Background: MicroRNAs are small non-protein-coding RNAs that inhibit target gene expression.
Results: p53 suppresses miR-520g expression, and miR-520g mediates drug resistance through down-regulation of p21 expression.
Conclusion: The p53/miR-520g/p21 signaling axis plays an important role in the response of colorectal cancer to chemotherapy.
Significance: Our study identifies miR-520g as a potential target against drug resistance in colorectal cancer, especially in patients with mutant p53.
Keywords: Apoptosis, Colorectal Cancer, Drug Resistance, MicroRNA (miRNA), p53, 5-FU, miR-520g, p21
Abstract
Development of drug resistance is one of the major causes of colorectal cancer recurrence, yet mechanistic understanding and therapeutic options remain limited. Here, we show that expression of microRNA (miR)-520g is correlated with drug resistance of colon cancer cells. Ectopic expression of miR-520g conferred resistance to 5-fluorouracil (5-FU)- or oxaliplatin-induced apoptosis in vitro and reduced the effectiveness of 5-FU in the inhibition of tumor growth in a mouse xenograft model in vivo. Further studies indicated that miR-520g mediated drug resistance through down-regulation of p21 expression. Moreover, p53 suppressed miR-520g expression, and deletion of p53 up-regulated miR-520g expression. Inhibition of miR-520g in p53−/− cells increased their sensitivity to 5-FU treatment. Importantly, studies of patient samples indicated that expression of miR-520g correlated with chemoresistance in colorectal cancer. These findings indicate that the p53/miR-520g/p21 signaling axis plays an important role in the response of colorectal cancer to chemotherapy. A major implication of our studies is that inhibition of miR-520g or restoration of p21 expression may have considerable therapeutic potential to overcome drug resistance in colorectal cancer patients, especially in those with mutant p53.
Introduction
Colorectal cancer is currently the third most common cancer diagnosed in the United States and the second leading cause of cancer-related deaths in both men and women. 5-Fluorouracil (5-FU)2 is one of the most commonly used drugs in colorectal cancer therapy. It acts principally as a thymidylate synthase inhibitor (1). Interrupting the action of this enzyme blocks synthesis of the pyrimidine thymidine, which is a nucleoside required for DNA replication. As a result, 5-FU is able to induce cell cycle arrest and/or apoptosis in cancer cells. Although adjuvant 5-FU treatment has a good success rate, recurrence of colorectal cancer is still frequent due to the development of drug resistance. Many mechanisms have been proposed to be responsible for drug resistance, including limitation of accumulation of drugs within cells by reducing uptake, enhancing efflux, or affecting membrane lipids; activation of detoxification; inhibition of apoptosis; and alterations in cell cycle and checkpoints (1–4). However, no significant advance has been made to identify targets against drug resistance (5). Therefore, it is important to better understand the mechanisms of drug resistance and uncover new targets to improve efficacy of chemotherapeutic treatment.
MicroRNAs (miRNAs) are a group of small non-protein-coding RNAs. They are evolutionarily conserved in the genomes of animals, plants, fungi, and viruses (6). miRNAs promote mRNA degradation or inhibit translation of many different target genes through sequence-specific interaction with their 3′-UTRs (7). It is estimated that miRNAs regulate more than one-third of human genes and the majority of genetic pathways (8, 9). Alterations in miRNA expression are associated with many human cancers (10). Therefore, miRNA profiling or signature in cancers has been proposed to be used to classify tumor subtypes, diagnose cancer, determine treatment plan, and predict patient outcome (11–13). In addition, miRNAs function as oncogenes or tumor suppressors depending on their target genes (10, 14). Much effort has been taken to understand the roles of miRNAs in cancer and the potential of manipulating miRNAs for cancer therapy. The pleiotropic nature of gene regulation by miRNAs suggests that some miRNAs may function as crucial mediators of drug resistance.
The p53 tumor suppressor is a transcriptional factor that can be activated by various stresses, including exposure to chemotherapeutic agents (15). Activation of p53 results in cell cycle arrest or apoptosis, mediated through enhanced expression of cell cycle-regulating proteins such as p21, an inhibitor of cyclin-dependent kinases, or of a number of pro-apoptotic proteins such as Bax (16). Although the role of p53 in the response of tumors to therapy is controversial (16–20), investigation of the mechanisms by which p53 mediates this response is important given that mutations inactivating p53 function are present in >50% of all human cancers. In this study, we discovered a novel p53/miR-520g/p21 signaling axis that mediates the response of colon cancer cells to 5-FU treatment. We show that p53 suppresses miR-520g expression and that deletion of p53 up-regulates miR-520g expression. Inhibition of miR-520g in p53−/− cells increased their sensitivity to 5-FU treatment. miR-520g conferred resistance to 5-FU-induced apoptosis through the inhibition of p21 expression, which is a direct target of miR-520g. Rescued expression of p21 in miR-520g-expressing colon cancer cells sensitized them to 5-FU-induced apoptosis. Importantly, experiments in tumor xenograft mouse models demonstrate that miR-520g reduced the effectiveness of 5-FU in the inhibition of tumor growth in vivo. Moreover, studies of colorectal cancer specimens indicate a positive correlation between miR-520g expression and chemoresistance. Our studies have identified miR-520g as a potential target for drug resistance in colorectal cancer.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
The human colon carcinoma RKO, HCT116, FET, and GEO cell lines were cultured in McCoy's 5A serum-free medium (Sigma) supplemented with 10 ng/ml epidermal growth factor, 20 μg/ml insulin, and 4 μg/ml transferrin (21). The HCT116 p53−/− cell line was cultured in McCoy's 5A serum-free medium supplemented with 10% fetal bovine serum (Life Technologies). Cells were maintained at 37 °C in a humidified incubator with 5% CO2. 5-FU and oxaliplatin were purchased from Sigma. Anti-hsa-miR-520g-3p miScript miRNA inhibitor was purchased from Qiagen (Valencia, CA). Antibodies for Western blot analyses were purchased as indicated: anti-cleaved poly(ADP-ribose) polymerase (PARP), Cell Signaling Technology (Beverly, MA); anti-p21 and anti-p53, Santa Cruz Biotechnology (Santa Cruz, CA); and anti-actin, Sigma.
Cell Viability and Apoptosis Assays
Colon cancer cells were plated in 96-well plates and treated with 5-FU or oxaliplatin for the indicated times. Cells were stained for 2 h with thiazolyl blue tetrazolium bromide (MTT; Sigma). The absorbance at 570 nm was read on an ELx808 absorbance microplate reader (BioTek, Winooski, VT) after the purple precipitates were dissolved with MTT detergent reagent. Cell viability was calculated as the ratio of the absorbance values of drug-treated samples to those of controls. Apoptosis was detected using a DNA fragmentation ELISA kit (Roche Applied Science).
Western Blot Analysis and Real-time Quantitative PCR (qPCR)
Whole cell lysates were prepared in radioimmune precipitation assay buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 10 mm Tris-HCl (pH 7.5), 5 mm EDTA, and protease inhibitor mixture (Sigma)). Equivalent amounts of protein were separated by SDS-PAGE and transferred to a PVDF membrane (Millipore, Billerica, MA). Proteins were detected using an enhanced chemiluminescence system (Amersham Biosciences).
Expression of miR-520g-3p was determined by miScript primer assays with an miScript SYBR Green PCR kit (Qiagen). RNU6-2 was used as an endogenous reference gene.
Luciferase Assays
The reverse complementary sequence of miR-520g-3p was synthesized and cloned into a Promega psiCHECK2 vector downstream of the Renilla reporter gene to generate psiCHECK2-520g. The 3′-UTR of p21 containing miR-520g-binding sites was amplified and cloned into the same vector to generate psiCHECK2-p21. The psiCHECK2 vector contains a second reporter gene (firefly luciferase) and is designed for end point lytic assays. The reporter was transfected into cells using Lipofectamine LTX (Life Technologies). Luciferase activity was measured 48 h later using the Dual-Luciferase reporter assay (Promega). Values were normalized with firefly luciferase activity.
Plasmid Construction and Lentiviral Infection
cDNA encoding human miR-520g precursor (∼300 bp) was cloned into the pCDH-CMV lentiviral vector (System Biosciences, Mountain View, CA). shRNAs targeting p21 were constructed by cloning annealed oligonucleotides into the FSIPPW lentiviral vector. The targeting sequences of p21 shRNA are GTGGACAGCGAGCAGCTGA and CTTCGACTTTGTCACCGAG. 293 packaging cells were cotransfected with pPACKH1 packaging plasmid mixture (System Biosciences) and the lentiviral vectors using FuGENE HD (Promega). Viruses were harvested 48 h later and used to infect target cells.
In Vivo Xenograft Model
Experiments involving animals were approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee. HCT116 cells (2 × 106) expressing miR-520g or an empty vector were injected into the flanks of male athymic nude mice (4–5 weeks old). One week after injection, 5-FU (40 mg/kg/day) or carrier was administered by intraperitoneal injection for 5 consecutive days/week for 2 weeks (22). Tumor volumes were measured at the beginning of the treatment and every other day after that until the mice were terminated. The estimated tumor volumes (V) were calculated by the formula V = W2 × L × 0.5, where W represents the largest tumor diameter in centimeters, and L represents the next largest tumor diameter. The relative tumor volumes (RTV) were calculated by RTV = Vx/V0, where Vx is the volume in cubic millimeters at a given time, and V0 is the volume at the beginning of the treatment (22). Tumors were dissected out and weighed.
TUNEL and Ki67 Staining
Formalin-fixed paraffin-embedded tissue blocks of tumors were stained for TUNEL and Ki67 using the procedure described previously (23). Three tumors from each group were analyzed. Ten histologically similar fields were randomly selected from each slide for analysis. Apoptosis and proliferation of tumor cells were determined quantitatively by counting the numbers and calculating the percentage of positively stained cells for TUNEL and Ki67 at ×20 magnification, respectively.
Determination of miR-520g Expression in Human Tissue Samples
Formalin-fixed paraffin-embedded blocks of human colorectal adenocarcinomas were obtained from files of the Department of Pathology and Microbiology at the University of Nebraska Medical Center. The ages of all patients (including both men and women) were between 55 and 85 years. The cancer patients received neoadjuvant chemoradiotherapy prior to surgical removal of the tumors. The study was performed with the approval of the ethics committee of the Institutional Review Board.
The formalin-fixed paraffin-embedded slides were deparaffinized using Histo-Clear and rehydrated following hematoxylin staining to identify the tumor areas under a microscope. The tumor areas were then scrapped with a fine scalpel and collected in an Eppendorf tube. RNA was extracted using an miRNeasy FFPE kit (Qiagen). Expression of miR-520g-3p was determined by qPCR as described above.
Statistical Analysis
Statistical analyses were performed using two-way analysis of variance or Student's t test.
RESULTS
miR-520g Confers Resistance to 5-FU-induced Apoptosis in Colon Cancer Cells in Vitro
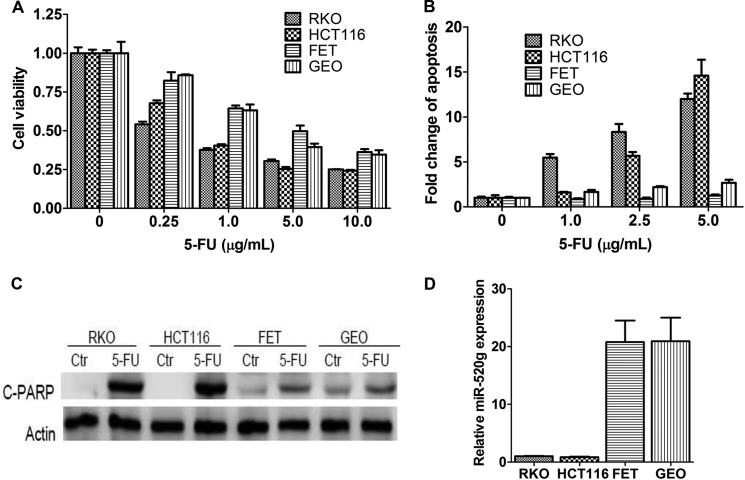
5-FU is one of the most commonly used chemotherapeutic agents for colorectal cancer. However, the lack of response due to drug resistance has been a main problem that affects the outcome of cancer therapy. To better understand the mechanisms of drug resistance, we examined a panel of colon cancer cell lines for their response to 5-FU treatment. Among the cell lines tested, RKO and HCT116 cells were more sensitive to 5-FU treatment compared with FET and GEO cells (Fig. 1A). Their differential response to 5-FU was attributed mainly to different sensitivities to 5-FU-induced apoptosis as reflected by DNA fragmentation assays (Fig. 1B) and PARP cleavage (Fig. 1C). To characterize the molecular determinants of these differences, we investigated the contribution of miRNAs. We identified potential miRNAs whose expression conferred 5-FU resistance in a functional screening of an miRNA library.3 Among those miRNAs identified, expression of miR-520g was positively correlated with 5-FU resistance in colon cancer cell lines. As shown in Fig. 1D, RKO and HCT116 cells expressed much lower levels of miR-520g compared with FET and GEO cells, suggesting that miR-520g may play a role in 5-FU resistance in colon cancer cells.
FIGURE 1.
Expression of miR-520g is correlated with resistance to 5-FU-induced apoptosis in colon cancer cells. A and B, RKO, HCT116, FET, and GEO cells were treated with different concentrations of 5-FU for 72 h. MTT assays (A) and DNA fragmentation assays (B) showed different sensitivity of these cells to 5-FU treatment. C, Western blot analysis showed that cleaved PARP (C-PARP) was higher in RKO and HCT116 cells than in FET and GEO cells after treatment with 10 μg/ml 5-FU for 72 h. Ctr, control. D, FET and GEO cells expressed much higher levels of miR-520g compared with RKO and HCT116 cells as determined by real-time qPCR analysis. The data are presented as the mean ± S.D. of triplicate experiments.
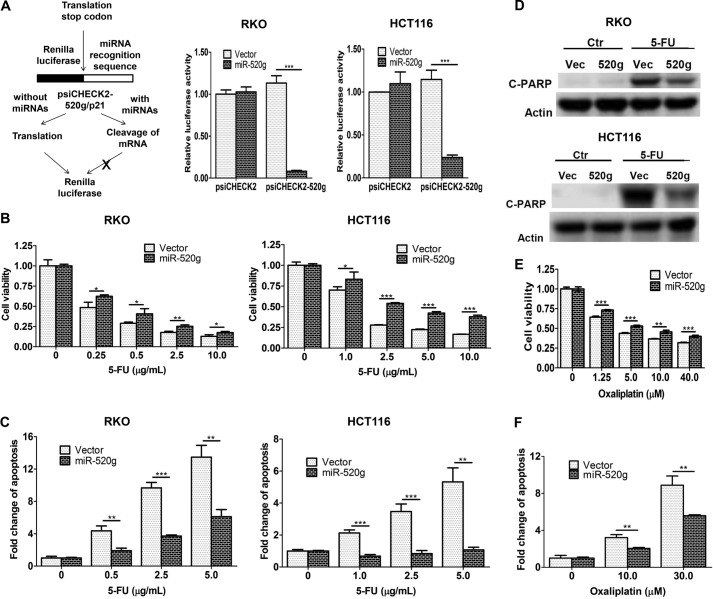
To determine whether miR-520g confers resistance to 5-FU, it was ectopically expressed in RKO and HCT116 cells. We used luciferase reporter assays to determine the function of ectopically expressed miR-520g. The reporter plasmid psiCHECK2-520g contains the miR-520g recognition element in the 3′-UTR of the Renilla luciferase gene. In the absence of miR-520g, luciferase will be expressed, whereas in the presence of miR-520g, luciferase mRNA will be degraded (Fig. 2A, left panel). Luciferase assays revealed that the luciferase activity of psiCHECK2-520g was decreased significantly in miR-520g-expressing cells compared with vector control cells, whereas there was little change in the luciferase activity of the control plasmid psiCHECK2 (Fig. 2A, middle and right panels). This indicated that ectopically expressed miR-520g was functional in RKO and HCT116 cells.
FIGURE 2.
miR-520g induces resistance to 5-FU treatment in colon cancer cells in vitro. A, the left panel shows a diagram elucidating how the luciferase reporter assays work. The reporter plasmid psiCHECK2-520g contains the miR-520g recognition element in the 3′-UTR of the Renilla luciferase gene. In the absence of miR-520g, luciferase will be expressed, whereas in the presence of miR-520g, luciferase mRNA will be degraded. The middle and right panels show the Dual-Luciferase assays performed in RKO and HCT116 cells, in which Renilla luciferase activity was determined and normalized to firefly luciferase activity. The luciferase activity of psiCHECK2-520g was reduced in miR-520g-expressing cells compared with vector control cells, whereas there was little change in the luciferase activity of the control plasmid psiCHECK2. B and C, miR-520g- and vector-expressing cells were treated with different concentrations of 5-FU for 72 h. MTT assays (B) and DNA fragmentation assays (C) showed increased resistance to 5-FU-induced apoptosis in miR-520g-expressing cells compared with the control cells. D, Western blot analysis showed that cleaved PARP (C-PARP) was lower in miR-520g-expressing cells (520g) than in vector control cells (Vec) after treatment with 10 μg/ml 5-FU for 72 h. Ctr, control. E and F, miR-520g- and vector-expressing cells were treated with different concentrations of oxaliplatin for 72 h. MTT assays (E) and DNA fragmentation assays (F) showed increased resistance to oxaliplatin-induced apoptosis in miR-520g-expressing cells. The data are presented as the mean ± S.D. of triplicate experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
After exposure to 5-FU at different concentrations, miR-520g-expressing cells displayed increased cell viability (Fig. 2B) and significantly reduced apoptosis (Fig. 2C) compared with vector control cells. Consistent with DNA fragmentation assays in Fig. 2C, induction of PARP cleavage by 5-FU was smaller in miR-520g-expressing cells than in vector control cells (Fig. 2D). These results indicated that miR-520g protected RKO and HCT116 cells from 5-FU-induced apoptosis.
To determine whether miR-520g plays a protective role in the resistance to other chemotherapeutic drugs, HCT116 cells were treated with oxaliplatin, an alkylating agent commonly used in combination with 5-FU for treating advanced colon cancer (24, 25). miR-520g-expressing cells were also more resistant to oxaliplatin treatment than vector control cells as reflected by increased cell viability (Fig. 2E) and reduced apoptosis (Fig. 2F). Although 5-FU and oxaliplatin function through different targets, they may share a similar mechanism for drug resistance. Therefore, these data suggest that increased miR-520g expression may be a common mechanism for drug resistance of different chemotherapeutic agents in colon cancer cells.
miR-520g Reduces the Effectiveness of 5-FU in Tumor Growth Inhibition in a Colon Cancer Xenograft Model in Vivo
To test whether miR-520g-mediated resistance to 5-FU-induced apoptosis in vitro translates to drug resistance in vivo, we established a colon tumor xenograft model by subcutaneously inoculating athymic nude mice with 2 million HCT116 cells expressing vector or miR-520g. After 7 days, mice were randomly divided into two groups. One group was treated with 5-FU (40 mg/kg/day) administered by intraperitoneal injection, and the other was treated with the carrier. The treatment was for 5 consecutive days/week for 2 weeks (22). Tumor growth and therapeutic sensitivity were monitored during the course of 5-FU treatment.
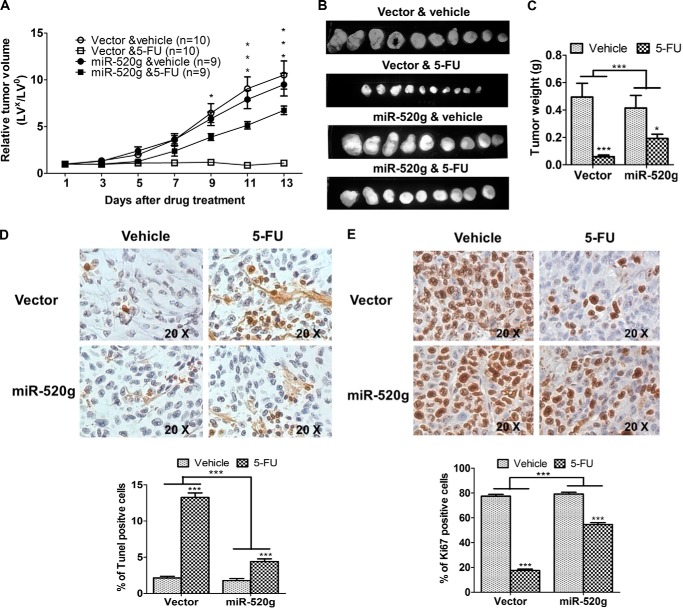
Xenograft tumor growth curves showed that tumors grew at similar rates independent of miR-520g expression. However, there was a marked difference in the response of tumors to 5-FU treatment. Tumors with vector-expressing cells (designated as vector tumors) did not grow during drug treatment, whereas tumors with miR-520g-expressing cells (designated as miR-520g tumors) continued to grow at a steady rate (Fig. 3A). In addition, miR-520g tumors showed significantly smaller reduction of tumor size (Fig. 3B) and tumor weight (Fig. 3C) after 5-FU treatment compared with vector tumors. These results indicated that 5-FU treatment was less effective in inhibiting the growth of miR-520g tumors than that of vector tumors. To determine whether miR-520g-mediated resistance to apoptosis in vitro is associated with reduced 5-FU effect in vivo, TUNEL assays were performed to determine the apoptotic index of tumors. TUNEL staining showed that, although the percentage of apoptotic cells was similar in miR-520g and vector tumors, the increase in apoptotic cells induced by 5-FU treatment was significantly lower in miR-520g tumors than in vector tumors (2.5-fold versus 6.2-fold) (Fig. 3D). In addition, Ki67 staining showed that 5-FU treatment decreased the percentage of proliferative cells in miR-520g tumors by 31% and in vector tumors by 77% (Fig. 3E). These studies indicated that the effect of miR-520g on drug resistance was a combined result of its resistance to 5-FU-induced apoptosis and inhibition of proliferation. Taken together, these in vitro and in vivo results demonstrate an important role of miR-520g in drug resistance of colon cancer cells.
FIGURE 3.
miR-520g reduces the effectiveness of 5-FU in inhibition of tumor growth in vivo. Xenograft tumor growth curves (A), images of tumors taken on the same scale (B), and tumor weights (C) are shown. D and E, images of TUNEL (D; ×20 magnification) and Ki67 (E; ×20 magnification) staining of tumors are shown in the upper panels. The images are representative of multiple fields of tumor sections from each group. The percentage of positive TUNEL (D) and Ki67 (E) staining cells was determined as described under “Experimental Procedures” (lower panels). The data are presented as the mean ± S.E. *, p < 0.05; ***, p < 0.001.
miR-520g Increases Drug Resistance by Reducing the Expression of Its Target Gene p21
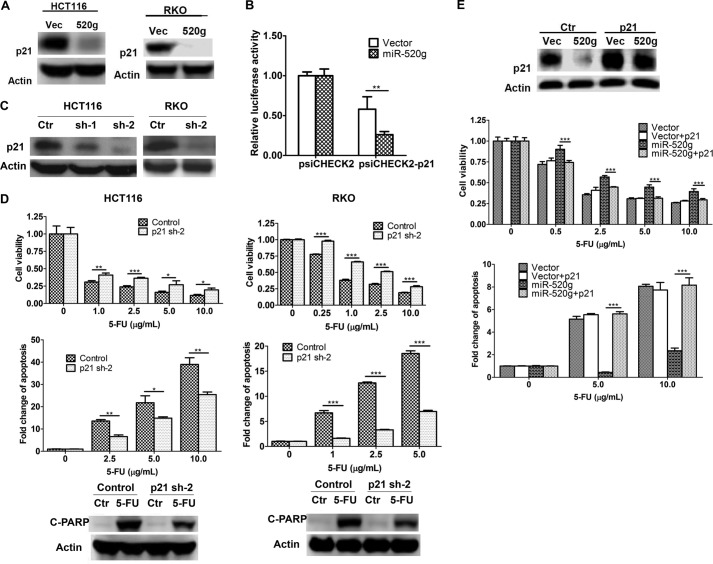
The ability of miR-520g to confer resistance to 5-FU-induced apoptosis is attributed to its ability to regulate expression of its target genes. To identify target genes of miR-520g, we used several algorithms that predict the mRNA targets of miRNAs: TargetScan (26), PicTar (27), and miRanda-mirSVR (28). Based on the representation of miR-520g recognition sites in their 3′-UTRs, candidate target genes were predicted. Among those tested, p21 showed reduced expression in HCT116 and RKO cells expressing miR-520g compared with vector cells (Fig. 4A). To determine whether p21 is a direct target of miR-520g, the 3′-UTR of p21 containing the potential miR-520g recognition element was cloned into the luciferase reporter construct psiCHECK2-p21 (Fig. 2A, left panel). Luciferase assays showed that the luciferase activity of psiCHECK2-p21 was decreased in miR-520g-expressing cells compared with vector control cells, whereas that of the control plasmid psiCHECK2 remained unchanged (Fig. 4B). These results indicated that miR-520g repressed the 3′-UTR of p21 and that expression of p21 was directly regulated by miR-520g.
FIGURE 4.
miR-520g increases drug resistance by reducing p21 expression. A, p21 expression was reduced by miR-520g expression in HCT116 and RKO cells as determined by Western blot analysis. B, the luciferase construct with the 3′-UTR of p21 (psiCHECK2-p21) was transfected into HCT116 cells. Dual-Luciferase assays were performed as described for Fig. 2A. The luciferase activity of psiCHECK2-p21 was reduced in miR-520g-expressing cells compared with vector control cells, whereas there was little change in the luciferase activity of the control plasmid psiCHECK2. C, expression of p21 was knocked down by shRNAs (sh) in HCT116 and RKO cells. Ctr, control. D, p21 knockdown and control cells were treated with different concentrations of 5-FU for 72 h. MTT assays (upper panels), DNA fragmentation assays (middle panels), and Western blot analysis of cleaved PARP (C-PARP; lower panels) were performed, all of which indicated that knockdown of p21 expression conferred resistance to 5-FU-induced apoptosis. E, p21 was ectopically expressed in HCT116 vector-expressing (Vec) and miR-520g-expressing (520g) cells. Western blot analysis was performed to confirm the rescued expression of p21 in miR-520g-expressing cells (upper panel). Cells were treated with different concentrations of 5-FU for 72 h. MTT assays (middle panel) and DNA fragmentation assays (lower panel) showed that restoration of p21 expression resensitized miR-520g-expressing cells to 5-FU treatment. The data are presented as the mean ± S.D. of triplicate experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
p21 has been reported to have both pro- and anti-apoptotic effects (29–34). To determine whether p21 plays a role in 5-FU-induced apoptosis, its expression was knocked down by two different shRNAs. shRNA-2 showed a better knockdown effect than shRNA-1, resulting in >90% reduction of p21 expression in HCT116 and RKO cells (Fig. 4C). Knockdown of p21 expression enhanced the resistance of HCT116 and RKO cells to 5-FU-induced apoptosis as reflected by increased cell viability (Fig. 4D, upper panel) and reduced apoptosis (middle and lower panels). These results indicated that p21 was required for 5-FU-induced apoptosis. We next determined whether miR-520g-mediated resistance to 5-FU-induced apoptosis could be reversed by restoration of p21 expression. p21 cDNA was introduced into miR-520g-expressing HCT116 cells. Ectopically expressed p21 is resistant to down-regulation mediated by miR-520g due to the lack of 3′-UTRs. Restoration of p21 expression almost completely restored the sensitivity of HCT116 cells to 5-FU-induced apoptosis (Fig. 4E). These results indicated that miR-520g promoted cell survival through the down-regulation of p21 expression, which further confirmed that p21 was required for 5-FU-induced apoptosis.
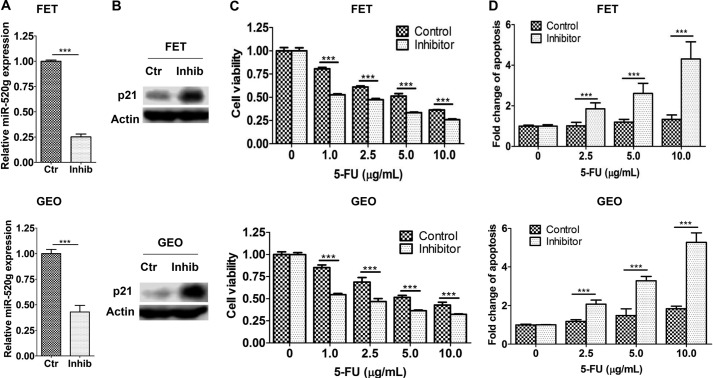
As a complementary approach, an anti-hsa-miR-520g miScript miRNA inhibitor was used to inhibit miR-520g expression and function. The miScript miRNA inhibitors from Qiagen are chemically synthesized, single-stranded, modified RNAs that specifically decrease endogenous miRNA levels and suppress their function when transfected into cells. We chose FET and GEO cells, which express high levels of endogenous miR-520g (Fig. 1D), for the inhibitor study. As shown in Fig. 5A, the inhibitor decreased miR-520g expression effectively in both FET and GEO cells. As a result, p21 expression was increased (Fig. 5B). The inhibitor sensitized FET and GEO cells to 5-FU-induced apoptosis as reflected by reduced cell viability (Fig. 5C) and increased apoptosis (Fig. 5D). Taken together, these results demonstrated that miR-520g conferred drug resistance in colon cancer cells and that it functioned through repressing p21 expression.
FIGURE 5.
A miR-520g inhibitor sensitizes FET and GEO cells to 5-FU-induced apoptosis. A chemically synthesized miR-520g inhibitor (Inhib) was transfected into FET and GEO cells, resulting in reduced miR-520g expression as determined by real-time qPCR assays (A) and increased p21 expression as determined by Western blot analysis (B). Ctr, control. The miR-520g inhibitor-transfected cells were treated with different concentrations of 5-FU for 72 h. MTT assays (C) and DNA fragmentation assays (D) showed that the inhibitor sensitized FET and GEO cells to 5-FU-induced apoptosis. The data are presented as the mean ± S.D. of triplicate experiments. ***, p < 0.001.
p53 Suppresses Expression of miR-520g in Colon Cancer Cells
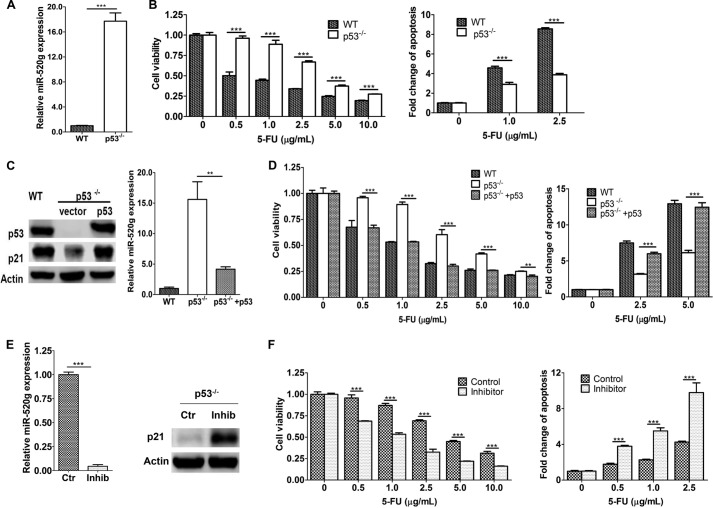
Based on in vitro and in vivo studies described above, miR-520g contributes to drug resistance of colon cancer cells. Therefore, it is important to determine how its expression is regulated, which would provide valuable information on the potential of inhibiting its expression to increase efficacy of drug treatment. As shown in Fig. 1D, miR-520g expression was high in FET cells with mutated p53 (35) and low in HCT116 and RKO cells with WT p53 (35, 36). p53 has been implicated to contribute to 5-FU- and other chemotherapeutic drug-induced apoptosis, which involves the DNA damage response (18, 37). We therefore hypothesized that p53 suppresses miR-520g expression in colon cancer cells. To test this hypothesis, we compared miR-520g expression in HCT116 WT and p53-null (p53−/−) cells. Expression of miR-520g was 16.7-fold higher in p53−/− cells than in WT cells (Fig. 6A). As a result, p53−/− cells were much more resistant to 5-FU-induced apoptosis compared with WT cells (Fig. 6B). Re-expression of p53 in p53−/− cells reduced miR-520g expression significantly (Fig. 6C), indicating that p53 inhibited miR-520g expression. Of note, expression of p21 was increased in p53-re-expressing cells (Fig. 6C, left panel). Consequently, restoration of p53 expression in HCT116 p53−/− cells sensitized them to 5-FU treatment (Fig. 6D).
FIGURE 6.
p53 suppresses miR-520g expression. A, miR-520g expression was higher in HCT116 p53−/− cells compared with HCT116 WT cells as determined by real-time qPCR assays. B, HCT116 WT and p53−/− cells were treated with different concentrations of 5-FU for 72 h. MTT assays (left panel) and DNA fragmentation assays (right panel) showed that p53−/− cells were more resistant to 5-FU-induced apoptosis than WT cells. C, p53 was ectopically expressed in HCT116 p53−/− cells. Western blot analysis showed restored expression of p53 and p21 (left panel). Real-time qPCR assays showed that restored p53 expression suppressed miR-520g expression (right panel). D, cells were treated with different concentrations of 5-FU for 72 h. MTT assays (left panel) and DNA fragmentation assays (right panel) showed that restored expression of p53 resensitized p53−/− cells to 5-FU-induced apoptosis. E, the miR-520g inhibitor (Inhib) was transfected into HCT116 p53−/− cells. As a result, miR-520g expression was suppressed as determined by real-time qPCR assays (left panel), and p21 expression was increased as determined by Western blot assays (right panel). Ctr, control. F, the miR-520g inhibitor-transfected cells were treated with different concentrations of 5-FU for 72 h. MTT assays (left panel) and DNA fragmentation assays (right panel) showed that the inhibitor sensitized p53−/− cells to 5-FU treatment. The data are presented as the mean ± S.D. of triplicate experiments. **, p < 0.01; ***, p < 0.001.
To determine whether miR-520g is responsible for the resistance of p53−/− cells to 5-FU-induced apoptosis, the miR-520g inhibitor described above was transfected into p53−/− cells. The inhibitor reduced miR-520g expression significantly in p53−/− cells as shown by real-time qPCR (Fig. 6E, left panel), which led to up-regulation of p21 expression (right panel). Inhibition of miR-520g effectively increased sensitivity to 5-FU-induced apoptosis in p53−/− cells (Fig. 6F). These results indicated that one of the mechanisms by which p53 mediated 5-FU-induced apoptosis was through the suppression of miR-520g expression and that elevated miR-520g expression contributed to 5-FU resistance in p53−/− cells.
Expression of miR-520g Correlates with Chemoresistance in Patient Samples
Although it has been shown that expression of miR-520g is increased in colorectal cancer compared with normal controls (Gene Expression Omnibus (GEO) Database), it is not clear whether this is relevant to patients' responses to chemotherapy. To demonstrate the clinical relevance of miR-520g expression in chemoresistance in cancer patients, we extended our analyses by assaying miR-520g expression in human colorectal adenocarcinoma specimens. RNA was isolated from sections prepared from 20 patients who had received neoadjuvant chemoradiotherapy. Among them, 10 patients had moderate responses, and 10 had no or poor responses. As shown in Fig. 7A, real-time qPCR assays demonstrated that expression of miR-520g in non- or poor responders was higher than that in moderate responders (*, p < 0.05). Expression of miR-520g in individual tissue samples is shown in Fig. 7B. These results indicated that expression of miR-520g correlated with chemoresistance in colorectal cancer patients. Taken together with in vitro and in vivo results, our studies demonstrate that miR-520g plays an important role in drug resistance of colorectal cancer.
FIGURE 7.
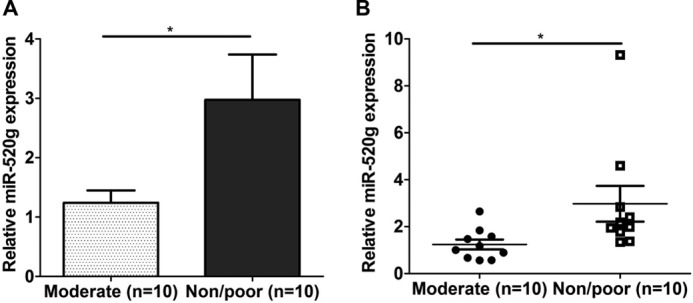
Expression of miR-520g correlates with chemoresistance in human colorectal cancer specimens. A, RNA samples prepared from 10 moderately responding and 10 non-responding or poorly responding colorectal tumors were used to determine miR-520g expression as described under “Experimental Procedures.” The data are presented as the mean ± S.E. B, expression of miR-520g in individual tumor samples is shown. Bars indicate the mean values of each group of samples. *, p < 0.05.
DISCUSSION
We have identified in this study a novel p53/miR-520g/p21 signaling axis that regulates the response of colon cancer cells to chemotherapeutic agents, including 5-FU and oxaliplatin (Fig. 8). In WT p53 cells, expression of miR-520g is suppressed and 5-FU treatment induces apoptosis whereas, in p53−/− cells, expression of miR-520g is elevated, and cells become resistant to 5-FU. A miR-520g inhibitor sensitizes p53−/− cells to 5-FU-induced apoptosis. Ectopic expression of miR-520g induces resistance to 5-FU treatment in vitro and in tumor xenografts in vivo. miR-520g confers drug resistance by inhibition of p21 expression, and p21 is required for 5-FU-induced apoptosis. Because resistance to 5-FU treatment is one of the major causes for chemotherapy failure in treating advanced colorectal cancer (38, 39), the discovery of miR-520g as a contributing factor of drug resistance and the identification of the regulatory p53/miR-520g/p21 signaling axis would help design strategies to increase the efficacy of chemotherapeutic treatment of colorectal cancer.
FIGURE 8.
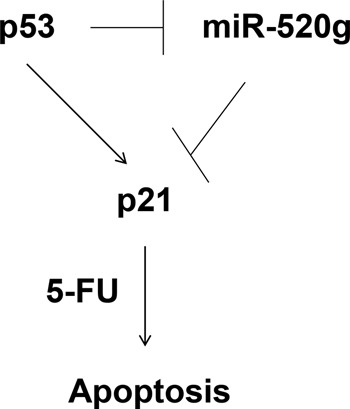
Proposed p53/miR-520g/p21 signaling axis that regulates 5-FU effect in colorectal cancer.
It is well known that p21 is a transcriptional target of p53. We have shown here that p21 is also a direct target of miR-520g in colon cancer cells. Because p53 suppresses miR-520g expression, p53 likely regulates p21 expression by two different mechanisms: regulating its promoter activity through transcriptional factors and its 3′-UTR through miR-520g (Fig. 8). Given that cancer cells often evade normal regulation of cell proliferation and survival, the multilevel regulation of p21 expression by p53 ensures that p53 induces p21 expression under stress conditions, including drug treatment, which would trigger cell cycle arrest and/or apoptosis in cancer cells. Of note, p53/p21 does not seem to affect cancer cell growth under normal conditions.
Although re-expression of p21 almost completely restored the sensitivity of miR-520g-expressing cells to 5-FU-induced apoptosis, miR-520g may also function through other targets and pathways. We found that expression of XIAP and survivin, two prosurvival proteins, was increased by miR-520g in HCT116 and RKO cells (data not shown). It has been shown that XIAP and survivin promote cell survival under stress conditions in colon cancer cells (40, 41) and that knockdown of their expression increases chemosensitivity of pancreatic cancer cells to 5-FU and gemcitabine treatment (42). We do not know the mechanisms by which miR-520g increases expression of XIAP and survivin. However, it has been recently reported that the TGFβ/PKA signaling transduceome negatively regulates expression of XIAP and survivin in colon cancer cells (40). We are currently investigating whether miR-520g affects TGFβ/PKA signaling.
There are contradictory reports on the relationship of p53, p21, and drug sensitivity in different cancer cells. Some studies show that drug-induced apoptosis is independent of p53, and others indicate that p53 is required (17–19, 43). In addition, p21 has been reported to have a dual role in apoptosis (29–34). Our studies show that both p53 and p21 are required for 5-FU-induced apoptosis in colon cancer cells. The differing results could be attributed to different cell context, experimental conditions, or assay approaches used in the various studies. In addition, it is still not clear how p21 mediates 5-FU-induced apoptosis in colon cancer cells. It has been shown that p21 promotes ceramide-induced apoptosis by enhancing Bax expression in human hepatocarcinoma cells (44). However, our results indicate that knockdown of p21 expression has little effect on expression of Bax (data not shown).
Several miRNAs have been shown to mediate 5-FU response by targeting different genes in colon cancer cells. For example, miR-21 induces resistance to 5-FU by down-regulating expression of human MSH2 (22). miR-143 increases sensitivity to 5-FU by reducing expression of ERK5, NF-κB, and Bcl-2 (45). Our discovery of miR-520g as a mediator of drug response in colorectal cancer provides another potential therapeutic target. Inhibition of miR-520g expression or activity could increase chemosensitivity in colorectal cancer treatment. Clinical data from the GEO Database show that expression of miR-520g is increased in colorectal cancer compared with normal controls. Because mutations inactivating p53 function occur frequently in colorectal cancer (46), it is probably one of the mechanisms responsible for up-regulated expression of miR-520g. p53 has been shown to regulate transcription and processing of miRNAs (47). The mechanism by which p53 suppresses miR-520g expression remains to be determined. Our studies suggest that miR-520g may be a promising target against drug resistance in colorectal cancer treatment, especially in patients with mutant p53.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA140988-01 from NCI. This work was also supported by Nebraska Department of Health and Human Services Grant LB506 (2014-40; to J. W.).
Y. Zhang and J. Wang, unpublished data.
- 5-FU
- 5-fluorouracil
- miRNA/miR
- microRNA
- PARP
- poly(ADP-ribose) polymerase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- qPCR
- quantitative PCR
- XIAP
- X-linked inhibitor of apoptosis.
REFERENCES
- 1. Longley D. B., Harkin D. P., Johnston P. G. (2003) 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3, 330–338 [DOI] [PubMed] [Google Scholar]
- 2. Liu Y.-Y., Han T.-Y., Giuliano A. E., Cabot M. C. (2001) Ceramide glycosylation potentiates cellular multidrug resistance. FASEB J. 15, 719–730 [DOI] [PubMed] [Google Scholar]
- 3. Longley D. B., Johnston P. G. (2005) Molecular mechanisms of drug resistance. J. Pathol. 205, 275–292 [DOI] [PubMed] [Google Scholar]
- 4. Synold T. W., Dussault I., Forman B. M. (2001) The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat. Med. 7, 584–590 [DOI] [PubMed] [Google Scholar]
- 5. Gottesman M. M. (2002) Mechanisms of cancer drug resistance. Annu. Rev. Med. 53, 615–627 [DOI] [PubMed] [Google Scholar]
- 6. Du T., Zamore P. D. (2005) microPrimer: the biogenesis and function of microRNA. Development 132, 4645–4652 [DOI] [PubMed] [Google Scholar]
- 7. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hwang H. W., Mendell J. T. (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 94, 776–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 10. Zhang B., Pan X., Cobb G. P., Anderson T. A. (2007) MicroRNAs as oncogenes and tumor suppressors. Dev. Biol. 302, 1–12 [DOI] [PubMed] [Google Scholar]
- 11. Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K. (2002) Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 99, 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calin G. A., Liu C.-G., Sevignani C., Ferracin M., Felli N., Dumitru C. D., Shimizu M., Cimmino A., Zupo S., Dono M., Dell'Aquila M. L., Alder H., Rassenti L., Kipps T. J., Bullrich F., Negrini M., Croce C. M. (2004) MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. U.S.A. 101, 11755–11760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Calin G. A., Croce C. M. (2006) MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 14. Calin G. A., Croce C. M. (2006) MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene 25, 6202–6210 [DOI] [PubMed] [Google Scholar]
- 15. Sun X.-X., Dai M.-S., Lu H. (2007) 5-Fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J. Biol. Chem. 282, 8052–8059 [DOI] [PubMed] [Google Scholar]
- 16. Miyashita T., Reed J. C. (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80, 293–299 [DOI] [PubMed] [Google Scholar]
- 17. Kim C. W., Lu J. N., Go S.-I., Jung J. H., Yi S. M., Jeong J.-H., Hah Y.-S., Han M. S., Park J. W., Lee W. S., Min Y. J. (2013) p53 restoration can overcome cisplatin resistance through inhibition of Akt as well as induction of Bax. Int. J. Oncol. 43, 1495–1502 [DOI] [PubMed] [Google Scholar]
- 18. Chakraborty S., Mazumdar M., Mukherjee S., Bhattacharjee P., Adhikary A., Manna A., Chakraborty S., Khan P., Sen A., Das T. (2014) Restoration of p53/miR-34a regulatory axis decreases survival advantage and ensures Bax-dependent apoptosis of non-small cell lung carcinoma cells. FEBS Lett. 588, 549–559 [DOI] [PubMed] [Google Scholar]
- 19. Metzinger D. S., Taylor D. D., Gercel-Taylor C. (2006) Induction of p53 and drug resistance following treatment with cisplatin or paclitaxel in ovarian cancer cell lines. Cancer Lett. 236, 302–308 [DOI] [PubMed] [Google Scholar]
- 20. Stoklosa T., Slupianek A., Datta M., Nieborowska-Skorska M., Nowicki M. O., Koptyra M., Skorski T. (2004) Report BCR/ABL recruits p53 tumor suppressor protein to induce drug resistance. Cell Cycle 3, 1463–1472 [DOI] [PubMed] [Google Scholar]
- 21. Boyd D. D., Levine A. E., Brattain D. E., McKnight M. K., Brattain M. G. (1988) Comparison of growth requirements of two human intratumoral colon carcinoma cell lines in monolayer and soft agarose. Cancer Res. 48, 2469–2474 [PubMed] [Google Scholar]
- 22. Valeri N., Gasparini P., Braconi C., Paone A., Lovat F., Fabbri M., Sumani K. M., Alder H., Amadori D., Patel T. (2010) MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl. Acad. Sci. U.S.A. 107, 21098–21103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geng L., Chaudhuri A., Talmon G., Wisecarver J. L., Are C., Brattain M., Wang J. (2014) MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene 33, 5332–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jun L., Haiping Z., Beibei Y. (2009) Genetic polymorphisms of GSTP1 related to response to 5-FU-oxaliplatin-based chemotherapy and clinical outcome in advanced colorectal cancer patients. Swiss Med. Wkly.139, 724–728 [DOI] [PubMed] [Google Scholar]
- 25. Kim S.-H., Kwon H.-C., Oh S. Y., Lee D. M., Lee S., Lee J.-H., Roh M.-S., Kim D.-C., Park K.-J., Choi H.-J., Kim H.-J. (2009) Prognostic value of ERCC1, thymidylate synthase, and glutathione S-transferase π for 5-FU/oxaliplatin chemotherapy in advanced colorectal cancer. Am. J. Clin. Oncol. 32, 38–43 [DOI] [PubMed] [Google Scholar]
- 26. Grimson A., Farh K. K.-H., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 28. Betel D., Koppal A., Agius P., Sander C., Leslie C. (2010) Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11, R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galmarini C. M., Bouchet B. P., Audoynaud C., Lamblot C., Falette N., Bertholon J., Wang Q., Beghin A., Dumontet C., Puisieux A. (2006) A p21/WAF1 mutation favors the appearance of drug resistance to paclitaxel in human noncancerous epithelial mammary cells. Int. J. Cancer 119, 60–66 [DOI] [PubMed] [Google Scholar]
- 30. Abukhdeir A. M., Vitolo M. I., Argani P., De Marzo A. M., Karakas B., Konishi H., Gustin J. P., Lauring J., Garay J. P., Pendleton C. (2008) Tamoxifen-stimulated growth of breast cancer due to p21 loss. Proc. Natl. Acad. Sci. U.S.A. 105, 288–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aneja R., Ghaleb A. M., Zhou J., Yang V. W., Joshi H. C. (2007) p53 and p21 determine the sensitivity of noscapine-induced apoptosis in colon cancer cells. Cancer Res. 67, 3862–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koster R., di Pietro A., Timmer-Bosscha H., Gibcus J. H., van den Berg A., Suurmeijer A. J., Bischoff R., Gietema J. A., de Jong S. (2010) Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer. J. Clin. Invest. 120, 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nefedova Y., Cheng P., Alsina M., Dalton W. S., Gabrilovich D. I. (2004) Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood 103, 3503–3510 [DOI] [PubMed] [Google Scholar]
- 34. Ferrandiz N., Caraballo J. M., Albajar M., Gomez-Casares M. T., Lopez-Jorge C. E., Blanco R., Delgado M. D., Leon J. (2010) p21Cip1 confers resistance to imatinib in human chronic myeloid leukemia cells. Cancer Lett. 292, 133–139 [DOI] [PubMed] [Google Scholar]
- 35. Gayet J., Zhou X.-P., Duval A., Rolland S., Hoang J.-M., Cottu P., Hamelin R. (2001) Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines. Oncogene 20, 5025–5032 [DOI] [PubMed] [Google Scholar]
- 36. Lim H. K., Bae W., Lee H.-S., Jung J. (2014) Anticancer activity of marine sponge Hyrtios sp. extract in human colorectal carcinoma RKO cells with different p53 status. BioMed Res. Int. 27, 413575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grassilli E., Narloch R., Federzoni E., Ianzano L., Pisano F., Giovannoni R., Romano G., Masiero L., Leone B. E., Bonin S. (2013) Inhibition of GSK3B bypass drug resistance of p53-null colon carcinomas by enabling necroptosis in response to chemotherapy. Clin. Cancer Res. 19, 3820–3831 [DOI] [PubMed] [Google Scholar]
- 38. Moertel C. G. (1978) Chemotherapy of gastrointestinal cancer. N. Engl. J. Med. 299, 1049–1052 [DOI] [PubMed] [Google Scholar]
- 39. Rougier P., Van Cutsem E., Bajetta E., Niederle N., Possinger K., Labianca R., Navarro M., Morant R., Bleiberg H., Wils J. (1998) Randomised trial of irinotecan versus fluorouracil by continuous infusion after fluorouracil failure in patients with metastatic colorectal cancer. Lancet 352, 1407–1412 [DOI] [PubMed] [Google Scholar]
- 40. Chowdhury S., Howell G. M., Rajput A., Teggart C. A., Brattain L. E., Weber H. R., Chowdhury A., Brattain M. G. (2011) Identification of a novel TGFβ/PKA signaling transduceome in mediating control of cell survival and metastasis in colon cancer. PLoS ONE 6, e19335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J., Yang L., Yang J., Kuropatwinski K., Wang W., Liu X.-Q., Hauser J., Brattain M. G. (2008) Transforming growth factor β induces apoptosis through repressing the phosphoinositide 3-kinase/AKT/survivin pathway in colon cancer cells. Cancer Res. 68, 3152–3160 [DOI] [PubMed] [Google Scholar]
- 42. Yang J., Ouyang J., Ouyang L., Ouyang L., Chen Y. (2013) Inhibition of cell proliferation and increase of chemosensitivity by simultaneous knockdown of XIAP and survivin in pancreatic carcinoma cells. Oncol. Res. 21, 43–50 [DOI] [PubMed] [Google Scholar]
- 43. Slichenmyer W. J., Nelson W. G., Slebos R. J., Kastan M. B. (1993) Loss of a p53-associated G1 checkpoint does not decrease cell survival following DNA damage. Cancer Res. 53, 4164–4168 [PubMed] [Google Scholar]
- 44. Kang K. H., Kim W. H., Choi K. H. (1999) p21 promotes ceramide-induced apoptosis and antagonizes the antideath effect of Bcl-2 in human hepatocarcinoma cells. Exp. Cell Res. 253, 403–412 [DOI] [PubMed] [Google Scholar]
- 45. Borralho P. M., Kren B. T., Castro R. E., Moreira da Silva I. B., Steer C. J., Rodrigues C. M. (2009) MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 276, 6689–6700 [DOI] [PubMed] [Google Scholar]
- 46. Fearon E. R., Vogelstein B. (1990) A genetic model for colorectal tumorigenesis. Cell 61, 759–767 [DOI] [PubMed] [Google Scholar]
- 47. Krell J., Frampton A. E., Colombo T., Gall T. M., De Giorgio A., Harding V., Stebbing J., Castellano L. (2013) The p53 miRNA interactome and its potential role in the cancer clinic. Epigenomics 5, 417–428 [DOI] [PubMed] [Google Scholar]