Background: Cell cycle-arrested cancer cells are resistant to conventional chemotherapy.
Results: Microarray analyses showed that expression of RFPL4A was significantly up-regulated in these G1-retained cells.
Conclusion: RFPL4A is a novel factor inducing G1 retention and reduced sensitivity to chemotherapy.
Significance: Combination therapy using RFPL4A inhibition and conventional anti-cancer drugs may represent a promising therapeutic approach for intractable cancer patients.
Keywords: Anticancer Drug, Cell Cycle, Colon Cancer, E3 Ubiquitin Ligase, Microarray, G1 Maintenance, RFPL4A
Abstract
Cell cycle-arrested cancer cells are resistant to conventional chemotherapy that acts on the mitotic phases of the cell cycle, although the molecular mechanisms involved in halting cell cycle progression remain unclear. Here, we demonstrated that RFPL4A, an uncharacterized ubiquitin ligase, induced G1 retention and thus conferred decreased sensitivity to chemotherapy in the human colorectal cancer cell line, HCT116. Long term time lapse observations in HCT116 cells bearing a “fluorescence ubiquitin-based cell cycle indicator” identified a characteristic population that is viable but remains in the G1 phase for an extended period of time (up to 56 h). Microarray analyses showed that expression of RFPL4A was significantly up-regulated in these G1-arrested cells, not only in HCT116 cells but also in other cancer cell lines, and overexpression of RFPL4A increased the G1 population and decreased sensitivity to chemotherapy. However, knockdown of RFPL4A expression caused the cells to resume mitosis and induced their susceptibility to anti-cancer drugs in vitro and in vivo. These results indicate that RFPL4A is a novel factor that increases the G1 population and decreases sensitivity to chemotherapy and thus may be a promising therapeutic target for refractory tumor conditions.
Introduction
During normal proliferation, mammalian cells withdraw from the cell cycle only when deprived of growth factors or exposed to growth inhibition signals during the early to middle G1 phase, before passing the restriction point controlled by the Rb/E2F transcription factor. Once past that point, cells are committed to undergo one round of DNA replication and cell division (1). However, genotoxic stresses, such as irradiation and reactive oxygen species (ROS),3 can transiently halt cell cycle progression at the G1, S, or G2 check point. The G1 check point represents the major machinery for responding to DNA damage and is regulated by the p53/MDM2-p21 pathway, which is capable of inducing sustained and sometimes permanent G1 arrest (2). The G1 checkpoint involves two critical tumor suppressors governed by p53 and RB, both of which represent major mechanisms commonly deregulated in many human cancers (3, 4).
In contrast, cell cycle-arrested cancer cells, such as cancer stem cells (CSCs), constitute a minor population of cancer cells that cause latency and the initiation of cancers, with several characteristic properties, including a self-renewal capacity, pluripotency, and quiescence (5). Recent evidence suggests a hierarchical model for cancer development, with a characteristic fraction of dormant CSCs remaining in the G0/G1 phase of the cell cycle that are resistant to conventional chemotherapy (6–8). Thus, past research has investigated novel cancer treatments targeting these dormant cells. For example, novel therapies are being tested for their ability to inhibit the action of Fbxw7, a key factor that maintains quiescence of leukemia-initiating “stem” cells, leading to an increase in sensitivity to chemotherapy (9, 10).
These previous studies shed light on the importance of cell cycle regulation in cancer cells, especially in the control of G1 arrest. Using long term time lapse observation of human colorectal cancer cells, we noticed a characteristic population that was viable but did not enter the cell cycle, remaining in a sustained G1 phase; these cells could be regarded as similar to so-called CSCs. Exhaustive expression analyses showed that RFPL4A was significantly up-regulated in this population, and this molecule induced G1 retention, resulting in decreased sensitivity to chemotherapy.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
The human colon cancer cell lines HCT116, HT29, and DLD1 were obtained from the American Type Culture Collection (ATCC, Manassas, VA). These cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin at 37 °C in a humidified incubator with 5% CO2 in air.
Establishment of Fluorescence Ubiquitin-based Cell Cycle Indicator (Fucci) Cell Lines
The mKO2-hCdt1(30/120) cells (provided by Dr. Miyawaki, RIKEN Brain Science Institute, Wako, Japan) were cloned into the CSII-EF-MCS vector (provided by Dr. Miyoshi, RIKEN BioResource Center, Tsukuba, Japan) and transfected into HEK293T cells with packaging plasmids (11). Stable transformants were selected using a FACSAria cell sorter (BD Biosciences). Fucci green (mAG) and Fucci red (mKO2) were excited using a 488-nm laser, and their emissions were detected using 530/30 and 585/42 band pass filters, respectively.
Time Lapse Imaging
Time lapse imaging was performed every 1 h using a confocal A1 microscope system (Nikon, Tokyo, Japan) equipped with a humidified imaging chamber (Nikon) at 37 °C, 5% CO2 in air. Time lapse images were analyzed using the NIS-Elements software (Nikon).
Flow Cytometry
Both mKO2 and mAG were excited by a 488-nm laser. Fluorescence signals were collected at 530 nm (530/28 band pass) for mAG and at 575 nm (575/26 band pass) for mKO2. Cell data were analyzed using the FACSCanto II or FlowJo software (Tree Star, Ashland, OR) and isolated using a FACSAria II cell sorter equipped with the FACS Diva software (BD Biosciences).
Sphere Formation Assay
We cultivated cells as hanging drops at a density of 1 × 103/100 μl in 6-well ultra-low attachment plates (MD6 with lid, low cell binding; Thermo Fisher Scientific). Cells were grown in serum-free medium containing 2 mm l-glutamine and 4.5% glucose and supplemented with B27 (MACS), N2 (Wako), 10 ng/ml EGF (Invitrogen), and 10 ng/ml basic fibroblast growth factor (Funakoshi). Sphere formation was assessed 14 days after seeding. Spheres were defined as cell clusters with diameters greater than 100 μm. In each of the 3–6 wells, the number of formed spheres was calculated.
ROS Assay
The ROS assay was performed as described previously (12). Intracellular ROS levels were determined by incubating the cells for 30 min at 37 °C with 5 μm CellROX Deep Red reagent (Invitrogen) in complete medium, followed by flow cytometry.
Quantitative Real-time RT-PCR and Protein Analysis
Total RNA was isolated using the Maxwell 16 LEV simplyRNA purification kit (Promega). Quantitative real-time PCR, preparation of cell lysates, and Western blotting were performed according to standard protocols. The primers used are listed in Table 1. The antibodies used for Western blotting are listed in Table 2.
TABLE 1.
Primer pairs used for real-time qPCR for verifying Fucci microarray data
The expected molecular size in bp is indicated.
| Forward (5′–3′) | Reverse (5′–3′) | bp | |
|---|---|---|---|
| RFPL4A | TGAACCGACAGGGGAAGATTG | CAGGGCTCGCAAACAGGAAT | 864 |
| Gapdh | TGTTGCCATCAATGACCCCTT | CTCCACGACGTACTCAGCG | 202 |
TABLE 2.
Antibodies and their applications
| Primary antibody | Company | Catalog ID | Application(s) and working dilution(s) |
|---|---|---|---|
| Anti-RFPL4A | Abcam | ab138017 | Western blot, 1:500; immunocytochemistry, 1:400 |
| Anti-β-actin | Santa Cruz Biotechnology, Inc. | sc-47778 | Western blot, 1:1000 |
Fucci Signal-based Microarray Analysis
Microarray analysis was performed as described previously (13). Briefly, the microarray platforms used in this study were Whole Human Genome Microarray 4x44K version 2 (G4845A) (Agilent Technologies, Santa Clara, CA). In total, 100 ng of total RNA derived from the 1.0–2.0 × 105 Fucci red cells were extracted using an RNeasy kit (Qiagen, Venlo, Netherlands) and then reverse transcribed and labeled using a low input Quick Amp labeling kit (Agilent) following the manufacturer's instructions. Raw data were imported into GeneSpring GX 11.0 (Agilent) for database management and quality control. The -fold change values were calculated as the ratio of the normalized value between the red-fluorescing retained (RR) and the red-fluorescing G1 (R) cells. The data have been deposited in GEO as GSE61088.
Immunocytochemistry
Cells were grown in incubation chambers coated with BioCoat collagen I (BD Biosciences). Immunocytochemistry was performed as described previously (14). All images were obtained using an A1 confocal microscope (Nikon). In all cases, the mean RFPL4A fluorescence intensity over the whole cell area was measured using the commercially available software NIS-Elements (Nikon).
Establishment of Overexpressing Cell Lines
A full-length cDNA of the human RFPL4A, obtained from the Kazusa DNA Research Institute (Chiba, Japan), was inserted in front of the intraribosomal entry site (IRES) of the retroviral vector pMX-IRES-Puro, via the EcoRI and XhoI sites, to obtain pMX-RFPL4A. Replication-defective retroviruses were generated by transient transfection of pMX-RFPL4A or pMX-IRES-Puro (control) vectors into PLAT-A-cells using FuGene 6 reagent (Promega, Tokyo, Japan) (15). HCT116 cells were transduced with the resulting retroviruses as described previously (16), positively selected, and expanded in the presence of 5 μg/ml puromycin.
Establishment of Knockdown Cell Lines
MISSION shRNA plasmids for human RFPL4A (Table 3), a scrambled control shRNA (17), and lentiviral packaging mix were purchased from Sigma. Lentivirus-mediated transfection of shRNAs was performed according to the manufacturer's instructions.
TABLE 3.
Sequences of shRNAs obtained from Sigma-Aldrich
| shRNA target | Sequence strand (5′–3′) | Catalog ID |
|---|---|---|
| RFPL4A 1 | CCGGACTAGAGCCCAAGCTGAAATCCTCGAGGATTTCAGCTTGGGCTCTAGTTTTTTG | TRCN0000230973 |
| RFPL4A 2 | CCGGCCAACAACTATCTCATCATTTCTCGAGAAATGATGAGATAGTTGTTGGTTTTTG | TRCN0000230974 |
| Control shRNA | CCGGGCTTCTAACACCGGAGGTCTTCTCGAGAAGACCTCCGGTGTTAGAAGCTTTTTG |
Cell Proliferation Assay
Isolated HCT116 cells were added to 96-well plates at a density of 5 × 102/well in 100 μl of DMEM supplemented with 10% FBS and incubated for 5 days. Cell proliferation was assessed using the Cell Counting Kit-8 incorporating WST-8 (Dojindo Molecular Technologies, Kumamoto, Japan) following the manufacturer's instructions and a plate reader (model 680XR; Bio-Rad).
In Vivo Cell Proliferation
Female NOD/SCID mice (6–8 weeks old; Charles River) were assigned randomly. HCT116 cells (OE-NC, RFPL4A-OE, sh-NC, RFPL4A-sh1, or RFPL4A-sh2) (2 × 106 cells) in 0.2 ml of PBS were injected into subcutaneous tissues.
In Vivo Chemotherapy
Female NOD/SCID mice (6–8 weeks old; Charles River) were assigned randomly. HCT116 cells (OE-NC, RFPL4A-OE, sh-NC, RFPL4A-sh1, or RFPL4A-sh2) (3 × 106 cells) in 0.2 ml of DMEM were injected into subcutaneous tissues. When xenografts reached a volume of 50–200 mm3 (∼12–14 days after injection), 5-FU (30 mg/kg/day) dissolved in 0.2 ml of PBS was administered by intraperitoneal injection for 2 consecutive days per week for 2 weeks (18–20). The estimated tumor volume (V) was calculated by the formula (21), V = W2 × L × 0.5, where W represents the largest tumor diameter (in cm), and L represents the next largest tumor diameter. The individual relative tumor volume (RTV) was calculated with the equation, RTV = Vx/V1, where Vx is the volume (in mm3) at a given time, and V1 is the volume at the start of treatment. Results are expressed as the mean daily percentage change in tumor volume for each group of mice.
In Vivo siRNA Treatment
HCT116 cells (5 × 106) were injected into subcutaneous tissues, and the resulting tumors were injected with siRNAs targeting RFPL4A (Table 4) or with a scrambled control siRNA, together with atelocollagen (AteloGene, Koken, Japan) 1 week after implantation. A 0.2-ml volume of siRNA solution (30 μmol/liter in 0.5% (v/v) atelocollagen) was injected directly into the tumors. Injected siRNAs were shown to remain stable in vivo for at least 1 week when supported by atelocollagen (22) (23). 5-FU (30 mg/kg/day) dissolved in 0.2 ml of PBS was administered by intraperitoneal injection for 2 consecutive days per week for 2 weeks.
TABLE 4.
Sequences of siRNA duplexes
| siRNA target | Sense (5′-3′) | Antisense (5′-3′) |
|---|---|---|
| RFPL4A 1 | ACTAGAGCCCAAGCTGAAATCTT | GATTTCAGCTTGGGCTCTAGTT |
| RFPL4A 2 | CCAACAACTATCTCATCATTTTT | AAATGATGAGATAGTTGTTGGTT |
| Scrambled | AUCCGCGCGAUAGUACGUATT | UACGUACUAUCGCGCGGAUTT |
Statistical Analyses
Differences between the control and treated groups were assessed by an unpaired Student's t test or a Mann Whitney U test and considered to be significant at p < 0.05 (*, p < 0.05; **, p < 0.01; ***, p < 0.005). Values are presented as means ± S.E. Statistical analyses were performed using the GraphPad Prism software (version 6.0; GraphPad Software). Image processing, reconstruction, analyses, and displays were performed using Imaris version 6.3 and 7.4 (Bitplane). A receiver operating characteristic (ROC) curve was used to obtain the optimal cut-off value.
RESULTS
Identification of G1-retained Cells Using Long Term Time Lapse Imaging
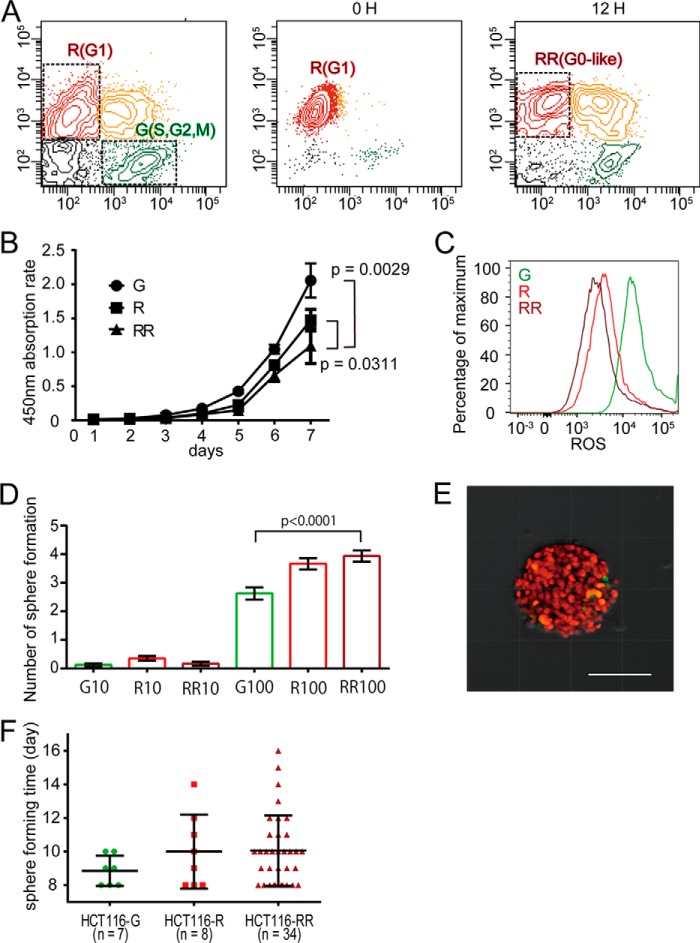
Cancer cells are heterogeneous in terms of their proliferative activity. To examine the cell division status in different cells, we used time lapse confocal microscopy with a Fucci probe to detect the cell cycle status of living cells (14). Using this method, geminin and Cdt1, nuclear proteins enriched in the S/G2/M and G1 phases, are marked as green and red fluorescing proteins, respectively. We generated Fucci-expressing HCT116 human colon cancer cell lines (24) and observed their proliferative time courses by confocal time lapse microscopy. The doubling time of HCT116 cells has been reported to be ∼21 h (25), although long term observations, up to 56 h, detected a minor population that was viable but remained in a “red” G1 state without entering the cell cycle (Fig. 1A and supplemental Video 1). We also collected these red G1 cells by sorting and cultured them for an extended period of time, confirming the presence of cells remaining in the G1 phase (Fig. 1, B and C). We assumed that the red-fluorescing population would consist of G1-retained cells.
FIGURE 1.
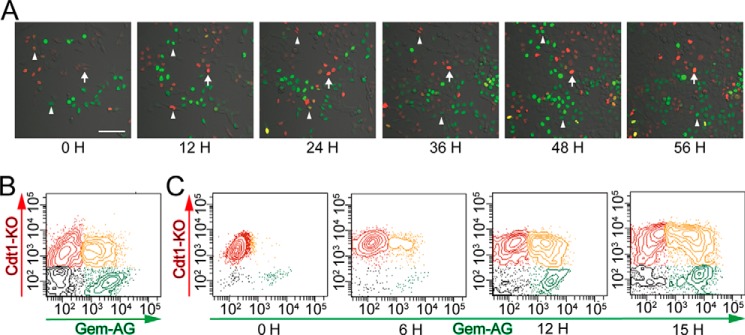
Identification of G1-retained cells using long term time lapse imaging. A, representative images of cell cycle changes in Fucci-expressing HCT116 cells, as visualized by confocal time lapse imaging. Red fluorescence (Cdt1-KO) and green fluorescence (Gem-AG) indicate the G0/G1 and S/G2/M phases of the cell cycle, respectively. The arrows indicate G1 (red) retained cells, and arrowheads indicate dividing cells. Scale bars, 100 μm. B, cell cycle analysis of Fucci-expressing HCT116 cells by flow cytometry. Shown is quantification of four subpopulations: Cdt1-KO+ (red), Gem-AG+ (green), double-positive (yellow), and double-negative (gray). C, Fucci red (KO)-positive HCT116 cells were sorted using a FACS Aria cell sorter and were cultured for the indicated periods of time.
Characterization of the Red-fluorescing “G1-retained” Cells and Identification of RFPL4A as a G1-determining Factor
To characterize the long term red-fluorescing G1-retained cells, we attempted to enrich this fraction (Fig. 2A). Fucci-introduced HCT116 cells were first separated into R and green-fluorescing S/G2/M (G) fractions. The R fraction was cultured for a further 12 h, from which a red-fluorescing G1 population (the RR population) was collected by cell sorting. R cells were cultured for an additional 12 h only because dividing S/G2/M cells re-enter a red-fluorescing G1 state and merge with the long term G1-retained cells on further incubation (Fig. 1C). The RR fraction was thus considered to be an enriched fraction of G1-arrested cells. Compared with the G and R fractions, the RR cells exhibited significantly delayed proliferation (Fig. 2B). Furthermore, RR cells had lower levels of ROS (Fig. 2C) and higher sphere-initiating capacity (Fig. 2, D and E), suggesting stem cell-like properties of RR cells (26–28). The spheres of RR cells were formed at ∼10 days (Fig. 2F).
FIGURE 2.
Characterization of red-fluorescing “G1-retained” cells. A, cell cycle analysis of Fucci-expressing HCT116 cells by flow cytometry. Shown are sorted red cells at 0 h and after culturing for 12 h. B, absorption rate of G, R, and RR HCT116 cells isolated by flow cytometry was examined using a cell counting kit. The statistical significance of differences among the G, R, and RR HCT116 cells was determined by a two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). C, intracellular ROS levels measured using CellROX-labeled allophycocyanin and flow cytometry in HCT116 cells. D, sphere formation results for G, R, and RR HCT116 cells isolated by flow cytometry. Sphere numbers formed within 14 days after seeding 10 or 100 cells/well are shown. Each bar represents the mean ± S.E. (error bars) of 48 independent experiments (n = 48). E, representative sphere formation for HCT116 cells expressing Fucci 14 days after seeding, as visualized by confocal microscopy. Scale bar, 100 μm. F, the sphere was defined when the diameter was 100 μm. Each bar represents the mean ± S.D. (error bars).
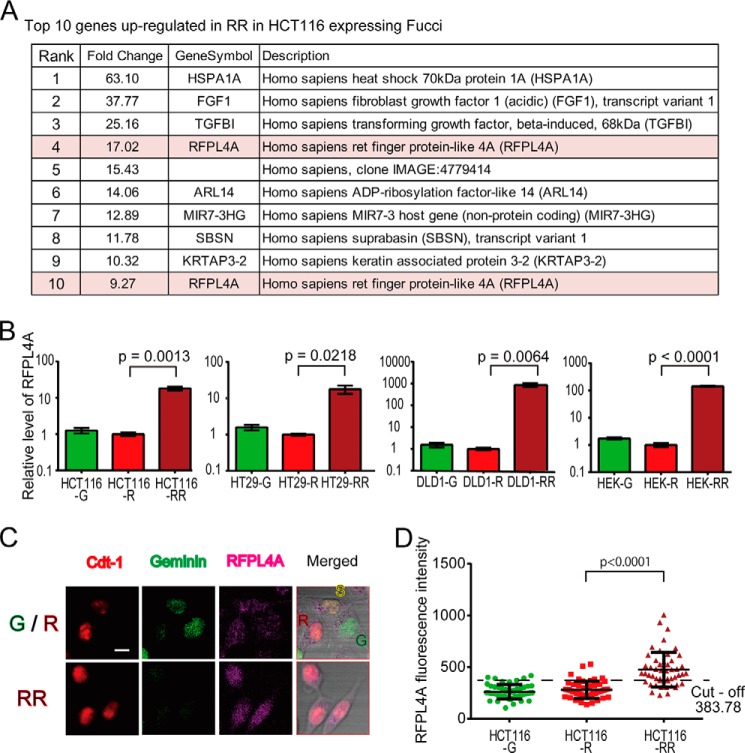
To determine the molecular basis of the G1 arrest/maintenance mechanism, we conducted cDNA microarray-based comparative analyses between the RR and R fractions. In the analysis, 518 of 34,127 probes showed greater than 2-fold changes in expression (Fig. 3A and Table 5). Among them, we noticed that a poorly characterized molecule, RFPL4A (Ret finger protein-like 4A), was significantly up-regulated in the RR versus the R fraction. Two probes for the RFPL4A gene were both ranked highly (4th and 10th) among the 518 probes (Fig. 3A and Table 5). The preferential expression of RFPL4A in RR cells was confirmed by quantitative RT-PCR analyses in several colon cancer cell lines, such as HCT116, HT29, and DLD1, and in non-cancer cell lines, such as HEK293 (Fig. 3B), suggesting a general mechanism for this characteristic regulation of RFPL4A expression among diverse cell types. To investigate the RFPL4A levels of individual cells, HCT116 cells expressing Fucci were immunostained by RFPL4A, and the intensity was compared (Fig. 3, C and D). According to the ROC curve for the intensity (RR versus R), the optimal cut-off value was 383.78. This cut-off value corresponded to a sensitivity of 94% and a specificity of 70%. The area under the ROC curve was 0.8852. The ratio of high RFPL4A in RR was 70% (35 of 50 cells).
FIGURE 3.
The identification of RFPL4A as a G1 maintenance factor. A, in total, the expression of 518 genes was up-regulated over 2-fold (Table 5; p < 0. 05). Of these, RFPL4A ranked highly, with two probes for this gene among the top 10. B, RFPL4A expression in various human colon cancer cell lines (HCT116, DLD1, and HT29) and a normal cell line (HEK293). In all of the cell lines, RFPL4A expression was significantly higher in RR. Each bar represents the mean ± S.E. (error bars) of three independent experiments (n = 3). C, the p21 HCT116 cells were visualized using immunofluorescence. Shown are representative images of Fucci-expressing HCT116 cells immunostained with RFPL4A. Top, R, G1 phase; S, early S phase; G, S/G2/M phase. Bottom, RR, G1-retained cells at 12 h after sorted G1 phase cells. Scale bar, 10 μm. D, 50 cells at G, R, and RR. Error bars, means ± S.D. The cut-off point was defined according to the ROC curve for intensity (RR versus R).
TABLE 5.
Top 50 genes up-regulated in RR in HCT116 cells expressing Fucci
| Rank | Change | Symbol | Description |
|---|---|---|---|
| -fold | |||
| 1 | 63.10 | HSPA1A | Homo sapiens heat shock 70-kDa protein 1A (HSPA1A) |
| 2 | 37.77 | FGF1 | H. sapiens fibroblast growth factor 1 (FGF1), transcript variant 1 |
| 3 | 25.16 | TGFBI | H. sapiens transforming growth factor, β-induced (TGFBI) |
| 4 | 17.02 | RFPL4A | H. sapiens Ret finger protein-like 4A (RFPL4A) |
| 5 | 15.43 | H. sapiens, clone IMAGE:4779414, mRNA | |
| 6 | 14.06 | ARL14 | H. sapiens ADP-ribosylation factor-like 14 (ARL14) |
| 7 | 12.89 | MIR7-3HG | H. sapiens MIR7–3 host gene (non-protein-coding) (MIR7-3HG) |
| 8 | 11.78 | SBSN | H. sapiens suprabasin (SBSN), transcript variant 1 |
| 9 | 10.32 | KRTAP3-2 | H. sapiens keratin associated protein 3-2 (KRTAP3-2) |
| 10 | 9.27 | RFPL4A | H. sapiens Ret finger protein-like 4A (RFPL4A) |
| 11 | 8.97 | C6orf222 | H. sapiens chromosome 6 open reading frame 222 (C6orf222) |
| 12 | 8.17 | PADI1 | H. sapiens peptidyl arginine deiminase, type I (PADI1) |
| 13 | 8.01 | GADD45G | H. sapiens growth arrest and DNA damage-inducible, γ (GADD45G) |
| 14 | 7.80 | MYPN | H. sapiens myopalladin (MYPN) |
| 15 | 7.02 | MAFB | H. sapiens v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (MAFB) |
| 16 | 7.00 | SNAI2 | H. sapiens snail homolog 2 (Drosophila) (SNAI2) |
| 17 | 6.86 | Q96IU8_HUMAN (Q96IU8) CSAG family, member 3A, partial (42%) | |
| 18 | 6.84 | KPNA7 | H. sapiens karyopherin α7 (importin α8) (KPNA7) |
| 19 | 6.82 | MIR7-3HG | H. sapiens MIR7-3 host gene (non-protein-coding) (MIR7-3HG) |
| 20 | 6.29 | DNAH17 | Dynein, axonemal, heavy chain 17 (source: HGNC Symbol;Acc:2946) |
| 21 | 6.28 | SCGN | H. sapiens secretagogin, EF-hand calcium binding protein (SCGN) |
| 22 | 6.16 | KRTAP4-1 | H. sapiens keratin-associated protein 4-1 (KRTAP4-1) |
| 23 | 6.13 | CPA4 | H. sapiens carboxypeptidase A4 (CPA4), transcript variant 1 |
| 24 | 6.06 | CRYAB | H. sapiens crystallin, αB (CRYAB) |
| 25 | 5.80 | TNF | H. sapiens tumor necrosis factor (TNF) |
| 26 | 5.67 | C8orf46 | H. sapiens chromosome 8 open reading frame 46 (C8orf46) |
| 27 | 5.61 | HMOX1 | H. sapiens heme oxygenase (decycling) 1 (HMOX1) |
| 28 | 5.52 | PMEPA1 | H. sapiens prostate transmembrane protein, androgen-induced 1 (PMEPA1), transcript variant 1 |
| 29 | 5.51 | CTGF | H. sapiens connective tissue growth factor (CTGF) |
| 30 | 5.46 | DKFZp451A211 | PREDICTED: H. sapiens DKFZp451A211 protein (DKFZp451A211) |
| 31 | 5.44 | FGF1 | H. sapiens fibroblast growth factor 1 (acidic) (FGF1), transcript variant 1 |
| 32 | 5.36 | PHEX | H. sapiens phosphate-regulating endopeptidase homolog, X-linked (PHEX) |
| 33 | 5.33 | INHBA | H. sapiens inhibin, βA (INHBA) |
| 34 | 5.32 | GADD45G | H. sapiens growth arrest and DNA-damage-inducible, gamma (GADD45G) |
| 35 | 5.24 | NR4A3 | H. sapiens nuclear receptor subfamily 4, group A, member 3 (NR4A3), transcript variant 3 |
| 36 | 5.19 | C9orf53 | H. sapiens chromosome 9 open reading frame 53 (C9orf53) |
| 37 | 5.15 | DIO3 | H. sapiens deiodinase, iodothyronine, type III (DIO3) |
| 38 | 5.11 | DLL3 | H. sapiens δ-like 3 (Drosophila) (DLL3), transcript variant 2 |
| 39 | 5.08 | C2orf54 | H. sapiens chromosome 2 open reading frame 54 (C2orf54), transcript variant 2 |
| 40 | 5.07 | LOC286059 | H. sapiens tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain pseudogene (LOC286059) |
| 41 | 5.07 | KBTBD5 | H. sapiens kelch repeat and BTB (POZ) domain-containing 5 (KBTBD5) |
| 42 | 5.07 | CDH16 | H. sapiens cadherin 16, KSP-cadherin (CDH16), transcript variant 1 |
| 43 | 5.05 | CXCL1 | H. sapiens chemokine (CXC motif) ligand 1 (melanoma growth-stimulating activity, α) (CXCL1) |
| 44 | 4.95 | TNFRSF9 | H. sapiens tumor necrosis factor receptor superfamily, member 9 (TNFRSF9) |
| 45 | 4.91 | SCEL | H. sapiens sciellin (SCEL), transcript variant 1 |
| 46 | 4.88 | Q9I2C8_PSEAE (Q9I2C8) probable two-component sensor, partial (6%) | |
| 47 | 4.85 | CD36 | H. sapiens CD36 molecule (thrombospondin receptor) (CD36), transcript variant 2 |
| 48 | 4.83 | Unknown | |
| 49 | 4.82 | CDO1 | H. sapiens cysteine dioxygenase, type I (CDO1) |
| 50 | 4.79 | KPNA7 | H. sapiens karyopherin α7 (importin α8) (KPNA7) |
Overexpression of RFLP4A Increases G1 Population and Inhibits Proliferation
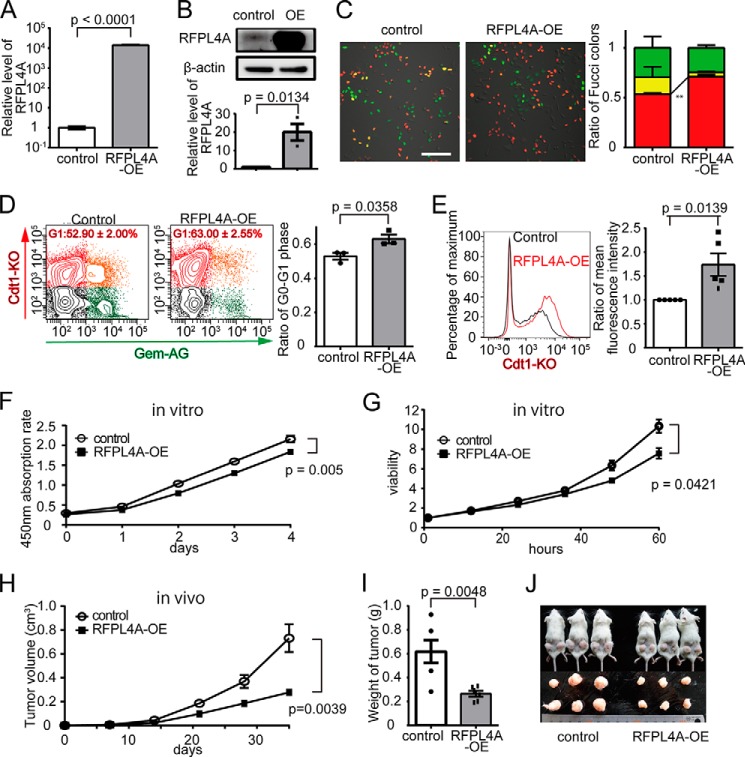
To investigate the function of RFPL4A, we first established an HCT116 cell line overexpressing RFPL4A (RFPL4A-OE). Abundant expression of RFPL4A in RFPL4A-OE cells was confirmed at both the mRNA (Fig. 4A) and protein (Fig. 4B) levels. The expression level of RFPL4A in OE cells (∼1,000-fold) was markedly higher than that in the control RR cells (∼10-fold), although it was still within the physiological range, because DLD1-RR cells expressed RFPL4A at ∼1,000-fold higher levels compared with DLD1-R/G cells (Fig. 3B). Microscopic examination revealed that the proportion of the red G0/G1 phase population was increased in RFPL4A-OE cells (control, 53.47 ± 1.14%, n = 3; RFPL4A-OE, 71.12 ± 2.03%, n = 3; p = 0.0016; Fig. 3C). Flow cytometric analyses also showed that the proportion of the red G0/G1 fraction in RFPL4A-OE cells was increased (control, 52.90 ± 2.00%, n = 3; RFPL4A-OE, 63.00 ± 2.55%, n = 3; p = 0.0358; Fig. 3D). Prolongation of the G0/G1 phase has been reported to result in an increase in red fluorescence intensity because of the accumulation of Cdt1-KO (29). Consistently, the red fluorescence intensity in RFPL4A-OE cells was significantly higher than that of the control (Fig. 4E). Additionally, RFPL4A-OE cells were found to exhibit significantly lower proliferative activity in vitro (Fig. 4, F and G) and in vivo (Fig. 4, H–J), also consistent with an extended cell cycle G1 arrest. Cell proliferation (tumor progression) would also be tracked in vivo for a longer period of time (∼30 days), which would make the differences more prominent.
FIGURE 4.
Overexpression of RFLP4A increases G1 population and inhibits proliferation. A, establishment of RFPL4A-overexpressing cells (RFPL4A-OE). Increased RFPL4A mRNA expression was confirmed by qPCR. B, protein expression was assessed by Western blotting and quantified by ImageJ. C, representative images of Fucci-expressing control and RFPL4A-overexpressing HCT116 cells. Scale bar, 100 μm. The ratios of Fucci colors counted by IMARIS are shown. Each bar represents the mean ± S.E. (error bars) of three independent experiments (n = 3). Total cell count was 744. D, cell cycle by flow cytometric analysis of Fucci-expressing control and RFPL4A-overexpressing HCT116 cells. Red, Cdt1-KO+; green, Gem-AG+; yellow, double-positive; gray, double-negative. The ratios of Cdt1-KO+ (red) are shown. Each bar represents the mean ± S.E. (error bars) of three independent experiments (n = 3). Total cell count was 1.2 × 105. E, red fluorescence intensity quantified by flow cytometry. Mean fluorescence intensity ratios were compared. Each bar represents the mean ± S.E. (error bars) of five individual experiments (n = 5 for each). F, the absorption rate of HCT116 cells was examined using a cell counting kit. The statistical significance of the difference between the control and RFPL4A-expressing HCT116 cells was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). G, viability of HCT116 cells was examined by time lapse imaging confocal microscopy, and the ratios were counted by IMARIS. Cell viability is presented as the ratio of HCT116 cells at each time relative to zero time. The statistical significance of the difference between the control and RFPL4A-expressing HCT116 cells was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). H, in vivo tumor expansion of HCT116 cells. Control and RFPL4A-overexpressing HCT116 cells are shown. Cancer cells (2 × 106/200 μl of PBS) were inoculated into subcutaneous tissue. Tumor sizes were measured every week for 5 weeks after inoculation. The statistical significance of the difference between the control and RFPL4A-expressing HCT116 cells was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of six individual experiments (n = 6 for each). I, comparison of the tumor weight in control versus RFPL4A-overexpressing cells at 35 days. Each bar represents the mean ± S.E. (error bars) of six independent experiments (n = 6 each). J, images of tumors excised on day 35.
Overexpression of RFLP4A Is Associated with a Stemlike Nature and Decreased Sensitivity to Chemotherapy
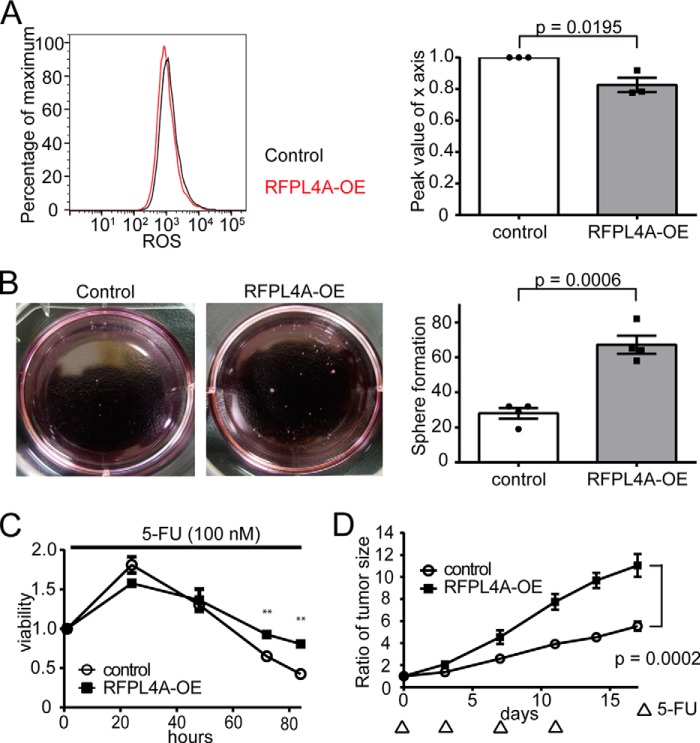
Next, we examined the effect of RFLP4A overexpression on the “stemness” and resistance to chemotherapy of the cells. We found that RFPL4A-OE cells had higher sphere-initiating capacity and lower reactive oxygen species levels compared with controls (Fig. 5, A and B), indicating stem cell-like properties of the RFPL4A-OE cells. Additionally, RFPL4A-OE cells showed higher viability in the presence of 5-FU in vitro (100 μm), which was statistically significant at 72 h after application of 5-FU (Fig. 5C). The chemotherapy resistance of RFPL4A-OE cells was also confirmed in a xenograft model, in which RFPL4A-OE cells could proliferate more effectively than control tumor cells under 5-FU treatment (30 mg/kg × 4 days; Fig. 5D). These results demonstrate that RFPL4A expression is closely associated with G1 retention and inhibited proliferation of cancer cells, resulting in induction of stemness and resistance to chemotherapy, which are clearly undesirable properties in terms of cancer treatments.
FIGURE 5.
Overexpression of RFLP4A is associated with a stemlike nature and decreased sensitivity to chemotherapy. A, intracellular ROS levels, measured using CellROX-labeled allophycocyanin and flow cytometry, in HCT116 cells (control and RFPL4A-OE). Peak values of the x axis ratio were compared. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). B, representative sphere formation of HCT116 expressing Fucci 14 days after seeding. Sphere formation results in HCT116 cells (control and RFPL4A-OE). Sphere numbers that formed within 14 days after seeding 2,000 cells/well are shown. Each bar represents the mean ± S.E. (error bars) of four independent experiments (n = 4). C, viability of HCT116 cells (control and RFPL4A overexpression) was examined by time lapse imaging using confocal microscopy, and the ratios were counted by IMARIS after treatment with 100 nm 5-FU for 84 h. Cell viability is presented as the ratio of HCT116 cells at each time relative to time zero. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). *, p < 0.05; **, p < 0.01; ***, p < 0.001. D, effect of 5-FU therapy in NOD/SCID mice (volume of tumor (V) = L × W2/2). HCT116 cells (control and RFPL4A-OE) were injected subcutaneously into NOD/SCID mice (n = 6 in each group). Cancer cells (3 × 106/200 μl of PBS) were inoculated into subcutaneous tissue. Treatment was initiated when tumors grew to a size of 5–7 mm in diameter (∼14 days after injecting cells). NOD/SCID mice were treated by intraperitoneal injection of 5-FU (30 mg/kg) at days 0, 3, 7, and 11. Data represent the mean volumes of the tumors, calculated by the formula, (longest diameter) × (shortest diameter)2 × 0.5. Statistical significance of the difference between the control and RFPL4A-expressing HCT116 cells was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of six individual experiments (n = 6 for each).
Knockdown of RFLP4A Decreased the G1 Population and Increased Proliferation
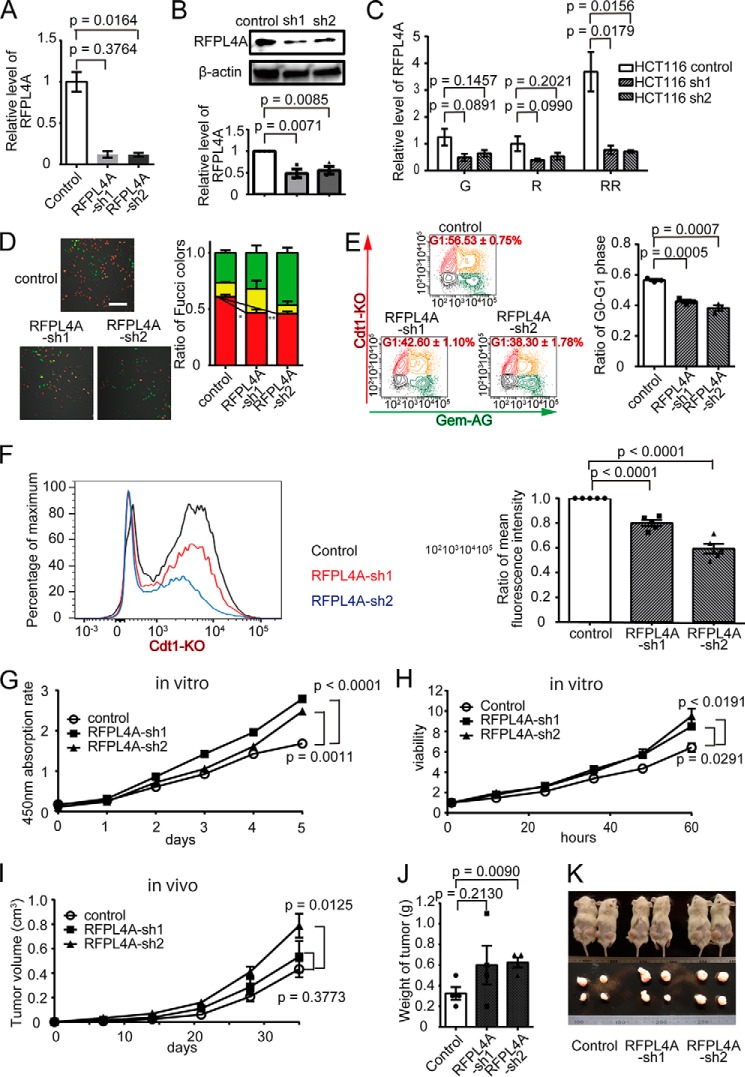
Next, we generated RFPL4A knockdown HCT116 cell lines (RFPL4A-sh1 and -sh2). The reduced expression of RFPL4A in RFPL4A-sh cells was confirmed at both the mRNA (Fig. 6A) and protein levels (Fig. 6B). The reduction of RFPL4A was prominent in the RFPL4A high RR fraction but not significantly reduced in the basal RFPL4A levels in the G and R fractions (Fig. 6C). The proportion of the red G0/G1 phase population was decreased significantly in RFPL4A-sh cells (control, 60.78 ± 0.02%, n = 3; RFPL4A-sh1, 46.59 ± 0.03%, n = 3, p = 0.0209; RFPL4A-sh2, 45.96 ± 0.02, n = 3, p = 0.0077), suggesting their proliferative tendencies (Fig. 6D). Flow cytometric analyses also showed that the proportion of the red G0/G1 fraction in the RFPL4A-sh cells was decreased (control, 56.53 ± 0.75%, n = 3; RFPL4A-sh1, 42.60 ± 1.10%, n = 3, p = 0.0005; RFPL4A-sh2, 38.30 ± 1.78, n = 3, p = 0.0007; Fig. 6E). Furthermore, the red fluorescence intensity of RFPL4A-sh cells was significantly lower than that of the control (Fig. 6F). Concordantly, RFPL4A-sh cells demonstrated significantly higher proliferative activity in vitro (Fig. 6, G and H) and in vivo (Fig. 6, I–K).
FIGURE 6.
Knockdown of RFLP4A decreased the G1 population and increased proliferation. A, establishment of RFPL4A knockdown HCT116 cell lines (RFPL4A-sh1 and -sh2). Decreased RFPL4A mRNA expression was confirmed by qPCR. B, protein expression was assessed by Western blotting and quantified by ImageJ. C, RFPL4A knockdown HCT116 cell lines (RFPL4A-sh1 and -sh2) were sorted to G, R, and RR (assessed by qPCR). Each bar represents the mean ± S.E. (error bars) of three independent experiments (n = 3). D, representative images of Fucci-expressing control and RFPL4A knockdown HCT116 cells (sh1 and sh2). Scale bar, 100 μm. The ratios of the Fucci colors, counted by IMARIS, are shown. Each bar represents the mean ± S.E. (error bars) of three independent experiments (n = 3). Total cell count was 829. E, cell cycle analysis of Fucci-expressing control and RFPL4A knockdown HCT116 cells (sh1 and sh2) by flow cytometry. Red, Cdt1-KO+; green, Gem-AG+; yellow, double-positive; gray, double-negative. The ratios of Cdt1-KO+ (red) are shown. Each bar represents the mean ± S.E. (error bars) of three independent experiments (n = 3). Total cell count was 1.8 × 105. F, red fluorescence intensity quantified by flow cytometry. G, the absorption rate of HCT116 cells was examined using a cell counting kit. Statistical significance of the difference between the control and RFPL4A knockdown (sh1 and sh2) HCT116 cells was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). H, viability of HCT116 cells was examined by time lapse imaging using confocal microscopy, and ratios were counted by IMARIS. Cell viability is presented as the ratio of HCT116 cells at each time relative to time zero. Statistical significance of the difference between control and RFPL4A knockdown HCT116 cells (sh1 and sh2) was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). I, in vivo tumor expansion of HCT116 cells. HCT116 cells treated with RFPL4A control and knockdown shRNAs (sh1 and sh2) are shown. Cancer cells (2 × 106 cells/200 μl of PBS) were inoculated into subcutaneous tissue. Tumor sizes were measured every week for 5 weeks after inoculation. The statistical significance of the difference between the control and RFPL4A-expressing HCT116 cells was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of four individual experiments (n = 4 each). J, comparison of the tumor weight in control versus RFPL4A knockdown cells (sh1 and sh2) at 35 days. Each bar represents the mean ± S.E. (error bars) of four independent experiments (n = 4 each). K, images of tumors excised on day 35.
Knockdown of RFLP4A Decreased Its Stemlike Nature and Increased Sensitivity to Chemotherapy
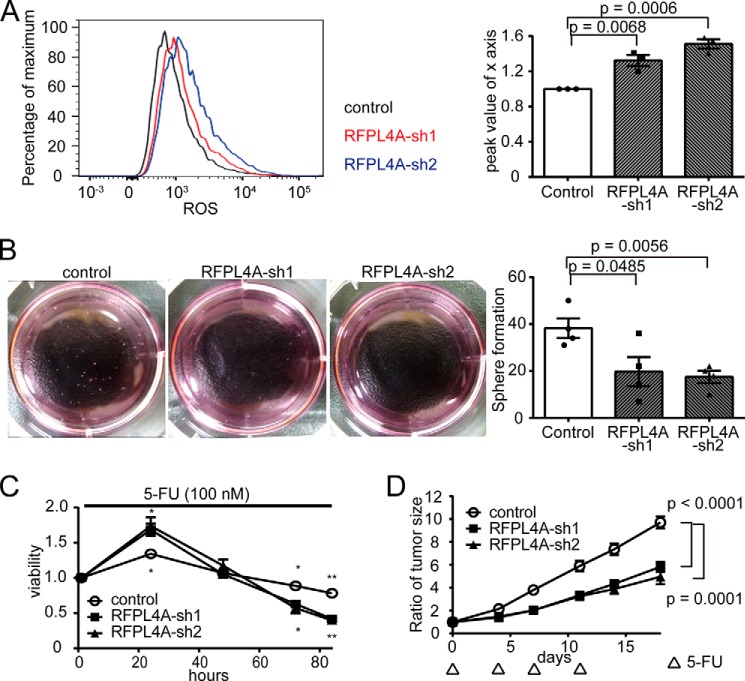
We also examined the effect of down-regulating RFLP4A on the stemness and chemotherapy resistance of the cells. RFPL4A-sh cells showed lower sphere-forming capacity and higher reactive oxygen species levels than did controls (Fig. 7, A and B). Additionally, RFPL4A-sh cells showed lower viability in the presence of 5-FU in vitro (100 nm), which was statistically significant at time points longer than 72 h after application of 5-FU (Fig. 7C). Increased susceptibility to chemotherapy of the RFPL4A-sh cells was also evident in a xenograft model, in which RFPL4A-sh cells proliferated less under periodic 5-FU treatment (30 mg/kg × 4 days) compared with controls (Fig. 7D). These results demonstrated that suppression of RFPL4A expression increased the susceptibility of cancer cells to chemotherapy; this could be useful for radical treatment against intractable cancers.
FIGURE 7.
Knockdown of RFLP4A decreased its stemlike nature and increased sensitivity to chemotherapy. A, intracellular ROS levels were measured using CellROX-labeled allophycocyanin and flow cytometry in HCT116 cells (control and RFPL4A-OE). Peak values of the x axis ratio were compared. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). B, representative sphere formation of HCT116 cells expressing Fucci 14 days after seeding. Sphere formation results in HCT116 cells (control and RFPL4A-sh1/-sh2 knockdown). The sphere numbers that formed within 14 days after seeding 2,000 cells/well are shown. Each bar represents the mean ± S.E. (error bars) of four independent experiments (n = 4). C, viability of HCT116 cells (control and RFPL4A-sh1/-sh2 knockdown) was examined by time lapse imaging using confocal microscopy, with the ratios counted by IMARIS, after treatment with 100 nm 5-FU for 84 h. Cell viability is presented as the ratio of HCT116 cells at each time relative to time 0. Each bar represents the mean ± S.E. (error bars) of three individual experiments (n = 3 for each). *, p < 0.05; **, p < 0.01; and ***, p < 0.001. Top, control versus sh1; bottom, represents control versus sh2. D, effect of 5-FU therapy in NOD/SCID mice (volume of tumor (V) = L × W2/2). HCT116 cells (control and RFPL4A-sh1/-sh2 knockdown) were injected subcutaneously into NOD/SCID mice (n = 4 in each group). Cancer cells (3 × 106 cells/200 μl of PBS) were inoculated into subcutaneous tissue. Treatment was initiated when tumors grew to a size of 5–7 mm in diameter (∼12 days after injection of cells). NOD/SCID mice were treated by intraperitoneal injection of 5-FU (30 mg/kg) at days 0, 4, 7, and 11. Data represent the mean volumes of tumors, calculated using the formula, (longest diameter) × (shortest diameter)2 × 0.5. Statistical significance of the difference between the control and RFPL4A-expressing HCT116 cells was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of four individual experiments (n = 4 each).
RNAi-based Inhibition of RFPL4A Expression Increased Susceptibility to Chemotherapy in Vivo
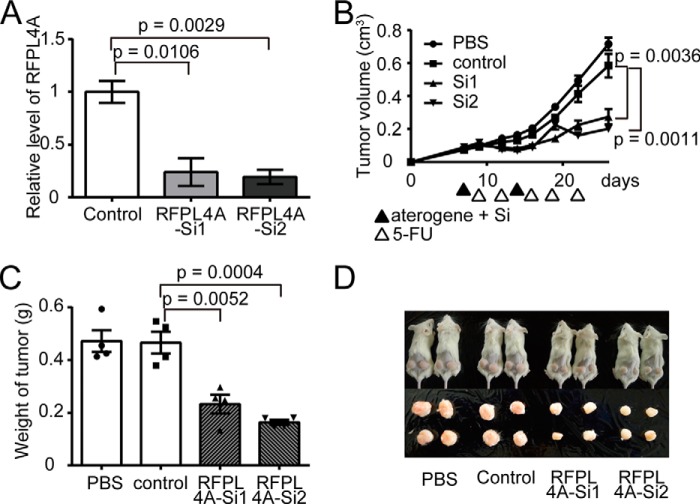
Thus, we have demonstrated that RFPL4A is a G0/G1 maintenance factor and endows cancer cells with a stemlike nature and reduced sensitivity to chemotherapy. Next, we examined the potential of this factor as a novel therapeutic target for treating chemotherapy-resistant cancers. Subcutaneously inoculated HCT116 tumors were subjected to local injection of siRNAs targeting RFPL4A or of a scrambled control siRNA (Fig. 8A). Injected siRNAs were preincubated with atelocollagen to facilitate their introduction into target cells (22, 23). The mice displaying inoculated tumors were then treated with 5-FU (30 mg/kg × 5 days). Treatment of RFPL4A-siRNAs concurrent with 5-FU reduced tumor development significantly (Fig. 8, B and D), suggesting that blocking RFPL4A may a promising adjuvant therapy for intractable cancers.
FIGURE 8.
RNAi-based inhibition of RFPL4A increased the sensitivity of HCT116 to chemotherapy in vivo. A, decreased expression of RFPL4A in HCT116 cells treated with siRNAs targeting RFPL4A (assessed by qPCR). Each bar represents the mean ± S.E. (error bars) of three independent experiments (n = 3). B, in vivo siRNA treatment of HCT116 tumors. Control HCT116 cells (5 × 106) were implanted in NOD/SCID mice and, on days 7 and 14, were treated with PBS, scrambled control siRNA plus atelocollagen, or two siRNAs (Si1 and Si2) against RFPL4A plus atelocollagen. NOD/SCID mice were administered intraperitoneal injection of 5-FU (30 mg/kg) at days 9, 12, 16, 19, and 22. Data represent the mean tumor volumes, calculated using the formula, (longest diameter) × (shortest diameter)2 × 0.5. The statistical significance of the difference between the control and RFPL4A-expressing HCT116 cells was determined by two-way ANOVA. Each bar represents the mean ± S.E. (error bars) of four individual experiments (n = 4 for each). C, comparison of the tumor weights after treatment with atelocollagen and 5-FU at 26 days. Each bar represents the mean ± S.E. (error bars) of four independent experiments (n = 4 for each). D, images of tumors excised on day 35.
DISCUSSION
Taking advantage of time lapse confocal microscopic imaging using a Fucci reporter system (14), we identified a characteristic population of cancer cells maintained long term in the G0/G1 phase, defined as the Fucci red-retained “RR” fraction. Because it possessed stem cell-like properties, such as high sphere-initiating capacity and low reactive oxygen species production, the RR fraction may be regarded as similar to CSCs. Additionally, in microarray analyses comparing the RR and R fractions (Fig. 3A and Table 5), we found dominant expression of several CSC signature molecules, such as FGF1, TGFβ1, snal2, and DLL3, supporting our theory that the RR fraction contains CSCs.
Here, we showed that RFPL4A was expressed preferentially in RR cells, the enriched fraction of CSC-like cells, in colorectal cancer cell lines. Expression of RFPL4A mRNA was found to be increased only in RR cells, with no differences between G and R cells, indicating that RFPL4A is up-regulated specifically in the G0 phase but not in the G1 phase. RFPL4A (Ret finger protein-like 4A) was first identified by an in silico screening for germ cell-specific genes (30) and is a unique RFPL family member of the RING finger-containing proteins that do not have a histidine in their RING finger motif (31). It has been demonstrated that RFPL4A targets cyclin B1 for proteasomal degradation in oocytes, a key process in meiosis (30, 32), suggesting a general role for this molecule in cell cycle control. This notion may be supported by the observation that preferential expression of RFPL4A in RR cells was detected not only in colorectal cancer cell lines, such as HCT116, HT29, and DLD1, but also in non-cancer cell lines, such as HEK293 cells (Fig. 3B). Some of the changes induced by RFPL4A up/down-regulation were relatively small, presumably because the stemlike property in RR states may be influenced by RFPL4A as well as several other factors (33, 34).
Many researchers have sought to develop novel cancer therapies targeting the supposed “dormant” cancer cells. For example, Fbxw7 has been shown to be required for the maintenance of quiescence in leukemia-initiating cells (9) (35), and Fbxw7-deficient leukemia-initiating cells are sensitive to anti-cancer drugs. Therefore, a combination therapy of Fbxw7 ablation and an anti-cancer drug was shown to be relatively effective for the eradication of leukemia-initiating cells, resulting in a survival advantage over treatment with anti-cancer drugs alone (9). The present data also indicate that inhibition of RFPL4A during chemotherapy eliminates colon cancer cells more effectively by preventing quiescence, compared with chemotherapy alone, a concept similar to Fbxw7-targeted therapy. Although RFPL4A was also expressed substantially in non-cancer cells, such as HEK293, and its inhibition might have some adverse effects, combination therapy using RFPL4A inhibition and conventional anti-cancer drugs may represent a promising therapeutic approach for intractable cancer patients in the future.
Acknowledgments
We are grateful to Drs. A. Miyawaki and H. Miyoshi (RIKEN) for providing expression plasmids and advice.
This work was funded by Grant-in-aid for Scientific Research on Innovative Areas 22113007 and by Grant-in-aid for Scientific Research (A) 25253070 from the Ministry of Education, Science, Sports and Culture of Japan; by International Human Frontier Science Program Grants RGY0077/2011; and by grants from the Takeda Science Foundation and Uehara Memorial Foundation.

This article contains supplemental Video 1.
- ROS
- reactive oxygen species
- CSC
- cancer stem cell
- 5-FU
- 5-fluoroacil
- ROC
- receiver operating characteristic
- Fucci
- fluorescence ubiquitin-based cell cycle indicator
- ANOVA
- analysis of variance
- qPCR
- quantitative PCR
- G fraction
- green-fluorescing S/G2/M fraction
- R fraction
- red-fluorescing G1 fraction
- RR fraction
- red-fluorescing retained fraction.
REFERENCES
- 1. Sherr C. J., McCormick F. (2002) The RB and p53 pathways in cancer. Cancer Cell 2, 103–112 [DOI] [PubMed] [Google Scholar]
- 2. Wahl G. M., Carr A. M. (2001) The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nat. Cell Biol. 3, E277–E286 [DOI] [PubMed] [Google Scholar]
- 3. Kastan M. B., Bartek J. (2004) Cell-cycle checkpoints and cancer. Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- 4. Shimada H., Nagata M., Cho A., Takiguchi N., Kainuma O., Soda H., Ikeda A., Nabeya Y., Yajima S., Yamamoto H., Sugiyama T., Itami M. (2014) Long-term monitoring of serum p53 antibody after neoadjuvant chemotherapy and surgery for esophageal adenocarcinoma: report of a case. Surg. Today 44, 1957–1961 [DOI] [PubMed] [Google Scholar]
- 5. Clevers H. (2011) The cancer stem cell: premises, promises and challenges. Nat. Med. 17, 313–319 [DOI] [PubMed] [Google Scholar]
- 6. Wicha M. S., Liu S., Dontu G. (2006) Cancer stem cells: an old idea: a paradigm shift. Cancer Res. 66, 1883–1890; discussion 1895–1886 [DOI] [PubMed] [Google Scholar]
- 7. Visvader J. E., Lindeman G. J. (2008) Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer 8, 755–768 [DOI] [PubMed] [Google Scholar]
- 8. Todaro M., Francipane M. G., Medema J. P., Stassi G. (2010) Colon cancer stem cells: promise of targeted therapy. Gastroenterology 138, 2151–2162 [DOI] [PubMed] [Google Scholar]
- 9. Takeishi S., Matsumoto A., Onoyama I., Naka K., Hirao A., Nakayama K. I. (2013) Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell 23, 347–361 [DOI] [PubMed] [Google Scholar]
- 10. Iwahashi S., Utsunomiya T., Shimada M., Saito Y., Morine Y., Imura S., Ikemoto T., Mori H., Hanaoka J., Bando Y. (2013) High expression of cancer stem cell markers in cholangiolocellular carcinoma. Surg. Today 43, 654–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyoshi H., Blömer U., Takahashi M., Gage F. H., Verma I. M. (1998) Development of a self-inactivating lentivirus vector. J. Virol. 72, 8150–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haraguchi N., Ishii H., Mimori K., Tanaka F., Ohkuma M., Kim H. M., Akita H., Takiuchi D., Hatano H., Nagano H., Barnard G. F., Doki Y., Mori M. (2010) CD13 is a therapeutic target in human liver cancer stem cells. J. Clin. Invest. 120, 3326–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Funato Y., Terabayashi T., Sakamoto R., Okuzaki D., Ichise H., Nojima H., Yoshida N., Miki H. (2010) Nucleoredoxin sustains Wnt/β-catenin signaling by retaining a pool of inactive dishevelled protein. Curr. Biol. 20, 1945–1952 [DOI] [PubMed] [Google Scholar]
- 14. Kagawa Y., Matsumoto S., Kamioka Y., Mimori K., Naito Y., Ishii T., Okuzaki D., Nishida N., Maeda S., Naito A., Kikuta J., Nishikawa K., Nishimura J., Haraguchi N., Takemasa I., Mizushima T., Ikeda M., Yamamoto H., Sekimoto M., Ishii H., Doki Y., Matsuda M., Kikuchi A., Mori M., Ishii M. (2013) Cell cycle-dependent Rho GTPase activity dynamically regulates cancer cell motility and invasion in vivo. PloS One 8, e83629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morita S., Kojima T., Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 16. Nishikawa K., Nakashima T., Hayashi M., Fukunaga T., Kato S., Kodama T., Takahashi S., Calame K., Takayanagi H. (2010) Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W., Gao X., Aldape K., Lu Z. (2011) Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bras-Gonçalves R. A., Pocard M., Formento J. L., Poirson-Bichat F., De Pinieux G., Pandrea I., Arvelo F., Ronco G., Villa P., Coquelle A., Milano G., Lesuffleur T., Dutrillaux B., Poupon M. F. (2001) Synergistic efficacy of 3n-butyrate and 5-fluorouracil in human colorectal cancer xenografts via modulation of DNA synthesis. Gastroenterology 120, 874–888 [DOI] [PubMed] [Google Scholar]
- 19. Sohn K. J., Croxford R., Yates Z., Lucock M., Kim Y. I. (2004) Effect of the methylenetetrahydrofolate reductase C677T polymorphism on chemosensitivity of colon and breast cancer cells to 5-fluorouracil and methotrexate. J. Natl. Cancer Inst. 96, 134–144 [DOI] [PubMed] [Google Scholar]
- 20. Wada H., Nagano H., Yamamoto H., Arai I., Ota H., Nakamura M., Damdinsuren B., Noda T., Marubashi S., Miyamoto A., Takeda Y., Umeshita K., Doki Y., Dono K., Nakamori S., Sakon M., Monden M. (2007) Combination therapy of interferon-α and 5-fluorouracil inhibits tumor angiogenesis in human hepatocellular carcinoma cells by regulating vascular endothelial growth factor and angiopoietins. Oncol. Rep. 18, 801–809 [PubMed] [Google Scholar]
- 21. Bras-Gonçalves R. A., Rosty C., Laurent-Puig P., Soulié P., Dutrillaux B., Poupon M. F. (2000) Sensitivity to CPT-11 of xenografted human colorectal cancers as a function of microsatellite instability and p53 status. Br. J. Cancer 82, 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takeshita F., Minakuchi Y., Nagahara S., Honma K., Sasaki H., Hirai K., Teratani T., Namatame N., Yamamoto Y., Hanai K., Kato T., Sano A., Ochiya T. (2005) Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 12177–12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Satow R., Shitashige M., Jigami T., Honda K., Ono M., Hirohashi S., Yamada T. (2010) Traf2- and Nck-interacting kinase is essential for canonical Wnt signaling in Xenopus axis formation. J. Biol. Chem. 285, 26289–26294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., Imamura T., Ogawa M., Masai H., Miyawaki A. (2008) Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 [DOI] [PubMed] [Google Scholar]
- 25. Brattain M. G., Fine W. D., Khaled F. M., Thompson J., Brattain D. E. (1981) Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 41, 1751–1756 [PubMed] [Google Scholar]
- 26. Hermann P. C., Huber S. L., Herrler T., Aicher A., Ellwart J. W., Guba M., Bruns C. J., Heeschen C. (2007) Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323 [DOI] [PubMed] [Google Scholar]
- 27. Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., Wicha M. S. (2003) In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. (2004) Identification of human brain tumour initiating cells. Nature 432, 396–401 [DOI] [PubMed] [Google Scholar]
- 29. Roccio M., Schmitter D., Knobloch M., Okawa Y., Sage D., Lutolf M. P. (2013) Predicting stem cell fate changes by differential cell cycle progression patterns. Development 140, 459–470 [DOI] [PubMed] [Google Scholar]
- 30. Rajkovic A., Lee J. H., Yan C., Matzuk M. M. (2002) The ret finger protein-like 4 gene, Rfpl4, encodes a putative E3 ubiquitin-protein ligase expressed in adult germ cells. Mech. Dev. 112, 173–177 [DOI] [PubMed] [Google Scholar]
- 31. Bonnefont J., Nikolaev S. I., Perrier A. L., Guo S., Cartier L., Sorce S., Laforge T., Aubry L., Khaitovich P., Peschanski M., Antonarakis S. E., Krause K. H. (2008) Evolutionary forces shape the human RFPL1,2,3 genes toward a role in neocortex development. Am. J. Hum. Genet. 83, 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Suzumori N., Burns K. H., Yan W., Matzuk M. M. (2003) RFPL4 interacts with oocyte proteins of the ubiquitin-proteasome degradation pathway. Proc. Natl. Acad. Sci. U.S.A. 100, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salcido C. D., Larochelle A., Taylor B. J., Dunbar C. E., Varticovski L. (2010) Molecular characterisation of side population cells with cancer stem cell-like characteristics in small-cell lung cancer. Br. J. Cancer 102, 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaminska B., Kocyk M., Kijewska M. (2013) TGF β signaling and its role in glioma pathogenesis. Adv. Exp. Med. Biol. 986, 171–187 [DOI] [PubMed] [Google Scholar]
- 35. Takeishi S., Nakayama K. I. (2014) Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br. J. Cancer 111, 1054–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]