Background: The six human SET1 family core complexes catalyze methylation of histone H3 lysine 4 (H3K4).
Results: Different SET1 family core complexes catalyze different levels of H3K4 methylation.
Conclusion: Product specificity of the SET1 family is correlated with evolutionary lineage.
Significance: Core complex subunits differentially regulate the product specificity of different SET1 family members.
Keywords: Cancer, Epigenetics, Histone Methylation, Leukemia, Phylogenetics, Ash2L, MLL, Product Specificity, SET1, WDR5
Abstract
Mixed lineage leukemia protein-1 (MLL1) is a member of the SET1 family of histone H3 lysine 4 (H3K4) methyltransferases that are required for metazoan development. MLL1 is the best characterized human SET1 family member, which includes MLL1–4 and SETd1A/B. MLL1 assembles with WDR5, RBBP5, ASH2L, DPY-30 (WRAD) to form the MLL1 core complex, which is required for H3K4 dimethylation and transcriptional activation. Because all SET1 family proteins interact with WRAD in vivo, it is hypothesized they are regulated by similar mechanisms. However, recent evidence suggests differences among family members that may reflect unique regulatory inputs in the cell. Missing is an understanding of the intrinsic enzymatic activities of different SET1 family complexes under standard conditions. In this investigation, we reconstituted each human SET1 family core complex and compared subunit assembly and enzymatic activities. We found that in the absence of WRAD, all but one SET domain catalyzes at least weak H3K4 monomethylation. In the presence of WRAD, all SET1 family members showed stimulated monomethyltransferase activity but differed in their di- and trimethylation activities. We found that these differences are correlated with evolutionary lineage, suggesting these enzyme complexes have evolved to accomplish unique tasks within metazoan genomes. To understand the structural basis for these differences, we employed a “phylogenetic scanning mutagenesis” assay and identified a cluster of amino acid substitutions that confer a WRAD-dependent gain-of-function dimethylation activity on complexes assembled with the MLL3 or Drosophila trithorax proteins. These results form the basis for understanding how WRAD differentially regulates SET1 family complexes in vivo.
Introduction
Methylation of histone H3 at lysine 4 (H3K4) is required for the epigenetic maintenance of transcriptionally active forms of chromatin in eukaryotes (1). H3K4 can be mono-, di-, or trimethylated, with each state associated with distinct genomic locations and functional outcomes (2). For example, H3K4 trimethylation (H3K4me3) is enriched in the promoters of active genes, whereas H3K4me2 is localized throughout open reading frames (3, 4). H3K4me1 is enriched in active enhancer elements (5) and is associated with gene silencing in lower eukaryotes (6–9). How cells achieve and maintain different H3K4 methylation states at distinct genomic regions is an important unsolved question in biology. This is particularly important given that genetic alteration of the genes that encode the enzymes that regulate H3K4 methylation are associated with a number of human developmental disorders and cancers (10–17).
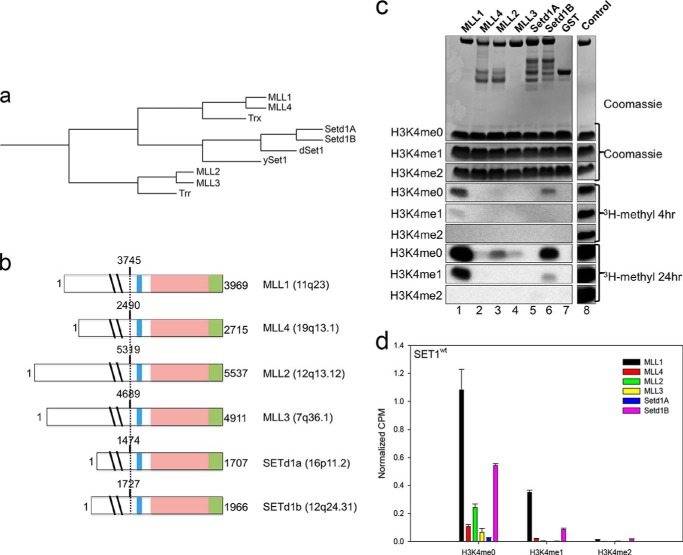
In humans, Mixed lineage leukemia protein-1 (MLL1, also known as HRX, ALL1, and KMT2A) catalyzes methyl group transfer from S-adenosylmethionine (AdoMet)2 to the ϵ-amino group of H3K4 (18), and it is frequently altered in poor-prognosis acute leukemias (19–21). MLL1 is a member of the SET1 family of H3K4 methyltransferases, which is conserved from yeast to humans. It has been suggested that in budding yeast Set1p is the sole H3K4 methyltransferase, as deletion of the set1 gene results in global reduction of H3K4 methylation (6). Drosophila melanogaster has three SET1 family members: Trithorax (Trx), trithorax-related (Trr), and dSet1 (22), whereas humans have six related SET1 family members: MLL1–4 (23–26) and SETd1A/B (27, 28). Phylogenetic analysis shows that the human SET1 family reduces to three distinct clades with MLL1 and MLL4 (also known as MLL2, Wbp7, and KMT2B) being most closely related to Trx, MLL2 (MLL4, ALR, and KMT2D) and MLL3 (KMT2C) being most closely related to Trr, and SETd1A and SETd1B being most closely related to Drosophila dSet1 and yeast Set1p (Fig. 1, a and b) (29, 30).
FIGURE 1.
Human SET1 family members predominantly catalyze monomethylation of H3K4. a, phylogenetic cluster analysis (Clustal Omega (66)) of SET1 family members using full-length protein sequences from Saccharomyces cerevisiae (ySet1), Drosophila (dSet1, Trx, and Trr), and humans (MLL1–4, SETd1A/B). b, schematic representation of full-length human SET1 family proteins. The catalytic SET domain is shown in light pink; the post-SET region is shown in green, and the WDR5 interaction (Win) motif is shown in blue. Dotted lines represent the N terminus of the recombinant constructs used in this study, beginning with the residues noted above each construct. c, comparison of histone methyltransferase activity among human SET1 family SET domains. Upper panels show Coomassie Blue-stained SDS-polyacrylamide gels, and the lower panels shows [3H]methyl incorporation by fluorography after a 4-h exposure (middle panels), and after a 24-h exposure (lowest panels). The control lane shows the activity of the MLL1 SET domain on 100 μm unmodified H3 peptide, which is included on each gel. The control lanes are from the same gel at the same exposure but were cropped for clarity. d, quantification of radioactivity from excised histone H3 bands by LSC. Data are normalized to the activity level of the control lane on each gel. Error bars represent the S.E. of measurement between three independent experiments.
SET1 family members share the properties that they all catalyze H3K4 methylation using the evolutionarily conserved suppressor of variegation, enhancer of zeste, trithorax (SET), domain (31), and all interact with an evolutionarily conserved subcomplex called WRAD (WD-40 repeat protein 5 (WDR5), Retinoblastoma-binding protein 5 (RBBP5), Absent small homeotic-2-like (ASH2L), and Dumpy-30 (DPY-30)) (32–35). MLL1 interacts with WRAD to form the MLL1 core complex, which is required for multiple H3K4 methylation in vitro and in vivo (32, 36). In vitro biochemical studies have shown that the isolated MLL1 SET domain catalyzes predominantly weak H3K4 monomethyltransferase activity (36). However, when in complex with WRAD, the rates of H3K4 mono- and dimethylation are markedly increased (36). The molecular mechanisms for how WRAD increases these activities are not well understood.
Although MLL1 has served as a paradigm for the mechanism of action of human SET1 family enzymes, several recent reports suggest different family members are controlled by distinct regulatory mechanisms. For example, although the MLL1 core complex has predominantly mono- and dimethyltransferase activity in vitro (36), a similar complex assembled with MLL3 shows only monomethylation activity (37, 38). Indeed, MLL2/3 complexes are linked to H3K4 monomethylation at active enhancers (39). In contrast, it has been suggested that SETd1A/B complexes catalyze the bulk of H3K4 trimethylation in cells (40, 41), but it is dependent on other factors that are unique to SETd1A/B complexes, such as WDR82 (Swd2 in yeast) (40, 41) and CFP1 (Spp1 in yeast) (8, 42). A further complication comes from the observation that histone H2B monoubiquitination is required for H3K4 trimethylation in a potential cross-talk mechanism (43–46). In yeast, this effect appears to be mediated by Spp1 and the N-SET domain of SET1p (42, 45), but it may also be a context-dependent phenomenon (47). It is unclear whether these enzymatic activities are intrinsic to SET1 family complexes or whether they are the result of direct or indirect regulation of SET1 family SET domains within cells. Lacking is a rigorous biochemical comparison of all human SET1 family core complexes under standard conditions.
In this investigation, we reconstituted each human SET1 family core complex from individual subunits and compared complex assembly and enzymatic activity using well defined standard in vitro assays. We found that in the absence of WRAD, all but one SET domain catalyzes at least weak H3K4 monomethylation. The SETd1A SET domain is inactive in the absence of WRAD, likely due to a disordered AdoMet binding pocket. We also found that all SET1 family SET domains assemble with WRAD but that the requirement for WDR5 for complex assembly and enzymatic activity differs among family members. In the presence of WRAD, all SET1 family SET domains showed stimulated monomethyltransferase activity but differed in their abilities to catalyze H3K4 di- and trimethylation. We found that these differences in product specificity are correlated with evolutionary lineage, suggesting these enzyme complexes have evolved to accomplish unique tasks within metazoan genomes. To begin to understand the structural basis for differences in product specificity, we employed a phylogenetic scanning mutagenesis assay and identified a nonactive site SET domain surface that is required for WRAD-dependent H3K4 dimethylation by SET1 family core complexes. These results form the basis for our understanding of how the incorporation of H3K4 methylation in mammalian genomes may be regulated.
EXPERIMENTAL PROCEDURES
Materials
WDR5 antibody was obtained from Abcam (ab22512). RBBP5 and ASH2L antibodies were obtained from Bethyl (A300-109A and A300-489A, respectively). An HRP-conjugated donkey anti-rabbit antibody was obtained from GE Healthcare. A polyclonal anti-GST antibody was obtained from GE Healthcare (27257701). Histone H3 peptides were synthesized by GenScript and contained residues 1–20 followed by GGK-biotin and were either unmodified, mono-methylated, or di-methylated at H3K4. All peptides were purified to greater than 95% purity. Furthermore, all peptides were blocked by amidation of the C terminus. MCF-7 cell extracts were obtained from Santa Cruz Biotechnology (sc-24793).
Protein Expression/Purification
Human SET1 family constructs consisting of residues MLL1(3745–3969) (UniProtKB ID Q03164), MLL2(5319–5537) (UniProtKB ID O14686), MLL3(4689–4911) (UniProtKB ID Q8NEZ4), MLL4(2490–2715) (UniProtKB ID Q9UMN6), SETd1A(1474–1708) (UniProtKB ID O15047), and SETd1B(1684–1924) (UniProtKB ID Q9UPS6) were subcloned into pGST parallel expression vectors (48), individually expressed in Escherichia coli (Rosetta 2 (DE3) pLysS; Novagen), then purified over a GSTrap-FF column (GE Healthcare), and eluted with a gradient of 0–10 mm reduced glutathione. Pooled fractions were dialyzed with three changes into Buffer 1 containing 50 mm Tris (pH 7.5), 300 mm NaCl, 10% glycerol, 3 mm dithiothreitol (DTT), and 1 μm ZnCl2. Full-length WRAD constructs in pHis parallel vectors (48) were individually expressed in E. coli (Rosetta 2 (DE3) pLysS; Novagen) and purified as described previously (49). WRAD components were further purified and buffer exchanged by gel filtration chromatography (Superdex 200, GE Healthcare) pre-equilibrated with 20 mm Tris (pH 7.5), 300 mm NaCl, 1 mm Tris(2-carboxyethyl)phosphine, and 1 μm ZnCl2 (Buffer 2). SET1 family mutants were prepared by subjecting DNA constructs to site-directed mutagenesis (QuikChange II XL, Agilent), expressed, and purified as described above.
Methyltransferase Activity Assays
Histone H3 methyltransferase assays were performed by incubating GST-tagged SET1 family members with a stoichiometric amount of WRAD (3 μm), 1 μCi of [3H]AdoMet (PerkinElmer Life Sciences), and 100 μm histone H3 peptides that were unmodified or previously mono- or di-methylated at H3K4. Reactions were incubated at 15 °C for 6 h. 15 °C was chosen as the incubation temperature due to SET domain instability at higher temperatures. Isolated SET1 family SET domains (5 μm) were assayed by incubating with 1 μCi of [3H]AdoMet (PerkinElmer Life Sciences) and 100 μm histone H3 peptides that were unmodified or previously mono- or di-methylated at H3K4. These reactions were incubated at 15 °C for 8 h. All reactions were quenched with SDS-loading buffer and separated by SDS-PAGE using a 4–12% BisTris gel (Invitrogen) run at 200 V for 30 min. The gels were enhanced for 30 min (Enlightning, PerkinElmer Life Sciences) and then dried for 2.5 h at 72 °C under constant vacuum. The dried gels were exposed to film (Kodak Biomax MS Film) for 4–24 h.
Liquid scintillation counting (LSC) was performed by excising bands corresponding to histone H3 peptides, which were dissolved in 750 μl of Solvable (PerkinElmer Life Sciences) and incubated at room temperature for 30 min followed by incubation at 50 °C for 3 h. The solubilized volume of each sample was transferred to liquid scintillation vials containing 10 ml of Ultima Gold XL liquid scintillation mixture (PerkinElmer Life Sciences). Samples were dark adapted for 1 h and then counted for 5 min each with a two-σ error cutoff using an all purpose scintillation counter (Beckman Coulter).
MALDI-TOF mass spectrometry methyltransferase assays were performed by incubating 7 μm SET1 family SET domain with 7 μm WRAD, 250 μm AdoMet (Cayman Chemicals), and 10 μm of H3(1–20) peptide (unmodified) at 15 °C for 24 h. The reactions were quenched with 0.5% TFA and then mixed 1:5 with α-cyano-4-hydroxycinnamic acid and shot on a Bruker Autoflex III mass spectrometer (State University of New York College of Environmental Sciences and Forestry, Syracuse, NY) in reflectron mode. Final shots were averaged from 200 shots per spot at five different positions. Relative methylation levels were quantitated using mMass (50). Reaction progress curves were globally fitted to irreversible consecutive reaction models using Dynafit (51).
[3H]AdoMet Cross-linking Assays
Purified GST-tagged SET1 family proteins (wild type and mutant) alone or assembled with WRAD (3 μm) were incubated with 1 μCi of [3H]AdoMet (PerkinElmer Life Sciences) at 15 °C for 3 h and then incubated on ice for 1 h. Half the volume of each sample was quenched with SDS loading buffer, and the other half was exposed to UV light (254 nm) in a Stratalinker oven at a distance of ∼15 cm for 30 min on ice. The UV-exposed samples were quenched with SDS loading buffer, and samples were separated by SDS-PAGE using a 4–12% BisTris gel (Invitrogen) run at 200 V for 30 min. The gels were enhanced for 30 min (Enlightning, PerkinElmer Life Sciences) and then dried for 2.5 h at 72 °C under constant vacuum. The dried gels were exposed to film (Kodak Biomax MS Film) for 3 days. Bands corresponding to SET domains were excised and counted by LSC as described above.
GST Pulldowns/Immunoblots
GST-tagged SET1 proteins were preincubated with a stoichiometric amount of purified WRAD components (3 μm) for 1 h at 4 °C before being added to pre-washed agarose beads coated with glutathione (Thermo Fisher) and incubated for an additional 2 h at 4 °C. The beads were washed three times with Buffer 2 supplemented with 0.05% Triton X-100 and 0.05% sodium deoxycholate. The complexes were eluted from the beads by boiling the samples at 95 °C in SDS-loading buffer for 10 min. Samples of the supernatant were run on a 4–12% BisTris gel (Invitrogen) and either stained with Coomassie Brilliant Blue or transferred to a PVDF membrane (Invitrogen) at 30 V for 1 h. PVDF membranes were blocked for 1 h with a 5% nonfat milk solution and then incubated with primary antibody (1:12,000) for 1 h at room temperature. Blots were washed four times and then incubated with an HRP-conjugated anti-rabbit secondary antibody (1:10,000) for 1 h at room temperature. Blots were washed an additional four times and then visualized by chemiluminescence (Clarity Western, Bio-Rad) on a Bio-Rad ChemiDoc MP Imager using the chemiluminescence setting.
For pulldown assays using cell extracts, a 3 μm concentration of GST-tagged SET domains was incubated with 100 μg of MCF-7 cell extracts for 16 h at 4 °C. Following the initial incubation, 20 μl of a 50:50 slurry of glutathione-agarose beads was added to each sample and incubated for an additional 2 h at 4 °C. Samples were washed three times in radioimmunoprecipitation (RIPA) buffer. Samples were eluted from beads by boiling the samples at 95 °C in SDS-loading buffer for 10 min. Samples of the supernatant were separated on a 4–12% BisTris gel (Invitrogen) and Western blotted as described above. Primary antibodies were used at a dilution of 1:2000, and the secondary antibody was used at a dilution of 1:5000.
Phylogenetic Scanning Mutagenesis Assays
Mutant and wild type constructs (GST-tagged) were expressed in E. coli (Rosetta 2 (DE3) pLysS; Novagen) in 5-ml cultures by inducing with 1 mm IPTG (MLL1/MLL3) or 750 μm IPTG (Trx) and growing for 24 h at 16 °C. Cells were harvested by centrifugation and lysed by resuspending pellets in Buffer 1 supplemented with a cOmplete protease inhibitor EDTA-free tablet (Roche Applied Science), 1× BugBuster (Novagen), and 0.25 mg/ml DNase A at 4 °C for 3 h with rotation. Lysates were harvested by centrifugation. The expression of each mutant was assessed by running samples of the lysate on a 4–12% BisTris gel (Invitrogen) and then Western blotted as described above. The primary α-GST antibody (GE Healthcare) was used at a dilution of 1:10,000 (MLL3 mutants) or 1:7500 (Trx mutants), and the secondary antibody (Jackson ImmunoResearch) was used at a dilution of 1:10,000.
Methyltransferase assays were performed by incubating each lysate with 3 μm WRAD, 250 μm H3 peptide (unmodified or mono-methylated), and 1 μCi (Trx) or 2 μCi (MLL3) [3H]AdoMet (PerkinElmer Life Sciences). Samples were incubated at 15 °C for 6 h (Trx) or 15 h (MLL3). The reactions were quenched with SDS-loading buffer and separated by SDS-PAGE using a 4–12% BisTris gel (Invitrogen) run at 200 V for 30 min. The gels were enhanced for 30 min (Enlightning, PerkinElmer Life Sciences) and then dried for 2.5 h at 72 °C under constant vacuum. The dried gels were exposed to film (Kodak Biomax MS Film) for 24 h (Trx) or 2 days (MLL3). Bands corresponding to H3 peptides were excised and counted by LSC as described above.
RESULTS
Human SET1 Family SET Domains Are Predominantly Monomethyltransferases
Previous structural studies have shown that the product specificity of SET domain enzymes depends on the presence of Phe or Tyr residue at a specific position in the SET domain active site, called the Phe/Tyr switch position (52–56). SET1 family SET domains all possess tyrosine at the switch position, which limits active site volume and predicts monomethyltransferase activity. We previously demonstrated that the isolated MLL1 SET domain is predominantly an H3K4 monomethyltransferase and that replacement of Tyr-3942 with Phe converts it into a processive trimethyltransferase (36). To determine whether the other members of the human SET1 family catalyze a similar degree of methylation, we purified each recombinant human SET1 family SET domain as a GST fusion protein from E. coli and compared histone methyltransferase activity and complex assembly in the presence and absence of WRAD. GST-SET domain constructs contained the SET and post-SET domains as well as the conserved WDR5 interaction (Win) motif (Fig. 1b), which we and others previously showed is crucial for MLL1 core complex assembly in vitro and in vivo (49, 57–59).
To determine the product specificity of the isolated human SET1 family constructs, we compared the enzymatic activity of each SET domain on histone H3 peptides that were unmodified or previously mono- or di-methylated at H3K4. When [3H]AdoMet and unmodified histone H3 peptide (H3K4me0) were incubated with each GST-SET domain protein, only MLL1 and SETd1B showed activity in the fluorogram after a 4-h exposure to film (Fig. 1c). After a 24-h exposure, weak activity was detected for MLL2, MLL3, and MLL4 but not the SETd1A protein (Fig. 1c). With the H3K4me0 peptide substrate, MLL1 showed the greatest amount of activity, followed by SETd1B, MLL2, MLL3, and MLL4 (Fig. 1d). A similar pattern of activity was observed upon liquid scintillation counting of excised peptide bands (Fig. 1d). When H3K4me1 was used as a substrate, weak activity was observed with the MLL1 and SETd1B enzymes (Fig. 1c). No activity was observed when H3K4me2 was the substrate. These results are consistent with previous experiments suggesting that SET1 family SET domains preferentially catalyze H3K4 monomethylation.
Human SET1 Family Core Complexes Display Different Product Specificities
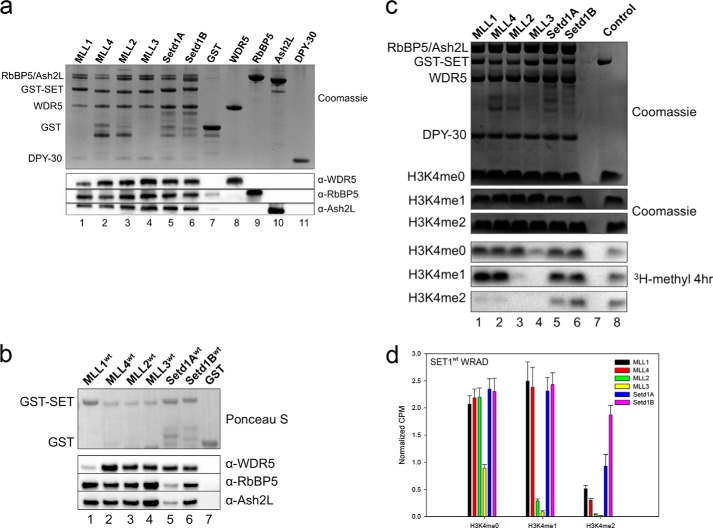
We previously showed that when MLL1 is assembled with WRAD under single turnover conditions, significant H3K4 mono- and dimethylation activities are observed with trace amounts of trimethylation after 24 h (36). To determine whether all human SET1 family core complexes show a similar product specificity, we compared complex formation and enzymatic activity among all six human SET1 family core complexes. GST pulldown experiments were used to compare the ability of each GST-SET domain to pull down recombinant WRAD subunits in vitro. As shown in Fig. 2a, all GST-SET1 family fusion proteins enriched all WRAD components (lanes 1–6), compared with the GST-only control (lane 7). Similarly, each GST-SET1 family fusion protein enriched endogenous WRAD subunits after incubation with MCF-7 cell extracts (Fig. 2b). These results demonstrate that all SET1 family SET domains form complexes with WRAD in vitro.
FIGURE 2.
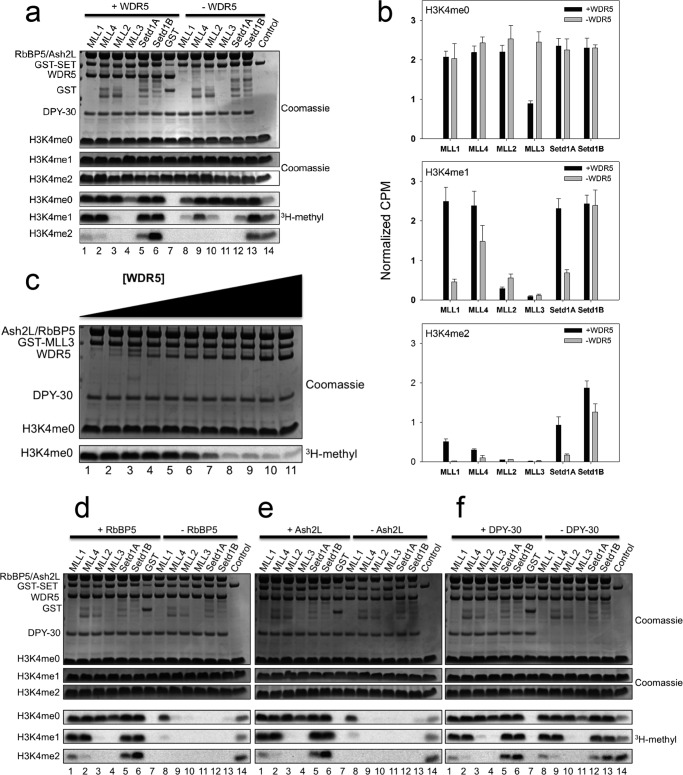
SET1 family core complexes catalyze different levels of H3K4 methylation. a, comparison of core complex assembly with each human SET1 family member by in vitro GST pulldown assays from purified components. Individual GST-tagged SET domains were incubated with purified WRAD components and glutathione-coated agarose beads. The upper panel shows a Coomassie Blue-stained gel, and the lower panels show the Western blot. Purified GST is used as a negative control (lane 7). Purified individual WRAD subunits were run on the gel (lanes 8–11) to compare with the migration of WRAD components from the pulldown lanes (lanes 1–8). b, comparison of core complex assembly with each human SET1 family member by in vitro pulldown experiments from MCF-7 breast cancer cell extracts. Individual GST-tagged SET domains were incubated with cell extracts and pulled down with glutathione-agarose beads. WRA components were detected by Western blotting. The upper panel shows a Ponceau S-stained PVDF membrane, and the lower panels show the Western blot. c, comparison of core complex methyltransferase activities among SET1 family members in complex with WRAD. The upper panels show Coomassie Blue-stained SDS-polyacrylamide gels, and the lower panels show [3H]methyl incorporation after 4 h as shown by fluorography. The control lane shows the activity of the MLL1 SET domain on 100 μm unmodified H3 peptide, which is included on each gel. d, quantification of radioactivity from excised histone H3 bands by LSC. Data are normalized to the activity level of the control lane on each gel. Error bars represent the S.E. of measurement between five independent experiments.
We next compared enzymatic activity of each complex using H3 peptides that were either unmodified or previously mono- or dimethylated at Lys-4. As shown in previous studies (36), the MLL1 core complex showed significant activity with H3K4me0 and H3K4me1 peptides, with trace amounts of activity with the H3K4me2 peptide (Fig. 2c, lane 1, and d). A similar activity profile was observed when GST-MLL4 was mixed with WRAD (Fig. 2c, lane 2). These results suggest that MLL1 and MLL4 core complexes are predominantly H3K4 mono- and dimethyltransferases. In contrast, the complexes assembled with the other human SET1 family SET domains showed striking differences. For example, although MLL2 and MLL3 complexes showed WRAD stimulated activity with H3K4me0 peptides, they showed little or no activity with H3K4me1 and H3K4me2 peptides (Fig. 2c, lanes 3 and 4). These results suggest that MLL2 and MLL3 core complexes are predominantly monomethyltransferases. In contrast, although we were not able to detect activity with SETd1A without WRAD (Fig. 1c), with WRAD it displayed activity with all three peptides (Fig. 2c, lane 5). A similar pattern was observed with the SETd1B enzyme (Fig. 2c, lane 6). These results suggest that SETd1A/B complexes catalyze mono-, di-, and trimethylation of H3K4.
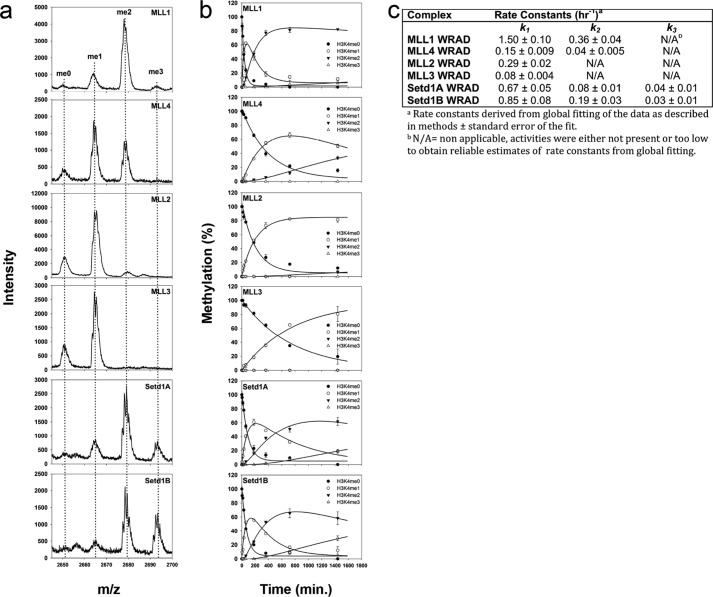
To confirm the observed product specificities using a different assay, we incubated each enzyme complex with the H3K4me0 peptide under single turnover conditions and monitored methylation over time by quantitative MALDI-TOF mass spectrometry. As shown in Fig. 3a (top panel), the MLL1 core complex converts most of the H3 peptide into the me2 form, with a small amount converted into the me3 form at 24 h. The complex assembled with MLL4 showed similar product specificity but with a slower overall rate (Fig. 3, a–c). MLL2 and MLL3 complexes, in contrast, showed predominantly monomethyltransferase activity after 24 h, whereas SETd1A/B complexes showed di- and trimethyltransferase activities (Fig. 3, a and b). For enzyme complexes catalyzing multiple methylation (MLL1, MLL4, and SETd1A/B), fitting of reaction progress curves to irreversible consecutive reactions models showed significant accumulation of the H3K4me1 and H3K4me2 intermediates (Fig. 3b), with rate constants for monomethylation (k1) that were ∼4–8-fold greater than that for dimethylation (k2) (Fig. 3, b and c). In addition, the rates constants for dimethylation by the SETd1A/B complexes were ∼2–6-fold greater than that for trimethylation (k3) (Fig. 3, b and c). These results are consistent with nonprocessive methylation.
FIGURE 3.
SET1 family core complex single turnover kinetics. a, MALDI-TOF mass spectrometry showing histone methylation of an unmodified H3 peptide after 24 h for each human SET1 family core complex. b, reaction progress curves globally fitted to irreversible consecutive reaction models using DynaFit. Each time point represents the mean percentage of total integrated area for each species in MALDI-TOF reactions. Error bars represent ± S.D. from duplicate measurements. c, summary of rate constants derived from global fitting of reaction progress curves as described under “Experimental Procedures.”
Taken together, these results are in agreement with that of the radiometric assays, and they suggest that different SET1 family core complexes catalyze different degrees of H3K4 methylation.
WRAD Increases AdoMet Binding in Several SET1 Family Members
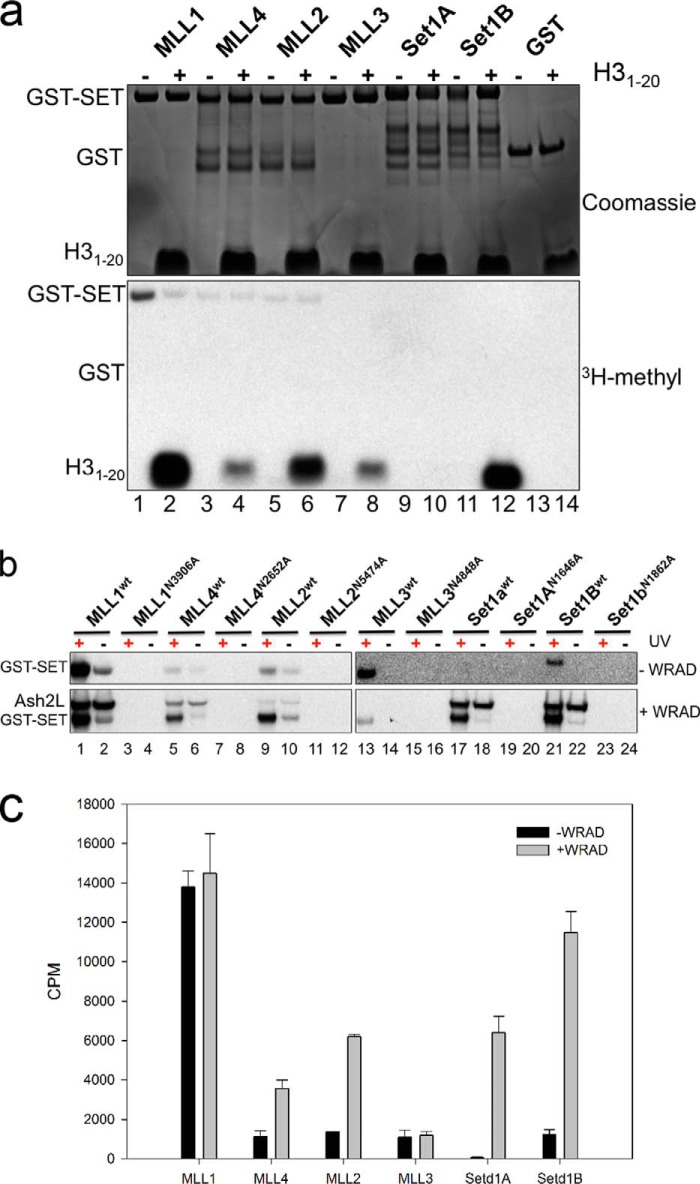
To begin to understand why WRAD increases the H3K4 monomethylation activity of all SET1 family complexes, we performed UV-dependent AdoMet cross-linking studies in the presence and absence of WRAD. A complicating feature of these studies is that we previously demonstrated that the isolated MLL1 SET domain undergoes a robust intramolecular automethylation activity in the absence of UV light (60). Indeed, in this investigation, we observed that the isolated MLL4 and MLL2 SET domains also undergo weaker automethylation reactions compared with that of MLL1 (Fig. 4a). In addition, when MLL1 is assembled with WRAD, we previously found that MLL1 methylates ASH2L in an intramolecular (intra-complex) manner (60). In this investigation, we observed a similar ASH2L methylation reaction in complexes assembled with MLL1, MLL4, MLL2, SETd1A, and SETd1B proteins in the absence of UV light (Fig. 4b, lower panels, lanes 2, 6, 10, 18, and 22, respectively). No ASH2L methylation was detected in the complex assembled with the MLL3 SET domain (Fig. 4b, lower panel, lane 14). Despite these activities, we found that UV-dependent cross-linking could be used to interrogate AdoMet binding in the presence and absence of WRAD. For example, all isolated GST-SET proteins except SETd1A showed an increase in radioactivity in the presence of UV light compared with that observed in the absence of UV light (Fig. 4b, upper panels). AdoMet cross-linking was abolished in each SET domain variant in which the AdoMet-binding asparagine (equivalent to N3906 in MLL1) was replaced with alanine (Fig. 4b, MLL1N3906A, MLL4N2652A, MLL2N5474A, MLL3N4848A, SETd1AN1646A, and SETd1BN1862A). These results suggest that all SET domain proteins can bind AdoMet, with the exception of the SETd1A protein.
FIGURE 4.
AdoMet cross-linking studies of the human SET1 family members. a, subset of SET1 family members undergoing auto-methylation. Isolated SET domains were incubated with [3H]AdoMet in the presence (+) or absence (−) of 250 μm unmodified H3 peptide. The upper panel shows a Coomassie Blue-stained SDS-polyacrylamide gel, and the lower panel shows [3H]methyl incorporation by fluorography after a 24-h exposure to film. b, UV exposure was used to cross-link [3H]AdoMet to isolated SET1 family SET domains (upper panels) or SET1 family core complexes (lower panels). All panels represent [3H]methyl incorporation by fluorography after a 3-day exposure to film. A single asparagine to alanine mutation in the AdoMet binding pocket of each SET domain abolishes [3H]AdoMet cross-linking. c, quantification of [3H]AdoMet cross-linking in the presence and absence of WRAD. Bands corresponding to the SET domains of each wild type SET1 family member were excised and quantified by liquid scintillation counting as described under “Experimental Procedures.” Error bars represent the S.E. of measurement from three independent experiments.
When similar assays were performed in the presence of WRAD, all SET domain proteins showed UV-dependent AdoMet cross-linking (Fig. 4b, lower panels). The SETd1A protein, which did not show evidence of cross-linking in the absence of WRAD, showed robust UV-dependent cross-linking in the presence of WRAD (Fig. 4, lane 17). These results suggest that WRAD rescues the AdoMet-binding defect on the GST-SETd1A protein. Indeed, WRAD significantly increased SET domain AdoMet cross-linking in MLL4, MLL2, SETd1A, and SETd1B complexes when excised bands were quantitated by liquid scintillation counting (Fig. 4c). In contrast, ASH2L methylation was unaffected in the presence or absence of UV light, indicating that it is the result of an enzymatic reaction that is catalyzed by the SET domain subunit, as suggested previously for MLL1 (60). The only complex that did not display ASH2L methylation was the MLL3 core complex (Fig. 4b, lane 15). These results suggest that WRAD induces a conformational change in SET1 family SET domains that results in increased binding of AdoMet for the majority of SET1 family members.
Functions of WRAD Subunits in Human SET1 Family Core Complexes
From the results presented above, WRAD stimulates H3K4 monomethylation in all SET1 family core complexes and is required for the di- and trimethylation activity of a subset of complexes. To further dissect the roles of WRAD subunits, we compared enzymatic activities in SET1 family complexes in which each WRAD subunit was systematically deleted. When the WDR5 subunit was omitted, we noticed that all SET1 family core complexes retained significant activity with the H3K4me0 substrate (Fig. 5a, lanes 8–13, and b). Interestingly, in the absence of WDR5, the MLL3 core complex displayed a significant stimulation in enzymatic activity with the H3K4me0 substrate (Fig. 5, compare lanes 4 and 11). These results suggest that WDR5 partially inhibits the H3K4 monomethyltransferase activity of the MLL3 core complex. To characterize WDR5 inhibition further, we titrated WDR5 into the MLL3 core complex and found that inhibition occurs only when the concentration of WDR5 approaches the concentration of the other subunits (Fig. 5c). Therefore, a near- or super-stoichiometric concentration of WDR5 partially inhibits the enzymatic activity of the MLL3 core complex.
FIGURE 5.
Functions of WRAD components in the SET1 family of core complexes. a, comparison of H3K4 methylation activity by SET1 family core complexes assembled with and without WDR5. The upper panels show Coomassie Blue-stained SDS-polyacrylamide gels, and the lower panels show [3H]methyl incorporation after a 4-h exposure to film. b, quantification of methyltransferase activity among SET1 family core complexes assembled with and without WDR5 by liquid scintillation counting. Error bars represent the S.E. of measurement from three to five independent experiments. c, titration of WDR5 into the MLL3-RAD complex. A 3 μm MLL3-RAD complex was assembled with increasing amounts of WRD5 and tested for methyltransferase activity when an unmodified H3 peptide was the substrate. WDR5 was titrated in 0.5 μm increments in lanes 1–9 and in 1 μm increments for lanes 10 and 11 (range is from 0 to 6 μm). d–f, comparison of H3K4 methylation activity by SET1 family core complexes assembled with and without RBBP5 (d), ASH2L (e), and DPY-30 (f). The upper panels show Coomassie Blue-stained SDS-polyacrylamide gels, and the lower panels show [3H]methyl incorporation after 4 h as shown by fluorography. All gels contain the activity of the isolated MLL1 SET domain on 100 μm unmodified H3 peptide as a control.
When the H3K4me1 or H3K4me2 peptides were used as substrates, significant differences among SET1 family members were observed upon omission of WDR5. H3K4 dimethylation activity is significantly reduced with MLL1 (Fig. 5a, compare lanes 1 and 8, and b) and SETd1A complexes (Fig. 5a, lanes 5 and 12). In contrast, little or no differences were observed with MLL4, MLL2, and SETd1B complexes (Fig. 5a and b). Despite stimulation of the monomethylation activity of the MLL3 complex upon omission of WDR5, little activity was observed when H3K4me1 or H3K4me2 peptides were used as substrates (Fig. 5a, lane 11). Taken together, these results suggest that MLL1 and Setd1A complexes depend on WDR5 for their complete di- and trimethylation activities. WDR5 appears to be somewhat dispensable for the enzymatic activity of the MLL2, MLL3, MLL4, and Setd1B complexes.
In contrast to that of WDR5, omission of RBBP5 or ASH2L subunits completely abolished WRAD stimulated effects on all human SET1 family core complexes (Fig. 5, d and e). The activity of each complex resembles that of the activity of the isolated SET domain in the absence of WRAD on all three substrates (Fig. 1c). These results suggest that RBBP5 and ASH2L are critical for the enzymatic activities of human SET1 family core complexes. Omission of DPY-30, conversely, did not significantly affect the activities of SET1 family core complexes on all three substrates (Fig. 5f).
Phylogenetic Scanning Mutagenesis Reveals SET Domain Surface Involved in WRAD-dependent Product Specificity Regulation
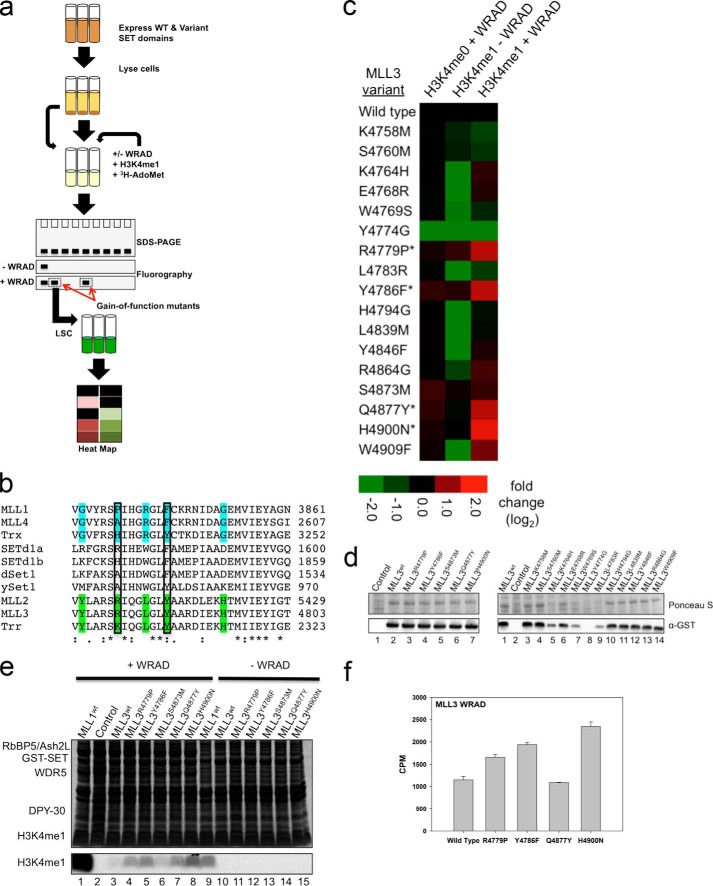
To begin to identify amino acids responsible for differences in product specificity among SET1 family complexes, we developed a high throughput “phylogenetic-scanning mutagenesis” assay, where systematic amino acid substitutions were made between SET domain paralogs or orthologs and assayed for gain-of-function activities. The assay works by expressing wild type or variant GST-SET domain proteins in 5-ml cultures of E. coli, followed by Western blotting with anti-GST antibodies to compare and normalize expression levels (Fig. 6a). The extracts were then incubated with [3H]AdoMet and the H3K4me0/1 peptide substrates and assayed directly for H3K4 dimethyltransferase activity in the presence and absence of purified WRAD. Gain-of-function substitutions are defined as those variants showing significantly increased H3K4 dimethylation activity (p ≤ 0.01) compared with that of the wild type enzyme.
FIGURE 6.
Phylogenetic scanning mutagenesis reveals a cluster of mutations that enhance the dimethylation activity of the MLL3 core complex. a, schematic of the phylogenetic scanning mutagenesis assay. Mutant constructs were expressed in 5-ml cultures, and lysates were analyzed for protein expression. Purified WRAD components along with [3H]AdoMet and H3 peptides (unmodified or previously mono-methylated at H3K4) were incubated with the lysates. Fluorography and liquid scintillation counting were used to analyze activity. b, representative sequence alignment of SET1 family SET domains (Clustal Omega). The MLL1 homologs are highlighted in blue, and the MLL3 homologs are highlighted in green. The two boxed positions represent gain-of-function hits from the screen. c, heat map of methyltransferase activities of wild type and mutant MLL3 constructs. The data represent the log2-fold change (mutant/wild type) in activity of MLL3 constructs with the indicated peptide. Asterisk represents significant increase in dimethylation activity (p < 0.01). d, assessment of the expression level of mutant MLL3 constructs. Lysate samples from mutant and wild type constructs were separated by SDS-PAGE, transferred to PVDF membranes, and blotted with an α-GST antibody. The upper panels show Ponceau S-stained PVDF membranes, and the lower panels show the Western blots. The control represents an untransformed E. coli lysate that was induced with 1 mm IPTG. e, representative gel of gain-of-function MLL3 mutants. The upper panel depicts a Coomassie Blue-stained SDS-polyacrylamide gel, and the lower panel shows [3H]methyl incorporation after 2 days as shown by fluorography. f, UV exposure was used to cross-link [3H]AdoMet to wild type or MLL3 variant core complexes. [3H]AdoMet cross-linking was quantitated by LSC of excised SET domain bands. Error bars represent S.E. of measurement from three independent experiments.
We used phylogenetic scanning mutagenesis to identify residues that confer a gain-of-function dimethyltransferase activity on the MLL3 core complex, which is predominantly an H3K4 monomethyltransferase (Fig. 2c). We performed a multiple sequence alignment with MLL1 and MLL3 orthologs and identified at least 17 positions that were conserved or semi-conserved within each phylogenetic clade but different between clades (Fig. 6b). We then systematically replaced each position in MLL3 with the corresponding amino acid in MLL1 and assayed for enzymatic activity using the H3K4me0 and H3K4me1 peptides as substrates. The resulting heat map shows that when H3K4me0 was the substrate there was very little variation in activity among MLL3 variants with the exception of Y4774G, which was significantly reduced compared with that of the complex assembled with wild type MLL3 (Fig. 6c). Indeed Western blotting revealed that the Y4774G GST fusion protein did not express in E. coli, explaining why no activity was observed (Fig. 6d, right panel, lane 8). In contrast, when H3K4me1 was used as the substrate, four of the 17 variants (R4779P, Y4786F, Q4877Y, and H4900N) showed a modest WRAD-dependent increase in dimethylation activity compared with that of wild type MLL3 (Fig. 6, c and e). These results cannot be explained by differences in expression levels as Western blotting revealed that each gain-of-function variant expressed at a level similar to that of wild type MLL3 in E. coli (Fig. 6d, left panel). In addition, there was little variation in activity when the H3K4me0 peptide was used as substrate, suggesting that similar amounts of folded protein were assayed (Fig. 6c).
To determine whether differences in AdoMet binding account for the gain-of-function activities, we purified the gain-of-function variants and compared AdoMet UV cross-linking in the presence of WRAD with that of the wild type MLL3 core complex. The results showed little changes in AdoMet cross-linking (0.9–2-fold) suggesting that differences in AdoMet binding alone do not account for increased dimethylation activity (Fig. 6f).
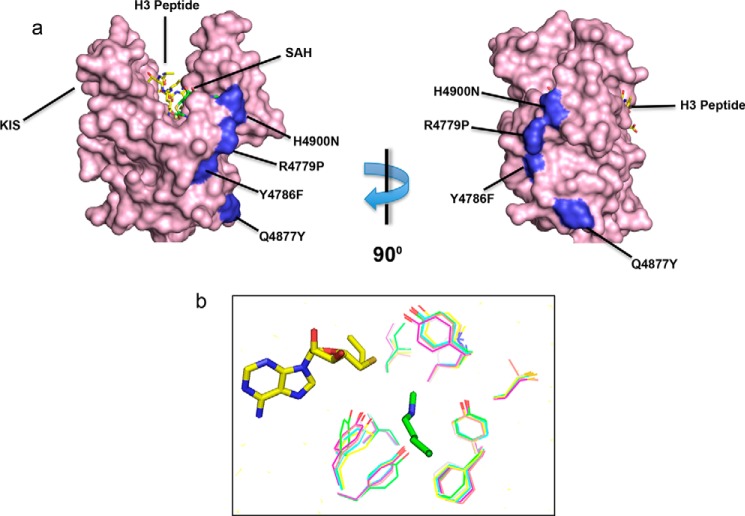
We then mapped the gain-of-function mutations on the three-dimensional structure of the MLL1 SET domain, and we noticed that all gain-of-function amino acid positions cluster around a solvent-exposed concave surface that is distinct from the SET domain active site cleft (Fig. 7). Modeling reveals that each gain-of-function position tolerates multiple rotomers of each mutant side chain without steric clash, making it likely that the mutations do not significantly alter the overall structure of the SET domain. These results suggest that this surface is involved in the WRAD-dependent dimethylation activity of the MLL1 core complex.
FIGURE 7.
a, surface representation of the crystal structure of the MLL1 SET domain bound to histone H3 peptide (yellow) and S-adenosyl-l-homocysteine (SAH) (green) (Protein Data Bank code 2W5Z) (67). The position of the four gain-of-function mutants are highlighted in blue and noted with their MLL3 numbering. The location of the previously identified Kabuki interaction surface (KIS) is noted. b, homology modeling of the conserved active site residues in the human SET1 family predicts that they adopt similar three-dimensional positions. Models were generated in Modeler (68) using the MLL1 structure (PDB code 2W5Z) as a template.
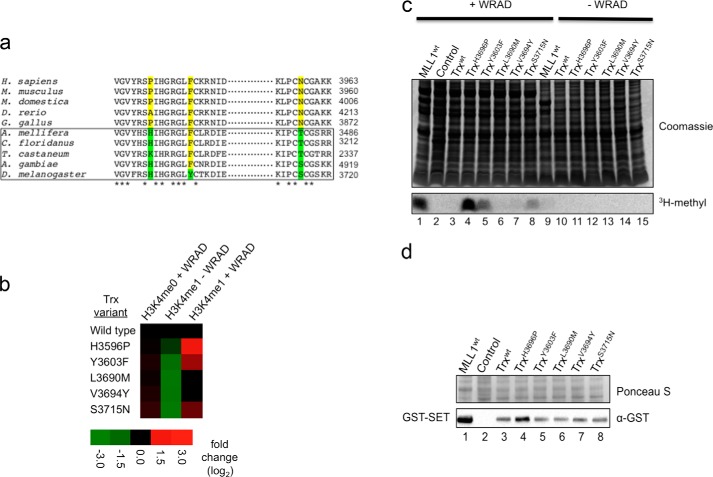
We next asked whether phylogenetic scanning mutagenesis could be used to determine whether variation at the same amino acid positions can explain why the Drosophila Trx ortholog of MLL1 does not catalyze H3K4 dimethylation when mixed with human WRAD. We previously found that like the human MLL1 SET domain, the Trx SET domain forms a complex with human WRAD and displays stimulated monomethyltransferase activity (61). However, unlike that of MLL1, Trx does not catalyze appreciable H3K4 dimethylation when assembled with human WRAD (61). Interestingly, several of the gain-of-function amino acid positions identified in the MLL3 screen showed an evolutionary split between vertebrate and invertebrate MLL1 orthologs (which includes Trx) (Fig. 8a). We replaced the four gain-of-function and one non gain-of-function amino acid positions in the Trx SET domain with the corresponding amino acids in MLL1 and assayed for H3K4 dimethyltransferase activity in the presence and absence of WRAD. The results revealed that three of the four amino acid substitutions showed a WRAD-dependent gain-of-function dimethylation activity in the Trx SET domain (Fig. 8, b–d). These results suggest that this surface is involved in the WRAD-dependent product specificity differences among SET1 family core complexes.
FIGURE 8.
Gain-of-function positions identified in MLL3 enhance WRAD-dependent dimethylation by the complex assembled with Drosophila Trx. a, representative alignment of MLL1 orthologs reveals amino acid positions that show a split between invertebrates (boxed) and vertebrates. b, heat map of methyltransferase activities of wild type and mutant Trx constructs. The data represent the log2-fold change (mutant/wild type) in activity of Trx constructs with the indicated peptide. c, representative gel of gain-of-function Trx mutants. The upper panel shows a Coomassie Blue-stained SDS-polyacrylamide gel, and the lower panel shows [3H]methyl incorporation after 24 h of fluorography. d, assessment of the expression level of mutant Trx constructs. Lysate samples from mutant and wild type constructs were separated by SDS-PAGE, transferred to PVDF membranes, and blotted with an α-GST antibody. The upper panel shows a Ponceau S-stained PVDF membrane, and the lower panel shows the Western blot. The control represents an untransformed E. coli lysate that was induced with 0.75 mm IPTG.
DISCUSSION
In this investigation, we have reconstituted and characterized the biochemical properties of all six human SET1 family core complexes under standard conditions in vitro. We found that all but one SET1 family SET domain catalyzes predominantly H3K4 monomethylation, which is stimulated by interaction with WRAD. In addition, we found that the SET domains interact with WRAD with similar stoichiometries in vitro but that the requirement for individual WRAD subunits for enzymatic activity differed. Although all SET1 family complexes require RBBP5 and ASH2L for full activity, the MLL3 core complex does not require WDR5 for enzymatic activity, and it appears to be inhibited by WDR5 when in stoichiometric excess. We found that WDR5 is required for the assembly of MLL1 and SETd1A core complexes but not for the assembly of MLL2–4 and SETd1B. Interestingly, these differences are inversely correlated with the differences in affinity observed between WDR5 and Win motif peptides derived from each SET1 family member. Paradoxically, MLL1 and SETd1A Win motif peptide sequences bind to WDR5 with the weakest affinity (0.5–2 μm compared with 30–100 nm for MLL2–4, SET1d1b) (57) yet require WDR5 for complex formation and multiple H3K4 methylation. Understanding the structural and functional basis for these differences will be important for translational efforts to target individual SET1 family members in diseased cells. Indeed, it has been shown that the MLL1-WDR5 interaction can be targeted for inhibition by peptides and small molecules in vitro and in mammalian cells (38, 49, 57). Even though WDR5 is not required for the enzymatic activity of all SET1 family complexes, it has been retained in each complex for some additional reason, possibly for gene targeting. For example, it has been shown that WDR5 binds to a long noncoding RNA called HOTTIP and is involved in recruitment of MLL family complexes to specific genomic loci (62, 63).
The major difference we observed among SET1 family complexes is that product specificities varied in a manner correlated with evolutionary lineage, which is surprising given their high degree of amino acid conservation. Because WRAD subunits were identical, these results indicate that SET domain amino acid sequence variation accounts for the observed product specificities. However, SET domain active site residues are strictly conserved and are predicted to adopt similar positions in three-dimensional models (Fig. 7b), implying that product specificity differences are due to amino acid sequence variation outside of the active site cleft. Indeed, we recently described the characterization of a nonactive site SET domain surface that is mutated in MLL2 in human Kabuki syndrome and in non-Hodgkin lymphomas (64). We found that this surface, called the Kabuki interaction surface, is required for the interaction of MLL1 with the RBBP5-ASH2L heterodimer and for H3K4 dimethylation by the MLL1 core complex (64). However, these residues are strictly conserved among SET1 family members and therefore cannot be responsible for the product specificity differences observed in this investigation.
We therefore developed a high throughput phylogenetic scanning mutagenesis assay to systematically switch amino acid residues between orthologs and paralogs, and we found a group of four amino acid positions that cluster on a common surface and appear to be involved in the H3K4 dimethylation reaction of the MLL1 core complexes in a WRAD-dependent manner. Interestingly, these amino positions are highly conserved among vertebrate MLL1 orthologs but are different among invertebrate orthologs. These residues map to a solvent-exposed surface that is opposite the Kabuki interaction surface cluster and distinct from the SET domain active site cleft. We propose that amino acid differences in this SET domain surface result in subtly different interactions with WRAD and “fine-tunes” the number of methyl groups that can be transferred to the histone substrate. The molecular mechanisms for how this works are unknown. One possibility is that different interactions between each SET domain and WRAD cause different allosteric alterations of SET domain active site residues, resulting in different product specificities. This mechanism is consistent with the “one-active site” model for multiple methylation, where mono-, di-, and trimethylation all occur within the SET domain active site. However, it is also possible that the amino acid sequence variation alters a surface that is involved in formation of a second active site, one that is created at the interface between WRAD and a nonactive site SET domain surface (36). This so-called “two-active site” model is supported by the demonstration that WRAD possesses an intrinsic H3K4 methyltransferase activity that is independent of the enzymatic activity of the MLL1 SET domain (36, 65). Further experiments will be required to distinguish between these mechanisms.
In summary, we have established that different SET1 family complexes catalyze varying degrees of H3K4 methylation when compared under identical conditions in vitro. These differences appear to be due in part to amino acid variation outside of the SET domain active site cleft. We suggest that variation in this SET domain surface regulates product specificity either through allosteric changes in SET domain active site residues or through modulation of the activity of a second active site. This information will facilitate future experiments that will distinguish these possibilities. In addition, this work forms the basis for understanding how additional regulatory inputs alter the enzymatic activities of human SET1 family core complexes in cells.
Acknowledgments
We thank Dr. David Skalnik for the Setd1A and Setd1B plasmids and Dr. Peter Harte for the Trx plasmid. We are grateful to Vicky Lyle at the DNA Sequencing Core Facility at SUNY Upstate Medical University. We thank Dr. Dave Amberg and Dr. Brian Haarer for the anti-GST and HRP-conjugated anti-goat antibody. In addition, we thank Dr. Nilda Alicea-Velazquez and Kevin Namitz for helpful comments on this manuscript. We also thank Dr. Nilda Alicea-Velazquez for technical assistance with the UV cross-linking experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01CA140522 (to M. S. C.).
- AdoMet
- S-adenosylmethionine
- Trx
- trithorax
- Trr
- trithorax-related
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- SET
- suppressor of variegation, enhancer of zeste, trithorax
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- LSC
- liquid scintillation counting.
REFERENCES
- 1. Strahl B. D., Ohba R., Cook R. G., Allis C. D. (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 96, 14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 3. Liu C. L., Kaplan T., Kim M., Buratowski S., Schreiber S. L., Friedman N., Rando O. J. (2005) Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3, e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 5. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 6. Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., Dent S. Y., Winston F., Allis C. D. (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nislow C., Ray E., Pillus L. (1997) SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8, 2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schneider J., Wood A., Lee J. S., Schuster R., Dueker J., Maguire C., Swanson S. K., Florens L., Washburn M. P., Shilatifard A. (2005) Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell 19, 849–856 [DOI] [PubMed] [Google Scholar]
- 9. van Dijk K., Marley K. E., Jeong B. R., Xu J., Hesson J., Cerny R. L., Waterborg J. H., Cerutti H. (2005) Monomethyl histone H3 lysine 4 as an epigenetic mark for silenced euchromatin in Chlamydomonas. Plant Cell 17, 2439–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morin R. D., Mendez-Lago M., Mungall A. J., Goya R., Mungall K. L., Corbett R. D., Johnson N. A., Severson T. M., Chiu R., Field M., Jackman S., Krzywinski M., Scott D. W., Trinh D. L., Tamura-Wells J., Li S., Firme M. R., Rogic S., Griffith M., Chan S., Yakovenko O., Meyer I. M., Zhao E. Y., Smailus D., Moksa M., Chittaranjan S., Rimsza L., Brooks-Wilson A., Spinelli J. J., Ben-Neriah S., Meissner B., Woolcock B., Boyle M., McDonald H., Tam A., Zhao Y., Delaney A., Zeng T., Tse K., Butterfield Y., Birol I., Holt R., Schein J., Horsman D. E., Moore R., Jones S. J., Connors J. M., Hirst M., Gascoyne R. D., Marra M. A. (2011) Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ng S. B., Bigham A. W., Buckingham K. J., Hannibal M. C., McMillin M. J., Gildersleeve H. I., Beck A. E., Tabor H. K., Cooper G. M., Mefford H. C., Lee C., Turner E. H., Smith J. D., Rieder M. J., Yoshiura K., Matsumoto N., Ohta T., Niikawa N., Nickerson D. A., Bamshad M. J., Shendure J. (2010) Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 790–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banka S., Howard E., Bunstone S., Chandler K. E., Kerr B., Lachlan K., McKee S., Mehta S. G., Tavares A. L., Tolmie J., Donnai D. (2013) MLL2 mosaic mutations and intragenic deletion-duplications in patients with Kabuki syndrome. Clin. Genet. 83, 467–471 [DOI] [PubMed] [Google Scholar]
- 13. Jones W. D., Dafou D., McEntagart M., Woollard W. J., Elmslie F. V., Holder-Espinasse M., Irving M., Saggar A. K., Smithson S., Trembath R. C., Deshpande C., Simpson M. A. (2012) De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 91, 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kleefstra T., Kramer J. M., Neveling K., Willemsen M. H., Koemans T. S., Vissers L. E., Wissink-Lindhout W., Fenckova M., van den Akker W. M., Kasri N. N., Nillesen W. M., Prescott T., Clark R. D., Devriendt K., van Reeuwijk J., de Brouwer A. P., Gilissen C., Zhou H., Brunner H. G., Veltman J. A., Schenck A., van Bokhoven H. (2012) Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am. J. Hum. Genet. 91, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pasqualucci L., Trifonov V., Fabbri G., Ma J., Rossi D., Chiarenza A., Wells V. A., Grunn A., Messina M., Elliot O., Chan J., Bhagat G., Chadburn A., Gaidano G., Mullighan C. G., Rabadan R., Dalla-Favera R. (2011) Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43, 830–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pugh T. J., Weeraratne S. D., Archer T. C., Pomeranz Krummel D. A., Auclair D., Bochicchio J., Carneiro M. O., Carter S. L., Cibulskis K., Erlich R. L., Greulich H., Lawrence M. S., Lennon N. J., McKenna A., Meldrim J., Ramos A. H., Ross M. G., Russ C., Shefler E., Sivachenko A., Sogoloff B., Stojanov P., Tamayo P., Mesirov J. P., Amani V., Teider N., Sengupta S., Francois J. P., Northcott P. A., Taylor M. D., Yu F., Crabtree G. R., Kautzman A. G., Gabriel S. B., Getz G., Jäger N., Jones D. T., Lichter P., Pfister S. M., Roberts T. M., Meyerson M., Pomeroy S. L., Cho Y. J. (2012) Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones D. T., Jäger N., Kool M., Zichner T., Hutter B., Sultan M., Cho Y. J., Pugh T. J., Hovestadt V., Stütz A. M., Rausch T., Warnatz H. J., Ryzhova M., Bender S., Sturm D., Pleier S., Cin H., Pfaff E., Sieber L., Wittmann A., Remke M., Witt H., Hutter S., Tzaridis T., Weischenfeldt J., Raeder B., Avci M., Amstislavskiy V., Zapatka M., Weber U. D., Wang Q., Lasitschka B., Bartholomae C. C., Schmidt M., von Kalle C., Ast V., Lawerenz C., Eils J., Kabbe R., Benes V., van Sluis P., Koster J., Volckmann R., Shih D., Betts M. J., Russell R. B., Coco S., Tonini G. P., Schüller U., Hans V., Graf N., Kim Y. J., Monoranu C., Roggendorf W., Unterberg A., Herold-Mende C., Milde T., Külozik A. E., von Deimling A., Witt O., Maass E., Rössler J., Ebinger M., Schuhmann M. U., Fruhwald M. C., Hasselblatt M., Jabado N., Rutkowski S., von Bueren A. O., Williamson D., Clifford S. C., McCabe M. G., Collins V. P., Wolf S., Wiemann S., Lehrach H., Brors B., Scheurlen W., Felsberg J., Reifenberger G., Northcott P. A., Taylor M. D., Meyerson M., Pomeroy S. L., Yaspo M. L., Korbel J. O., Korshunov A., Eils R., Pfister S. M., Lichter P. (2012) Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Milne T. A., Briggs S. D., Brock H. W., Martin M. E., Gibbs D., Allis C. D., Hess J. L. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 19. Djabali M., Selleri L., Parry P., Bower M., Young B. D., Evans G. A. (1992) A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat. Genet. 2, 113–118 [DOI] [PubMed] [Google Scholar]
- 20. Tkachuk D. C., Kohler S., Cleary M. L. (1992) Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71, 691–700 [DOI] [PubMed] [Google Scholar]
- 21. Ziemin-van der Poel S., McCabe N. R., Gill H. J., Espinosa R., 3rd, Patel Y., Harden A., Rubinelli P., Smith S. D., LeBeau M. M., Rowley J. D. (1991) Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl. Acad. Sci. U.S.A. 88, 10735–10739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eissenberg J. C., Shilatifard A. (2010) Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura T., Mori T., Tada S., Krajewski W., Rozovskaia T., Wassell R., Dubois G., Mazo A., Croce C. M., Canaani E. (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 24. Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L., Roeder R. G. (2005) Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121, 873–885 [DOI] [PubMed] [Google Scholar]
- 25. Hughes C. M., Rozenblatt-Rosen O., Milne T. A., Copeland T. D., Levine S. S., Lee J. C., Hayes D. N., Shanmugam K. S., Bhattacharjee A., Biondi C. A., Kay G. F., Hayward N. K., Hess J. L., Meyerson M. (2004) Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell 13, 587–597 [DOI] [PubMed] [Google Scholar]
- 26. Wysocka J., Myers M. P., Laherty C. D., Eisenman R. N., Herr W. (2003) Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17, 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee J. H., Skalnik D. G. (2005) CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 280, 41725–41731 [DOI] [PubMed] [Google Scholar]
- 28. Lee J. H., Tate C. M., You J. S., Skalnik D. G. (2007) Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem. 282, 13419–13428 [DOI] [PubMed] [Google Scholar]
- 29. Hubert A., Henderson J. M., Ross K. G., Cowles M. W., Torres J., Zayas R. M. (2013) Epigenetic regulation of planarian stem cells by the SET1/MLL family of histone methyltransferases. Epigenetics 8, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu H., Min J., Lunin V. V., Antoshenko T., Dombrovski L., Zeng H., Allali-Hassani A., Campagna-Slater V., Vedadi M., Arrowsmith C. H., Plotnikov A. N., Schapira M. (2010) Structural biology of human H3K9 methyltransferases. PLoS One 5, e8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z. W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D., Jenuwein T. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 [DOI] [PubMed] [Google Scholar]
- 32. Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D., Roeder R. G. (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713–719 [DOI] [PubMed] [Google Scholar]
- 33. Steward M. M., Lee J. S., O'Donovan A., Wyatt M., Bernstein B. E., Shilatifard A. (2006) Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 13, 852–854 [DOI] [PubMed] [Google Scholar]
- 34. Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K. (2007) PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282, 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Nuland R., Smits A. H., Pallaki P., Jansen P. W., Vermeulen M., Timmers H. T. (2013) Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell. Biol. 33, 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel A., Dharmarajan V., Vought V. E., Cosgrove M. S. (2009) On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 284, 24242–24256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu L., Lee S. Y., Zhou B., Nguyen U. T., Muir T. W., Tan S., Dou Y. (2013) ASH2L regulates ubiquitylation signaling to MLL: trans-regulation of H3 K4 methylation in higher eukaryotes. Mol. Cell 49, 1108–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao F., Townsend E. C., Karatas H., Xu J., Li L., Lee S., Liu L., Chen Y., Ouillette P., Zhu J., Hess J. L., Atadja P., Lei M., Qin Z. S., Malek S., Wang S., Dou Y. (2014) Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell 53, 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu D., Gao X., Morgan M. A., Herz H. M., Smith E. R., Shilatifard A. (2013) The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu M., Wang P. F., Lee J. S., Martin-Brown S., Florens L., Washburn M., Shilatifard A. (2008) Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 28, 7337–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J. H., Skalnik D. G. (2008) Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol. Cell. Biol. 28, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takahashi Y. H., Lee J. S., Swanson S. K., Saraf A., Florens L., Washburn M. P., Trievel R. C., Shilatifard A. (2009) Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol. Cell. Biol. 29, 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., Shilatifard A. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277, 28368–28371 [DOI] [PubMed] [Google Scholar]
- 44. Lee J. S., Shukla A., Schneider J., Swanson S. K., Washburn M. P., Florens L., Bhaumik S. R., Shilatifard A. (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131, 1084–1096 [DOI] [PubMed] [Google Scholar]
- 45. Kim J., Kim J. A., McGinty R. K., Nguyen U. T., Muir T. W., Allis C. D., Roeder R. G. (2013) The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol. Cell 49, 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun Z. W., Allis C. D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 47. Thornton J. L., Westfield G. H., Takahashi Y. H., Cook M., Gao X., Woodfin A. R., Lee J. S., Morgan M. A., Jackson J., Smith E. R., Couture J. F., Skiniotis G., Shilatifard A. (2014) Context dependency of Set1/COMPASS-mediated histone H3 Lys4 trimethylation. Genes Dev. 28, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sheffield P., Garrard S., Derewenda Z. (1999) Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 49. Patel A., Vought V. E., Dharmarajan V., Cosgrove M. S. (2008) A conserved arginine containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J. Biol. Chem. 283, 32162–32175 [DOI] [PubMed] [Google Scholar]
- 50. Niedermeyer T. H., Strohalm M. (2012) mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS One 7, e44913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 52. Qian C., Wang X., Manzur K., Sachchidanand, Farooq A., Zeng L., Wang R., Zhou M. M. (2006) Structural insights of the specificity and catalysis of a viral histone H3 lysine 27 methyltransferase. J. Mol. Biol. 359, 86–96 [DOI] [PubMed] [Google Scholar]
- 53. Trievel R. C., Flynn E. M., Houtz R. L., Hurley J. H. (2003) Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat. Struct. Biol. 10, 545–552 [DOI] [PubMed] [Google Scholar]
- 54. Xiao B., Jing C., Kelly G., Walker P. A., Muskett F. W., Frenkiel T. A., Martin S. R., Sarma K., Reinberg D., Gamblin S. J., Wilson J. R. (2005) Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 19, 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiao B., Jing C., Wilson J. R., Walker P. A., Vasisht N., Kelly G., Howell S., Taylor I. A., Blackburn G. M., Gamblin S. J. (2003) Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421, 652–656 [DOI] [PubMed] [Google Scholar]
- 56. Zhang X., Yang Z., Khan S. I., Horton J. R., Tamaru H., Selker E. U., Cheng X. (2003) Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell 12, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dharmarajan V., Lee J. H., Patel A., Skalnik D. G., Cosgrove M. S. (2012) Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J. Biol. Chem. 287, 27275–27289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Patel A., Dharmarajan V., Cosgrove M. S. (2008) Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J. Biol. Chem. 283, 32158–32161 [DOI] [PubMed] [Google Scholar]
- 59. Song J. J., Kingston R. E. (2008) WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 283, 35258–35264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patel A., Vought V. E., Swatkoski S., Viggiano S., Howard B., Dharmarajan V., Monteith K. E., Kupakuwana G., Namitz K. E., Shinsky S. A., Cotter R. J., Cosgrove M. S. (2014) Automethylation activities within the mixed lineage leukemia-1 (MLL1) core complex reveal evidence supporting a “two-active site” model for multiple histone H3 lysine 4 methylation. J. Biol. Chem. 289, 868–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tie F., Banerjee R., Saiakhova A. R., Howard B., Monteith K. E., Scacheri P. C., Cosgrove M. S., Harte P. J. (2014) Trithorax monomethylates H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize Polycomb silencing. Development 141, 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang Y. W., Flynn R. A., Chen Y., Qu K., Wan B., Wang K. C., Lei M., Chang H. Y. (2014) Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife 3, e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang K. C., Yang Y. W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B. R., Protacio A., Flynn R. A., Gupta R. A., Wysocka J., Lei M., Dekker J., Helms J. A., Chang H. Y. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shinsky S. A., Hu M., Vought V. E., Ng S. B., Bamshad M. J., Shendure J., Cosgrove M. S. (2014) A nonactive site SET domain surface crucial for the interaction of MLL1 and the RbBP5/Ash2L heterodimer within MLL family core complexes. J. Mol. Biol. 426, 2283–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Patel A., Vought V. E., Dharmarajan V., Cosgrove M. S. (2011) A novel non-SET domain multi-subunit methyltransferase required for sequential nucleosomal histone H3 methylation by the mixed lineage leukemia protein-1 (MLL1) core complex. J. Biol. Chem. 286, 3359–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Southall S. M., Wong P. S., Odho Z., Roe S. M., Wilson J. R. (2009) Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol. Cell 33, 181–191 [DOI] [PubMed] [Google Scholar]
- 68. Sali A., Blundell T. L. (1993) Comparative protein structure modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]