895; TetR AmpR | From − 20 of XCC2888 to stop of XCC2895 | This study |
| pC-nixE | pCZ1016-XCC2888; TetR AmpR | From − 20 to stop | This study |
| pC-nixF | pCZ1016-XCC2889; TetR AmpR | From − 30 to stop | Ref. 22 |
| pC-nixG | pCZ1016-XCC28908004; TetR AmpR | From − 30 to stop | Ref. 22 |
| pC-nixH | pCZ917-XCC2891; TetR AmpR | From − 30 to stop | This study |
| pC-nixI | pCZ1016-XCC2892; TetR AmpR | From − 30 to stop | This study |
| pC-nixJ | pCZ1016-XCC2893; TetR AmpR | From − 25 to stop | This study |
| pC-nixK | pCZ1016-XCC2894; TetR AmpR | From − 30 to stop | This study |
| pC-nixL | pCZ1016-XCC2895; TetR AmpR | From − 26 to stop | This study |
| pC-nixE-8His | pCZ1016-XCC2888-8His; TetR AmpR | From − 20 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pC-nixF-8His | pCZ1016-XCC2889-8His; TetR AmpR | From − 30 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pC-nixG-8His | pCZ1016-XCC28908004-8His; TetR AmpR | From − 30 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pC-nixH-8His | pCZ917-XCC2891-8His; TetR AmpR | From − 30 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pC-nixI-8His | pCZ1016-XCC2892-8His; TetR AmpR | From − 30 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pC-nixJ-8His | pCZ1016-XCC2893-8His; TetR AmpR | From − 25 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pC-nixK-8His | pCZ1016-XCC2894-8His; TetR AmpR | From − 30 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pC-nixL-8His | pCZ1016-XCC2895-8His TetR AmpR | From − 26 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pC-pghAxc-8His | pCZ1016-XCC3459-8His; TetR AmpR | From − 30 to stop with sequence coding for eight histidines upstream of the stop codon | This study |
| pMAL-nixE | pMAL-c2-XCC2888; AmpR | From + 117 to stop | This study |
| pMAL-nixF | pMAL-c2-XCC2889; AmpR | From + 69 to stop | This study |
| pMAL-nixG | pMAL-c2-XCC28908004; AmpR | From + 102 to stop | This study |
| pMAL-nixH | pMAL-c2-XCC2891; AmpR | From + 84 to stop | This study |
| pMAL-nixI | pMAL-c2-XCC2892; AmpR | From + 84 to stop | This study |
| pMAL-nixJ | pMAL-c2-XCC2893; AmpR | From + 153 to stop | This study |
| pMAL-nixK | pMAL-c2-XCC2894; AmpR | From + 69 to stop | This study |
| pMAL-nixL | pMAL-c2-XCC2895; AmpR | From + 69 to stop | This study |
a AmpR, KanR, RifR, and TetR indicate resistance to ampicillin, kanamycin, rifampicin, and tetracyclin, respectively.
Plasmid Constructions
DNA manipulations followed the procedures described by Sambrook et al. (25). All constructs were verified by sequencing.
nixE to nixL single genes were amplified by PCR using appropriate oligonucleotide primers. In Xcc ATCC33913 strain, insertion of one nucleotide five codons upstream of the stop codon of nixG has led to a frameshift with an extension of 281 amino acids as compared with NixG of all Xcc strains sequenced to date including the most related one, Xcc strain 8004 (26). Therefore, for this gene, we constructed plasmids expressing NixG8004 (named NixG under “Results”).
For complementations, PCR products were cloned into the multiple cloning site of the complementation plasmid pCZ1016 (27) (or pCZ917 (28) for nixH because we were not able to introduce pCZ1016-nixH in Xcc). Plasmids were named pC-nixE to pC-nixL, respectively (Table 1). The nixE-nixL cluster was subcloned into the multiple cloning site of the pCZ1016 plasmid from the pCZ1034-nixE-nixL (22).
For protein expression, we cloned nixE to nixL without the sequence encoding the N-terminal signal sequence into pMAL-c2 in frame with malE, which encodes the maltose-binding protein (MBP, translational fusion at the N-terminal end of Nix proteins; New England Biolabs, Inc.). The plasmids obtained were named pMAL-nixE to pMAL-nixL (Table 1). No activity could be detected for MBP-fused NixH, NixI, and NixK expressed in E. coli. Therefore, in a second set of experiments, we cloned nixE to nixL genes into pCZ1016 (or pCZ917 for nixH; see above) with a sequence coding for eight histidines at the 3′ end of the gene upstream of the stop codon. Plasmids were named pC-nixE-8His to pC-nixL-8His, respectively (Table 1), and proteins were expressed in Xcc. Plasmids were introduced into E. coli by electroporation and into Xcc strains by triparental conjugation as described by Turner et al. (29).
Construction of Xcc Deletion Mutants
Deletion mutants were constructed using the sacB system (30). PCR amplicons of 700 bp corresponding to regions located upstream and downstream of the region to delete were cloned into the pK18mobsacB plasmid. The deleted regions are indicated in Table 1.
Enzymatic Activity Analyses Using para-Nitrophenyl (pNP) Glycosides on Xcc Total Extracts
pNP-α-l-fucopyranoside (pNP-α-Fuc), pNP-N-acetyl-β-d-glucosaminide (pNP-β-GlcNAc), pNP-N-acetyl-β-d-galactosaminide (pNP-β-GalNAc), pNP-β-d-mannopyranoside (pNP-β-Man), pNP-β-d-xylopyranoside (pNP-β-Xyl), pNP-α-d-mannopyranoside (pNP-α-Man), and pNP-β-d-galactopyranoside (pNP-β-Gal) were obtained from Sigma-Aldrich. All these substrates were diluted in dimethylformamide at 100 mg·ml−1 except pNP-β-GalNAc, which was diluted at 25 mg·ml−1.
Enzymatic activities in Xcc were analyzed as previously described for β-glucuronidase assays (31). Briefly, after overnight precultures in MOKA-rich medium, bacterial cells were harvested by centrifugation, washed, and resuspended in MME minimal medium. 5 ml of MME minimal medium were inoculated at an A600 of 0.2, and 5 ml of MME supplemented with 10 mm GlcNAc or MOKA media were inoculated at an A600 of 0.1. After 6 h of growth at 28 °C, cells were centrifuged 5 min at 9000 × g and resuspended in water, and A600 was measured. For all the substrates except pNP-α-Man, 40 μl of 5× β-glucuronidase extraction buffer (250 mm NaPi, pH 7, 50 mm β-mercaptoethanol, 50 mm EDTA, pH 8, 0.5% Triton X-100, 0.5% sodium lauryl sarcosine (32)) were added to 160 μl of bacterial cells. For pNP-α-Man substrate, 160 μl of cells were centrifuged and resuspended in 200 μl of BugBuster Master Mix (Novagen®; EMD Chemicals Inc.). After 15 min at 37 °C, pNP substrates were added at a final concentration of 1 mg·ml−1. When the coloration turned to yellow, the reaction was stopped by addition of 100 μl of 1 m Na2CO3 and incubated 10 min on ice. After centrifugation for 5 min at 11,000 × g, A415 and A550 were measured using the supernatant. Activities were given as Miller units [1000 × (A415 − 1.75 × A550)/(timemin × volumeml × A600)] (31).
Subcellular Localization of Nix Enzymes
The localization of Nix enzymes was analyzed in Xcc strains carrying plasmids expressing His-tagged proteins. Cell lysate fractions, periplasmic-enriched fractions, and supernatant fractions were prepared from the same overnight culture. Cell lysate fractions were prepared using MagneHisTM purification system (Promega) from the pellet of 4 ml of culture. Proteins were eluted in 70 μl, and 10 μl were loaded on the gel.
Periplasmic-enriched fractions were prepared from the pellet of 2 ml of culture using chloroform extraction as described (33). His-tagged proteins were further purified as described above. Proteins were eluted in 50 μl, and 20 μl were loaded on the gel.
Supernatant fractions were prepared from the supernatant of 6 ml of culture. Remaining cells were removed by filtration on 0.45-μm filters (Millipore). Proteins were concentrated using Amicon® Ultracel-30K (Millipore) by centrifugation at 4000 × g to obtain a final volume of 100 μl. Ten μl were loaded on the gel. His-tagged proteins were detected by Western blot using anti-His antibody (GE Healthcare) after separation on 10% SDS-polyacrylamide gels and transfer to Immobilon-P (Millipore).
Recombinant Protein Production and Purification
For each NixX-8His enzymes, 100 ml of MOKA-rich medium were inoculated at an A600 of 0.15 from overnight precultures of Xcc WT strains containing pC-nixX-8His plasmids (Table 1). After about 6 h at 28 °C (i.e. to an A600 of 0.6), cultures were centrifuged at 4000 × g for 20 min at 4 °C. Pellets were stored at −20 °C prior to enzyme purification. Pellets (corresponding to 25 ml of cultures) were resuspended in 2 ml of 1× β-glucuronidase extraction buffer (see above) and incubated for 15 min at 37 °C. Supernatants obtained after centrifugation at 9000 × g for 15 min at 4 °C corresponded to crude extracts (CEs). These CEs were dialyzed against 50 mm sodium acetate buffer (pH 5) using MidiGeBAflex tubes (Gene Bio-Application) and used for deglycosylation tests of N-glycopeptides. Alternatively, CEs were used for affinity purification. Enzymes were purified either one by one or in mixture (from independent cultures), with a maximum of three proteins co-purified. Depending on the level of expression and on the concentration required for each purified protein, 1–4 ml of CE were incubated with 100–500 μl of MagneHisTM resin (Promega). Proteins purification was performed following supplier's instructions. The first elution fraction (200 μl to 1 ml) was dialyzed against optimal buffer (Table 2) using MidiGeBAflex tubes (Gene Bio-Application). Dialyzed fractions were used for enzyme assays, both for kinetic parameter determination and for N-glycopeptide degradation.
TABLE 2.
Enzymatic behavior of Xcc NixX-8His recombinant enzymes
Optimal conditions and apparent kinetic parameters were determined for hydrolysis of pNP synthetic substrates. Kinetic parameters were measured from three independent experiments. They were not measurable for NixF and NixJ enzymes because appropriated substrates are not available. Activity on N-glycopeptide observed in this study is reported. nd, not determined; ND, not detected.
| Protein | NixE-8His | NixF-8His | NixG-8His | NixH-8His | NixI-8His | NixJ-8His | NixK-8His | NixL-8His |
|---|---|---|---|---|---|---|---|---|
| Activity | α-1,3-Fucosidase | Endo-β-N-acetyl-glucosaminidase | β-N-Acetyl-glucosaminidase | β-1,4-Mannosidase | β-1,2-Xylosidase | α-1,6-Mannosidase | α-1,3-Mannosidase | β-Galactosidase |
| GH familya | GH29 | GH18 | GH20 | GH2 | GH3 | GH125 | GH92 | GH35 |
| Synthetic substrate used | pNP-α-Fuc | Not available | pNP-β-GlcNAc | pNP-β-Man | pNP-β-Xyl | Not available | pNP-α-Man | pNP-β-Gal |
| Optimal conditionsb | pH 5, 37 °C | pH 6, 37 °Cc | pH 6, 37 °C | pH 5, 37 °C | pH 5, 37 °C | ND | pH 5, 37 °C | pH 6, 37 °C |
| Km app (mm) | 0.7 ± 0.1 | nd | 0.4 ± 0.1 | 7.4 ± 2.6 | 0.47 ± 0.16 | ND | 1.2 ± 0.2 | 0.24 ± 0.01 |
| kcat app (s−1) | 6.1 ± 2.0 | nd | 4.2 ± 2.2 | 1552.3 ± 267.7 | 60.5 ± 17.2 | ND | 96.0 ± 12.9 | 42.0 ± 7.4 |
| kcat app/Km app (s−1. mm−1) | 8.71 | nd | 10.5 | 209.77 | 128.72 | ND | 80.0 | 175.0 |
| Activity on N-glycopeptided | Yes (4 or 5) | ND | ND | Yes (4 or 5) | Yes (2) | Yes (3) | Yes (1) | ND |
a According to CAZy.
b Buffers used were 50 mm sodium acetate and 50 mm MES for pH 5 and 6, respectively.
c Determined on chitooligosaccharides using MBP-NixF.
d The activity on N-glycopeptide is indicated, as well as the order (in parentheses) in which each GH is assumed to be involved.
MBP-fused Nix proteins were expressed in E. coli and purified by affinity chromatography (New England Biolabs, Inc). Elution fractions (50 μl) were dialyzed against optimal buffer (Table 2) using MidiGeBAflex tubes and used for enzymatic assays. Protein content was estimated at each step of the purification using the Bradford method (34), and proteins were analyzed on 12.5% SDS-polyacrylamide gels (35) stained with colloïdal blue using PageBlueTM Protein Staining Solution (Fermentas).
Determination of Enzymatic Parameters
Enzymatic parameters of NixX-8His proteins purified from Xcc and/or MBP-Nix proteins purified from E. coli were determined using pNP-glycosides corresponding to each enzymatic activity. To determine optimal conditions, assays were first performed from 4 to 60 °C and from pH 3.5 to 7 (sodium citrate, sodium acetate, sodium succinate, and MES buffers were tested) using substrates at a final concentration of 1 mg·ml−1 (i.e. from 3.3 to 3.7 mm) and purified enzymes. Enzymatic assays were carried out in triplicate at 37 °C in optimal buffer (50 mm sodium acetate, pH 5, or 50 mm MES, pH 6; Table 2). Kinetic parameters were determined using 3.7 to 12.9 μg·ml−1 (final concentrations) of NixX-8His purified enzyme by quantifying pNP release rate from pNP-glycosides at final concentrations ranging from 0.1 to 40 mm. Assays were achieved in microplates in 200 μl of final volume, and the A405 was measured at regular time intervals using TriStar LB 941 microplate reader (Berthold Technologies) for up to 16 h. The extinction coefficients of pNP were determined experimentally in 50 mm sodium acetate buffer (pH 5; 137.3 m−1·cm−1) and in 50 mm MES (pH 6; 739.9 m−1·cm−1), and were used to calculate pNP concentrations. The apparent kinetic parameters were determined by fitting the initial rates of pNP release to the Michaelis-Menten equation.
Thin Layer Chromatography
E. coli recombinant strains harboring pMAL-nixF and pMAL-nixG8004 constructs were harvested from overnight precultures and used to inoculate 100 ml of fresh LB medium at a final A600 of 0.1. Growth was subsequently allowed until A600 reached 0.5. Isopropyl β-d-thiogalactopyranoside was added at 200 μm final concentration, and cultures were continued for 2 h. Bacterial cells were recovered by centrifugation and resuspended in 25 ml of lysis buffer (20 mm Tris-HCl, pH 6, 200 mm NaCl, 1 mm EDTA, 10 mm β-mercaptoethanol). Crude enzymatic preparations were obtained after applying several cycles of French press and after centrifugation at 5000 × g for 5 min to eliminate the remaining debris. Total soluble protein concentration was estimated as described above. Chitooligosaccharides (Megazyme) were incubated at a final concentration of 10 mm with 1 μg of crude enzymatic preparation in reaction buffer (0.1 m phosphate buffer, pH 6, optimum pH). Reactions were performed at 37 °C for 24 h in 20 μl. Degradation products were separated on a TLC plate (SilicaGel 60; Merck) with a mobile phase consisting of a mix of butanol/water/acetic acid (2/2/1). Chitooligosaccharides and GlcNAc were revealed with the aniline/diphenylamine/o-phosphoric acid reagent used as a dipping solution (2 g of diphenylamine, 2 ml of aniline, 15 ml of o-phosphoric acid, 85 ml of acetone). After immersion in dipping solution for 2 s, the plate was heated at 100 °C for at least 10 min.
N-Glycopeptide Preparation
An N-glycopeptide was prepared from a commercial recombinant avidine from egg white produced in corn (Sigma-Aldrich). After solubilization in water, 2.5 mg of avidine were separated by SDS-PAGE. Bands of interest were excised from the gel following a slight coloration using Coomassie Blue R-250 at 0.003% (Sigma-Aldrich). Gel pieces were washed in 0.1 m acetonitrile/NH4HCO3 (50/50 v/v) and incubated for 45 min in 0.1 m DTT (Sigma-Aldrich) in 0.1 m NH4HCO3 at 56 °C. After DTT removal, proteins were treated with 55 mm iodoacetamine (Sigma-Aldrich) in 0.1 m NH4HCO3 for 20 min under gentle shaking at room temperature. Proteins were then digested in gel using trypsin (Promega; 1 μg for 40 μg of avidine) overnight at 37 °C. After trypsin denaturation for 10 min at 95 °C and supernatant removal, peptides were extracted from the gel by three successive washes using acetonitrile, 0.1 m NH4HCO3, and acetonitrile. The three supernatants were pooled and concentrated using SPD SpeedVac® (Thermo Scientific). The resulting sample was resuspended in 1.5 ml of concanavalin A (ConA) binding buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm each MgCl2/MnCl2/CaCl2) for subsequent purification of N-glycopeptide by affinity chromatography on ConA lectin. ConA chromatography was carried out using 0.5 ml of ConA-agarose matrix (Sigma-Aldrich) in batch mode by decantation. Before equilibration, resin was conditioned with 5 ml of prewash buffer (20 mm Tris-HCl, pH 7.4, 1 m NaCl, 3.33 mm each of MgCl2, MnCl2, and CaCl2) to remove unbound lectins. After resin equilibration with 5 ml of ConA binding buffer, avidine tryptic digest was put in contact with the matrix for 1 h at room temperature. After flow-through removal, the resin was washed three times by 0.5 ml of binding buffer. Glycopeptides were eluted with 3 × 0.25 ml of binding buffer supplemented with 1 m methyl-α-d-glucopyranose (Sigma-Aldrich). Elution fractions were concentrated three times using SPD SpeedVac® and stored at −20 °C before use. Elution fraction 2 was used for enzymatic assays.
N-Glycopeptide Degradation
NixX-8His purified enzymes were tested using N-glycopeptide as substrate, either alone or in mixture. For mixture assays, enzymes were introduced either all together at t0 or sequentially during kinetics. Substrate (ConA elution fraction 2, diluted 5×) was incubated with enzyme preparations (final concentration, 6–72 μg·ml−1) at 37 °C in 50 mm sodium acetate buffer (pH 5) supplemented with 2 mm CaCl2. Reactions were carried out in low volumes (10 μl) for up to 72 h. Samples (2 μl) were taken regularly during the kinetics, with highest frequency during the first 4 h following enzyme addition. Reactions were stopped by heating at 95 °C for 5 min. Samples were stored at −20 °C till their analysis by MALDI-TOF MS.
MALDI-TOF MS Analysis of N-Glycopeptides
MALDI-TOF MS analyses were performed using a Voyager-DE STR mass spectrometer (Applied Biosystems/MD SCIEX). An aliquot of 1 μl of each sample was spotted on the mass spectrometer plate (MALDI sample plate SS; Applied Biosystems) with 1 μl of the matrix solution (6 g·l−1 α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 0.1% trifluoroacetic acid). Spectra were acquired in positive reflectron mode with the following parameters: accelerating voltage, 20 kV; grid voltage, 68%; extraction delay time, 200 ns; and shoot number, 1000. Acquisition m/z range was between 750 and 3000. Mass calibration was performed using standard peptide mixture (Applied Biosystems). Spectra were treated using the Data Explorer software (Applied Biosystems).
RESULTS
The N-Glycan Cluster of Xcc Is Involved in N-Glycan Degradation
MALDI-TOF analysis of the enriched N-glycopeptide from avidine expressed in corn reveals a major peak of 3006.31 m/z corresponding to an N2M3FX N-glycan (two GlcNacs (N), three mannoses (M), one fucose (F), and one xylose (X); Fig. 1, A and B) attached to an asparagine residue (N underlined in the peptide sequence hereafter) on the WTNDLGSNMTIGAVNSR tryptic peptide. Minor peaks observable on the mass spectrum (Fig. 1B) may correspond to other glycopeptides and/or peptides present as trace in the N-glycopeptide preparation. The N2M3FX motif, which corresponds to one of the most widespread N-glycan in plants, should allow observation of α- and β-mannosidases, β-xylosidase, α-fucosidase, and β-N-acetylglucosaminidase activities.
FIGURE 1.
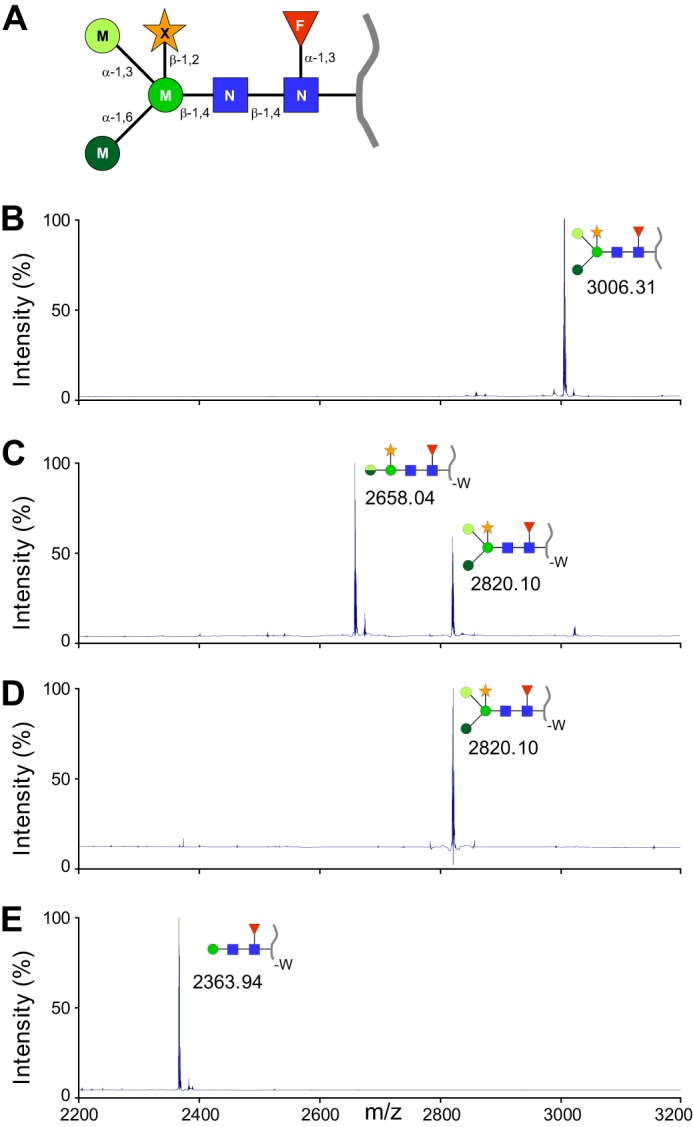
Nix enzymes encoded by Xcc are involved in the cleavage of the glycosyl moiety of a N-glycopeptide. A, schematic representation of the N-glycopeptide used in this study. The blue squares represent GlcNAc, the green circles represent mannose (with different color tones according to different glycosidic linkages), the red triangles represent fucose, and the orange stars represent xylose. B–E, N-glycopeptides were analyzed by MALDI-TOF MS. B, nontreated N-glycopeptide. C, N-glycopeptide incubated for 24 h in the presence of total extracts of Xcc wild-type strain (WT pC-0). For the 2658.04 m/z N-glycopeptide, the two half-circles represent the remaining mannose, which can be either an α-1,3-mannose (upper half-circle, light green) or an α-1,6-mannose (lower half-circle, dark green). D, N-glycopeptide incubated for 24 h in the presence of total extracts of Xcc ΔnixE-nixL pC-0 strain. E, N-glycopeptide incubated for 24 h in the presence of total extracts of the complemented ΔnixE-nixL pC-nixE-nixL strain. The peptide moiety of the N-glycopeptide (WTNDLGSNMTIGAVNSR, B–E) is represented by a gray line. -W, loss of the N-terminal tryptophan residue on the peptide moiety.
Enzymatic assays with Xcc total extracts were followed by MALDI-TOF MS analyses of the N-glycopeptide substrate and products. Three Xcc strains were used as enzyme sources: (i) the wild-type strain (WT pC-0), (ii) the strain deleted of the N-glycan cluster (ΔnixE-nixL pC-0), and (iii) the complemented strain (ΔnixE-nixL pC-nixE-nixL). Regarding the nontreated N-glycopeptide substrate profile (Fig. 1B), two products corresponding to peaks at 2820.10 and 2658.04 m/z were observed after 24 h of reaction in presence of dialyzed CEs of WT strain (Fig. 1C), indicating a disappearance of the substrate. We assume that the peak at 2820.10 m/z corresponds to the loss of the N-terminal tryptophan residue (−186.21 Da) of the peptide and the peak at 2658.04 m/z to the additional loss of a hexose residue (−162.06 Da). Degradation of the peptide indicates aminopeptidase activity, whereas the loss of a carbohydrate moiety attests of a mannosidase activity because the substrate only contains mannose hexoses. However, this mannosidase activity was not observed when using the deleted strain because we only observed the 2820.10 m/z peak (Fig. 1D), suggesting that the N-glycan cluster is responsible for mannose removal. Interestingly, the reaction carried out with CE of the complemented strain overexpressing NixE to NixL resulted in a more advanced degradation of the N-glycopeptide. A peak at 2363.94 m/z corresponding to the loss of a tryptophan (−186.21 Da), two mannoses (−2 × 162.06 Da), and one xylose (−132.04 Da) was observed after 24 h (Fig. 1E). This reveals two additional enzymatic activities, i.e. another mannosidase and a xylosidase, overexpressed in the complemented strain. These results indicate that at least three enzymes encoded by the N-glycan cluster of Xcc are involved in N-glycan degradation in vitro.
Validation of Predicted Nix Enzymatic Activities in Xcc
To study the enzymatic activities of Nix GHs encoded by the N-glycan cluster, we first analyzed the ability of Xcc strains to degrade synthetic substrates. Using synthetic pNP-glycoside substrates, α-fucosidase, β-N-acetylglucosaminidase, β-mannosidase, β-xylosidase, α-mannosidase, and β-galactosidase activities were detected in the Xcc WT strain and were reduced in the ΔnixE-nixL deleted strain (Fig. 2A). These activities were still detected in the deleted strain, suggesting that Xcc codes for other enzymes active on these pNP substrates that are not encoded by the N-glycan cluster. All the tested activities were considerably enhanced in the complemented ΔnixE-nixL pC-nixE-nixL strain, confirming that these enzymatic activities are encoded by the N-glycan cluster.
FIGURE 2.
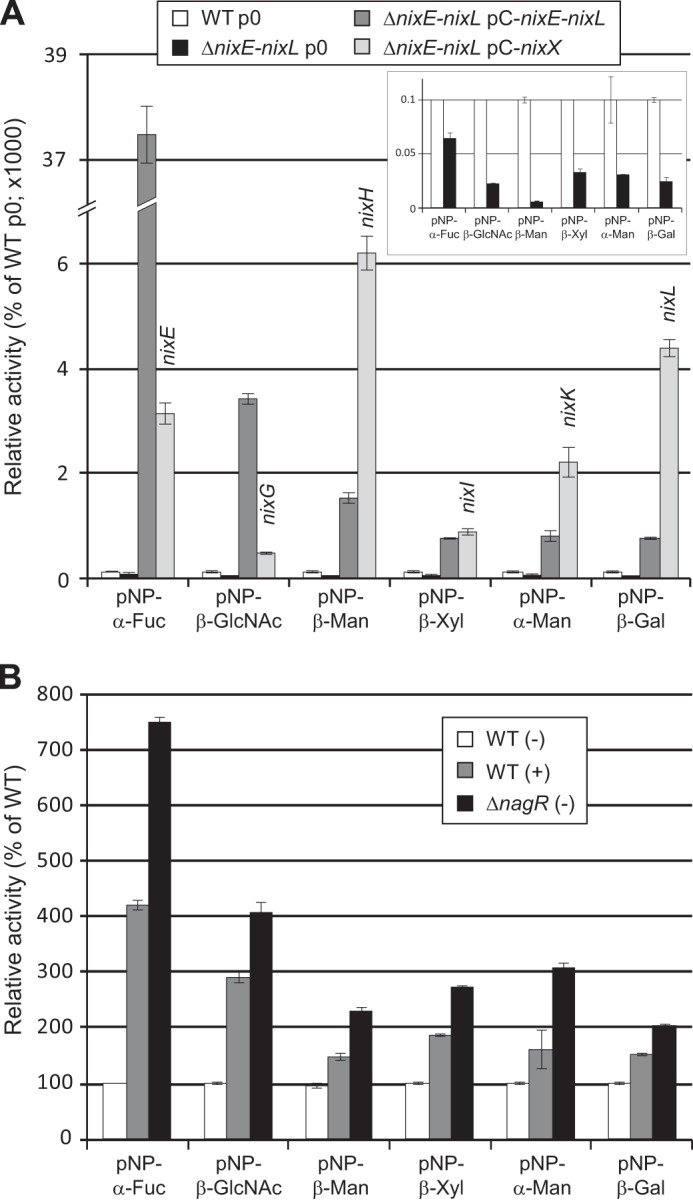
Xcc nix genes encode GHs active on synthetic pNP substrates. Enzymatic activities were analyzed on total cell lysates of different Xcc strains, and activities were calculated as Miller units. A, strains were cultivated for 6 h in MOKA-rich medium, and activities were represented as percentages of the wild-type strain (WT p0). For complemented strains with plasmids expressing one nix gene (ΔnixE-nixL pC-nixX strains), the gene expressed is indicated above the histogram bar. The inset corresponds to an enlargement of the histogram for WT p0 and ΔnixE-nixL p0 strains. B, strains were cultivated for 6 h in MME minimal medium supplemented (+) or not (−) with GlcNAc at 10 mm. Activities were represented as percentages of the WT strain. The error bars indicate the standard deviations obtained from three independent experiments.
Using plasmids expressing each Nix enzyme in the ΔnixE-nixL deleted strain, we confirmed the predicted activities of NixE (α-fucosidase, GH29), NixG (β-N-acetylglucosaminidase, GH20), NixH (β-mannosidase, GH2), NixK (α-mannosidase, GH92), and NixL (β-galactosidase, GH35) (Fig. 2A). Introduction of the plasmid pC-nixI complemented the enzymatic activity of the ΔnixE-nixL strain on pNP-β-Xyl, suggesting that NixI (annotated as a β-glucosidase, GH3) displays a β-xylosidase activity. The plasmid expressing nixF (predicted to code for an endo-β-N-acetylglucosaminidase, GH18) did not complement the activity of the ΔnixE-nixL strain on pNP-β-GlcNAc, consistent with the endo-activity predicted for this enzyme (data not shown). Similarly and as reported for enzymes belonging to the GH125 family (19), the plasmid expressing nixJ (predicted to code for an α-1,6-mannosidase, GH125) was not active on pNP-α-Man (data not shown). Activity on pNP-Fuc was 11-fold lower when NixE was expressed from pC-nixE as compared with expression from pC-nixE-nixL (Fig. 2A), although the 5′ ends of fragments cloned in both plasmids were identical. Because a similar profile was observed for NixG, this could either be due to a difference in stability of transcripts or to the fact that enzymes of the cluster form a complex that stabilizes these proteins. Conversely, activities observed for NixH, NixK, and NixL were higher when their translation start is located at the 5′ end of the transcript (expression from pC-nixX) as compared with its location in a polycistronic transcript (expression from pC-nixE-nixL).
Enzymatic activities encoded by the N-glycan cluster were all induced both when cells were cultured in presence of GlcNAc and in the ΔnagR strain as compared with medium without GlcNAc and WT strain, respectively (Fig. 2B). These results confirm that the entire N-glycan cluster is under the control of GlcNAc via NagR, even if we did not observe transcriptional regulation by NagR for nixI to nixL (22).
It should be noted that although NixG was active on pNP-β-GalNAc, the level of activity was at least 15-fold lower than when observed using pNP-β-GlcNAc (3 ± 0 compared with 57 ± 3 Miller units, respectively, in the WT strain, and 33 ± 2 compared with 550 ± 25 in the ΔnixE-nixL pC-nixG strain), suggesting that its preferred substrate is β-GlcNAc. This is consistent with a common feature of enzymes belonging to the EC 3.2.1.52 class (β-N-acetyl-hexosaminidase), which are active on both pNP-β-GlcNAc and pNP-β-GalNAc with substrate preference for pNP-GlcNAc (36).
Determination of Enzymatic Parameters of NixX-8His on pNP Synthetic Substrates
Kinetic parameters of NixX-8His recombinant proteins were determined on pNP synthetic substrates. The presence of the His-tag did not affect enzymatic activities of the corresponding proteins because activities on pNP synthetic substrates with total extracts of bacterial strains expressing either tagged and nontagged proteins were not significantly different (data not shown). NixX-8His proteins were purified by affinity chromatography using paramagnetic precharged nickel particles. For each of them, colloïdal blue staining of elution fractions after SDS-PAGE separation showed a unique band at the expected molecular mass (data not shown). Protein content of purified preparations was estimated between 6 and 36 μg·ml−1.
After the optimal buffer, pH and temperature were determined (Table 2), kinetic parameters were measured in the optimal conditions for each NixX-8His protein. Note that enzymatic parameters could not be determined for NixF- and NixJ-8His enzymes because no synthetic substrates were available for them. All the tested enzymes obey Michaelis-Menten kinetics. Vmax and Km were determined from Lineweaver and Burk linearization by plotting 1/Vmax versus 1/[substrate] at various concentrations of pNP-carbohydrates (Table 2). Km values are all in the mm order of magnitude (0.24–7.4 mm), whereas the kcat values are more disparate (6.1–1552 s−1). Catalytic efficiency (kcat/Km) ranged from 8.7 to 209.7 s−1·mm−1, with NixH and NixE as the most and the least efficient enzymes toward pNP-carbohydrates, respectively (Table 2).
NixF Is an Endo-β-N-acetylglucosaminidase and NixG Is an Exo-β-N-acetylglucosaminidase
NixF is annotated as an endo-β-N-acetylglucosaminidase. We therefore analyzed its ability to hydrolyze glycosidic bonds between two GlcNAc residues. For that purpose, chitooligosaccharides from polymerization degrees 3–6 (chitotriose to chitohexaose, respectively) were treated with total extracts of E. coli overexpressing NixF from plasmid pMAL and subsequently analyzed by TLC (Fig. 3). Chitohexaose and chitopentaose treated with total extracts of E. coli overexpressing MBP-NixF were degraded into chitotetraose + chitobiose and chitotriose + chitobiose, respectively. These results confirm that NixF is an endo-β-N-acetylglucosaminidase. Almost no degradation of chitotetraose or chitotriose by MBP-NixF was observed, suggesting that the minimal size of the substrate for this enzyme is a pentamer. Extracts of E. coli containing an empty pMAL were used as negative controls, and no degradation of chitooligosaccharides was observed. Similar reactions carried out using MBP-NixG led to the total degradation of all tested chitooligosaccharides into GlcNAc (Fig. 3), demonstrating that NixG is an exo-β-N-acetylglucosaminidase. This result corroborates the activity on pNP-GlcNAc demonstrated for NixG-8His (see above). This is also in agreement with its involvement in chitooligosaccharides utilization in Xcc (22) and with results obtained for its ortholog in the Xanthomonadaceae Stenotrophomonas maltophilia (Hex, 45.8% identity with NixG at the amino acid level) (37).
FIGURE 3.
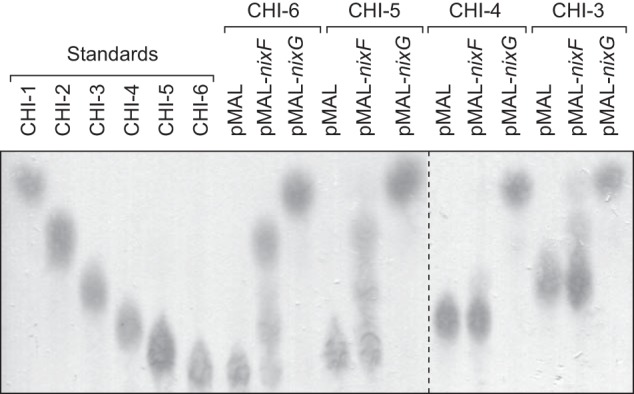
MBP-NixF protein of Xcc is an endo-β-N-acetylglucosaminidase, and MBP-NixG is an exo-β-N-acetylglucosaminidase. Chitooligosaccharides were analyzed by TLC after incubation during 24 h in presence of crude extracts of E. coli containing empty plasmid pMAL (pMAL) or pMAL expressing MBP-NixF (pMAL-nixF) or MBP-NixG (pMAL-nixG). CHI-1, N-acetylglucosamine; CHI-2, chitobiose; CHI-3, chitotriose; CHI-4, chitotetraose; CHI-5, chitopentaose; CHI-6, chitohexaose.
Recombinant NixX-8His Enzymes Are Active on an N-Glycopeptide Substrate
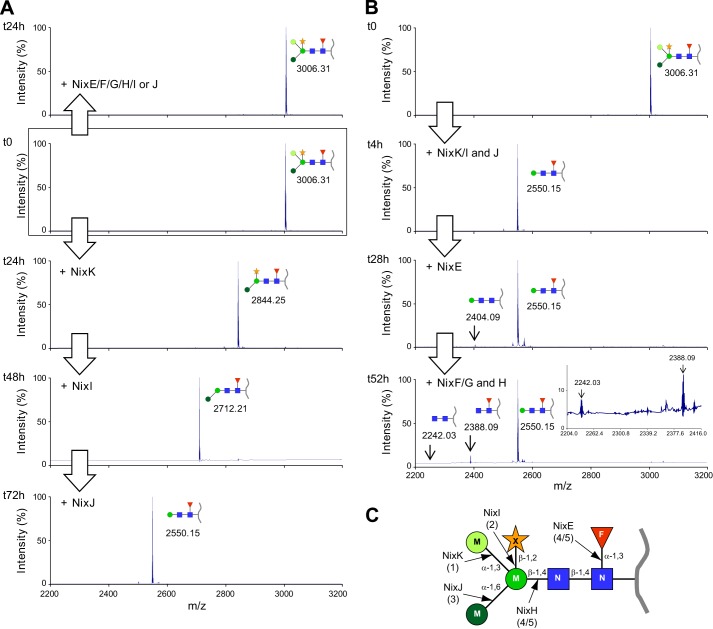
Following the enzymatic characterization of NixX-8His enzymes using synthetic substrates, we wanted to demonstrate their involvement into N-glycopeptide degradation. First, we performed assays in which the purified N-glycopeptide was incubated with a mixture of NixE-8His to NixK-8His enzymes purified individually. NixL, predicted as a β-galactosidase, was not included because the N-glycopeptide substrate does not contain a terminal galactose residue (Fig. 1A). Kinetic analyses of this reaction by MALDI-TOF MS allowed the detection of different degradation products of the N-glycopeptide (Fig. 4). It should be noted that the tryptophan moiety removal observed with crude Xcc extracts (see above, Fig. 1) was not observed here, indicating that NixX-8His enzymes were only involved in glycan degradation. After 30 min of reaction, the peak corresponding to the substrate (3006.31 m/z) almost disappeared, whereas two peaks at 2844.25 and 2712.21 m/z were detected (Fig. 4B). The first product corresponded to the loss of a hexose (−162.06 Da), and the second one corresponded to that of a pentose (−132.04 Da), namely mannose and xylose residues, respectively, according to the N-glycan structure (Fig. 1A). After 4 h, two additional products were detected, indicating that the substrate has been completely consumed. The peak detected at 2550.15 m/z corresponded to the loss of an additional mannose (−162.06 Da) and the peak at 2404.09 m/z to the additional loss of the fucose residue (−146.06 Da) (Fig. 4C). The major reaction product observed at the end of the reaction (peak at 2550.15 m/z described above) corresponds to the N2MF-glycopeptide. Shorter compounds (2242.03 and 2404.09 m/z) were only detected as traces. The smaller form corresponds to a glycopeptide with two GlcNAc residues (Fig. 4D). Altogether, our results demonstrate five enzymatic activities, i.e. two α-mannosidases (α-1,3 and α-1,6), a β-1,4-mannosidase, a β-1,2-xylosidase, and an α-1,3-fucosidase involved in the degradation of N-glycan (Fig. 1A). Considering results previously obtained with pNP substrates, these activities could be assigned to NixK (α-mannosidase), NixH (β-mannosidase), NixI (β-xylosidase), and NixE (α-fucosidase). In addition, the kinetic of formation of products suggested a sequential degradation of the glycopeptide.
FIGURE 4.
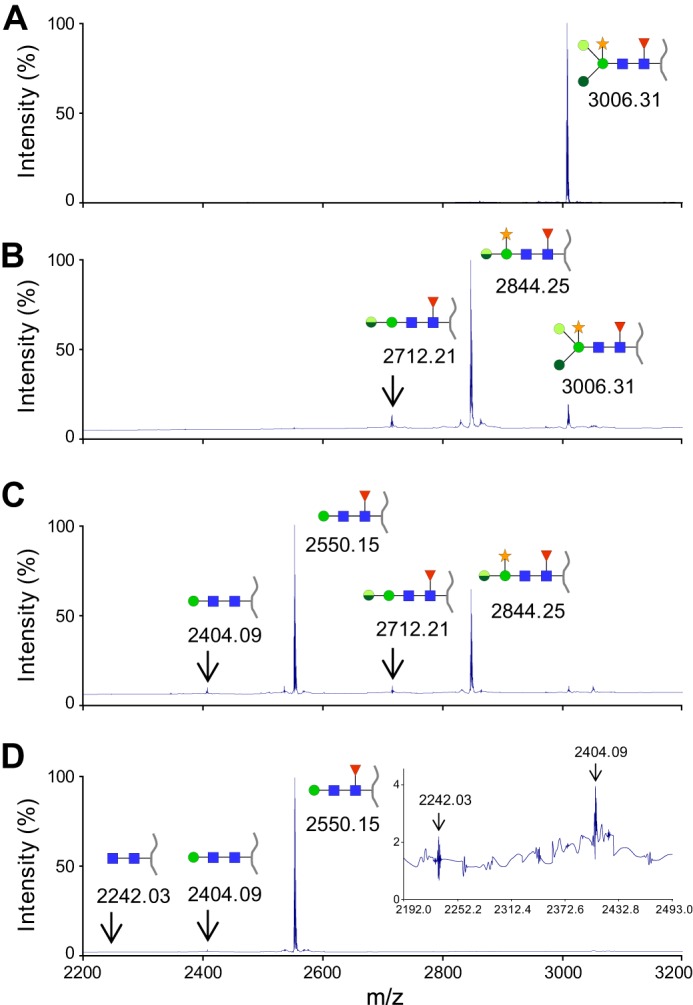
NixX-8His enzymes purified from Xcc are involved in N-glycopeptide degradation. N-Glycopeptides were analyzed by MALDI-TOF MS at different time points after addition of a mixture of purified His-tagged NixE-, NixF-, NixG-, NixH-, NixI-, NixJ-, and NixK-8His enzymes. A, nontreated N-glycopeptide. B, 30 min after addition of NixX-8His enzymes. C, 4 h after addition of NixX-8His enzymes. D, 48 h after addition of NixX-8His enzymes. The inset corresponds to an enlargement of the mass spectra showing peaks corresponding to N2 and N2M N-glycopeptides. The blue squares represent GlcNAc, the green circles represent mannose (with different color tones according to different glycosidic linkages), the red triangles represent fucose, and the orange stars represent xylose. The peptide moiety is represented by a gray line. For the 2844.25 and 2712.21 m/z N-glycopeptides, the two half-circles represent the remaining mannose, which can be either an α-1,3-mannose (upper half-circle, light green) or an α-1,6-mannose (lower half-circle, dark green).
The Degradation of N-Glycopeptide Is Sequential
To confirm the role of each enzyme and to go further into the mechanism of deglycosylation, we carried out assays by introducing each of the seven NixX-8His enzymes of the cluster (i.e. all except NixL-8His; see above) individually. NixK-8His was the only enzyme active on the N-glycopeptide substrate used in this study, indicating that NixK-8His initiates the deglycosylation, generating a product at 2844.25 m/z (Fig. 5A). Considering a possible sequential mechanism, we subsequently introduced in the reaction assay the NixI-8His enzyme, predicted to act on the NixK-8His product. A product at 2712.21 m/z was detected, confirming that NixI-8His hydrolyzed the xylose from the N-glycopeptide only after mannose removal (Fig. 5A). In the same way, sequential addition of NixJ-8His confirmed that it was able to remove a mannose residue from NixI-8His product. These data demonstrate that NixK-8His, NixI-8His, and NixJ-8His are responsible for the sequential degradation of the N-glycopeptide, leading to the product at 2550.15 m/z (peptide with N2MF glycan), which was shown to accumulate in our assay conditions. Interestingly, it should be noted that NixK-8His and NixJ-8His are both α-mannosidases but with different specificities because they are devoted to the removal of only one of the two α-mannose residues attached either by α-1,3 or α-1,6 linkages. Because NixJ was classified in the GH125 family containing only α-1,6-mannosidases (Carbohydrate-Active EnZymes database), we deduced that NixK-8His is an α-1,3-mannosidase. Assays carried out by adding subsequently other enzymes of the cluster (i.e. NixE-8His, NixF-8His, NixG-8His, and NixH-8His) were too diluted to detect significant MALDI-TOF MS signals (data not shown).
FIGURE 5.
Sequential degradation of the N-glycan from a N-glycopeptide by purified NixX-8His enzymes from Xcc. N-Glycopeptides were analyzed by MALDI-TOF MS at different time points after sequential addition of purified NixX-8His enzymes. A, identification of the three first cleavage steps of the N-glycan. When added individually, no cleavage was observed when either NixE-, NixF-, NixG-, NixH-, NixI-, or NixJ-8His was added (top panel). Deglycosylation of the N2M3FX N-glycopeptide was initiated by NixK-8His (α-1,3-mannosidase), then NixI-8His eliminated the xylose residue, and finally NixJ-8His removed the α-1,6-mannose residue. B, identification of final cleavage steps. NixK-, NixI-, and NixJ-8His were added simultaneously, and after 4 h, the product (N2MF) obtained was used as substrate for other NixX-8His enzymes. NixE-8His (α-fucosidase) and NixH-8His (β-mannosidase) were able to cleave fucose and mannose residues, respectively. The inset corresponds to an enlargement of the mass spectra showing peaks corresponding to N2 and N2F N-glycopeptides. C, schematic representation of the N-glycopeptide used in this study. Cleavages observed with purified Xcc NixX-8His enzymes are indicated. The numbers in parentheses indicate the order of action of enzymes. The blue squares represent GlcNAc (N), the green circles represent mannose (M, with different color tones according to different glycosidic linkages), the red triangles represent fucose (F), and the orange star represents xylose (X). The peptide moiety is represented by a gray line.
To overcome this limitation, we developed new assays by co-purifying NixK-8His, NixI-8His, and NixJ-8His from independent bacterial cultures. Enzyme concentrations were then high enough to observe a full conversion of the N-glycopeptide substrate to the 2550.15 m/z product after 4 h (Fig. 5B). NixE-8His, assumed to remove subsequently the fucose residue, was then introduced, and after 24 h of reaction, a product at 2404.09 m/z (peptide with N2M glycan) was detected as trace. This suggests that NixE-8His, although active on pNP-Fuc, is poorly efficient on the N-glycopeptide substrate at 2550.15 m/z, suggesting that another enzyme is needed prior to its activity. However, the addition of NixF-8His, NixG-8His, and NixH-8His, alone or in mixture, did not result in efficient degradation of the 2550.15 m/z substrate. Two new products were detected at 2388.09 and 2242.03 m/z. These correspond to the loss of a β-mannose residue (−162.06 Da) from the N2MF glycopeptide (at 2550.15 m/z) and of a fucose residue (−146.06 Da) from the N2F glycopeptide (at 2388.09 m/z), respectively (Fig. 5B). These data confirm that NixH-8His, predicted as a β-mannosidase, is a β-1,4-mannosidase and that NixE-8His is able to remove an α-1,3-fucose from NixH-8His product. No activities were observed for NixG-8His and NixF-8His enzymes.
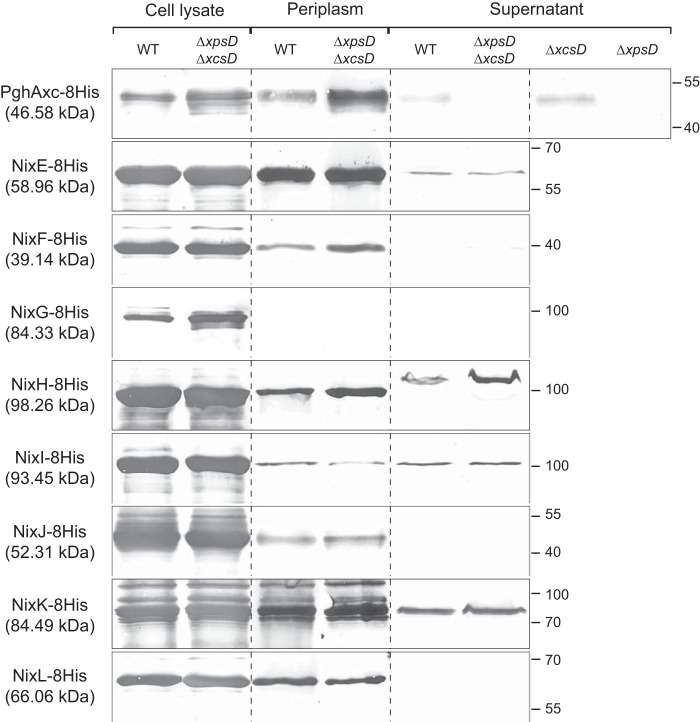
Subcellular Localization of Nix Enzymes
All Nix enzymes possess a signal peptide, suggesting that these proteins are translocated across the inner membrane to the periplasm (22). Xcc possesses two type II secretion systems (Xps and Xcs), the first one playing a major role in the secretion of plant cell wall degradative enzymes (38). Therefore, subcellular localization of Nix enzymes was analyzed in WT strain and strains with nonfunctional Xps and/or Xcs machinery (ΔxpsD, ΔxcsD and ΔxpsD ΔxcsD mutants). We performed Western analyses using different fractions (total cell lysate, periplasmic-enriched fraction, and concentrated supernatant) prepared from Xcc strains overexpressing 8His-tagged Nix proteins; for each strain, fractions loaded on the gel were normalized to correspond to the same number of bacterial cells. The polygalacturonase PghAxc (XCC3459) was used as a control. Indeed, in Xcc strain 8004, PghAxc is secreted via the type II secretion system in an Xps-dependent and Xcs-independent manner (39). In agreement with these observations in strain 8004, we detected PghAxc-8His from strain ATCC33913 in the supernatant fractions of both WT and ΔxcsD strains but not in ΔxpsD or ΔxpsD ΔxcsD mutants (Fig. 6). All NixX-8His enzymes were detected in total cell lysates. Except NixG-8His, all enzymes were also detected in the periplasmic enriched fraction of Xcc cells. NixE-, NixH-, NixI-, and NixK-8His were also detected in the culture supernatant of both WT and ΔxpsD ΔxcsD strains (Fig. 6), suggesting that these proteins are secreted in a type II secretion system-independent manner, as previously observed for amylase and cellulase activities in X. campestris pv. vesicatoria (40). NixF-8His was weakly detected in the culture supernatant of ΔxpsD ΔxcsD strain but not of the WT strain, which is probably due to the detection limits of the experiment. Finally, the absence of detection of NixG-, NixJ-, and NixL-8His in the supernatant and NixG-8His in periplasmic-enriched fractions of either WT or ΔxpsD ΔxcsD strains could be due to the detection limits of the experiment. Alternatively, NixG-8His could be cytoplasmic or associated with one membrane whereas NixJ- and NixL-8His would not be secreted in the supernatant and either remain in the periplasm or may be associated to the outer membrane. Surprisingly, NixH-8His was detected in the supernatant at an apparent molecular mass higher than that of the protein detected in the cell lysate and in the periplasmic enriched fraction (Fig. 6), suggesting that NixH-8His is modified during its translocation through the outer membrane.
FIGURE 6.
Subcellular localization of Xcc NixX-8His enzymes. Western blot analysis using anti-His antibody was carried out on total cell lysates, periplasmic enriched fractions, and cell-free supernatants of Xcc WT and type II secretion system mutants (ΔxpsD, ΔxcsD and/or ΔxpsD-ΔxcsD strains) overexpressing NixX-8His enzymes from replicative plasmids. PghAxc-8His was used as a control for protein secretion dependent on the Xps type II secretion system in Xcc. Molecular masses are indicated in parentheses beneath each protein name correspond to the predicted mass of the His-tagged protein without the signal peptide. Molecular mass markers (in kDa) are indicated on the right.
DISCUSSION
Xcc, the causal agent of black rot disease on Brassica plants, produces an extensive repertoire of GHs, lyases, and esterases (41) that target linkages present in plant cell wall polysaccharides such as cellulose, mannan, xylan, galacturonan, and pectin. In this paper, we characterized Nix GHs belonging to the Xcc GlcNAc utilization system on pNP synthetic substrates, and we demonstrated their ability to degrade a plant N-glycan. This is the first evidence of in vitro N-glycan degradation by a plant pathogen suggesting that N-glycans may by metabolized by phytopathogenic bacteria during infection. By using Xcc total extracts or His-tagged proteins purified from Xcc, we demonstrated that N-glycan processing is sequential. On the N2M3FX N-glycopeptide substrate, NixK initiates the deglycosylation by removing the α-1,3-mannose, NixI acts on the β-1,2-xylose in a second step, and then NixJ releases the α-1,6-mannose. NixE and NixH are assumed to act at a later stage, by removing the α-1,3-fucose and the β-1,4-mannose, respectively. This sequential mode of action corroborates literature data reporting that, in the context of fruit ripening, the removal of α-1,3-mannose from N-glycan is a prerequisite for hydrolysis of the β-1,2-xylose by a tomato xylosidase (42). In addition, in the human N-glycan degrading Firmicutes, a sequential model is proposed, with the removal of the α-1,3-mannose by a GH38 as a first step necessary for the activity of a GH125 subsequently involved in the removal of the α-1,6-mannose (19). Such common features suggest that N-glycan processing is of widespread importance, in particular for pathogens in a variety of host contexts.
In Xcc, although the N-glycan cluster potentially encodes all the enzymes required for complete degradation of the N2M3FX N-glycan, the N2MF product accumulates, suggesting that in our conditions, there is a limiting step for the N2MF glycopeptide degradation process. The reasons for this limiting step may be multiple. Indeed, it could be due to inappropriate condition assays or to the fact that the polypeptidic part of the glycopeptide provides a steric hindrance for NixE, NixF, and/or NixH activities. It is also possible that NixE, NixF, and/or NixH are not in a form active on N-glycans or that NixF is not able to accommodate in its active site the N-glycopeptide used in this study. Furthermore, we cannot exclude that other secreted critical enzyme(s), not encoded by the N-glycan cluster, are required for full N-glycan processing. Finally, this limiting step could be a strategy for the bacterium to partially deglycosylate plant N-glycoproteins. Indeed, in S. pneumoniae strain D39, although the genome codes for α- and β-mannosidases (belonging to GH2, GH38, GH92, and GH125 families; Carbohydrate-Active EnZymes database), deglycosylation of human complex-type N-glycoconjugates is partial with only removal of terminal sialic acid, galactose and GlcNAc residues. This led to the exposure of mannose residues on host glycoproteins, which plays a role in adherence of unencapsulated pneumococci on human cells (43).
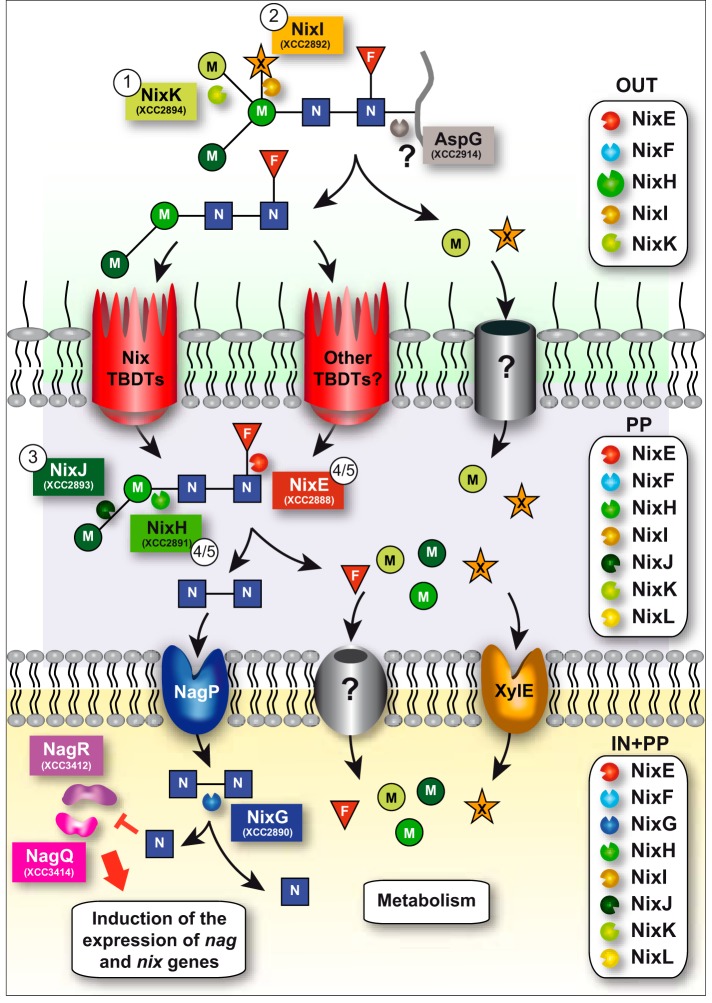
The results obtained here suggest that in Xcc, as observed in S. oralis (17) and S. pneumoniae (43), deglycosylation is sequential because of the action of exo-enzymes. However, in human pathogens such as E. faecalis (16), S. pyogenes (44), B. fragilis (15), and C. canimorsus (10, 21), and in infant gut-associated Bifidobacteria (45), the ability to release N-glycans from host proteins has been mainly associated with endo-β-N-acetylglucosaminidase activity. In these bacteria, it seems that deglycosylation is initiated by the cleavage of the N-glycan between the two GlcNAc residues common to all N-glycans. Because the two first enzymes acting on the N2M3FX substrate (i.e. NixK and NixI) in Xcc were detected in the supernatant-enriched fraction of bacterial cultures, we can propose that degradation of the N2M3FX N-glycan is initiated outside the cell (Fig. 7). Possibly after cleavage of the peptide by the asparaginase AspG (XCC2914, proposed in the KEGG pathway database to cleave the N-glycan from the asparagine residue of N-glycoproteins), uptake of partially degraded N-glycans through the outer membrane could be achieved by Nix TonB-dependent transporters. Indeed, we have recently observed that these active transporters belonging to the GlcNAc utilization system are involved in the uptake of GlcNAc-containing complex molecules (22). This is in agreement with the proposed role of the TonB-dependent transporter GpdC of C. canimorsus in the uptake of the N-glycan moiety after cleavage by the endo-β-N-acetylglucosaminidase GpdG (21). Cleavage could continue in the periplasm because all Nix enzymes except NixG were detected in this cell compartment. The resulting chitobiose could be imported inside the cell through the MFS transporter NagP (28), whereas uptake of xylose through the inner membrane would be achieved via XylE (27). NixG would further degrade chitobiose into GlcNAc, which would subsequently directly or indirectly inhibit repressors NagR and NagQ, leading to the induction of the expression of nag and nix genes (22, 28).
FIGURE 7.
Model of the N2M3FX N-glycopeptide degradation and utilization in Xcc ATCC33913 strain. NixK and NixI initiate deglycosylation of the N-glycopeptide outside the cell. We propose that AspG (XCC2914) would cleave the peptide moiety, and the resulting N2M2F N-glycan would be transported through Nix and/or other TonB-dependent transporters to the periplasm where NixJ, NixH, and NixE continue its degradation. Uptake of chitobiose (N2) through the inner membrane occurs via NagP, and NixG would be responsible for its degradation inside the cell. Free GlcNAc would then directly or indirectly inhibit repressors NagR and NagQ, leading to the induction of the expression of all nag and nix genes. Uptake of xylose through the inner membrane is performed by XylE. Transporters responsible for the uptake of xylose and mannose through the outer membrane and fucose and mannose through the inner membrane are not known. Free monosaccharides (fucose, mannose, GlcNAc, and xylose) would then be utilized for the metabolism of the bacterium. Numbers in circles indicate the order of action of Nix enzymes as determined in this paper. The boxes on the right indicate NixX-8His enzymes detected in total cell lysates (IN+PP), in periplasmic-enriched fraction (PP) and in the supernatant (OUT) of Xcc. F, fucose; M, mannose; N, GlcNAc; X, xylose; Nix, N-acetylglucosamine induced in Xanthomonas; TBDT, TonB-dependent transporter.
In Xcc, the genes involved in N-glycan degradation are clustered and transcribed polycistronically (22), suggesting that they may act synergistically. A similar genetic organization has been recently described in Bacteroides species (10, 46) and the corresponding cluster, important for growth on mucins in B. fragilis (46), has been proposed to be involved in sialoglycoconjugate utilization (sgu locus). However, some enzymes encoded by the sgu locus are different from those encoded by the Xcc N-glycan cluster. Indeed, the sgu locus codes for a sialidase (neuraminidase; GH33) and two other enzymes involved in sialic acid metabolism, whereas the Xcc N-glycan cluster codes for a β-xylosidase (GH3). These differences reflect structural specificities of mammal N-glycans as compared with plant N-glycans, respectively, and therefore suggest functional convergence to adapt these clusters to the ecological niches encountered by both bacteria.
It has previously been described that glycans of N-glycoproteins can serve as nutrients for human pathogenic bacteria aiding in survival or persistence in vivo (16, 17, 20, 21, 47–49). We have recently observed that in Xcc, the N-glycan cluster participates in the degradation of GlcNAc-containing molecules of plant origin during infection (22), suggesting that deglycosylation probably occurs in planta during infection. Although N-glycans are widely distributed, these compounds represent only a small fraction of carbohydrates found in plants. This could explain the absence of phenotype in planta of the mutant deleted from the entire N-glycan cluster when inoculated into plants by piercing of the central vein (22). Although the oligosaccharide moiety could be the nutrient source, N-glycan degradative enzymes could also help bacterial proteases gain access to the peptide portion of the glycoprotein, resulting in an optimized nutrient supply. Indeed, glycoproteins become more sensitive to proteases after deglycosylation of N-linked oligosaccharides (50). In addition to their function in nutrition, the degradation of plant cell wall glycoproteins may lead to weakening of the cell wall, and better accessibility of cell wall polymers to degradative enzymes. Finally, this cluster could have an impact in signaling by activating or inactivating the function of proteins from the host. Indeed, in animals, many of the proteins involved in adaptive and innate immunity are glycosylated. Interestingly, the S. pyogenes, E. faecalis, and C. canimorsus human pathogens were shown to cleave the conserved N-glycan from human IgG (21, 51, 52), which, in addition to the potential role in nutrition, might impact immunity. In S. pyogenes, the EndoS protein that cleaves the chitobiose core of N-glycans (44) interferes with the adaptive immune response and promotes bacterial growth in human blood (47). Similarly, plant pattern recognition receptors involved in innate immunity have N-glycosylations that are important for their function in pathogen-associated molecular pattern perception (53). Further investigations are now being undertaken to decipher on the role of the N-glycan cluster in Xcc during its life cycle, i.e. epiphytic life, entry into the leaf through hydathodes, infection, and survival on plant debris.
Acknowledgments
We are grateful to Patrice Lerouge (Université de Rouen) for stimulating discussion and for welcoming S. D. in his laboratory. We also thank François Lemauff and Coralie Boyer for technical assistance. We gratefully acknowledge Deborah Hinton and Nicolas Denancé for critical reading of the manuscript.
This work was supported by French Agence Nationale de la Recherche Grant ANR-08-BLAN-0193-01, the local Fédération de Recherche Agrobiosciences, Interactions et Biodiversité, the Université Paul Sabatier, the Centre National de la Recherche Scientifique, the Institut National de la Recherche Agronomique, and the French Ministry of Research and Technology (to A.B.).
- GH
- glycoside hydrolase
- Xcc
- Xanthomonas campestris pv. campestris
- MME
- minimal medium for hrp gene expression
- MBP
- maltose-binding protein
- pNP
- para-nitrophenyl
- pNP-α-Fuc
- pNP-α-l-fucopyranoside
- pNP-β-GlcNAc
- pNP-N-acetyl-β-d-glucosaminide
- pNP-β-GalNAc
- pNP-N-acetyl-β-d-galactosaminide
- pNP-β-Man
- pNP-β-d-mannopyranoside
- pNP-β-Xyl
- pNP-β-d-xylopyranoside
- pNP-α-Man
- pNP-α-d-mannopyranoside
- pNP-β-Gal
- pNP-β-d-galactopyranoside
- CE
- crude extract
- ConA
- concanavalin A.
REFERENCES
- 1. Spiro R. G. (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12, 43R–56R [DOI] [PubMed] [Google Scholar]
- 2. Messner P., Allmaier G., Schäffer C., Wugeditsch T., Lortal S., König H., Niemetz R., Dorner M. (1997) Biochemistry of S-layers. FEMS Microbiol. Rev. 20, 25–46 [DOI] [PubMed] [Google Scholar]
- 3. Jarrell K. F., Ding Y., Meyer B. H., Albers S. V., Kaminski L., Eichler J. (2014) N-Linked glycosylation in archaea: a structural, functional, and genetic analysis. Microbiol. Mol. Biol. Rev. 78, 304–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Faye L., Boulaflous A., Benchabane M., Gomord V., Michaud D. (2005) Protein modifications in the plant secretory pathway: current status and practical implications in molecular pharming. Vaccine 23, 1770–1778 [DOI] [PubMed] [Google Scholar]
- 5. Mitra N., Sinha S., Ramya T. N., Surolia A. (2006) N-Linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci 31, 156–163 [DOI] [PubMed] [Google Scholar]
- 6. Rudd P. M., Elliott T., Cresswell P., Wilson I. A., Dwek R. A. (2001) Glycosylation and the immune system. Science 291, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 7. An H. J., Lebrilla C. B. (2010) A glycomics approach to the discovery of potential cancer biomarkers. Methods Mol. Biol. 600, 199–213 [DOI] [PubMed] [Google Scholar]
- 8. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ladevèze S., Tarquis L., Cecchini D. A., Bercovici J., André I., Topham C. M., Morel S., Laville E., Monsan P., Lombard V., Henrissat B., Potocki-Véronèse G. (2013) Role of glycoside phosphorylases in mannose foraging by human gut bacteria. J. Biol. Chem. 288, 32370–32383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nihira T., Suzuki E., Kitaoka M., Nishimoto M., Ohtsubo K., Nakai H. (2013) Discovery of β-1,4-d-mannosyl-N-acetyl-d-glucosamine phosphorylase involved in the metabolism of N-glycans. J. Biol. Chem. 288, 27366–27374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martens E. C., Koropatkin N. M., Smith T. J., Gordon J. I. (2009) Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284, 24673–24677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tailford L. E., Money V. A., Smith N. L., Dumon C., Davies G. J., Gilbert H. J. (2007) Mannose foraging by Bacteroides thetaiotaomicron: structure and specificity of the β-mannosidase, BtMan2A. J. Biol. Chem. 282, 11291–11299 [DOI] [PubMed] [Google Scholar]
- 13. Zhu Y., Suits M. D., Thompson A. J., Chavan S., Dinev Z., Dumon C., Smith N., Moremen K. W., Xiang Y., Siriwardena A., Williams S. J., Gilbert H. J., Davies G. J. (2010) Mechanistic insights into a Ca2+-dependent family of alpha-mannosidases in a human gut symbiont. Nat. Chem. Biol. 6, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thompson A. J., Williams R. J., Hakki Z., Alonzi D. S., Wennekes T., Gloster T. M., Songsrirote K., Thomas-Oates J. E., Wrodnigg T. M., Spreitz J., Stütz A. E., Butters T. D., Williams S. J., Davies G. J. (2012) Structural and mechanistic insight into N-glycan processing by endo-α-mannosidase. Proc. Natl. Acad. Sci. U.S.A. 109, 781–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao Y., Rocha E. R., Smith C. J. (2014) Efficient utilization of complex N-linked glycans is a selective advantage for Bacteroides fragilis in extraintestinal infections. Proc. Natl. Acad. Sci. U.S.A. 111, 12901–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roberts G., Tarelli E., Homer K. A., Philpott-Howard J., Beighton D. (2000) Production of an endo-β-N-acetylglucosaminidase activity mediates growth of Enterococcus faecalis on a high-mannose-type glycoprotein. J. Bacteriol. 182, 882–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Byers H. L., Tarelli E., Homer K. A., Beighton D. (1999) Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human α1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9, 469–479 [DOI] [PubMed] [Google Scholar]
- 18. Suits M. D., Zhu Y., Taylor E. J., Walton J., Zechel D. L., Gilbert H. J., Davies G. J. (2010) Structure and kinetic investigation of Streptococcus pyogenes family GH38 α-mannosidase. PLoS One 5, e9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gregg K. J., Zandberg W. F., Hehemann J. H., Whitworth G. E., Deng L., Vocadlo D. J., Boraston A. B. (2011) Analysis of a new family of widely distributed metal-independent alpha-mannosidases provides unique insight into the processing of N-linked glycans. J. Biol. Chem. 286, 15586–15596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mally M., Shin H., Paroz C., Landmann R., Cornelis G. R. (2008) Capnocytophaga canimorsus: a human pathogen feeding at the surface of epithelial cells and phagocytes. PLoS Pathog. 4, e1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Renzi F., Manfredi P., Mally M., Moes S., Jenö P., Cornelis G. R. (2011) The N-glycan glycoprotein deglycosylation complex (Gpd) from Capnocytophaga canimorsus deglycosylates human IgG. PLoS Pathog. 7, e1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boulanger A., Zischek C., Lautier M., Jamet S., Rival P., Carrère S., Arlat M., Lauber E. (2014) The plant pathogen Xanthomonas campestris pv. campestris exploits N-acetylglucosamine during infection. MBio 5, e01527–e01514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blanvillain S., Meyer D., Boulanger A., Lautier M., Guynet C., Denancé N., Vasse J., Lauber E., Arlat M. (2007) Plant carbohydrate scavenging through tonb-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2, e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arlat M., Gough C. L., Barber C. E., Boucher C., Daniels M. J. (1991) Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum. Mol. Plant Microbe Interact. 4, 593–601 [DOI] [PubMed] [Google Scholar]
- 25. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26. Qian W., Jia Y., Ren S. X., He Y. Q., Feng J. X., Lu L. F., Sun Q., Ying G., Tang D. J., Tang H., Wu W., Hao P., Wang L., Jiang B. L., Zeng S., Gu W. Y., Lu G., Rong L., Tian Y., Yao Z., Fu G., Chen B., Fang R., Qiang B., Chen Z., Zhao G. P., Tang J. L., He C. (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15, 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Déjean G., Blanvillain-Baufumé S., Boulanger A., Darrasse A., Dugé de Bernonville T., Girard A. L., Carrére S., Jamet S., Zischek C., Lautier M., Solé M., Büttner D., Jacques M. A., Lauber E., Arlat M. (2013) The xylan utilization system of the plant pathogen Xanthomonas campestris pv campestris controls epiphytic life and reveals common features with oligotrophic bacteria and animal gut symbionts. New Phytol. 198, 899–915 [DOI] [PubMed] [Google Scholar]
- 28. Boulanger A., Déjean G., Lautier M., Glories M., Zischek C., Arlat M., Lauber E. (2010) Identification and regulation of the N-acetylglucosamine utilization pathway of the plant pathogenic bacterium Xanthomonas campestris pv. campestris. J. Bacteriol. 192, 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turner P., Barber C. E., Daniels M. J. (1985) Evidence for clustered pathogenicity genes in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 199, 338–343 [Google Scholar]
- 30. Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 [DOI] [PubMed] [Google Scholar]
- 31. Miller J. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32. Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ames G. F., Prody C., Kustu S. (1984) Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J. Bacteriol. 160, 1181–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 35. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 36. Horsch M., Mayer C., Sennhauser U., Rast D. M. (1997) β-N-Acetylhexosaminidase: a target for the design of antifungal agents. Pharmacol. Ther. 76, 187–218 [DOI] [PubMed] [Google Scholar]
- 37. Katta S., Ankati S., Podile A. R. (2013) Chitooligosaccharides are converted to N-acetylglucosamine by N-acetyl-β-hexosaminidase from Stenotrophomonas maltophilia. FEMS Microbiol. Lett. 348, 19–25 [DOI] [PubMed] [Google Scholar]
- 38. Büttner D., Bonas U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133 [DOI] [PubMed] [Google Scholar]
- 39. Wang L., Rong W., He C. (2008) Two Xanthomonas extracellular polygalacturonases, PghAxc and PghBxc, are regulated by type III secretion regulators HrpX and HrpG and are required for virulence. Mol. Plant Microbe Interact. 21, 555–563 [DOI] [PubMed] [Google Scholar]
- 40. Szczesny R., Jordan M., Schramm C., Schulz S., Cogez V., Bonas U., Büttner D. (2010) Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. New Phytol. 187, 983–1002 [DOI] [PubMed] [Google Scholar]
- 41. da Silva A. C., Ferro J. A., Reinach F. C., Farah C. S., Furlan L. R., Quaggio R. B., Monteiro-Vitorello C. B., Van Sluys M. A., Almeida N. F., Alves L. M., do Amaral A. M., Bertolini M. C., Camargo L. E., Camarotte G., Cannavan F., Cardozo J., Chambergo F., Ciapina L. P., Cicarelli R. M., Coutinho L. L., Cursino-Santos J. R., El-Dorry H., Faria J. B., Ferreira A. J., Ferreira R. C., Ferro M. I., Formighieri E. F., Franco M. C., Greggio C. C., Gruber A., Katsuyama A. M., Kishi L. T., Leite R. P., Lemos E. G., Lemos M. V., Locali E. C., Machado M. A., Madeira A. M., Martinez-Rossi N. M., Martins E. C., Meidanis J., Menck C. F., Miyaki C. Y., Moon D. H., Moreira L. M., Novo M. T., Okura V. K., Oliveira M. C., Oliveira V. R., Pereira H. A., Rossi A., Sena J. A., Silva C., de Souza R. F., Spinola L. A., Takita M. A., Tamura R. E., Teixeira E. C., Tezza R. I., Trindade dos Santos M., Truffi D., Tsai S. M., White F. F., Setubal J. C., Kitajima J. P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417, 459–463 [DOI] [PubMed] [Google Scholar]
- 42. Yokouchi D., Ono N., Nakamura K., Maeda M., Kimura Y. (2013) Purification and characterization of β-xylosidase that is active for plant complex type N-glycans from tomato (Solanum lycopersicum): removal of core alpha1–3 mannosyl residue is prerequisite for hydrolysis of β1–2 xylosyl residue. Glycoconj. J. 30, 463–472 [DOI] [PubMed] [Google Scholar]
- 43. King S. J., Hippe K. R., Weiser J. N. (2006) Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59, 961–974 [DOI] [PubMed] [Google Scholar]
- 44. Collin M., Olsén A. (2001) EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 20, 3046–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garrido D., Nwosu C., Ruiz-Moyano S., Aldredge D., German J. B., Lebrilla C. B., Mills D. A. (2012) Endo-β-N-acetylglucosaminidases from infant gut-associated Bifidobacteria release complex N-glycans from human milk glycoproteins. Mol. Cell. Proteomics 11, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakayama-Imaohji H., Ichimura M., Iwasa T., Okada N., Ohnishi Y., Kuwahara T. (2012) Characterization of a gene cluster for sialoglycoconjugate utilization in Bacteroides fragilis. J. Med. Invest. 59, 79–94 [DOI] [PubMed] [Google Scholar]
- 47. Collin M., Svensson M. D., Sjöholm A. G., Jensenius J. C., Sjöbring U., Olsén A. (2002) EndoS and SpeB from Streptococcus pyogenes inhibit immunoglobulin-mediated opsonophagocytosis. Infect. Immun. 70, 6646–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burnaugh A. M., Frantz L. J., King S. J. (2008) Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garbe J., Sjögren J., Cosgrave E. F., Struwe W. B., Bober M., Olin A. I., Rudd P. M., Collin M. (2014) EndoE from Enterococcus faecalis hydrolyzes the glycans of the biofilm inhibiting protein lactoferrin and mediates growth. PLoS One 9, e91035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berger S., Menudier A., Julien R., Karamanos Y. (1995) Do de-N-glycosylation enzymes have an important role in plant cells? Biochimie 77, 751–760 [DOI] [PubMed] [Google Scholar]
- 51. Collin M., Olsén A. (2001) Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect. Immun. 69, 7187–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collin M., Fischetti V. A. (2004) A novel secreted endoglycosidase from Enterococcus faecalis with activity on human immunoglobulin G and ribonuclease B. J. Biol. Chem. 279, 22558–22570 [DOI] [PubMed] [Google Scholar]
- 53. Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010) Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285, 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meyer D., Lauber E., Roby D., Arlat M., Kroj T. (2005) Optimization of pathogenicity assays to study the Arabidopsis thaliana-Xanthomonas campestris pv. campestris pathosystem. Mol. Plant Pathol. 6, 327–333 [DOI] [PubMed] [Google Scholar]