Background: ClipR-59 is implicated in the regulation of muscle function.
Results: ClipR-59 knockdown in C2C12 cells blocks muscle differentiation. ClipR-59 interacts with Elmo2 to regulate Rac1 activation.
Conclusion: ClipR-59 is required for myoblast fusion.
Significance: Identification of ClipR-59 as a novel promoter of muscle differentiation through its ability to orchestrate Rac signaling.
Keywords: Differentiation, Muscle, Myogenesis, Protein-protein Interaction, Rho (Rho GTPase), Dock1
Abstract
Recent studies using ClipR-59 knock-out mice implicated this protein in the regulation of muscle function. In this report, we have examined the role of ClipR-59 in muscle differentiation and found that ClipR-59 knockdown in C2C12 cells suppressed myoblast fusion. To elucidate the molecular mechanism whereby ClipR-59 regulates myoblast fusion, we carried out a yeast two-hybrid screen using ClipR-59 as the bait and identified Elmo2, a member of the Engulfment and cell motility protein family, as a novel ClipR-59-associated protein. We showed that the interaction between ClipR-59 and Elmo2 was mediated by the atypical PH domain of Elmo2 and the Glu-Pro-rich domain of ClipR-59 and regulated by Rho-GTPase. We have examined the impact of ClipR-59 on Elmo2 downstream signaling and found that interaction of ClipR-59 with Elmo2 enhanced Rac1 activation. Collectively, our studies demonstrate that formation of an Elmo2·ClipR-59 complex plays an important role in myoblast fusion.
Introduction
Elmo (engulfment and cell motility) proteins were identified based on their homology to Caenorhabditis elegans Ced-12, a protein that is required for apoptotic cell engulfment and cell migration (1). Elmo proteins are characterized by the presence of a Ras GTPase-binding domain, a region that is present only in Elmo proteins and ElmoD protein (Elmo domain), an atypical PH domain (aPH)4 and a proline-rich region with a PXXP motif (2, 3). In addition, two regions in Elmo proteins were shown to mediate a closed and inactive conformation state, the Elmo inhibitory domain and the Elmo autoregulation domain (EAD) (4). The exact function of this autoregulation remains to be fully established. There are three isoforms of Elmos in mammals including Elmo1, Elmo2, and Elmo3. Elmo1 and Elmo2 have been most extensively studied and are closely related based on their sequences.
Elmos have no apparent enzymatic activity and exert their biologic activity through interacting with a panel of different proteins (5). To date, a number of proteins have been found to interact with Elmo. These include proteins associated with plasma membrane such as brain-specific angiogenesis inhibitors 1–3 (6, 7), Gαi2 (8), Gβγ (9), Nck-1 (10), ILK2 (11), and also cytosolic proteins such as the DOCK proteins (1, 12, 13), RhoG (14), Arl4A (15), ACF7 (16), and ERM (17, 18). In general, Elmo proteins are recruited to the plasma membrane through interacting proteins for efficient activation of downstream effectors. For instance, interaction of Elmo1 and -2 with brain-specific angiogenesis inhibitor recruits them onto the proximal membrane where the DOCK180·Elmo complex promotes activation of Rac1 (6, 7), a small GTPase that plays a critical role in a wide variety of biology processes including cytoskeleton rearrangement, cell migration, and muscle differentiation (19, 20).
ClipR-59 is a membrane-associated protein characterized by three distinct structural features including a Glu-Pro-rich domain, three ankryin repeats, and two putative CAP-Gly domains (21). CAP-Gly domains are the signature domain of the ClipR-170 protein family that binds microtubules and regulates microtubule dynamics (22, 23). ClipR-59 shows no microtubule-binding activity but instead associates with cellular membranes including plasma membrane and trans-Golgi membrane via its palmitoylated cysteine residue (24, 25). Both CAP-Gly domains and ankyrin repeats are protein-protein interaction modules suggesting that ClipR-59 functions as an adaptor to recruit specific factors to the membrane. Supporting this view, ClipR-59 interacts with Akt and modulates Akt plasma membrane compartmentalization in adipocytes, thereby promoting Glut4 membrane translocation (26, 27). ClipR-59 also interacts with TNF receptor to regulate TNFα signaling (28). Inactivation of ClipR-59 in mice resulted in perinatal lethality in part due to muscle malfunction, implying that this gene plays a crucial role in muscle development (29). However, how ClipR-59 affects muscle function remains unexplored.
In this report, we have examined the role of ClipR-59 in muscle differentiation and found that ClipR-59 is required for myoblast fusion. Moreover, we also found that ClipR-59 interacts with Elmo2 to enhance Rac1 activation.
EXPERIMENTAL PROCEDURES
Reagents
Insulin, DAPI, mouse monoclonal anti-FLAG antibody, HRP-conjugated anti-FLAG, and anti-HA antibodies were from Sigma. Mouse monoclonal anti-HA antibody was from Covance. Mouse monoclonal anti-Elmo2, anti-GST and anti-Myc antibodies, anti-myogenin, anti-myosin heavy chain, and anti-β-tubulin antibodies were from Santa Cruz. Rabbit monoclonal anti-Akt and phospho-Akt antibodies were from Cell Signaling. Rabbit anti-ClipR-59 antibody has been described previously (26). TnT kit was from Promga.
Plasmids and Virus Production
ClipR-59 and its mutants have been described (27). Myc-tagged Elmo expression vectors and FLAG-Dock180 have been described (6). FLAG-180 was a gift of Dr. Michyuki Matsuda of Kyoto University. GST-CRIB was a gift of Dr. Rachel Bushsbaum, Tufts Medical Center (30). Rac1 and RhoG expression vectors were a gift of Dr. Ralph Isberg, Tufts University School of Medicine (31). The sequence of ClipR-59 shRNA has been described (26). The lentiviral vectors and subcloning strategy to generate shRNA lentiviral expression vectors and produce the lentiviral particles have been described (32). These lentiviral vectors encode a GFP protein independent of shRNA expression so that the transduced cells can be seen through green fluorescence.
The deletion mutants of Elmo2 were generated by using convenient restriction sites in the Elmo2 cDNA. To generate mammalian GST-Elmo2 expression vectors, mouse Elmo2 cDNA was amplified by PCR with primers 5′-GGAGATCTATGCCGCCTCCGTCTGACA-3′ and 5′-cctctagatcagccatagtgatagac-3′ and cloned into BamHI and SpeI site of pEBG. To generate Elmo2aPH, the sequences after the PH domain were amplified with primers 5′-GGAATTCATGTCCAGCGAGCTAACC-3′, and 5′-CCCTCGAGTCAGCCATAGTGATAGAC-3′ and digested with EcoRI and XhoI. The full-length Elmo2 cDNA was amplified with primers 5′-CCTCTAGATCAGCCATAGTGATAGAC-3′ and 5′-CCCTCGAGTCAGCCATAGTGATAGAC-3′ and digested with BglII and MfeI. Then, the EcoRI-Xho fragment and BglII-MfeI fragments were together ligated to either pCMV-Myc vector or pCMV HA vectors. Finally, the EcoRI and XhoI fragment was also cloned into pC-FLAG and pCHA vectors to obtain HA and FLAG-tagged Elmo2, respectively. To generate GST expression vector that express the aPH domain of Elmo2, aPH (aa 534–677) were amplified with primer 5′-GGATCCCCTGAGATCCTGGAGCTG-3′ and 5′-CACTCGAGTCATAGCTCGCTGGACATATCC-3′ and digested with BamHI and XhoI and cloned into pEBG.
To generate retroviral expression vectors for ClipR-59 and its mutants, cDNA fragments were cloned into pMigR1 retroviral vector between HpaI and XhoI sites. The viral particles were produced as described (32). To generate GST-E/PClipR-59, the E/P domain was amplified with primers 5′-GTGGATCCATGACTAAGACAGATCCTG-3′ and 5′-CACTCGAGTCACCCGTCACGATCGTTCACATG-3′, digested with BamHI and XhoI, and cloned into pGEX 5-1 expression vector.
Cell Culture and Transfection
COS-7 cells were grown in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mm l-glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin (Invitrogen). C2C12 cells were grown in the same DMEM but supplemented with 20% bovine serum instead of FBS. The differentiation of C2C12 cell was as follows. Briefly, C2C12 cells were cultured to 60–80% confluence. Then, the medium was changed to differentiation medium (DMEM supplemented with 2% horse serum). 72–96 h later, myoblast formation was examined.
Cell Imaging
C2C12 cells transduced with the indicated viruses were differentiated for 72–96 h. Then the cells were fixed and stained with DAPI. The cells were mounted on a glass coverslide. The fluorescence imaging is captured with confocal microscopy (Olympus).
Quantification of Myoblast Fusion
The myoblast fusion is quantified as the ratio of differentiated cells and total cells, which is referred to as the fusion index. Briefly, after differentiation, C2C12 cells were fixed and stained with DAPI. Then, the total number of viral transduced cells (with green fluorescence) and the fluorescence cells that consist of more than three nuclei (based on DAPI staining) were counted, respectively. Then, the ratio of the number of fluorescence cells, which has more three nuclei, and total fluorescence cells are calculated as myoblast fusion index.
Immunoprecipitation Assays
Transfected COS-7 cells or C2C12 cells were extracted with immunoprecipitation buffer (150 mm NaCl, 25 mm Tris, pH 7.6, 0.5 mm EDTA, 10% glycerol, 0.5% Nonidet P-40 plus a protease inhibitor mixture). 500 μg of total proteins from the cell lysates were subjected to immunoprecipitation with the corresponding antibodies.
GST Pulldown Assay
GST-ClipR-59 fusion peptide was expressed in COS-7 or HEK293 cells from pEBG expression vector and purified with glutathione-Sepharose 4B according to the manufacturer's instruction. For examining Elmo2 and ClipR-59 interaction, purified GST-ClipR-59 or Elmo2 was mixed with the total cell lysate of COS-7 cells that had been transiently transfected with proper expression vectors in immunoprecipitation buffer and incubated for 4 h. Then, the beads were washed 3 times, and the proteins associated with GST beads were analyzed in Western blot with the corresponding antibodies. Also, before washing, 2% volume of the mixtures was taken out for Western blot to determine the input level of each component. In some cases, cells were cotransfected with PEBG expression vectors with the indicated Elmo2 or ClipR-59 expression constructs.
Rac1 Activation Assay
The assay to measure GTP-loaded Rac1 was carried out as previously described (30). Briefly, GST-CRIB was produced in Escherichia coli BL21 and purified on glutathione beads. Total lysates from COS-7 cells that were transiently transfected with proper expression vectors as indicated in the figure legends were incubated with GST-CRIB beads. After 4 h incubation, GST beads were washed extensively and analyzed in Western blot with the proper antibodies.
Western Blotting
After treatments, cells were washed twice with PBS and extracted with cell lysis buffer (20 mm Tris, pH 7.6, 150 mm NaCl, 0.5 mm EDTA, 0.5 mm DTT, 10% glycerol, protease and phosphatase inhibitors). For cellular fractionation experiments, the cellular fractions were directly dissolved into lysis buffer. Equal amounts of protein were subjected to SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). After blocking in 5% dry milk, the membranes were incubated with each primary antibody, followed by incubation with a horseradish peroxidase-conjugated secondary antibody. The protein bands were visualized using the ECL detection system (Pierce). The quantification of Western blot was determined with Image J software.
Yeast Two-hybrid Screen
The yeast two-hybrid screen was carried out with full-length ClipR-59 as the bait as described previously (33).
Statistical Analysis
Mean ± S.D. were calculated and statistically significant differences among groups were determined by one-way analysis of variance analysis followed by post hoc comparisons, or by two-tailed unpaired Student's t test between two groups as appropriate, with significance at p < 0.05.
RESULTS
ClipR-59 Is Involved in Myoblast Fusion
Recent studies revealed that ClipR-59 knock-out mice display embryonic lethality in part because of impaired muscle function (29). This implied that ClipR-59 could play a role in muscle development and prompted us to examine the role of ClipR-59 in muscle differentiation. First, we examined ClipR-59 expression during differentiation of C2C12 cells into myoblasts. This cell line is widely used to study muscle differentiation and is amenable to dissect myoblast fusion (34). As shown in Fig. 1A, expression of ClipR-59 was increased following C2C12 differentiation. This change in expression of ClipR-59 is correlated to muscle differentiation as the expression of myogenin (Fig. 1, panel ii) and myosin heavy chain (MHC) (panel iii), markers of muscle cell differentiation, were also increased. Together, these data demonstrate that ClipR-59 expression is induced during muscle cell differentiation. We next introduced a validated ClipR-59 shRNA into C2C12 cells via lentiviral gene transfer. As a control cells were transduced with a lentiviral expression vector that expresses an shRNA targeting luciferase. As shown in Fig. 1, B and C, interfering with ClipR-59 expression in C2C12 cells reduced the number of myoblasts (>3 nuclei in each fiber) by more than 80%.
FIGURE 1.
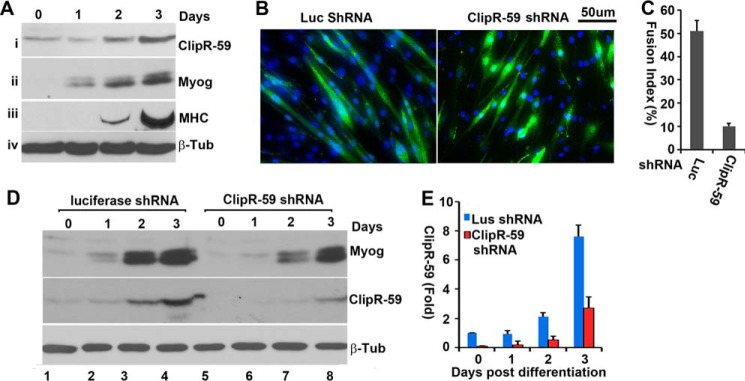
ClipR-59 modulates myoblast fusion. A, ClipR-59 expression during C2C12 muscle differentiation. Total cell lysates from C2C12 cells that had undergone differentiation at the indicated time points were analyzed in Western blot with anti-ClipR-59 (i), anti-myogenin (ii), anti-myosin heavy chain (MHC, iii), and anti-β-tubulin (iv) antibodies, respectively. B, ClipR-59 shRNA expression in C2C12 cells suppressed myoblast fusion. C2C12 cells were transduced with lentiviral vectors that expressed luciferase, and ClipR-59 shRNA, respectively. 48 h post-transduction, cells were switched into differentiation medium (DMEM supplemented with 2% horse serum). 72 h later, the cells were fixed and stained with DAPI. The scale bar represents 50 μm. C, quantification of C2C12 myoblast fusion. The fusion index is the ratio of GFP positive cells that contains more than 3 nuclei and total GFP cells. Bar graphs show mean ± S.D., n = 9 (fields). D, Western blot analysis of total cell lysates collected from the cells in B with anti-myogenin (MyoG), ClipR-59, and β-tubulin antibodies, respectively. E, densitometric analysis of ClipR-59 expression in D. The amount of ClipR-59 in luciferase shRNA expressing C2C12 cells before differentiation was set as 1 after normalized to β-tubulin. The bar graphs show ± S.D. (n = 3). In all cases, p < 0.05.
Next, we examined the expression of myogenin in control and ClipR-59 shRNA expressing C2C12 cells. As shown in Fig. 1D, a marginal difference in the expression of myogenin between control and ClipR-59 shRNA-expressing C2C12 cells was observed. In these experiments, we also examined the expression of ClipR-59. As expected, the expression of ClipR-59 is induced during muscle cell differentiation (Fig. 1, panel ii) and suppressed by ClipR-59 shRNA from 90 to 60% depending on the time of differentiation. Taken together, these data demonstrate that ClipR-59 plays a role in muscle differentiation at the myoblast fusion step.
ClipR-59 Interacts with Elmo2
ClipR-59 is a membrane-associated protein without enzymatic activity. To gain mechanistic insights on the role of ClipR-59 in muscle differentiation, we set out to identify its interacting proteins. Toward this end, we carried out a yeast two-hybrid screen using a murine embryonic fibroblast F422A-3T3 cDNA library using ClipR-59 as the bait. Several potential ClipR-59-interacting proteins were identified in this screen. One of them, Elmo2, was particularly interesting because it was recently reported to play an essential role in myoblast fusion (6).
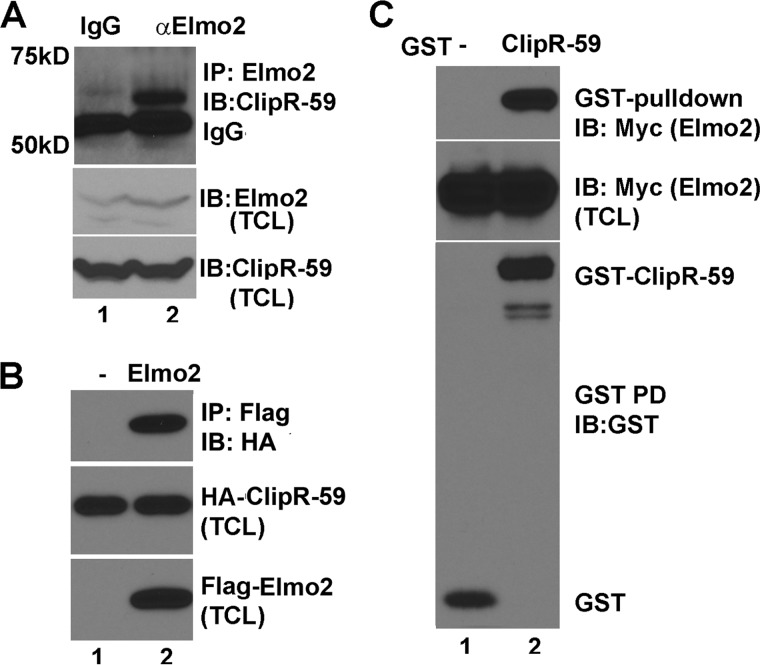
We next aimed to validate Elmo2 as a bona fide ClipR-59-interacting protein in mammalian cells. Toward this goal, a co-immunoprecipitation assay with differentiated C2C12 total lysates was carried out using an anti-Elmo2 monoclonal antibody. As shown in Fig. 2A, a 60-kDa anti-ClipR-59 immunoreactive band, just above IgG heavy chain, was detected in the anti-Elmo2 immunoprecipitate but not in the nonspecific IgG immunoprecipitate. The 60-kDa immunoreactive band to anti-ClipR-59 antibody was not due to sample variation as comparable levels of ClipR-59 (middle) and Elmo2 (bottom) were observed in each sample.
FIGURE 2.
ClipR-59 interacts with Elmo2. A, co-immunoprecipitation assay to show the interaction between endogenous ClipR-59 and Elmo2. Total cell lysates from differentiated C2C12 cells were immunoprecipitated with anti-Elmo2 antibody. The anti-Elmo2 immunoprecipitates were analyzed in Western blot with anti-ClipR-59 antibody (top). The middle and bottom panels show the levels of Elmo2 and ClipR-59 C2C12 total cell lysates. B, co-immunoprecipitation assay to show that transiently expressed Elmo2 and ClipR-59 interact. Total cell lysates from COS-7 cells transiently co-transfected with HA-tagged ClipR-59 expression vectors plus vectors expressing either FLAG-tag alone or FLAG-Elmo2 were subjected to immunoprecipitation with anti-FLAG antibody. The anti-FLAG immunoprecipitates were analyzed in Western blot with HRP-conjugated anti-HA antibody (top). The middle and bottom panels show the levels of FLAG-Elmo2 and HA-ClipR-59 in TCL. C, GST-pulldown assay with GST alone or GST-ClipR-59 purified from HEK293T cells and the total cell lysates from COS-7 cells that transiently expressed Myc-tagged Elmo2. The GST beads were analyzed in Western blot with anti-Myc (top) and anti-GST antibodies (bottom), respectively. The middle shows the total cellular levels of Myc-Elmo2. These experiments were repeated 2–4 times with similar results. IB, immunoblot.
To further validate our findings, we next carried out co-immunoprecipitation assays with FLAG-tagged Elmo2 and HA-tagged ClipR-59 transiently expressed in COS-7 cells. As shown in Fig. 2B, HA-ClipR-59 was detected in anti-FLAG immunoprecipitates from the cell lysates of COS-7 cells that were transiently co-transfected with Myc-Elmo2 and HA-ClipR-59 expression vectors, but not with empty FLAG expression vector. The middle and bottom panels of Fig. 2 represents the levels of HA-ClipR-59 and FLAG-Elmo2 in total cell lysates (TCL).
To further evaluate if the association of ClipR-59 and Elmo2 is direct, we carried out a GST pulldown assay. GST and GST-ClipR-59 were produced in 293T cells and were incubated with COS-7 cell lysates that expressed Myc-Elmo2. As shown in Fig. 2C, Myc-Elmo2 was robustly detected in GST-ClipR-59 beads. The middle and bottom panels represent the cellular levels of Myc-Elmo2 and GST fusion proteins, respectively. Collectively, these data demonstrate that ClipR-59 interacts specifically and directly with Elmo2 in mammalian cells.
ClipR-59 Preferentially Interacts with Elmo2 and Elmo3
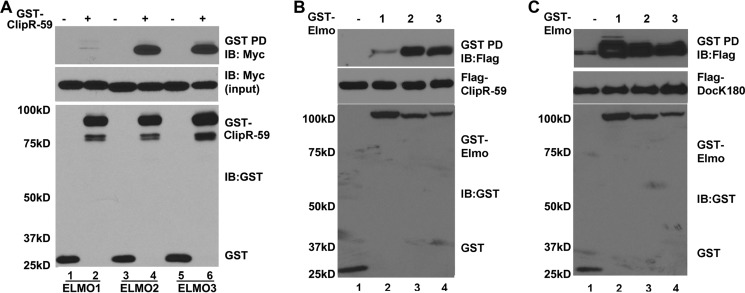
There are three Elmo proteins in mammals including Elmo1, Elmo2, and Elmo3 (5). We investigated whether ClipR-59 would also interact with other isoforms of Elmo proteins. To test this, a GST pulldown assay was carried out with purified GST-ClipR-59 isolated from HEK293T cell lysate and incubated with total cell lysates of COS-7 cells that were transiently transfected with expression vectors coding for Myc-tagged Elmo1, Elmo2, or Elmo3. As shown in Fig. 3A, all Elmo proteins were retained on GST-ClipR-59 beads, but the amount of Elmo2 and Elmo3 was markedly higher than that of Elmo1. The Elmo proteins retained on the beads were specific to GST-ClipR-59 as no detectable Elmo proteins were found on GST beads alone. The variation among Elmo proteins retained on GST ClipR-59 beads could not be ascribed to sample variations as comparable levels of Elmo proteins and GST proteins were observed.
FIGURE 3.
ClipR-59 preferentially interacts with Elmo2 and Elmo3. A, GST pulldown assay to examine interaction between GST-ClipR-59 and Elmo family proteins. GST-ClipR-59 or GST prepared from 239T cells was incubated with cell lysates from COS-7 cells that were transiently co-transfected with Myc-tagged Elmo expression vectors. The proteins associated with GST beads were analyzed in Western blot with anti-Myc antibody (top). The middle panel shows the levels of Myc-Elmo in TCL. The bottom panels show the levels of GST fusion proteins associated with GST beads. B, GST pulldown assay to examine the interaction between ClipR-59 and GST-Elmo proteins. Total cell lysates from COS-7 cells that were transiently co-transfected with FLAG-ClipR-59 expression vectors plus either pEBG (GST vector) or pEBG-Elmo (lanes 1–3) were subjected to a GST pulldown assay. The GST beads were analyzed in Western blot with anti-FLAG antibody (top). The middle panel shows the levels of FLAG-ClipR-59 in TCL. The bottom panels show the levels of GST fusion proteins associated with GST beads. C, similar to B except FLAG-tagged Dock180 was used. These experiments were repeated 3–4 times with similar results. IB, immunoblot.
To examine this differential association further, we carried out GST pulldown assays with GST-Elmo proteins and FLAG-tagged ClipR-59 expressed in COS-7. As shown in Fig. 3B, ClipR-59 was retained on all GST-Elmo beads. However, the amount of ClipR-59 on GST-Elmo2 and Elmo3 beads was 6-fold higher than that on GST-Elmo1 beads. The ClipR-59 retained on GST-Elmo was specific to Elmo proteins as no ClipR-59 was found on GST beads alone (lane 1, top panel) and comparable levels of ClipR-59 (middle) and GST fusion proteins were observed in each sample.
To rule out the possibility that the lower affinity of Elmo1 for ClipR-59 is due to non-functional Elmo1, we examined the association of Elmo1, Elmo2, and Elmo3 with Dock180, a known Elmo-interacting protein. As expected, all three Elmo proteins exhibited a comparable capability of interacting with Dock180 (Fig. 3C). Taken together, these data demonstrate that ClipR-59 preferentially interacts with Elmo2 and Elmo3.
The Interaction between ClipR-59 and Elmo2 Enhances the Levels of Active Rac1
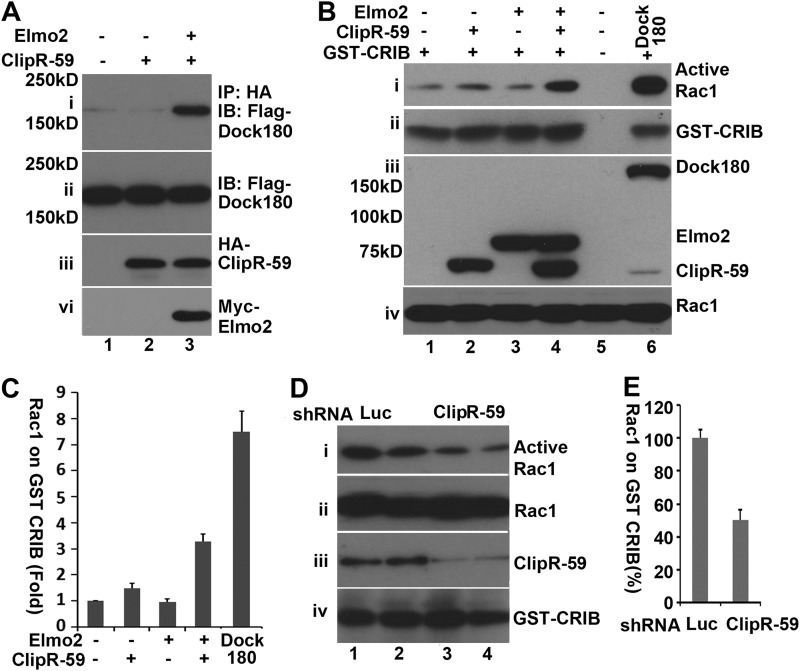
Elmo2 interacts with the Dock family protein to activate downstream targets including Rac1 (35). To determine whether Elmo2, ClipR-59, and Dock protein forms a tertiary complex, a co-immunoprecipitation assay was carried out with COS-7 cell lysates that express FLAG-Dock180 alone or HA-ClipR-59 plus FLAG-Dock2, or HA-ClipR-59 plus FLAG-Dock180 and Myc-Elmo2, with anti-FLAG antibody. As shown in Fig. 4A, FLAG-Dock180 was only significantly immunoprecipitated with anti-HA antibody in the presence of both HA-ClipR-59 and Myc-Elmo2, indicating that ClipR-59, Elmo2, and Dock180 are present in the same complex.
FIGURE 4.
The interaction between ClipR-59 and Elmo2 induces Rac1 activation. A, co-immunoprecipitation (IP) assay to show that Dock180 is present in the Elmo2·ClipR-59 complex. Total cell lysates from COS-7 cells that were transiently co-transfected with FLAG-Dock1 and HA-ClipR-59 or HA-ClipR59 plus Myc-Elmo2 expression vectors as indicated were subjected to immunoprecipitation with anti-HA antibody. The anti-HA immunoprecipitates were analyzed in a Western blot with HRP-conjugated anti-FLAG (i) or anti-HA-antibodies (iii), respectively. Panels ii and iv represent the cellular levels of FLAG-Dock180 and Myc-Elmo2. B, GST pulldown assay to show that interaction between ClipR-59 and Elmo2 increases the level of active Rac1. The total cell lysates from COS-7 cells that were cotransfected with FLAG-Rac1 and FLAG-ClipR-59, or FLAG-Elmo2 or FLAG-ClipR-59 plus FLAG-Elmo2 were subjected to a GST pulldown assay with recombinant GST-CRIB. The levels of Rac1 associated with GST-CRIB beads were analyzed by Western blot with anti-FLAG antibody (i). Meanwhile, total cellular levels of FLAG-Rac1, FLAG-Elmo2, and FLAG·ClipR-59 were also examined in Western blots with anti-FLAG antibody. The lane that expressed FLAG-Dock1 served as a control in this experiment. C, densitometry of the gel in B, the levels of Rac1 associated with GST-CRIB. The level of Rac1 associated with GST-CRIB in control cells was set as 1. The bar graphs show ±S.D. (n = 3). D, GST pulldown assay to show that ClipR-59 knockdown in C2C12 cells suppressed Rac activation. Total cell lysates from C2C12 cells that were transduced with either luciferase shRNA or expression lentivirus or different doses of ClipR-59 shRNA expressing lentiviruses. The levels of Rac1 associated with GST-CRIB were analyzed in Western blot with anti-Rac1 antibody. Total cellular levels of Rac1, ClipR-59, and β-tubulin were also examined in Western blots with anti-Rac1 (ii) anti-ClipR-59 (iii), and anti-β-tubulin (iv), respectively. E, densitometry of the gel in D, the levels of Rac1 associated with GST-CRIB. The level of Rac1 associated with GST-CRIB in control cells was set as 100%. The bar graphs show ± S.D. (n = 4). IB, immunoblot.
Binding of Elmo2 to Dock family proteins enhances the levels of active Rac1 (GTP-bound Rac1). Active Rac1, in turn, binds Pak1 and activates it (36). The notion that ClipR-59, Elmo2, and Dock180 are in the same complex raises the possibility that ClipR-59 enhances Rac1 activation. To test this, we carried out a GST pulldown assay to examine the impact of ClipR-59 and Elmo2 co-expression on Rac1 GTP loading using recombinant GST-CRIB. GST-CRIB is a GST fusion protein that consists of Cdc42/Rac1 interactive-binding (CRIB) domain of the p21-activated kinase 1 and only interacts with active Rac1 or Cdc42 (30). As shown in Fig. 4B, the levels of Rac1 associated with GST-CRIB under forced expression of either ClipR-59 or Elmo2 were not different from that of control cells. In marked contrast, the amount of Rac1 associated with GST-CRIB increased about 3-fold with ClipR-59 and Elmo2 co-expression (Fig. 4B, top panel). As a control, we also examined the association of Rac1 and GST-CRIB with Dock180 expression. As expected, forcing expression of Dock180 strongly enhanced the association of Rac1 and GST-CRIB.
To further verify the impact of ClipR-59 on Rac1 activation, we next examined the impact of ClipR-59 on Rac1 activation in C2C12 cells. As shown in Fig. 4D, active Rac1 were readily pulled down with GST-CRIB in luciferase shRNA expressing C2C12 cells (Fig. 4, panel i). In contrast, a 40% reduction of Rac1 pulled by GST-CRIB was observed in ClipR-59 shRNA expression cells (Fig. 4D, panel i, for quantification). As expected, ClipR-59 shRNA was effective to suppress ClipR-59 expression (Fig. 4D, panel iii). Together, these data demonstrate that interaction of ClipR-59 with Elmo2 enhances the cellular levels of active Rac1.
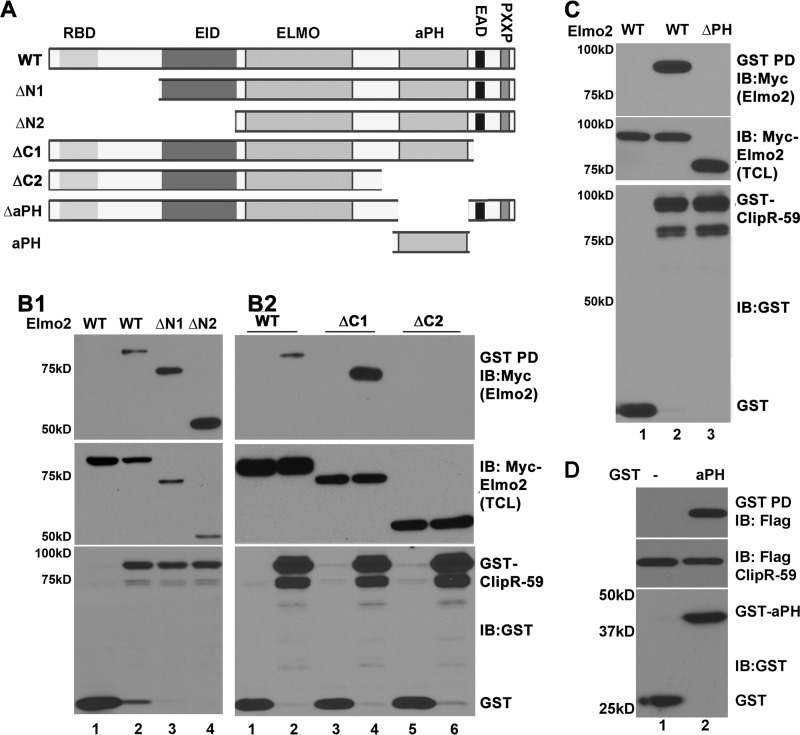
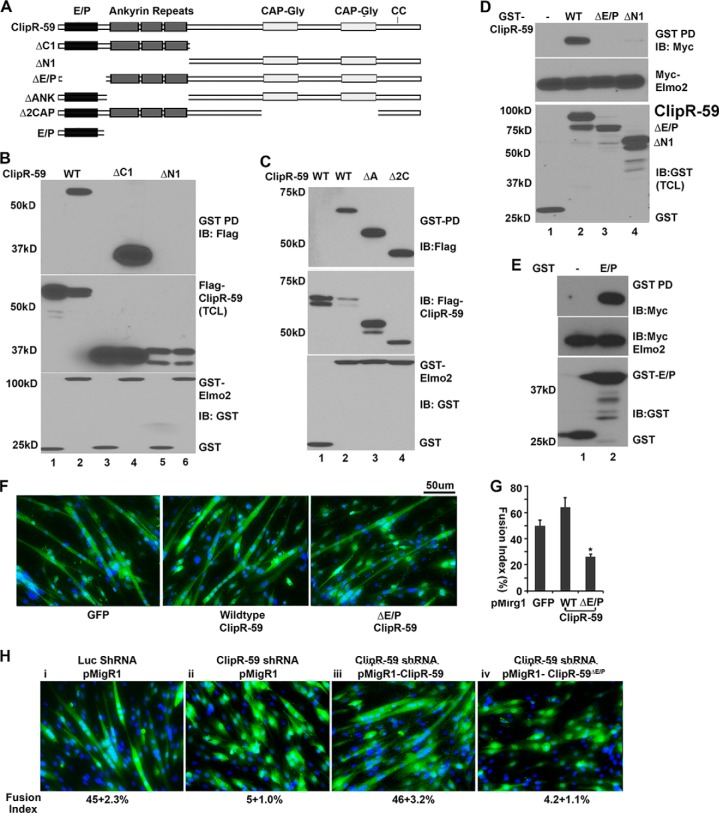
The aPH Domain of Elmo2 Is Required for Interaction between Elmo2 and ClipR-59
Among three Elmo proteins, Elmo1 and Elmo2 are the most studied. Because ClipR-59 preferentially interacts with Elmo2, we set out to characterize the interaction between ClipR-59 and Elmo2 in more detail. Toward this, we generated a series of Elmo2 mutants in which different portions of Elmo2 were deleted (Fig. 5A). We tested the ability of each mutant to interact with ClipR-59 in a GST pulldown assay. As shown in Fig. 5B1, removal of the first 107 (ΔN1) and 307 aa (ΔN2) from the amino terminus of Elmo2 did not prevent binding but actually increased the amount of Elmo2 retained on GST-ClipR-59 beads. Removal of the last 60 aa (ΔC1) from the carboxyl terminus of Elmo2 also increased the amount of Elmo2 retained on GST-ClipR-59 beads (Fig. 5B2, top panel, compare lanes 2 and 4). However, removal of the last 190 aa from the carboxyl terminus of Elmo2 (ΔC2) abolished Elmo2 precipitation by GST-ClipR-59 beads (Fig. 5B2, top panel, compare lanes 2 and 5). The failure to detect ΔC2 Elmo2 on GST-ClipR-59 beads was not because of sample variation as the levels of Elmo2 and Elmo2 mutants (Fig. 5B2, middle panel) as well as that of the GST protein (Fig. 5B2, bottom panel) were observed in each sample.
FIGURE 5.
aPH domain of Elmo2 mediates the interaction between Elmo2 and ClipR-59. A, the schematic representation of Elmo2 proteins and Elmo2 mutants used in this study. RBD, Rho-GTPase binding domain; EID, Elmo inhibitory domain; Elmo, Elmo domain; PXXP, SH3 domain binding motif. X represents any amino acid residue. B1 and B2, GST pulldown assay to show that deletion of the carboxyl-terminal sequence of Elmo2 abolished the interaction between ClipR-59 and Elmo2. Total cell lysates from COS-7 cells were transiently co-transfected with Myc-Elmo (wild-type and mutant) expression vectors plus either pEBG (GST vector) or pEBG-ClipR-59. 36 h post-transfection, the total cell lysates were prepared and subjected to pulldown with glutathione-agarose (GST beads). The proteins associated with GST beads were analyzed in a Western blot with anti-FLAG antibody (top). The middle panel shows the levels of Myc-Elmo2 and its mutant in TCL. The bottom panels show the levels of GST fusion proteins associated with GST beads. C, GST pulldown assay to show that the aPH domain of Elmo3 mediates the interaction between ClipR-59 and Elmo. D, GST pulldown assay to show that the aPH domain alone is sufficient to mediate the interaction between ClipR-59 and Elmo2. The experiments were carried out as in B except GST-aPHElmo2 and FLAG-ClipR-59 were used. These experiments were repeated 2–4 times with similar results. The result shown is representative. Depending on the individual assay, each experiment was repeated 3–4 times with similar results. IB, immunoblot.
The last 190 aa of Elmo2 consists of a putative PH domain, EAD, and a PXXP motif. EAD and PXXP motifs are localized in the last 60 amino acids. Because removal of the EAD and PXXP motifs had no effect on the interaction between Elmo2 and ClipR-59, we reasoned that the aPH domain of Elmo2 likely mediates the interaction of Elmo2 with ClipR-59. To test this, the aPH domain of Elmo2 was internally deleted and the resultant mutant (designated as Elmo2ΔPH) was tested for interaction with ClipR-59 in a GST pulldown assay. As shown in Fig. 5C, wild-type Elmo2 was readily detected in GST-ClipR-59 beads but in contrast no ΔaPH Elmo2 was found on GST-ClipR-59 beads (top panel) despite comparable levels of wild-type and Elmo2ΔPH (middle panel) as well as GST-ClipR-59 among these samples.
To confirm that the aPH domain of Elmo2 is sufficient to mediate the interaction between Elmo2 and ClipR-59, we next carried out a GST pulldown assay with GST-aPHElmo2 and ClipR-59 coexpressed in COS-7 cells. As shown in Fig. 5D, ClipR-59 was readily detected in GST-aPHelmo2 beads, but not in GST beads (top panel). As expected, removal of the first 300 aa from Elmo2 had no impact on the interaction between ClipR-59 and Elmo2 (top panel). Together, we conclude that the aPH domain of Elmo2 mediates the interaction between Elmo2 and ClipR-59. In addition, these data suggest that binding of Elmo2 to ClipR-59 is in part dependent on the Elmo conformation state.
ClipR-59 and Elmo2 Interaction Is Regulated by Rho GTPase
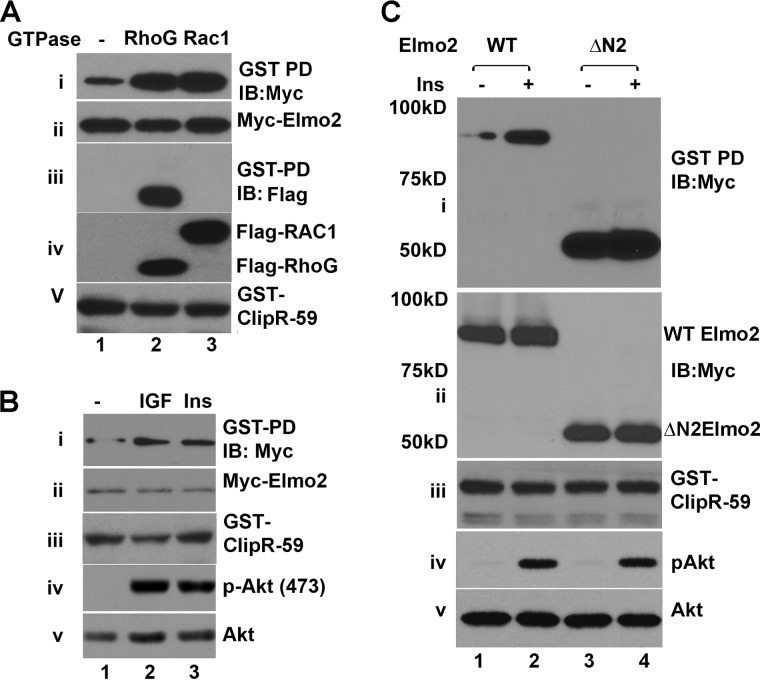
Elmo proteins exist as autoinhibited scaffolds under basal conditions through the intramolecular interaction between the Elmo inhibitory domain and EAD (4) (Fig. 5A). At present, how this inhibitory status is relieved remains elusive. It is postulated that binding of active RhoG GTPase to Elmo proteins targets the Elmo·Dock complex to the membrane, via the Ras GTPase-binding domain, to mediate Rac1 activation (4, 14). As described above, we noticed that the removal of the first 100 aa from the amino terminus as well as 60 aa from the carboxyl terminus enhances the interaction between ClipR-59 and Elmo2 (Fig. 5B), raising the possibility that the interaction between ClipR-59 and Elmo2 is subject to specific regulation. With this in mind, we tested the impact of active RhoG expression on ClipR-59 and Elmo2 in GST pulldown assays. As shown in Fig. 6A, exogenous expression of constitutively active RhoGG12V increased the amount of Elmo2 associated with GST-ClipR-59 by more than 3-fold (top panel). In these experiments, we also examined the impact of active Rac1G12V on the interaction between ClipR-59 and Elmo2 as Rac1 has been reportedly involved in the interaction between Dock180 and Elmo1 (37). Indeed, we observed that expression of constitutively active Rac1G12V also increased the association of ClipR-59 with Elmo2 (Fig. 6A, top panel). In these experiments, we also examined the presence of RhoG and Rac1 in GST-ClipR-59 beads. In agreement with the notion that RhoG is associated with Elmo2 (14), RhoG was detected in GST-ClipR-59 beads (panel iii). Panels ii, iv, and v show the cellular levels of Myc-Elmo2, GST-ClipR-59, and FLAG-RhoG and FLAG-Rac1, respectively.
FIGURE 6.
The interaction between ClipR-59 and Elmo2 is regulated by insulin. A, GST pulldown assay to show that active RhoG and Rac1 enhance ClipR-59 and Elmo2 interaction. The total cell lysates from COS-7 cells that were transiently co-transfected with Myc-Elmo2 expression vectors plus either pEBG (GST vector) or pEBG-ClipR-59 with either constitutive active RhoGG12V or Rac1G12V were subjected to a GST pulldown assay. The proteins associated with GST beads were analyzed in a Western blot with anti-FLAG antibody (i). Panels ii and iii show the levels of Myc-Elmo2 and GFP-tagged RhoG or Rac in TCL. Panel iv shows the levels of GST fusion proteins associated with GST beads. B, insulin and IGF enhances the interaction between Elmo2 and ClipR-59. COS-7 cells were co-transiently transfected with pEBG-ClipR-59 and Myc-Elmo2. 24 h post-transfection, cells were serum-deprived overnight. After treatment with insulin (100 nm) or IGF (5 ng/ml) for 15 min, the cell total lysates were prepared for a GST pulldown assay. The GST beads were analyzed in Western blot with anti-Myc antibody (i). The TCL were analyzed in Western blot with anti-Myc (ii), anti-phospho-Akt (iii), and anti-GST (iv) antibodies, respectively. C, similar to B, except ΔN2 Elmo2 was also used and the cells were only treated with insulin. All of these experiments were repeated three times with similar results. Where necessary, molecular mass markers are shown on the left of the panel. IB, immunoblot.
Rho-GTPases are activated by a range of extracellular stimuli including insulin and growth factors. With this in mind, we tested the impact of insulin and IGF on the interaction between ClipR-59 and Elmo2. As shown in Fig. 6B, treatment of cells with either IGF or insulin enhanced the interaction between ClipR-59 and Elmo2 (panel i, compare lanes 1, 2, and 3). The IGF and insulin used here are active as they induced Akt phosphorylation (panels iv and v). Panels ii and iii show the levels of Myc-Elmo2 and GST-ClipR-58 in total cell lysates, respectively.
To further evaluate the regulation of ClipR-59 and Elmo2 interaction by insulin, we next examined the impact of insulin on the interaction between ClipR-59 and ΔN2 Elmo2, which showed a stronger interaction with ClipR-59 (Fig. 5B). As shown in Fig. 6C, as expected, ΔN2 Elmo2 still showed a strong interaction with ClipR-59 (compare lanes 1 and 3, panel i). However, insulin treatment appeared to diminish the interaction between ClipR-59 and Elmo2, suggesting that insulin treatment relieved Elmo2 from an autoinhibitory status. Insulin treatment was effective as it induced Akt phosphorylation without an effect on total cellular levels of Akt (panels iv and v). Panels ii and iii show the total cellular levels of Myc-Elmo2 and GST-ClipR-59.
The Domain in ClipR-59 That Mediates the Interaction between Elmo2 and ClipR-59
To determine the domain in ClipR-59 that mediates the interaction between Elmo2 and ClipR-59, ClipR-59 was divided into portions 1–245 (designated as ΔC1) and 245–547 (designated as ΔN1) and the resulting ClipR-59 peptides were tested for their interaction with Elmo2 in a GST pulldown assay. As shown in Fig. 7B, ΔC1-ClipR-59, but not ΔN1-ClipR-59 was retained on GST-Elmo2 beads, suggesting that sequences within the first 245 aa residues of ClipR-5 mediate the interaction between ClipR-59 and Elmo2 interaction.
FIGURE 7.
The amino terminus of ClipR-59 mediates the interaction between ClipR-59 and Elmo2. A, schematic presentation of ClipR-59 proteins. The functional domains are indicated. E/P, Glu-Pro-rich domain; ankyrin, ankryin repeats; CC, palmitoylated Cys at 534 and 535. B, GST pulldown assay to show that the amino-terminal sequence of ClipR-59 mediates the interaction between ClipR-59 and Elmo2. Total cell lysates from COS-7 cells were transiently co-transfected with FLAG-ClipR-59 (wild-type and mutants) expression vectors plus either pEBG (GST vector) or pEBG-Elmo2 were subject to a GST pulldown assay. The proteins associated with GST beads were analyzed in a Western blot with anti-FLAG antibody (top). The middle panel shows the levels of FLAG-ClipR-59 and its mutant in TCL. The bottom panels show the levels of GST fusion proteins associated with GST beads. C, GST pulldown assay to show that ankyrin repeats of Elmo2 are not required for the interaction between ClipR-59 and Elmo2. The experiments were carried out similar to B, except FLAG-ΔANK-ClipR-59 (ΔA) and FLAG-Δ2CAP-ClipR59 (Δ2C) were used. D, GST pulldown assay to show that the E/P domain of ClipR-59 mediates the interaction between ClipR-59 and Elmo2. Total cell lysates from COS-7 cells that were transiently co-transfected with Myc-Elmo2 (wild-type) expression vectors plus either pEBG (GST vector) or pEBG-ClipR-59 or pEBG-ClipR-59ΔE/P subjected to GST pulldown assay. The proteins associated with GST beads were analyzed in Western blot with anti-FLAG antibody (top). The middle panel shows the levels of Myc-Elmo2 in TCL. The bottom panel shows the levels of GST fusion proteins associated with GST beads. These experiments were repeated 2–4 times with similar results. A representative result is shown. E, GST pulldown assay to show that the E/P domain of ClipR-59 mediates the interaction between Elmo2 and ClipR-59 with recombinant GST-E/PClipR-59 and in vitro translated FLAG-Elmo2. Top, GST beads associated with FLAG-Elmo2; middle, FLAG-Elmo2 input; bottom, GST fusion protein input. F, forcing expression of the Elmo2 interaction defective ClipR-59ΔE/P impaired C2C12 cell differentiation. C2C12 cells were transduced with pMigR1 (left, served as a control), pMigR-ClipR-59 (middle), and pMigR ClipR-59ΔE/P, respectively. 48 h post-transduction, cells were switched into differentiation medium (DMEM supplemented with 2% horse serum). 72 h later, the cells were fixed and stained with DAPI. G, quantification of C2C12 myoblast fusion. The fusion index is the ratio of GFP positive cells that contains more than 3 nuclei and total GFP cells. Bar graphs show mean ± S.D., n = 9 (fields). In all cases, p < 0.05. The scale bar is 50 μm. H, forcing expression of ClipR-59, but not ClipR-59ΔE/P in ClipR-59 shRNA expressing C2C12 cells rescues C2C12 differentiation. The C2C12 cells were co-transduced with lentiviral vectors that express ClipR-59 shRNA and pMigr1 vectors as indicated. Then, the cells were differentiated and examined as described in F. The numbers under each panel are the fusion index as mean ± S.D., n = 9 (fields). In all cases, p < 0.05. IB, immunoblot.
The first 254 aa of ClipR-59 consists of a Glu-Pro-rich domain and three ankyrin repeats. Both are protein-protein interaction modules. To determine whether ankyrin repeats play a role in ClipR-59 and Elmo2 interaction, we internally deleted ankyrin repeats from ClipR-59 (designated as ΔANK ClipR-59) and examined the interaction of ΔANK ClipR-59 with Elmo2 in a GST pulldown assay. As shown in Fig. 7C, ΔANK ClipR-59 showed a similar capacity to wild-type ClipR-59 in the interaction with Elmo2 (compare lanes 2 and 3, top panel), ruling out the role of ankyrin repeats in the interaction between ClipR-59 and Elmo2. In these experiments, we also examined interaction of the ClipR-59 mutant Δ2CAP ClipR-59 in which both CAP-Gly domains were deleted. No alteration of interaction between Elmo2 and ClipR-59 was observed when both CAP-Gly domains were deleted.
The notion that ankyrin repeats in ClipR-59 play no role in the interaction between ClipR-59 and Elmo2 implies that the first 100 amino acid residues of ClipR-59 mediate the interaction between ClipR-59 and Elmo2. To test this, we generated an additional GST-ClipR-59 fusion protein in which aa 7–107 (designated as ClipR-59ΔE/P) and the first 245 aa (ΔN1) were deleted, respectively. In GST pulldown assays, Elmo2 was unable to be detected in either GST-ClipR-59ΔE/P or GST-ΔN1 ClipR-59 beads (Fig. 7D).
To confirm that the E/P domain of ClipR-59 alone is sufficient to mediate the interaction between ClipR-59 and Elmo2, a GST pulldown assay with recombinant GST-E/PClipR-59 and Elmo2 protein from in vitro transcription and translations were carried out. As shown in Fig. 7E, Elmo2 were detected in the GST-E/PClipR-59 bead, but not GST beads alone (top). Collectively, these results demonstrated that the E/P of ClipR-59 mediate the direct interaction between ClipR-59 and Elmo2.
To determine whether the interaction between ClipR-59 and Elmo2 is required for ClipR-59 to modulate C2C12 differentiation, we introduced wild-type and Elmo2 interaction defective ΔN2 ClipR-59 into C2C12 cells and examined how forcing the expression of ClipR-59 affected C2C12 differentiation. As shown in Fig. 7E, forcing expression of ClipR-59 modestly enhanced C2C12 differentiation (50 versus 60%) presumably because there is a sufficient amount of endogenous ClipR-59 in the cells. However, in sharp contrast, forcing expression of ΔN2 ClipR-59 reduced C2C12 cell myoblast fusion by about 50% (Fig. 7F).
To further examine the requirement for the interaction between ClipR-59 in the role of ClipR-59 in C2C12 differentiation, we next co-transduced C2C12 cells with the lentiviral vector expressing ClipR-59 shRNA and pMigr1 expression vector that expresses ClipR-59 or ClipR-59ΔE/P and examined whether C2C12 differentiation suppressed by ClipR-59 shRNA could be rescued, respectively. Then, viral-transduced cells were subjected to differentiation. The control cells were co-transduced with lentiviral vector expressing ClipR-59 shRNA and pMigr1 empty vector, or lentiviral vector expressing luciferase shRNA and pMigr1 empty vector. As shown in Fig. 7H, ClipR-59 expression in ClipR-59 shRNA expressing cells clearly increased the level of differentiation (panels ii and iii), which was at a comparable level with luciferase shRNA expressing cells (panels i and iii). On the other hand, expression of ClipR-59ΔE/P has no such effect (panels iii and iv). Together, these data argue that the interaction between ClipR-59 and Elmo2 is required for ClipR-59 to modulate muscle differentiation.
DISCUSSION
In the present report, we have explored the role of ClipR-59 in muscle differentiation. Based on the view that ClipR-59 knockdown in C2C12 cells suppressed muscle differentiation, without affecting the expression of differentiation markers such as myogenin, we propose a novel function for ClipR-59 in regulating myoblast fusion (Fig. 1). Mechanistically, ClipR-59 is likely involved in myoblast fusion as its expression was induced in the later stage of C2C12 differentiation (Fig. 1A) and it interacted with Elmo2, which itself regulates myoblast fusion (6). This is also in agreement with the belief that ClipR-59 plays a role in the regulation of membrane dynamics (24).
To illustrate the mechanism by which ClipR-59 regulates muscle differentiation, we set to identify new ClipR-59-interacting proteins and identified Elmo2 using a yeast two-hybrid screening approach. Elmo2 is essential for muscle differentiation. Our data indicate that ClipR-59 and Elmo2 interact in vivo (Fig. 2) and regulate Rac1 activation (Fig. 4), indicating the possible mechanism by which ClipR-59 modulates muscle differentiation. There are three Elmo proteins in mammals. Given the sequence similarity, Elmo proteins are believed functionally exchangeable. Interestingly, we found that ClipR-59 preferably interacted with Elmo2 and Elmo3 among three mammalian Elmo proteins, indicating the possibility that each Elmo protein may have a specific function.
To better understand the interaction between ClipR-59 and Elmo2, we have characterized the sequences in ClipR-59 and Elmo2 that mediate the interaction between ClipR-59 and Elmo2. We found that the PH domain of Elmo2 and the Glu-Pro-rich domain of ClipR-59 mediated the interaction between Elmo2 and ClipR-59 (Figs. 4 and 7). The PH domain of Elmo2 has no binding activity toward phospholipid (38), therefore this domain is considered an atypical PH domain. The PH domain is often associated with an acidic motif (39). The Glu-Pro-rich domain of ClipR-59 is highly acidic. The finding that the Glu-Pro-rich domain ClipR-59 interacts with the PH domain of Elmo2 is in line with the notion.
The atypical PH domain of the Elmo protein was required for Elmo protein to interact with Dock protein. In our studies, we found that the PH domain was also required for Elmo2 to interact with ClipR-59. The question raised is how the same domain mediates the interaction of Elmo2 with both proteins? Studies of the interaction between Elmo and Dock protein revealed that the aPH domain does not directly contact with Dock, but provides a structural determinant in that the helice (αN and αC) before and after the PH domain along the PXXP motif were arranged to direct contact with Dock protein (35, 38). In our studies, we found that the aPH domain without these helices was sufficient to interact with ClipR-59 (Fig. 5D). These studies argue that the interaction of Elmo2 with ClipR-59 and Dock180 is through different sequences and unlikely mutually excluded. Further analysis of the structural requirement in the Elmo2 aPH domain is required to clarify this issue.
In the current study, we used overexpressed ClipR-59, Elmo2, as well as Dock180 to examine formation of the ClipR-59·Elmo2·Dock180 complex. Although these studies might not provide direct evidence that there is a complex consisting of these three proteins endogenously, the notion that knockdown of ClipR-59 reduced the levels of active Rac1 will argue that an endogenous complex consisting of these proteins is likely presented in cells (Fig. 4D). Further studies are required to clarify this issue.
Elmo2 exists in an autoinhibited status in which an intramolecular interaction between the Elmo inhibitory domain and EAD occurs (4). It is believed that the autoinhibitory status of Elmo2 is relieved following extracellular cues. In our studies, we found that the removal of either the Elmo inhibitory domain or the autoregulatory domain enhanced the interaction between ClipR-59. This suggests that the interaction between ClipR-59 and Elmo2 is subject to regulation. Supporting this notion, we found that expression of active Rho-GTPases such as RhoGG12V and Rac1G12V or cells stimulated with either IGF or insulin, enhanced the interaction between ClipR-59 and Elmo2 (Fig. 6, A and B). At present, the mechanism by which Rac1, IGF, and insulin promote interaction between Elmo2 and ClipR-59 requires further investigation. However, the ability of RhoG to enhance the interaction between ClipR-59 and Elmo2 could result from interaction between RhoG and Elmo2 as Elmo2 specifically interacts with active RhoG and was found in the ClipR-59·Elmo2 complex (Fig. 6A). Further studies are required to clarify these issues. Regardless, our studies here present the first experimental evidence that Elmo2 activity is regulated by extracellular stimuli.
To determine whether regulation of muscle differentiation by ClipR-59 required the interaction between ClipR-59 and Elmo2, we examined impact of the Elmo2 interacting defective ClipR-59 on C2C12 muscle differentiation and found that Elmo2 interacting defective ClipR-59 seemed to suppress C2C12 muscle cell differentiation (Fig. 7E), providing evidence that the interaction of ClipR-59 with Elmo2 is required for ClipR-59 to modulate muscle differentiation.
In summary, we have identified Elmo2 as a ClipR-59 interacting protein and showed that ClipR-59 regulates myoblast fusion. Overall, our studies suggest that by interacting with Elmo2, ClipR-59 facilitates Rac1 activation, and thereby muscle differentiation. Further examining the muscle development in ClipR-59 knock-out mice will be required to better define the role of ClipR-59.
Acknowledgments
We express our gratitude to Dr. Ralph Isberg for Rac and RhoG expression vectors, Dr. Michiyuki Matsuda (University of Kyoto) for FLAG-Dock180 expression plasmid, and Dr. Rachel Buchsbaum for GST-CRIB expression vector.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 DK084319 (to K. D.).
- aPH
- atypical plextrin homology domain
- EAD
- Elmo autoregulation domain
- TCL
- total cell lysates
- CRIB
- Cdc42/Rac1 interactive-binding
- Clip
- cytoplasmic linker protein
- ClipR-59
- Clip-related 59 kDa
- IGF
- insulin-like growth factor
- aa
- amino acid(s)
- Dock
- dedicator of cytokinesis
- CAP-Gly
- cytoskeleton-associated protein-Gly rich.
REFERENCES
- 1. Gumienny T. L., Brugnera E., Tosello-Trampont A. C., Kinchen J. M., Haney L. B., Nishiwaki K., Walk S. F., Nemergut M. E., Macara I. G., Francis R., Schedl T., Qin Y., Van Aelst L., Hengartner M. O., Ravichandran K. S. (2001) CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107, 27–41 [DOI] [PubMed] [Google Scholar]
- 2. Ravichandran K. S., Lorenz U. (2007) Engulfment of apoptotic cells: signals for a good meal. Nat. Rev. Immunol 7, 964–974 [DOI] [PubMed] [Google Scholar]
- 3. Laurin M., Côté J. F. (2014) Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev. 28, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel M., Margaron Y., Fradet N., Yang Q., Wilkes B., Bouvier M., Hofmann K., Côté J. F. (2010) An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr. Biol. 20, 2021–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patel M., Pelletier A., Côté J. F. (2011) Opening up on ELMO regulation: new insights into the control of Rac signaling by the DOCK180/ELMO complex. Small GTPases 2, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamoud N., Tran V., Croteau L. P., Kania A., Côté J. F. (2014) G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 111, 3745–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park D., Tosello-Trampont A. C., Elliott M. R., Lu M., Haney L. B., Ma Z., Klibanov A. L., Mandell J. W., Ravichandran K. S. (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434 [DOI] [PubMed] [Google Scholar]
- 8. Li H., Yang L., Fu H., Yan J., Wang Y., Guo H., Hao X., Xu X., Jin T., Zhang N. (2013) Association between Galphai2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat. Commun. 4, 1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fritsch R., de Krijger I., Fritsch K., George R., Reason B., Kumar M. S., Diefenbacher M., Stamp G., Downward J. (2013) RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell 153, 1050–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang G., Chen X., Qiu F., Zhu F., Lei W., Nie J. (2014) A novel interaction between SH2 domain of signaling adaptor protein Nck-1 and the upstream regulator of the Rho family GTPase Rac1-engulfment and cell motility 1 (ELMO1) promotes Rac1 activation and cell motility. J. Biol. Chem. 289, 23112–23122 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11. Ho E., Dagnino L. (2012) Emerging role of ILK and ELMO2 in the integration of adhesion and migration pathways. Cell Adh. Migr. 6, 168–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanui T., Inayoshi A., Noda M., Iwata E., Stein J. V., Sasazuki T., Fukui Y. (2003) DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood 102, 2948–2950 [DOI] [PubMed] [Google Scholar]
- 13. Hiramoto K., Negishi M., Katoh H. (2006) Dock4 is regulated by RhoG and promotes Rac-dependent cell migration. Exp. Cell Res. 312, 4205–4216 [DOI] [PubMed] [Google Scholar]
- 14. Katoh H., Negishi M. (2003) RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature 424, 461–464 [DOI] [PubMed] [Google Scholar]
- 15. Bowzard J. B., Cheng D., Peng J., Kahn R. A. (2007) ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J. Biol. Chem. 282, 17568–17580 [DOI] [PubMed] [Google Scholar]
- 16. Margaron Y., Fradet N., Côté J. F. (2013) ELMO recruits actin cross-linking family 7 (ACF7) at the cell membrane for microtubule capture and stabilization of cellular protrusions. J. Biol. Chem. 288, 1184–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grimsley C. M., Lu M., Haney L. B., Kinchen J. M., Ravichandran K. S. (2006) Characterization of a novel interaction between ELMO1 and ERM proteins. J. Biol. Chem. 281, 5928–5937 [DOI] [PubMed] [Google Scholar]
- 18. Hiramoto-Yamaki N., Takeuchi S., Ueda S., Harada K., Fujimoto S., Negishi M., Katoh H. (2010) Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J. Cell Biol. 190, 461–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duquette P. M., Lamarche-Vane N. (2014) Rho GTPases in embryonic development. Small GTPases 5, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Linseman D. A., Loucks F. A. (2008) Diverse roles of Rho family GTPases in neuronal development, survival, and death. Front. Biosci. 13, 657–676 [DOI] [PubMed] [Google Scholar]
- 21. Perez F., Pernet-Gallay K., Nizak C., Goodson H. V., Kreis T. E., Goud B. (2002) CLIPR-59, a new trans-Golgi/TGN cytoplasmic linker protein belonging to the CLIP-170 family. J. Cell Biol. 156, 631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maekawa H., Schiebel E. (2004) CLIP-170 family members: a motor-driven ride to microtubule plus ends. Dev. Cell 6, 746–748 [DOI] [PubMed] [Google Scholar]
- 23. Galjart N. (2005) CLIPs and CLASPs and cellular dynamics. Nat. Rev. Mol. Cell Biol. 6, 487–498 [DOI] [PubMed] [Google Scholar]
- 24. Lallemand-Breitenbach V., Quesnoit M., Braun V., El Marjou A., Poüs C., Goud B., Perez F. (2004) CLIPR-59 is a lipid raft-associated protein containing a cytoskeleton-associated protein glycine-rich domain (CAP-Gly) that perturbs microtubule dynamics. J. Biol. Chem. 279, 41168–41178 [DOI] [PubMed] [Google Scholar]
- 25. Ren W., Sun Y., Du K. (2013) DHHC17 palmitoylates ClipR-59 and modulates ClipR-59 association with the plasma membrane. Mol. Cell. Biol. 33, 4255–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ding J., Du K. (2009) ClipR-59 interacts with Akt and regulates Akt cellular compartmentalization. Mol. Cell. Biol. 29, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ren W., Cheema S., Du K. (2012) The association of ClipR-59 protein with AS160 modulates AS160 protein phosphorylation and adipocyte Glut4 protein membrane translocation. J. Biol. Chem. 287, 26890–26900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fujikura D., Ito M., Chiba S., Harada T., Perez F., Reed J. C., Uede T., Miyazaki T. (2012) CLIPR-59 regulates TNF-α-induced apoptosis by controlling ubiquitination of RIP1. Cell Death Dis. 3, e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Couesnon A., Offner N., Bernard V., Chaverot N., Backer S., Dimitrov A., Perez F., Molgó J., Bloch-Gallego E. (2013) CLIPR-59: a protein essential for neuromuscular junction stability during mouse late embryonic development. Development 140, 1583–1593 [DOI] [PubMed] [Google Scholar]
- 30. Buchsbaum R. J., Connolly B. A., Feig L. A. (2002) Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol. Cell. Biol. 22, 4073–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mohammadi S., Isberg R. R. (2009) Yersinia pseudotuberculosis virulence determinants invasin, YopE, and YopT modulate RhoG activity and localization. Infect. Immun. 77, 4771–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kato S., Du K. (2007) TRB3 modulates C2C12 differentiation by interfering with Akt activation. Biochem. Biophys. Res. Commun. 353, 933–938 [DOI] [PubMed] [Google Scholar]
- 33. Du K., Herzig S., Kulkarni R. N., Montminy M. (2003) TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 34. Miller J. B. (1990) Myogenic programs of mouse muscle cell lines: expression of myosin heavy chain isoforms, MyoD1, and myogenin. J. Cell Biol. 111, 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanawa-Suetsugu K., Kukimoto-Niino M., Mishima-Tsumagari C., Akasaka R., Ohsawa N., Sekine S., Ito T., Tochio N., Koshiba S., Kigawa T., Terada T., Shirouzu M., Nishikimi A., Uruno T., Katakai T., Kinashi T., Kohda D., Fukui Y., Yokoyama S. (2012) Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms. Proc. Natl. Acad. Sci. U.S.A. 109, 3305–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bagrodia S., Cerione R. A. (1999) Pak to the future. Trends Cell Biol. 9, 350–355 [DOI] [PubMed] [Google Scholar]
- 37. Lu M., Kinchen J. M., Rossman K. L., Grimsley C., deBakker C., Brugnera E., Tosello-Trampont A. C., Haney L. B., Klingele D., Sondek J., Hengartner M. O., Ravichandran K. S. (2004) PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat. Struct. Mol. Biol. 11, 756–762 [DOI] [PubMed] [Google Scholar]
- 38. Komander D., Patel M., Laurin M., Fradet N., Pelletier A., Barford D., Côté J. F. (2008) An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Mol. Biol. Cell 19, 4837–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burks D. J., Wang J., Towery H., Ishibashi O., Lowe D., Riedel H., White M. F. (1998) IRS pleckstrin homology domains bind to acidic motifs in proteins. J. Biol. Chem. 273, 31061–31067 [DOI] [PubMed] [Google Scholar]