Background: Some streptococci target host extracellular matrix glycosaminoglycans for infection.
Results: The streptococcal metabolic pathway of glycosaminoglycan-derived unsaturated uronates was identified. The crystal structures of its isomerase and dehydrogenase were determined.
Conclusion: Streptococci include an assembled genetic cluster for depolymerization, import, degradation, and metabolism of glycosaminoglycans.
Significance: This study contributes to understanding of the streptococcal degradation/metabolism mechanism of glycosaminoglycans.
Keywords: Bacterial Metabolism, Dehydrogenase, Glycosaminoglycan, Streptococcus, X-ray Crystallography
Abstract
Glycosaminoglycans in mammalian extracellular matrices are degraded to their constituents, unsaturated uronic (glucuronic/iduronic) acids and amino sugars, through successive reactions of bacterial polysaccharide lyase and unsaturated glucuronyl hydrolase. Genes coding for glycosaminoglycan-acting lyase, unsaturated glucuronyl hydrolase, and the phosphotransferase system are assembled into a cluster in the genome of pathogenic bacteria, such as streptococci and clostridia. Here, we studied the streptococcal metabolic pathway of unsaturated uronic acids and the structure/function relationship of its relevant isomerase and dehydrogenase. Two proteins (gbs1892 and gbs1891) of Streptococcus agalactiae strain NEM316 were overexpressed in Escherichia coli, purified, and characterized. 4-Deoxy-l-threo-5-hexosulose-uronate (Dhu) nonenzymatically generated from unsaturated uronic acids was converted to 2-keto-3-deoxy-d-gluconate via 3-deoxy-d-glycero-2,5-hexodiulosonate through successive reactions of gbs1892 isomerase (DhuI) and gbs1891 NADH-dependent reductase/dehydrogenase (DhuD). DhuI and DhuD enzymatically corresponded to 4-deoxy-l-threo-5-hexosulose-uronate ketol-isomerase (KduI) and 2-keto-3-deoxy-d-gluconate dehydrogenase (KduD), respectively, involved in pectin metabolism, although no or low sequence identity was observed between DhuI and KduI or between DhuD and KduD, respectively. Genes for DhuI and DhuD were found to be included in the streptococcal genetic cluster, whereas KduI and KduD are encoded in clostridia. Tertiary and quaternary structures of DhuI and DhuD were determined by x-ray crystallography. Distinct from KduI β-barrels, DhuI adopts an α/β/α-barrel structure as a basic scaffold similar to that of ribose 5-phosphate isomerase. The structure of DhuD is unable to accommodate the substrate/cofactor, suggesting that conformational changes are essential to trigger enzyme catalysis. This is the first report on the bacterial metabolism of glycosaminoglycan-derived unsaturated uronic acids by isomerase and dehydrogenase.
Introduction
Mammalian extracellular matrices are important for the connection of neighboring cells and protection of cells against physicochemical stresses, such as osmotic pressure or invasion by pathogenic microbes (1). Glycosaminoglycans, one of the major components of extracellular matrices, are highly negatively charged polysaccharides with a repeating disaccharide unit comprising a uronic acid residue (glucuronic (GlcUA)3 or iduronic acid (IdoUA)) and an amino sugar residue (glucosamine or galactosamine), frequently N-acetylated (2). Based on the sugar composition, mode of glycosidic bond, and sulfation level, glycosaminoglycans are classified as hyaluronan, chondroitin sulfate, dermatan sulfate, keratan sulfate, heparin, and heparan sulfate, which are widely present in human tissues, such as the arterial wall, cartilage, heart valve, intestinal mucosa, liver, lung, and skin (3, 4). Except for hyaluronan, most glycosaminoglycans are bound to core proteins as a proteoglycan (5).
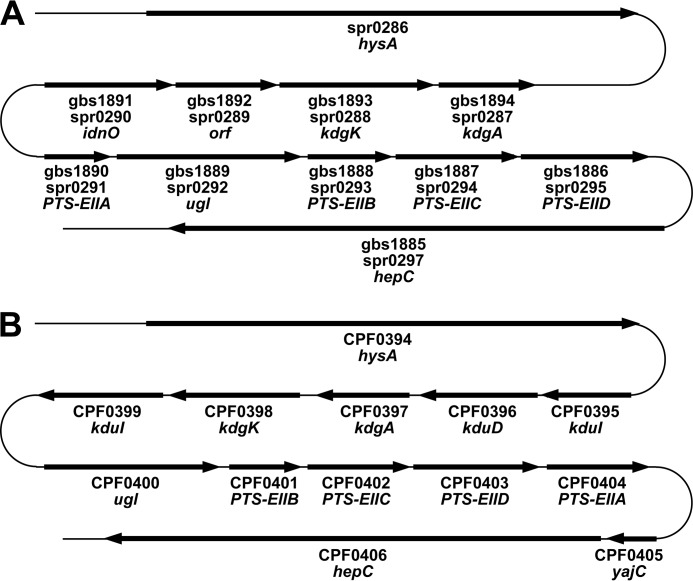
Some pathogenic bacteria are known to recognize host extracellular matrices (e.g. glycosaminoglycans) as targets for adhesion and/or degradation (6). A large number of streptococci depolymerize hyaluronan to unsaturated disaccharides by hyaluronate lyase (7) (Fig. 1). We have identified unsaturated glucuronyl hydrolase (UGL), which is essential for degrading unsaturated disaccharides to constituent monosaccharides (i.e. unsaturated uronic acids and amino sugars) in bacteria, including streptococci (8, 9), and found that a genome segment termed the UGL genetic cluster is responsible for the depolymerization, import, and degradation of glycosaminoglycans in streptococci, such as Streptococcus agalactiae, Streptococcus pneumoniae, and Streptococcus pyogenes (10) (see Fig. 2A). The UGL genetic cluster of S. agalacitae is inducibly transcribed in the presence of hyaluronan (10). Recently, the disruption of the UGL genetic cluster in S. pneumoniae has been demonstrated to reduce the bacterial attachment to host cells and the ability to cause infectious diseases (11). These reports indicate the significance of the UGL genetic cluster in bacterial infections.
FIGURE 1.
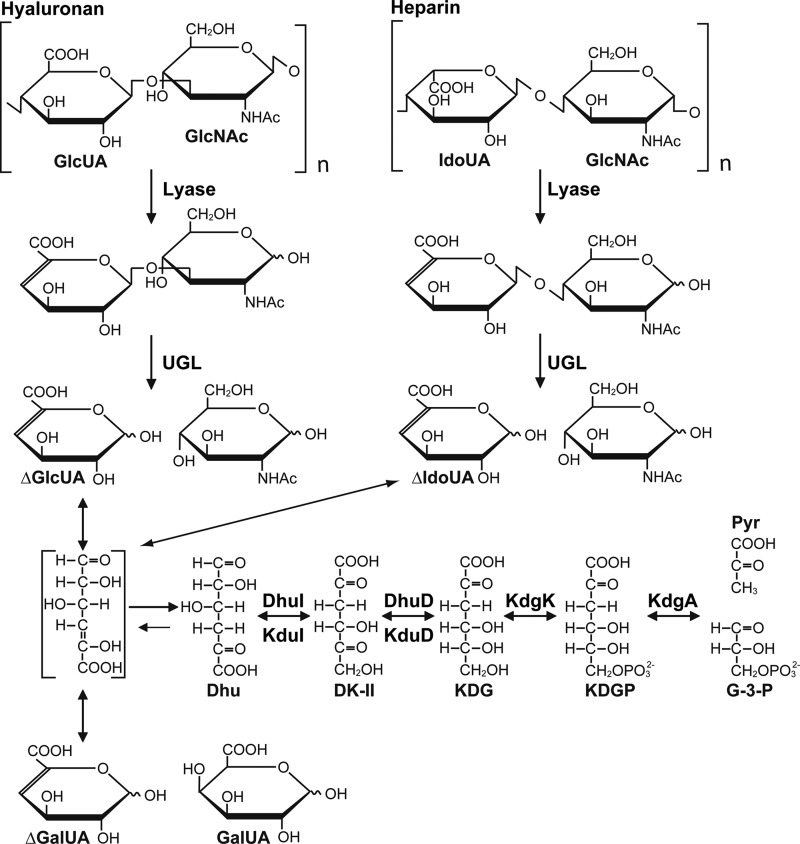
Bacterial degradation and metabolism of glycosaminoglycans. Glycosaminoglycans (e.g. hyaluronan and heparin) are depolymerized by lyase, and the resultant unsaturated disaccharides are subsequently degraded to unsaturated uronic acids and amino sugars by UGL. Unsaturated glucuronic/iduronic acids are nonenzymatically converted into Dhu. Dhu is metabolized to pyruvate (Pyr) and glyceraldehyde-3-phosphate (G-3-P) through the subsequent reactions of four enzymes (i.e. isomerase (DhuI or KduI), dehydrogenase (DhuD or KduD), kinase (KdgK), and aldolase (KdgA)). GlcNAc, N-acetylglucosamine; DK-II, 3-deoxy-d-glycero-2,5-hexodiulosonate; KDGP, 2-keto-3-deoxy-6-phosphogluconate. Dhu is also generated from pectin via unsaturated galacturonic acid (ΔGalUA).
FIGURE 2.
Genetic cluster for depolymerization, import, degradation, and metabolism of glycosaminoglycans. A, streptococcal genetic cluster. gbs1885–1894 and spr0286–0297, gene annotations of S. agalactiae strain NEM 316 and S. pneumoniae strain R6, respectively. PTS-EIIA-D, subunit domains of the phosphotransferase system for amino sugar import. B, clostridial genetic cluster. CPF0394–0404, gene annotations of C. perfringens ATCC 13124.
Two streptococci, S. pneumoniae and S. pyogenes, have been shown to grow on hyalruonan as the sole carbon source (11–13), but little knowledge about the bacterial assimilation mechanism of glycosaminoglycans has been accumulated. More recently, streptococcal UGL has been demonstrated to act on various unsaturated disaccharides, such as hyaluronan, chondroitin sulfate, heparin, and heparan sulfate, and to release unsaturated uronic acids and amino sugars (9, 14). Due to the lack of the hydroxyl group at C4, unsaturated glucuronic (ΔGlcUA) and iduronic acids (ΔIdoUA) are chemically identical (Fig. 1). The utilization pathway of amino sugars has already been identified in bacteria, such as Escherichia coli (15) and Streptococcus mutans (16). In contrast, the metabolism of unsaturated uronic acids from glycosaminoglycans remains to be clarified, although these unsaturated uronic acids are known to be nonenzymatically converted to 4-deoxy-l-threo-5-hexosulose uronic acid (Dhu) (Fig. 1). Our previous study (10) suggested that these unsaturated uronic acids are converted to 2-keto-3-deoxy-d-gluconate (KDG) because the streptococcal UGL genetic cluster includes two homologous genes coding for KDG kinase and aldolase.
The elucidation of the metabolic pathway of unsaturated uronic acids will facilitate the development of novel drugs against streptococcus-induced infectious diseases. The present study deals with the molecular identification of the streptococcal isomerase and dehydrogenase involved in the metabolism of unsaturated uronic acids from glycosaminoglycans, and the structures of these enzymes are also determined by x-ray crystallography.
EXPERIMENTAL PROCEDURES
Materials
Unsaturated chondroitin disaccharide was purchased from Seikagaku Biobusiness. Oligonucleotides used in this study were synthesized by Hokkaido System Science or GeneDesign, and their nucleotide sequences are listed in Table 1. Restriction endonucleases and DNA-modifying enzymes were purchased from Toyobo. All other analytical grade chemicals were obtained from commercial sources.
TABLE 1.
Oligonucleotides used in this study
Single and double underlines show restriction and mutation sites, respectively.
| Oligonucleotide | Sequence |
|---|---|
| gbs1892NdeI | 5′-GGCATATGAAAATTGCATTAATCAACGAAAATAGC-3′ |
| gbs1892XhoI | 5′-GGCTCGAGGTCTAAGACAGATTTCAAGTAGTCTGC-3′ |
| spr0289NdeI | 5′-GGCATATGAAAATCGCATTAATCAATGAAAATAGT-3′ |
| spr0289XhoI | 5′-GGCTCGAGCTTGGCTAATACTTCTTTCAAATAAGC-3′ |
| SPy0637NdeI | 5′-GGCATATGAAAATTGCATTAATTAATGAAAATAGC-3′ |
| SPy0637XhoI | 5′-GGCTCGAGCTGTTCAAGTACTGATTTCAAATAAGT-3′ |
| gbs1891NdeI | 5′-GGCATATGACTGAACAATTCTTAAAAGACAACTTT-3′ |
| gbs1891XhoI | 5′-GGCTCGAGTTGAGGTTGTTTACCGATGTAAGCTAA-3′ |
| spr0290NdeI | 5′-GGCATATGACAAATACATCATTCTCAATTGAGCAG-3′ |
| spr0290XhoI | 5′-GGCTCGAGCTCAGGTTGTTTTCCGATGTAGGCTAA-3′ |
| SPy0636NdeI | 5′-GGCATATGGAAAATATGTTTTCGTTACAAGGTAAG-3′ |
| SPy0636XhoI | 5′-GGCTCGAGAGGTTGTTTTCCAATATAAGCTAAAAT-3 |
| EcoKduINdeI | 5′-GGCATATGGACGTAAGACAGAGCATCCACAGTGCG-3′ |
| EcoKduIXhoI | 5′-CCCTCGAGGCGCAAATCTTTAACGGCCACATGGTC-3′ |
| gbs1892 C72A sense | 5′-GCAGATTTTGTTATTACAGGCGCTGGGACAGGAATTGGAGCGATG-3′ |
| gbs1892 C72A antisense | 5′-CATCGCTCCAATTCCTGTCCCAGCGCCTGTAATAACAAAATCTGC-3′ |
| gbs1892 T74A sense | 5′-GTTATTACAGGCTGTGGGGCAGGAATTGGAGCGATGCTTGC-3′ |
| gbs1892 T74A antisense | 5′-GCAAGCATCGCTCCAATTCCTGCCCCACAGCCTGTAATAAC-3′ |
| gbs1892 N106A sense | 5′-GCTTACTTATTCTCTCAAGTAGCCGGAGGAAATGCTCTTTCCC-3′ |
| gbs1892 N106A antisense | 5′-GGGAAAGAGCATTTCCTCCGGCTACTTGAGAGAATAAGTAAGC-3′ |
| gbs1891 S150A sense | 5′-GGTAAAATCATCAATATTTGCGCCATGATGAGTGAGCTTGGACGC-3′ |
| gbs1891 S150A antisense | 5′-GCGTCCAAGCTCACTCATCATGGCGCAAATATTGATGATTTTACC-3′ |
| gbs1891 Y163F sense | 5′-GGACGCGAAACAGTTGCTGCCTTTGCTGCTGCCAAAGGGGGACTT-3′ |
| gbs1891 Y163F antisense | 5′-AAGTCCCCCTTTGGCAGCAGCAAAGGCAGCAACTGTTTCGCGTCC-3′ |
| gbs1891 K167A sense | 5′-GTTGCTGCCTATGCTGCTGCCGCAGGGGGACTTAAAATGTTAACC-3′ |
| gbs1891 K167A antisense | 5′-GGTTAACATTTTAAGTCCCCCTGCGGCAGCAGCATAGGCAGCAAC-3′ |
Microorganisms and Culture Conditions
S. agalactiae strain NEM316 (CIP 82.45) was purchased from the Institut Pasteur. S. pneumoniae strain R6 (ATCC BAA-255) and S. pyogenes strain M1 GAS SF370 (ATCC 700294) were from the American Type Culture Collection. For isolating the genome DNAs, the three streptococci were statically grown at 30 °C under 5% (v/v) CO2 in tryptic soy broth (Difco) containing 5% (v/v) defibrinated sheep blood (Nippon Bio-Test Laboratories). As the host for plasmid amplification, E. coli strain DH5α (Toyobo) cells were routinely cultured at 37 °C in Luria-Bertani (LB) medium (1% (w/v) tryptone, 0.5% (w/v) yeast extract, and 1% (w/v) NaCl) containing sodium ampicillin (0.1 mg/ml). E. coli strain BL21 (DE3) pLysS (Novagen) was used as the host for the expression of streptococcal proteins. For expression in E. coli, cells were aerobically precultured at 30 °C in LB medium supplemented with sodium ampicillin (0.1 mg/ml). When the turbidity reached about 0.5 at 600 nm, isopropyl-β-d-thiogalactopyranoside was added to the culture at a final concentration of 0.1 mm, and the cells were further cultured at 16 °C for 44 h.
DNA Manipulations
Genomic DNA isolation, subcloning, transformation, and gel electrophoresis were performed as described previously (17). The nucleotide sequences of the streptococcal genes amplified by PCR or constructed through site-directed mutagenesis were determined by the dideoxy-chain termination method using the automated DNA sequencer model 3730xl (Applied Biosystems) (18).
Construction of the Overexpression System
Overexpression systems for streptococcal isomerases (DhuIs) (S. agalactiae gbs1892 (SagDhuI), S. pneumoniae spr0289 (SpnDhuI), S. pyogenes SPy0637 (SpyDhuI)) and dehydrogenases (DhuDs) (S. agalactiae gbs1891 (SagDhuD), S. pneumoniae spr0290 (SpnDhuD), S. pyogenes SPy0636 (SpyDhuD)) were constructed in E. coli as follows. To clone the streptococcal genes, PCR was conducted in a reaction mixture (10 μl) consisting of 0.2 units of KOD Plus polymerase (Toyobo), 40 ng of genomic DNA, 0.3 pmol of forward and reverse primers, 2 nmol of dNTPs, 10 nmol of MgCl2, 1 μl of dimethyl sulfoxide, and the commercial reaction buffer supplied with KOD Plus polymerase. A restriction site (NdeI or XhoI) was added to each of the 5′ regions of the forward and reverse primers. The PCR conditions were as follows: 94 °C for 2 min followed by 30 cycles at 94 °C for 15 s, 50 °C for 30 s, and 68 °C for 1 min. The PCR products were ligated with HincII-digested pUC119 (Takara Bio), and the resultant plasmids were digested with NdeI and XhoI to isolate the streptococcal genes. Further, DNA fragments of the streptococcal genes were ligated with NdeI- and XhoI-digested pET21b (Novagen). The pET21b vector was designed to express proteins with a hexahistidine-tagged sequence at the C terminus. An overexpression system for E. coli 4-deoxy-l-threo-5-hexosulose-uronate ketol-isomerase (KduI) (EcoKduI) was also constructed as described above.
Purification
Unless otherwise specified, all procedures were performed between 0 and 4 °C. E. coli cells harboring each plasmid were grown in 3.0 liters of LB medium (1.5 liters/flask), collected by centrifugation at 6,000 × g and 4 °C for 5 min, washed with 20 mm Tris-HCl (pH 7.5), and then resuspended in the same buffer. The cells were ultrasonically disrupted (Insonator Model 201M, Kubota) at 0 °C and 9 kHz for 20 min, and the clear solution obtained by centrifugation at 20,000 × g and 4 °C for 20 min was used as the cell extract. Recombinant streptococcal proteins and EcoKduI were purified to homogeneity from E. coli cells harboring each plasmid using column chromatography with three different separation media: affinity (TALON (Clontech, 1.0 × 10 cm)), anion exchange (Toyopearl SuperQ-650 M (Tosoh, 1.0 × 9.5 cm)), and gel filtration (Sephacryl S-200HR (GE Healthcare, 2.6 × 65 cm)). Protein purity was confirmed using SDS-PAGE (19). The fractions containing each protein were combined and dialyzed against 20 mm Tris-HCl (pH 7.5). The dialysate was then concentrated to about 10–30 mg/ml by ultrafiltration using a Centriprep (molecular weight cut-off, 10,000) (Millipore). The concentrate was used as the purified enzyme source. Pectobacterium carotovorum 2-keto-3-deoxy-d-gluconate dehydrogenase KduD (PcaKduD) was obtained from recombinant E. coli cells as described previously (53). The UGLs of S. agalactiae (SagUGL) and Bacillus sp. GL1 (BacillusUGL) were purified from recombinant E. coli cells as described previously (10).
Assays for Enzymes and Proteins
Because both Dhu and 3-deoxy-d-glycero-2,5-hexodiulosonate are commercially unavailable, Dhu was nonenzymatically obtained from ΔGlcUA prepared from 10 mm unsaturated chondroitin disaccharide sulfated at the C6 position of N-acetylgalactosamine residue through the reaction of SagUGL or from 10 mm unsaturated gellan tetraccharide (unsaturated glucuronyl-glucosyl-rhamnosyl-glucose) (20) after treatment with BacillusUGL. The standard dehydrogenase activity was assayed at 30 °C in a reaction mixture (0.5 ml) consisting of 0.2 mm Dhu, 0.2 mm cofactor (NADH or NADPH), 50 mm Tris-HCl (pH 7.5), and an appropriate amount of enzymes. The activity was measured by continuously monitoring the decrease in the absorbance at 340 nm, which corresponds to the oxidation of NADH or NADPH. One unit of enzymatic activity was defined as the amount of enzyme required to oxidize 1 μmol of cofactor per min at 30 °C. The kinetic parameters (Km and kcat) were determined using the Michaelis-Menten equation with the KaleidaGraph program (Synergy Software). In the case of the purified enzyme, the protein concentration was estimated by measuring the absorbance at 280 nm. The absorbance coefficient for 1 mg/ml of each protein with His tag is as follows: SagDhuI, 0.840; SpnDhuI, 0.888; SpyDhuI, 0.845; SagDhuD, 0.586; SpnDhuD, 0.636; SpyDhuD, 0.600.
Determination of Molecular Mass
In order to determine the molecular weight of streptococcal proteins, SDS-PAGE, blue native PAGE (Invitrogen), and gel filtration chromatography (Sephacryl S-200HR) were performed.
Optimal pH and Temperature and Thermal Stability
The substrate (3-deoxy-d-glycero-2,5-hexodiulosonate) for SagDhuD was prepared as follows. Dhu was reacted with the purified SagDhuI, and the resultant product was heated at 95 °C for 3 min to inactivate SagDhuI. Experiments were performed using the purified SagDhuD. For optimal pH, reactions were performed at 30 °C in the following 46 mm buffers: sodium acetate, potassium phosphate, Tris-HCl, and glycine-NaOH. For optimal temperature, reactions were performed at different temperatures in 50 mm Tris-HCl (pH 7.5). For thermal stability, after preincubating the enzyme at different temperatures for 5 min, residual activity was measured at 30 °C in 50 mm Tris-HCl (pH 7.5).
Site-directed Mutagenesis
Cys72, Thr74, and Asn106 of SagDhuI were substituted with Ala (C72A), Ala (T74A), and Ala (N106A), respectively, and Ser150, Tyr163, and Lys167 of SagDhuD were substituted with Ala (S150A), Phe (Y163F), and Ala (K167A), respectively, using the QuikChange site-directed mutagenesis kit (Stratagene) except that KOD Plus polymerase was used as the PCR enzyme. Site-directed mutagenesis was conducted using each expression plasmid as the template and synthetic oligonucleotides as sense and antisense primers (Table 1). The mutations in the resultant plasmids were confirmed by DNA sequencing. E. coli (i.e. BL21 (DE3) pLysS) was transformed with the plasmids. Moreover, the mutant enzymes were purified from the E. coli transformants approximately as described under “Purification.”
Crystallization and Structure Determination
To determine the three-dimensional structures of SagDhuI and SagDhuD, each purified enzyme was crystallized using the sitting drop vapor diffusion method. The SagDhuI T74A mutant (1 μl; 26.4 mg/ml protein in 20 mm Tris-HCl (pH 7.5) and 10 mm dithiothreitol) was mixed with an equal volume of reservoir solution (15% (w/v) polyethylene glycol 10,000, 2% (v/v) dioxane, and 0.1 m sodium citrate (pH 6.5)). The SagDhuI T74A mutant (1 μl) was also mixed with an equal volume of reservoir solution (2.0 m ammonium sulfate, 0.2 m potassium sodium tartrate, and 0.1 m sodium citrate (pH 5.6)). Furthermore, the SagDhuD S150A mutant (1 μl; 9.7 mg/ml protein in 20 mm Tris-HCl (pH 7.5)) was mixed with an equal volume of reservoir solution (20% (w/v) polyethylene glycol monomethyl ether 550, 0.1 m sodium chloride, and 0.1 m sodium N,N-bis(2-hydroxyethyl)glycine (pH 9.0)). Protein solutions were then incubated at 20 °C, and single crystals were allowed to grow for about 1 month. The crystals were flash-frozen under the cold nitrogen gas stream at −173 °C after soaking in the mother liquor containing 20% glycerol as the cryoprotectant.
X-ray diffraction data were collected at λ = 1.00 Å using a Quantum 210 or Quantum 315 CCD detector (Area Detector Systems Corp.) at BL38B1 station of SPring-8 (Hyogo, Japan). Data were processed and scaled up to 1.55 or 2.00 Å for SagDhuI and 2.90 Å for SagDhuD using the HKL2000 program (21). The structures were determined by molecular replacement using the Molrep program (22) supplied in the CCP4 program package (23) using the coordinates of the S. pneumoniae uncharacterized ribose 5-phosphate isomerase (Protein Data Bank (PDB) code 2PPW) as the initial model for SagDhuI and Streptococcus suis gluconate 5-dehydrogenase (Ga5DH) (PDB code 3CXR) as the initial model for SagDhuD. Structure refinement was conducted using the refmac5 program supplied in the CCP4 program package (24). Randomly selected 5% reflections were excluded from the refinement and used to calculate the Rfree. After each cycle of refinement, the model was manually adjusted using the winCoot program (25). Water molecules were incorporated into isolated electron densities exceeding 3σ in the Fo − Fc map and/or 1.2σ in the 2Fo − Fc map. Final model quality was checked with the PROCHECK program (26). Figures of protein structures were prepared using PyMOL (27). Coordinates used in this work were taken from the Protein Data Bank (28).
RESULTS AND DISCUSSION
Metabolism of Unsaturated Uronic Acids from Glycosaminoglycans
Streptococci such as S. agalactiae, S. pneumoniae, and S. pyogenes include the UGL genetic cluster responsible for the depolymerization (by lyases (HysA and/or HepC) for hyaluronan and/or heparan sulfate), import (by the phosphotransferase system; PTS), and degradation (by UGL) of glycosaminoglycans in the bacterial genomes (10, 11) (Fig. 2A). In addition to these glycosaminoglycan-related genes, the putative genes coding for 5-keto-d-gluconate dehydrogenase/reductase (IdnO), 2-keto-3-deoxygluconate kinase (KdgK), and 2-keto-3-deoxy-6-phosphogluconate aldolase (KdgA) involved in the metabolism of KDG are situated upstream of the UGL gene (29–31) (Fig. 2A). A gene for a hypothetical protein is inserted between idnO and kdgK in the streptococcal genomes. KDG has been demonstrated to convert to pyruvate and glyceraldehyde-3-phosphate through succeeding reactions by KdgK and KdgA (32) (Fig. 1).
The metabolic pathway of alginate-derived unsaturated mannuronic/guluronic acids and pectin-derived unsaturated galacturonic acid has been identified in bacteria (33, 34). α-Keto acid, 4-deoxy-l-erythro-5-hexoseulose uronic acid, from unsaturated mannuronic/guluronic acids is converted to KDG by NADPH-dependent reductase. We have recently identified the enzyme (A1-R) gene in an alginate-assimilating bacterium (35). In pectin-assimilating bacteria, such as Dickeya and Pectobacterium, formerly known as Erwinia, α-keto acid, Dhu, from unsaturated galacturonic acid is converted to KDG via 3-deoxy-d-glycero-2,5-hexodiulosonate through subsequent reactions of KduI and 2-keto-3-deoxy-d-gluconate dehydrogenase (KduD) (36) (Fig. 1). In E. coli, KduI and KduD are also demonstrated to be involved in the metabolism of galacturonate and GlcUA (37). To the best of our knowledge, no reports have been published on the metabolism of ΔGlcUA/ΔIdoUA derived from glycosaminoglycans, although unsaturated galacturonic acid as well as ΔGlcUA/ΔIdoUA are nonenzymatically converted to Dhu (Fig. 1).
The gene arrangement in the UGL genetic cluster and the metabolism of unsaturated uronic acids suggest that α-keto acid (Dhu) generated from ΔGlcUA/ΔIdoUA through the UGL reaction is also converted to KDG by certain enzymes. Thus, hypothetical proteins (i.e. gbs1892, spr0289, and SPy0637) and IdnO homologues (i.e. gbs1891, spr0290, and SPy0636) from S. agalactiae, S. pneumoniae, and S. pyogenes were expressed in recombinant E. coli cells and purified to homogeneity (Fig. 3, A–C). Purified proteins were subjected to enzyme assay using Dhu as the substrate. Three gbs1892 mutants (C72A, T74A, and N106A) with a mutation at a possible catalytic residue were also constructed based on the crystal structure of gbs1892 in complex with the substrate analog and its structural comparison with sugar isomerase, as described below. In the case of gbs1891, three mutants (S150A, Y163F, and K167A) with a mutation at the catalytic triad well conserved in the short-chain dehydrogenase/reductase (SDR) family were subjected to the enzyme assay and crystallization.
FIGURE 3.
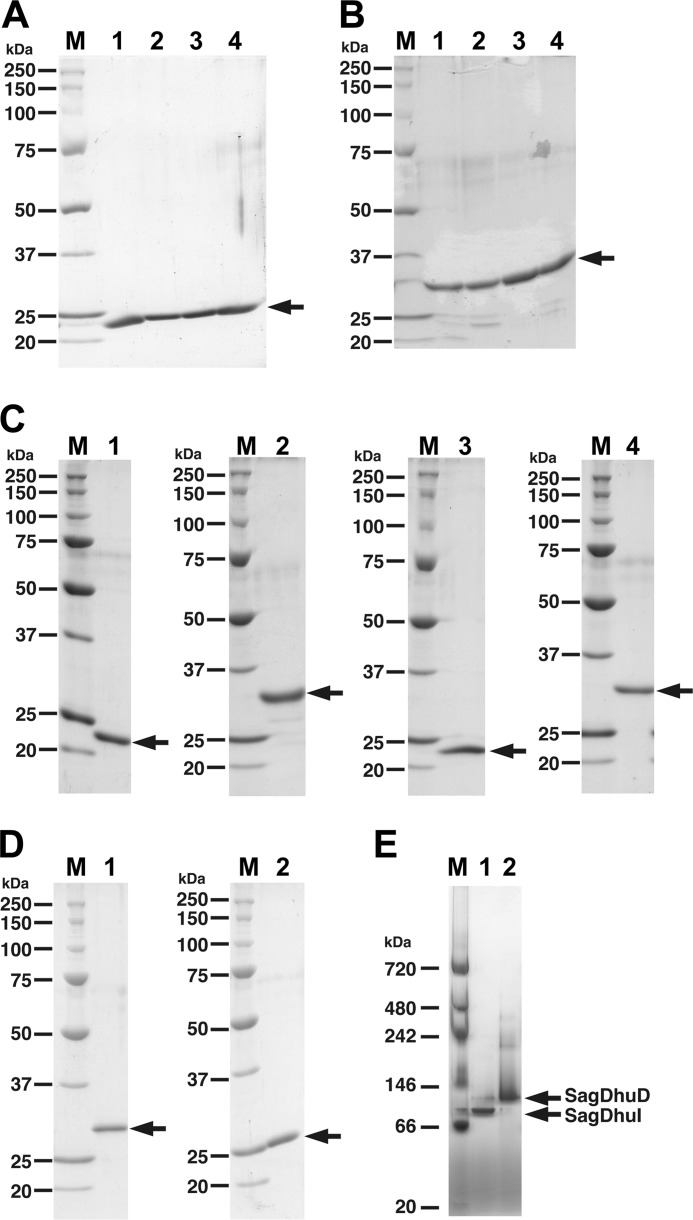
Electrophoretic profile of purified enzymes. A, SDS-PAGE profile of gbs1892 (SagDhuI). Lane 1, wild-type SagDhuI; lane 2, SagDhuI C72A; lane 3, SagDhuI T74A; lane 4, SagDhuI N106A. B, SDS-PAGE profile of gbs1891 (SagDhuD). Lane 1, wild-type SagDhuD; lane 2, SagDhuD S150A; lane 3, SagDhuD Y163F; lane 4, SagDhuD K167A. C, SDS-PAGE profile of spr0289 (SpnDhuI, lane 1), spr0290 (SpnDhuD, lane 2), SPy0637 (SpyDhuI, lane 3), and SPy0636 (SpyDhuD, lane 4). D, SDS-PAGE profile of EcoKduI (lane 1) and PcaKduD (lane 2). Lane M in A–D, molecular weight standards (from top): recombinant polypeptides with molecular weights of 250,000, 150,000, 100,000, 75,000, 50,000, 37,000, 25,000, and 20,000. E, blue native PAGE. Lane 1, gbs1892 (SagDhuI); lane 2, gbs1891 (SagDhuD). Lane M, molecular weight standards (from top): polypeptides with molecular weights of 720,000, 480,000, 242,000, 146,000, 66,000, and 20,000.
No oxidation of NADH or NADPH occurred in the presence of Dhu and IdnO homologues, whereas these proteins slightly reduced 5-keto-d-gluconate, depending on the presence of NADH. On the other hand, NADH was readily oxidized in the presence of Dhu and IdnO homologues as well as hypothetical proteins, indicating that Dhu was metabolized through subsequent reactions of the hypothetical protein and IdnO homologue. These reactions seem to correspond to those by KduI and KduD involved in the metabolism of unsaturated galacturonic acid (Fig. 1). The purified EcoKduI and PcaKduD with molecular masses of 31 and 27 kDa, respectively, were therefore used instead of the streptococcal enzymes (Fig. 3D). NADH was readily oxidized in the reaction mixtures of the combinations Dhu, EcoKduI, and PcaKduD; Dhu, EcoKduI, and gbs1891; and Dhu, gbs1892, and PcaKduD, demonstrating that the enzyme reactions by gbs1892 and gbs1891 are identical to those by EcoKduI and PcaKduD, respectively. These results indicated that Dhu was first converted to 3-deoxy-d-glycero-2,5-hexodiulosonate by gbs1892 (EC 5.3.1.17; 4-deoxy-l-threo-5-hexosulose-uronate ketol-isomerase) and subsequently to KDG by gbs1891 (EC 1.1.1.127; 2-keto-3-deoxy-d-gluconate dehydrogenase). The resultant KDG is considered metabolized to pyruvate and glyceraldehyde 3-phosphate through the subsequent reactions of KdgK and KdgA (Fig. 1). Because S. agalactiae genes encoding four enzymes are inducibly expressed in the presence of hyaluronan (10), these enzymes are suggested to be crucial for the metabolism of ΔGlcUA from the polysaccharide. In addition to gbs1892 and gbs1891, spr0289 and SPy0637 were found to enzymatically correspond to EcoKduI, and spr0290 and SPy0636 were found to correspond to PcaKduD. Therefore, streptococcal hypothetical proteins (i.e. gbs1892, spr0289, and SPy0637) and IdnO homologues (i.e. gbs1891, spr0290, and SPy0636) encoded in the UGL genetic cluster should be redefined as 4-deoxy-l-threo-5-hexosulose-uronate ketol-isomerase and 2-keto-3-deoxy-d-gluconate dehydrogenase, respectively.
Classification of the UGL Genetic Cluster
The primary structure of gbs1892 is significantly different from that of EcoKduI, and the sequence identity between gbs1891 and PcaKduD is also low (35.6%). The low sequence identity suggests that streptococcal genes coding for isomerase and dehydrogenase involved in the metabolism of glycosaminoglycans have been separately evolved from kduID, which is the operon for KduI and KduD, thus postulating that the streptococcal isomerase and dehydrogenase are designated as DhuI and DhuD, respectively, distinct from KduI and KduD. The operon kduID was first identified in Dickeya dadantii (36) and distributed in Escherichia, Klebsiella, Salmonella, Vibrio, and Yersinia (38). We also found kduID and kdgAK in the UGL genetic cluster in pathogenic Clostridium (39) and Enterococcus (40) (Fig. 2B), suggesting that these bacteria convert ΔGlcUA/ΔIdoUA to KDG through the successive reactions of KduI and KduD. Similar to clostridia and enterococci, most bacteria having the UGL genetic cluster include kduID, but not dhuDI. Based on the type of isomerase and dehydrogenase, bacterial UGL genetic clusters are classified into two groups (i.e. dhuDI and kduID), although dhuDI is rather specific to streptococci.
Enzyme Characteristics
Because, compared with the well characterized KduI (41) and KduD (42), the enzyme properties of DhuI and DhuD remain unclear, streptococcal enzymes, especially DhuD, were further characterized.
Molecular Weight
The molecular masses of DhuI (i.e. gbs1892, spr0289, and SPy0637) and DhuD (i.e. gbs1891, spr0290, and SPy0636) were determined to be 24 and 29 kDa, respectively, by SDS-PAGE (Fig. 3, A–C). These values were almost comparable with the theoretical ones deduced from the predicted amino acid sequences of the enzymes. Blue native PAGE indicated that SagDhuI (gbs1892) and SagDhuD (gbs1891) have molecular masses of 94 and 115 kDa, respectively (Fig. 3E), indicating that both SagDhuI and SagDhuD are tetrameric. This result was also supported by gel filtration chromatography on Sephacryl S-200HR (data not shown).
pH and Temperature
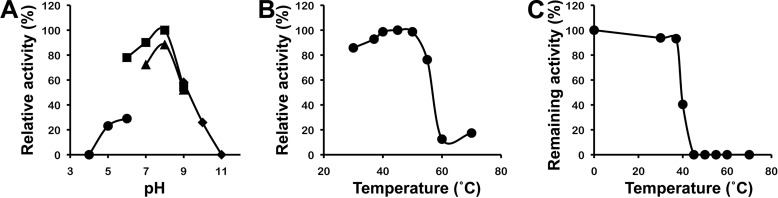
SagDhuD was most active at around pH 8.0 in potassium phosphate (Fig. 4A) at 45 °C (Fig. 4B). Over 50% of the enzyme activity was lost after preincubation at 40 °C for 5 min in 20 mm Tris-HCl (pH 7.5) (Fig. 4C).
FIGURE 4.
Optimal pH and temperature and thermal stability of SagDhuD. A, optimal pH. Experiments were performed at 30 °C using the following buffers: sodium acetate (circles), potassium phosphate (squares), Tris-HCl (triangles), and glycine-NaOH (diamonds). The maximum activity at pH 8.0 (potassium phosphate) was relatively taken as 100%. B, optimal temperature. Reactions were carried out at various temperatures in 50 mm Tris-HCl (pH 7.5). The maximum activity at 45 °C was relatively taken as 100%. C, thermal stability. After preincubation for 5 min at various temperatures, the residual activity was measured at 30 °C in 50 mm Tris-HCl (pH 7.5). The activity of the enzyme preincubated at 0 °C was taken as 100%.
Substrate Specificity
The substrate specificity of SagDhuD was investigated using 3-deoxy-d-glycero-2,5-hexodiulosonate, 5-keto-d-gluconate, 2-keto-d-gluconate, and gluconate. A low concentration (16.8 μm) of SagDhuD readily oxidized NADH when using about 0.2 mm 3-deoxy-d-glycero-2,5-hexodiulosonate as a substrate, whereas a high concentration (10 mm) of 5-keto-d-gluconate was reduced in the presence of NADH by a high concentration (1.68 mm) of SagDhuD. The specific activity (4.28 units/mg) of SagDhuD for 5-keto-d-gluconate was extremely low compared with that (175 units/mg) for 3-deoxy-d-glycero-2,5-hexodiulosonate. The intrinsic substrate (3-deoxy-d-glycero-2,5-hexodiulosonate) at more than 4 mm interfered with the measurement of absorbance at 340 nm, and determination of kinetic parameters for the substrate was difficult. On the other hand, Km and kcat for 5-keto-d-gluconate were determined to be 260 ± 51 mm and 0.521 ± 0.038 s−1, respectively. 2-Keto-d-gluconate and gluconate were inert substrates for SagDhuD. The reaction product (KDG) or its derivative (2-keto-3-deoxy-6-phosphogluconate) exhibited no inhibitory effect on the SagDhuD activity.
Cofactor Dependence
SagDhuD showed a preference for NADH (295 units/mg, 100%) rather than for NADPH (3.75 units/mg, 1.27%). Thus, the kinetic parameters (Km and kcat) of the enzyme for cofactor were measured using 3-deoxy-d-glycero-2,5-hexodiulosonate as the substrate. Km and kcat of the enzyme for NADH were determined to be 9.62 ± 1.05 μm and 2.67 ± 0.07 s−1, respectively. When 5-keto-d-gluconate was used as the substrate, Km and kcat for NADH were determined to be 26.3 ± 16.2 μm and 1.57 ± 0.30 s−1, respectively, whereas the kinetic parameters for NADPH could not be determined due to its low activity. This result indicated that SagDhuD is an NADH-dependent enzyme. There is a significant difference in the cofactor preference between DhuD and KduD, because KduD is known to use both NADH and NADPH (42).
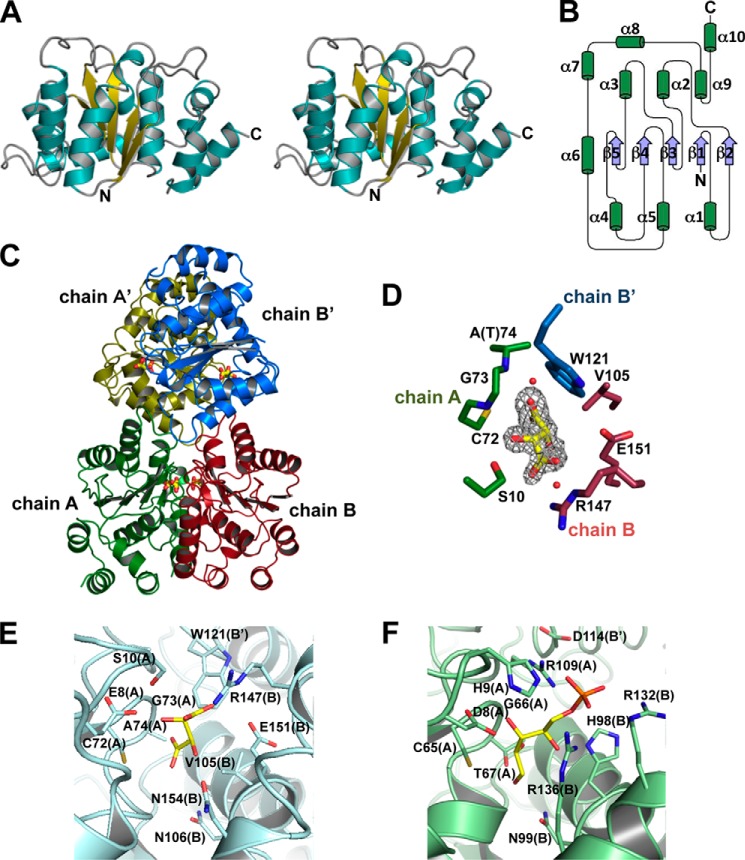
Crystal Structure of SagDhuI
To clarify the structure/function relationship, SagDhuI proteins (wild type and mutant (C72A, T74A, and N106A) enzymes) were subjected to x-ray crystallography. Although all of the wild type and mutants of SagDhuI were crystallized and subsequently employed for structure determination, we focused on the SagDhuI T74A mutants because their crystals diffracted to higher resolution.
Two types of SagDhuI T74A crystals formed under different crystallization conditions. The crystal in droplet A containing polyethylene glycol as the major precipitant showed a space group of I222, whereas the space group of the other crystal in droplet B with ammonium sulfate as the major precipitant was P41212. The structures of the SagDhuI crystals in droplets A and B were determined at 2.00 and 1.55 Å resolutions, respectively. Data collection and model refinement statistics are summarized in Table 2. Both crystal structures were identical except that the electron density map for tartrate included in the droplet B was also observed in the enzyme. SagDhuI adopts a Rossmann fold-like α/β/α-barrel structure as the basic scaffold (Fig. 5A), and a parallel β-sheet composed of five strands is sandwiched by α-helices (Fig. 5B). Four crystallographically symmetrical molecules of SagDhuI are assembled into a tetramer (Fig. 5C). There is no structural homology between SagDhuI and EcoKduI, although both catalyze the same reaction. EcoKduI is a homohexameric enzyme, each subunit of which consists of two β-barrels and shows structural similarity to the cupin superfamily (43).
TABLE 2.
Statistics for data collection and structure refinement
| SagDhuI T74A | SagDhuI T74A/tartrate | SagDhuD S150A | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 1.00 | 1.00 | 1.00 |
| Resolution range (Å) | 50.00–2.00 (2.07–2.00)a | 50.00–1.55 (1.61–1.55) | 50.00–2.90 (2.95–2.90) |
| Space group | I222 | P41212 | P6222 |
| Unit cell parameters (Å) | a = 53.23, b = 87.97, c = 90.30 | a = b = 92.96, c = 129.35 | a = b = 83.94, c = 181.10 |
| Total observations | 88,502 | 615,489 | 201,293 |
| Unique reflections | 14,717 | 82,052 | 8964 |
| Completeness (%) | 100 (100) | 99.4 (95.7) | 99.8 (100) |
| I/σ(I) | 28.6 (5.3) | 47.3 (4.8) | 78.5 (13.1) |
| Rmerge | 0.148 (0.443) | 0.084 (0.385) | 0.069 (0.414) |
| Refinement | |||
| Rcryst | 0.183 (0.282) | 0.198 (0.256) | 0.211 (0.323) |
| Rfree | 0.227 (0.255) | 0.211 (0.268) | 0.288 (0.449) |
| No. of subunits per ASUb | 1 | 2 | 1 |
| No. of non-hydrogen atoms | |||
| Proteins | 1670 | 3381 | 1994 |
| Water molecules | 121 | 525 | 5 |
| Ligand atoms | 20 | ||
| Average B-factor (Å2) | 27.31 | 17.40 | 76.08 |
| RMSDc from ideal | |||
| Bond length (Å) | 0.0057 | 0.0045 | 0.0073 |
| Bond angle (degrees) | 0.971 | 0.932 | 1.195 |
| Ramachandran plot (%) | |||
| Most favored | 96.2 | 95.1 | 89.9 |
| Additionally allowed | 3.8 | 4.9 | 10.1 |
a Data for the highest shells is given in parenthesis.
b Asymmetric unit.
c Root mean square deviation.
FIGURE 5.
Structure of sugar isomerases. A, overall structure of monomeric SagDhuI (stereo diagram). B, topology of monomeric SagDhuI. C, quaternary structure of SagDhuI. The four monomeric subunits are colored in green (chain A), red (chain B), yellow (chain A′), and blue (chain B′). Stick models indicate tartrate molecules. D, tartrate-binding residues in SagDhuI T74A. Shown is an omit map (Fo − Fc) of tartrate (gray mesh with a contour of 3σ). E, active site of SagDhuI T74A. F, ribose 5-phosphate-bound active site of clostridial RpiB (PDB code 3HEE). Subunit molecules (chains) are given in parenthesis. Atoms of carbon, nitrogen, oxygen, and phosphorus are colored in yellow, blue, red, and orange, respectively.
The structural homologues of SagDhuI were sought in the PDB using the Structure navigator program. The top seven proteins having the Rossmann fold displayed structural homology with SagDhuI (Table 3). In the Pfam database, these proteins belong to the RpiB/LacAB family (44), although the structural homologues of SagDhuI are divided into groups I and II based on sequence identity and amino acid length. The RpiB/LacAB family includes sugar isomerases, and its typical members are ribose 5-phosphate isomerase B (RpiB) and galactose isomerase subunits A and B (LacAB). Due to the high sequence identity, the overall structure of SagDhuI was highly similar to three group I protein structures from S. pneumoniae (PDB code 2PPW), Novosphingobium aromaticivorans (PDB code 3C5Y), and Vibrio parahaemeolyticus (PDB code 3ONO). These proteins, composed of ∼200 residues, are assigned to RpiB based on the Pfam motif, although their enzyme characteristics and crystal structures have not yet been published. In contrast, group II proteins (PDB codes 1NN4, 3K8C, 3HEE, and 3SDW), which consist of ∼150 residues, moderately resemble the tertiary structure of SagDhuI, although little sequence identity is observed. Distinct from group I proteins, group II proteins are experimentally identified as RpiB, which are involved in the pentose phosphate pathway (45–48). Group I proteins, including SagDhuI, have four additional α-helices at the C terminus. Genes coding for group I proteins from S. pneumoniae and V. parahaemeolyticus are located at the genetic clusters for the metabolism of glycosaminoglycan and pectin, respectively, in the bacterial genomes. These structural and genetic features suggest that group I proteins should be reclassified as DhuI.
TABLE 3.
Structural homologs of SagDhuI
| PDB entry | Organism | No. of residues | RMSDa to SagDhuI (no. of matching Cα atoms) | Sequence identity with SagDhuI | References |
|---|---|---|---|---|---|
| Å | % | ||||
| 2PPW | S. pneumoniae | 213 | 0.823 (192) | 74 | Unpublished |
| 3C5Y | N. aromaticivoraus | 212 | 0.887 (192) | 57 | Unpublished |
| 3ONO | V. parahaemolyticus | 211 | 1.031 (198) | 52 | Unpublished |
| 1NN4 | E. coli | 149 | 2.045 (115) | 21 | Ref. 44 |
| 3K8C | T. cruzi | 159 | 1.823 (124) | 17 | Ref. 45 |
| 3HEE | C. thermocellum | 149 | 2.315 (112) | 21 | Ref. 46 |
| 3SDW | C. immitis | 163 | 2.191 (117) | 14 | Ref. 47 |
a Root mean square deviation.
Active Site of SagDhuI
Four molecules of tartrate are bound to the homotetrameric SagDhuI (Fig. 5C), and each tartrate is accommodated at the interface formed by three subunits of SagDhuI (Fig. 5, D and E). The binding mode of tartrate to SagDhuI through hydrogen bond formation and van der Waals contacts is listed in Table 4. This binding site corresponds to the active site of RpiB (Fig. 5F), suggesting that tartrate was bound to SagDhuI as a substrate analog. Thus, three mutants (i.e. SagDhuI C72A, T74A, and N106A) carrying a mutation at the binding site were constructed based on the binding mode of tartrate and structural comparison between SagDhuI and RpiB (Fig. 3A). In the conjugation assay of SagDhuI and SagDhuD, three mutants caused a significant reduction of SagDhuD activity due to their low substrate production. When the enzyme activity using wild-type SagDhuI was taken as 100%, the relative activities using SagDhuI C72A, T74A, and N106A were determined to be 4.15, 0.474, and 6.29%, respectively. This site-directed mutagenesis indicated that tartrate was bound to the active site of SagDhuI. In fact, the catalytically important residues in group II RpiBs are also conserved in group I proteins, whereas phosphate-recognizing residues are specific to group II proteins (Fig. 6), thus suggesting that group I proteins are defined as DhuI but not as RpiB.
TABLE 4.
Interactions between SagDhuI T74A and tartrate
| Tartrate atom | Protein/Water | Chain | Atom | Distancea |
|---|---|---|---|---|
| Å | ||||
| Hydrogen bonds | ||||
| O1A | Wat44 | 2.9 | ||
| Arg147 | B | NH2 | 2.8 | |
| O1B | Arg147 | B | NE | 2.8 |
| Arg147 | B | NH2 | 3.0 | |
| O2 | Ser10 | A | OG | 3.0 |
| Gly73 | A | N | 2.9 | |
| O3 | Wat44 | 2.6 | ||
| O4A | Wat66 | 2.8 | ||
| Cys72 | A | SG | 3.3 | |
| O4B | Wat66 | 3.1 | ||
| Ala74 (Thr) | A | N | 2.9 | |
| C-C contacts | ||||
| C1 | Ser10 | A | CB | 3.7 |
| Arg147 | B | CZ | 3.9 | |
| C2 | Ser10 | A | CB | 4.4 |
| Gly73 | A | CA | 4.2 | |
| Trp121 | B′ | CH2 | 4.3 | |
| C3 | Val105 | B | OG1 | 3.8 |
| Glu151 | B | CG | 4.2 | |
| Trp121 | B′ | CH2 | 4.2 | |
| C4 | Val105 | B | CG1 | 3.7 |
| Cys72 | A | CB | 4.1 | |
| Gly73 | A | CA | 4.4 | |
a For hydrogen bonds, the distance was <3.3 Å, and for C-C contacts, the distance was <4.4 Å.
FIGURE 6.

Sequence alignment of SagDhuI and its structural homologues. DhuI, SagDhuI; 2PPW, S. pneumoniae uncharacterized RpiB; 3C5Y, N. aromaticivorans putative RpiB; 3ONO, V. parahaemeolyticus RpiB; 1NN4, E. coli RpiB; 3PH4, Clostridium thermocellum RpiB; 3K8C, Trypanosoma cruzi RpiB; 3SDW, Coccidioides immitis RpiB. Solid lined box, catalytically important residues; broken lined box, phosphate group-recognizing residues in RpiB.
Crystal Structure of SagDhuD
The crystal structure of SagDhuD S150A was determined at 2.90 Å resolution. The data collection and model refinement statistics are summarized in Table 2. Similar to SagDhuI, SagDhuD adopts a Rossmann fold-like α/β/α-barrel structure as the basic scaffold (Fig. 7, A and B), and a parallel β-sheet composed of seven strands is surrounded by α-helices (Fig. 7B). Four crystallographically symmetrical molecules of SagDhuD are assembled into a tetramer (Fig. 7C).
FIGURE 7.
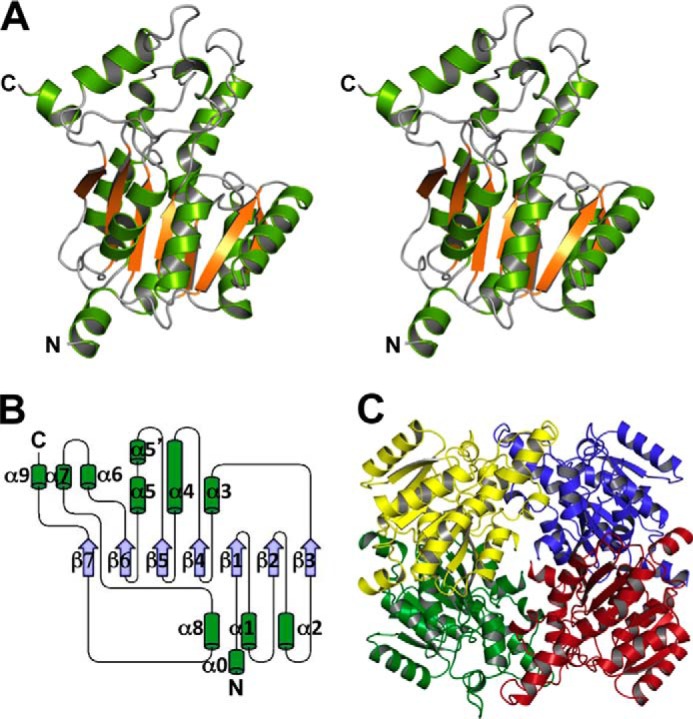
Structure of SagDhuD. A, overall structure of monomeric SagDhuD (stereo diagram). B, topology of monomeric SagDhuD. C, quaternary structure of SagDhuD. The four monomeric subunits are colored in green, red, yellow, and blue.
Based on the primary structure, SagDhuD belongs to the SDR family (49). A large number of SDR enzymes have been structurally analyzed, and their structure/function relationships have been well documented (50). Catalytic triads (Ser, Tyr, and Lys) are identified to be crucial for the enzyme reaction. Ser150, Tyr163, and Lys167 are also conserved in wild-type SagDhuD, and the SagDhuD mutants (S150A, Y163F, and K167A) constructed in this study (Fig. 3B) exhibited extremely lower activity (wild type, 100%; S150A, 0.240%; Y163F, 0.0581%; K167A, 0.0682%). Although three mutants were subjected to the kinetics analysis, their kinetic parameters for 5-keto-d-gluconate could not be determined due to their extremely low activity. The crystal structure of S. suis Ga5DH, one of the SDR enzymes, has been determined (51), although no description regarding its activity as dehydrogenase is included in the literature. Based on its gene location in the bacterial genome (the UGL genetic cluster) and high sequence identity (83%) with SagDhuD, the enzyme is considered to function as DhuD.
Structural Comparison in the SDR Family
Interesting structural features specific to SagDhuD were found. Compared with Ga5DH (PDB code 3CXR), SagDhuD has a bent α-helix 4 (Fig. 8A) and a short α-helix 5 (Fig. 8B). Instead of the long α-helix 5 of Ga5DH, the N-terminal half portion of the helix is loosened to a loop and a short α-helix 5′ (Fig. 8B). The loop between the β-strand 5 and α-helix 5′, which is crucial for the formation of the substrate and cofactor-binding sites in most SDR enzymes, is subjected to conformational changes to cover the binding sites. This structural characteristic in the crystal of SagDhuD is probably unsuitable for binding substrate and/or cofactor. Generally, no conformational change occurs in the corresponding loop through substrate/cofactor binding in SDR enzymes, except in E. coli β-ketoacyl-acyl carrier protein reductase (FabG). A cofactor-induced conformational change has been observed in the loop of FabG (52), although this change is small compared with the drastic change in SagDhuD. The bending of the α-helix 4 is probably caused by the destruction of the hydrogen bonds in the main chain due to the presence of Pro126 at the center of the helix (Fig. 8A). Two neighboring glycine residues (Gly168-Gly169) possibly contribute to the conformational change of the shorter α-helix 5 because the highly flexible glycine residue is entropically unsuitable for the construction of α-helices. The lack of the amino group in Pro193 in the β-strand 6 causes no hydrogen bond formation with Cys149 (Fig. 8B), thus resulting in no attendance of Cys149 to the formation of the β-strand 5 and determining its high flexibility. These structural features were also confirmed in other wild-type and mutant SagDhuD enzymes crystallized under different conditions.
FIGURE 8.
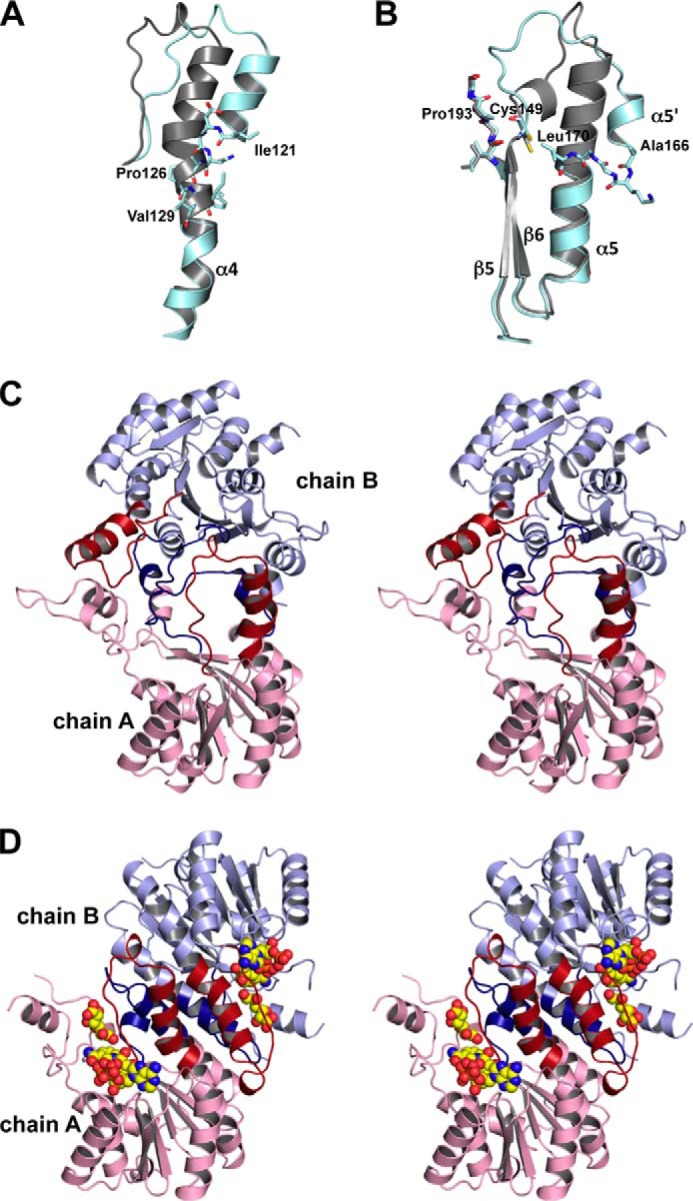
Structural comparison. A, the bent helix 4 in SagDhuD. Cyan, SagDhuD; gray, Ga5DH (PDB code 3CXR). The residues Ile121 and Val129 are indicated by stick models. B, part of helix 5 of SagDhuD (cyan) is loosened distinct from the long helix 5 of Ga5DH (gray). The residues Cys149, Ala166–Leu170, and Ile191–Gly194 are indicated by stick models. C, interaction between two monomeric subunits (chains A and B) (stereo diagram). The bent and shortened helices of SagDhuD are colored red and blue, respectively. D, subunit interaction of substrate/cofactor-bound Ga5DH (PDB code 3O03) (stereo diagram). Helices 4 and 5 are colored in red and blue, respectively. Substrate and cofactor are indicated by sphere models. The orientation of chain A is identical to that of chain A in C.
On the other hand, Ga5DH, which has a straight α-helix 4 and long α-helix 5, accepts the cofactor (51), although all of the above-mentioned SagDhuD residues, such as Pro126, Cys149, Gly168, Gly169, and Pro193, are completely conserved in Ga5DH. In this study, SagDhuD was experimentally demonstrated to exhibit an NADH-dependent reductase/dehydrogenase activity, suggesting that SagDhuD also adopts a Ga5DH-like structure through conformational changes caused by the substrate/cofactor binding and that two structural conformations (i.e. active and inactive forms) are mutually converted through the binding and release of the substrate/cofactor. The quaternary structure of SagDhuD (Fig. 8C) is different from that of Ga5DH (Fig. 8D). The possible conformational change in SagDhuD may affect the assembly of two subunits in the tetramer (chains A and B) (Fig. 8C), distinct from FabG, in which a slight conformational change occurs in the tertiary structure but not in the quaternary structure. To demonstrate the conformational change in SagDhuD, we are attempting to obtain the substrate/cofactor-binding crystals of the enzyme.
In conclusion, the metabolic pathway of unsaturated glucuronic/iduronic acids from glycosaminoglycans and its relevant enzymes/genes were elucidated for the first time in streptococci. The isomerase DhuI and dehydrogenase DhuD were specific mainly to streptococci, although they enzymatically correspond to KduI and KduD, which are involved in the metabolism of unsaturated galacturonic acid from pectin. X-ray crystallography revealed that SagDhuI and SagDhuD include the structural characteristics essential for the enzyme reaction.
Acknowledgments
We thank Drs. S. Baba and N. Mizuno of the Japan Synchrotron Radiation Research Institute (JASRI) for kind help with data collection. Diffraction data for crystals were collected at the BL-38B1 station of SPring-8 (Hyogo, Japan) with the approval of JASRI (Projects 2010B1149, 2011A1186, and 2011B2055). We also thank C. Tokunaga and A. Matsunami for excellent technical assistance.
Note Added in Proof
In the version of this article that was published as a Paper in Press on January 20, 2015, two identical descriptions for “CPF0404” were found in Fig. 2. “CPF0404” on hepC should have said “CPF0406”. In addition, the sequence for spr0290XhoI was missing from Table 1. The correct Fig. 2 and Table 1 are now shown.
This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (to K. M. and W. H.); the Targeted Proteins Research Program (to W. H.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; and a research grant (to W. H.) from the Mizutani Foundation for Glycoscience.
The atomic coordinates and structure factors (codes 4U8E, 4U8F, and 4U8G) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GlcUA
- d-glucuronic acid
- IdoUA
- l-iduronic acid
- UGL
- unsaturated glucuronyl hydrolase
- ΔGlcUA
- unsaturated GlcUA
- ΔIdoUA
- unsaturated IdoUA
- KDG
- 2-keto-3-deoxy-d-gluconate
- Dhu
- 4-deoxy-l-threo-5-hexosulose uronic acid
- DhuI
- 4-deoxy-l-threo-5-hexosulose-uronate ketol-isomerase
- DhuD
- 2-keto-3-deoxy-d-gluconate dehydrogenase
- SagDhuI
- S. agalactiae gbs1892
- SpnDhuI
- S. pneumoniae spr0289
- SpyDhuI
- S. pyogenes SPy0637
- SagDhuD
- S. agalactiae gbs1891
- SpnDhuD
- S. pneumoniae spr0290
- SpyDhuD
- S. pyogenes SPy0636
- KduI
- 4-deoxy-l-threo-5-hexosulose-uronate ketol-isomerase
- EcoKduI
- E. coli KduI
- KduD
- 2-keto-3-deoxy-d-gluconate dehydrogenase
- PcaKduD
- P. carotovorum KduD
- SagUGL
- S. agalactiae UGL
- BacillusUGL
- Bacillus sp. GL1 UGL
- Ga5DH
- S. suis gluconate 5-dehydrogenase
- PDB
- Protein Data Bank
- PTS
- phosphotransferase system
- IdnO
- 5-keto-d-gluconate dehydrogenase/reductase
- KdgK
- 2-keto-3-deoxygluconate kinase
- KdgA
- 2-keto-3-deoxy-6-phosphogluconate aldolase
- SDR
- short-chain dehydrogenase/reductase
- RpiB
- ribose 5-phosphate isomerase B
- FabG
- E. coli β-ketoacyl-acyl carrier protein reductase.
REFERENCES
- 1. Sawitzky D. (1996) Protein-glycosaminoglycan interactions: infectiological aspects. Med. Microbiol. Immunol. 184, 155–161 [DOI] [PubMed] [Google Scholar]
- 2. Imberty A., Lortat-Jacob H., Pérez S. (2007) Structural view of glycosaminoglycan-protein interactions. Carbohydr. Res. 342, 430–439 [DOI] [PubMed] [Google Scholar]
- 3. Lindahl U., Höök M. (1978) Glycosaminoglycans and their binding to biological macromolecules. Annu. Rev. Biochem. 47, 385–417 [DOI] [PubMed] [Google Scholar]
- 4. Couchman J. R., Pataki C. A. (2012) An introduction to proteoglycans and their localization. J. Histochem. Cytochem. 60, 885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gandhi N. S., Mancera R. L. (2008) The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 72, 455–482 [DOI] [PubMed] [Google Scholar]
- 6. Ernst S., Langer R., Cooney C. L., Sasisekharan R. (1995) Enzymatic degradation of glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 30, 387–444 [DOI] [PubMed] [Google Scholar]
- 7. Li S., Kelly S. J., Lamani E., Ferraroni M., Jedrzejas M. J. (2000) Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. EMBO J. 19, 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hashimoto W., Kobayashi E., Nankai H., Sato N., Miya T., Kawai S., Murata K. (1999) Unsaturated glucuronyl hydrolase of Bacillus sp. GL1: novel enzyme prerequisite for metabolism of unsaturated oligosaccharides produced by polysaccharide lyases. Arch. Biochem. Biophys. 368, 367–374 [DOI] [PubMed] [Google Scholar]
- 9. Nakamichi Y., Maruyama Y., Mikami B., Hashimoto W., Murata K. (2011) Structural determinants in streptococcal unsaturated glucuronyl hydrolase for recognition of glycosaminoglycan sulfate groups. J. Biol. Chem. 286, 6262–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maruyama Y., Nakamichi Y., Itoh T., Mikami B., Hashimoto W., Murata K. (2009) Substrate specificity of streptococcal unsaturated glucuronyl hydrolases for sulfated glycosaminoglycan. J. Biol. Chem. 284, 18059–18069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marion C., Stewart J. M., Tazi M. F., Burnaugh A. M., Linke C. M., Woodiga S. A., King S. J. (2012) Streptococcus pneumoniae can utilize multiple sources of hyaluronic acid for growth. Infect. Immun. 80, 1390–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bidossi A., Mulas L., Decorosi F., Colomba L., Ricci S., Pozzi G., Deutscher J., Viti C., Oggioni M. R. (2012) A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7, e33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Starr C. R., Engleberg N. C. (2006) Role of hyaluronidase in subcutaneous spread and growth of group A streptococcus. Infect. Immun. 74, 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakamichi Y., Mikami B., Murata K., Hashimoto W. (2014) Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin. J. Biol. Chem. 289, 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brinkkötter A., Klöss H., Alpert C., Lengeler J. W. (2000) Pathways for the utilization of N-acetyl-galactosamine and galactosamine in Escherichia coli. Mol. Microbiol. 37, 125–135 [DOI] [PubMed] [Google Scholar]
- 16. Kawada-Matsuo M., Mazda Y., Oogai Y., Kajiya M., Kawai T., Yamada S., Miyawaki S., Oho T., Komatsuzawa H. (2012) GlmS and NagB regulate amino sugar metabolism in opposing directions and affect Streptococcus mutans virulence. PLoS One 7, e33382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 18. Sanger F., Nicklen S., Coulson A. R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 20. Hashimoto W., Maesaka K., Sato N., Kimura S., Yamamoto K., Kumagai H., Murata K. (1997) Microbial system for polysaccharide depolymerization: enzymatic route for gellan depolymerization by Bacillus sp. GL1. Arch. Biochem. Biophys. 339, 17–23 [DOI] [PubMed] [Google Scholar]
- 21. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22. Vagin A. A., Isupov M. N. (2001) Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 57, 1451–1456 [DOI] [PubMed] [Google Scholar]
- 23. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 27. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
- 28. Berman H., Henrick K., Nakamura H., Markley J. L. (2007) The Worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 35, D301–D303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Glaser P., Rusniok C., Buchrieser C., Chevalier F., Frangeul L., Msadek T., Zouine M., Couvé E., Lalioui L., Poyart C., Trieu-Cuot P., Kunst F. (2002) Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45, 1499–1513 [DOI] [PubMed] [Google Scholar]
- 30. Hoskins J., Alborn W. E., Jr., Arnold J., Blaszczak L. C., Burgett S., DeHoff B. S., Estrem S. T., Fritz L., Fu D. J., Fuller W., Geringer C., Gilmour R., Glass J. S., Khoja H., Kraft A. R., Lagace R. E., LeBlanc D. J., Lee L. N., Lefkowitz E. J., Lu J., Matsushima P., McAhren S. M., McHenney M., McLeaster K., Mundy C. W., Nicas T. I., Norris F. H., O'Gara M., Peery R. B., Robertson G. T., Rockey P., Sun P. M., Winkler M. E., Yang Y., Young-Bellido M., Zhao G., Zook C. A., Baltz R. H., Jaskunas S. R., Rosteck P. R., Jr., Skatrud P. L., Glass J. I. (2001) Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183, 5709–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferretti J. J., McShan W. M., Ajdic D., Savic D. J., Savic G., Lyon K., Primeaux C., Sezate S., Suvorov A. N., Kenton S., Lai H. S., Lin S. P., Qian Y., Jia H. G., Najar F. Z., Ren Q., Zhu H., Song L., White J., Yuan X., Clifton S. W., Roe B. A., McLaughlin R. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U.S.A. 98, 4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. (1994) Molecular analysis of the Erwinia chrysanthemi region containing the kdgA and zwf genes. Mol. Microbiol. 11, 67–75 [DOI] [PubMed] [Google Scholar]
- 33. Preiss J., Ashwell G. (1962) Alginic acid metabolism in bacteria. II. The enzymatic reduction of 4-deoxy-L-erythro-5-hexoseulose uronic acid to 2-keto-3-deoxy-d-gluconic acid. J. Biol. Chem. 237, 317–321 [PubMed] [Google Scholar]
- 34. Preiss J., Ashwell G. (1963) Polygalacturonic acid metabolism in bacteria. II. Formation and metabolism of 3-deoxy-d-glycero-2,5-hexodiulosonic acid. J. Biol. Chem. 238, 1577–1583 [PubMed] [Google Scholar]
- 35. Takase R., Ochiai A., Mikami B., Hashimoto W., Murata K. (2010) Molecular identification of unsaturated uronate reductase prerequisite for alginate metabolism in Sphingomonas sp. A1. Biochim. Biophys. Acta 1804, 1925–1936 [DOI] [PubMed] [Google Scholar]
- 36. Condemine G., Robert-Baudouy J. (1991) Analysis of an Erwinia chrysanthemi gene cluster involved in pectin degradation. Mol. Microbiol. 5, 2191–2202 [DOI] [PubMed] [Google Scholar]
- 37. Rothe M., Alpert C., Loh G., Blaut M. (2013) Novel insights into E. coli's hexuronate metabolism: KduI facilitates the conversion of galacturonate and glucuronate under osmotic stress conditions. PLoS One 8, e56906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodionov D. A., Gelfand M. S., Hugouvieux-Cotte-Pattat N. (2004) Comparative genomics of the KdgR regulon in Erwinia chrysanthemi 3937 and other γ-proteobacteria. Microbiology 150, 3571–3590 [DOI] [PubMed] [Google Scholar]
- 39. Myers G. S., Rasko D. A., Cheung J. K., Ravel J., Seshadri R., DeBoy R. T., Ren Q., Varga J., Awad M. M., Brinkac L. M., Daugherty S. C., Haft D. H., Dodson R. J., Madupu R., Nelson W. C., Rosovitz M. J., Sullivan S. A., Khouri H., Dimitrov G. I., Watkins K. L., Mulligan S., Benton J., Radune D., Fisher D. J., Atkins H. S., Hiscox T., Jost B. H., Billington S. J., Songer J. G., McClane B. A., Titball R. W., Rood J. I., Melville S. B., Paulsen I. T. (2006) Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16, 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paulsen I. T., Banerjei L., Myers G. S., Nelson K. E., Seshadri R., Read T. D., Fouts D. E., Eisen J. A., Gill S. R., Heidelberg J. F., Tettelin H., Dodson R. J., Umayam L., Brinkac L., Beanan M., Daugherty S., DeBoy R. T., Durkin S., Kolonay J., Madupu R., Nelson W., Vamathevan J., Tran B., Upton J., Hansen T., Shetty J., Khouri H., Utterback T., Radune D., Ketchum K. A., Dougherty B. A., Fraser C. M. (2003) Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299, 2071–2074 [DOI] [PubMed] [Google Scholar]
- 41. Preiss J. (1966) 4-Deoxy-l-threo-5-hexosulose uronic acid isomerase. Methods Enzymol. 10.1016/0076-6879(66)09121-3 [DOI] [Google Scholar]
- 42. Condemine G., Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. (1984) An enzyme in the pectinolytic pathway of Erwinia chrysanthemi: 2-keto-3-deoxygluconate oxidoreductase. J. Gen. Microbiol. 10.1099/00221287-130-11-2839 [DOI] [Google Scholar]
- 43. Crowther R. L., Georgiadis M. M. (2005) The crystal structure of 5-keto-4-deoxyuronate isomerase from Escherichia coli. Proteins 61, 680–684 [DOI] [PubMed] [Google Scholar]
- 44. Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L., Tate J., Punta M. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang R. G., Andersson C. E., Skarina T., Evdokimova E., Edwards A. M., Joachimiak A., Savchenko A., Mowbray S. L. (2003) The 2.2 Å resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction. J. Mol. Biol. 332, 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stern A. L., Naworyta A., Cazzulo J. J., Mowbray S. L. (2011) Structures of type B ribose 5-phosphate isomerase from Trypanosoma cruzi shed light on the determinants of sugar specificity in the structural family. FEBS J. 278, 793–808 [DOI] [PubMed] [Google Scholar]
- 47. Jung J., Kim J. K., Yeom S. J., Ahn Y. J., Oh D. K., Kang L. W. (2011) Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics. Appl. Microbiol. Biotechnol. 90, 517–527 [DOI] [PubMed] [Google Scholar]
- 48. Edwards T. E., Abramov A. B., Smith E. R., Baydo R. O., Leonard J. T., Leibly D. J., Thompkins K. B., Clifton M. C., Gardberg A. S., Staker B. L., Van Voorhis W. C., Myler P. J., Stewart L. J. (2011) Structural characterization of a ribose-5-phosphate isomerase B from the pathogenic fungus Coccidioides immitis. BMC Struct. Biol. 11, 39–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Persson B., Kallberg Y. (2013) Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs). Chem. Biol. Interact. 202, 111–115 [DOI] [PubMed] [Google Scholar]
- 50. Kavanagh K. L., Jörnvall H., Persson B., Oppermann U. (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65, 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Q., Peng H., Gao F., Liu Y., Cheng H., Thompson J., Gao G. F. (2009) Structural insight into the catalytic mechanism of gluconate 5-dehydrogenase from Streptococcus suis: crystal structures of the substrate-free and quaternary complex enzymes. Protein Sci. 18, 294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Price A. C., Zhang Y. M., Rock C. O., White S. W. (2004) Cofactor-induced conformational rearrangements establish a catalytically competent active site and a proton relay conduit in FabG. Structure 12, 417–428 [DOI] [PubMed] [Google Scholar]
- 53. Takase R., Mikami B., Hashimoto W., Murata K. (2012) X-ray crystallography of bacterial α-keto acid reductases responsible for polyuronate metabolism. Abstract paper of the Annual Meeting of the Japan Society for Bioscience, Biotechnology, and Agrochemistry, Kyoto, Japan, March 25, 2012, 4C10a08, 110 [Google Scholar]