Background: RNA editing in plant organelles requires at least one of nine MORF proteins.
Results: MORF proteins connect in specific homo- and heteromeric protein-protein interactions.
Conclusion: The observed homo- and heteromeric combinations of MORF proteins explain why full editing can require two MORFs but one MORF can sustain basal editing levels.
Significance: These findings will help to better understand RNA editing in plant organelles.
Keywords: Arabidopsis thaliana, Chloroplast, Mitochondria, Protein-Protein Interaction, RNA Editing, MORF Proteins
Abstract
RNA editing in plastids and mitochondria of flowering plants requires pentatricopeptide repeat proteins (PPR proteins) for site recognition and proteins of the multiple organellar RNA editing factor (MORF) family as cofactors. Two MORF proteins, MORF5 and MORF8, are dual-targeted to plastids and mitochondria; two are targeted to plastids, and five are targeted to mitochondria. Pulldown assays from Arabidopsis thaliana tissue culture extracts with the mitochondrial MORF1 and the plastid MORF2 proteins, respectively, both identify the dual-targeted MORF8 protein, showing that these complexes can assemble in the organelles. We have now determined the scope of potential interactions between the various MORF proteins by yeast two-hybrid, in vitro pulldown, and bimolecular fluorescence complementation assays. The resulting MORF-MORF interactome identifies specific heteromeric MORF protein interactions in plastids and in mitochondria. Heteromers are observed for MORF protein combinations affecting a common site, suggesting their functional relevance. Most MORF proteins also undergo homomeric interactions. Submolecular analysis of the MORF1 protein reveals that the MORF-MORF protein connections require the C-terminal region of the central conserved MORF box. This domain has no similarity to known protein modules and may form a novel surface for protein-protein interactions.
Introduction
RNA editing in flowering plants converts 400–500 cytidines to uridines in transcripts in mitochondria and 30–40 cytidines in plastid mRNAs (1, 2). In some non-vascular plants, more than a thousand editing events have been documented in mitochondria, and hundreds have been documented in plastids (3). Each editing site is recognized by a trans-acting protein that addresses a specific target sequence located several nucleotides 5′ of the edited cytidine (4–8).
These site-specific proteins belong to a subgroup of the pentatricopeptide repeat (PPR)2 protein family with similar structural compositions (9, 10). A variable number of elements of 34–37 amino acids connect to the RNA, each element binding to one specific nucleotide as identified by statistical analyses and crystal structures (8, 11–15). The combination of several elements recognizes one or very few RNA sequence motifs that define the target sites. The ∼200 RNA editing specificity factors in Arabidopsis thaliana are all C-terminally extended by an extension (E) domain, and roughly half contain an additional region, often with the C-terminal amino acid triplet DYW (9, 10–12). The enzymatic reaction deaminating a C to a U may be performed by the DYW domain either in an RNA-binding DYW-PPR protein or in an additional PPR protein containing this region (12, 16, 17). Alternatively, a separate enzyme could be recruited to the PPR protein attached to the target editing site.
The recently identified group of multiple organellar RNA editing factor (MORF, also called RIP) proteins may provide the link between the RNA-binding PPR protein and the protein contributing the enzymatic activity (18, 19). In addition, another protein group with at least one degenerated central MORF sequence seems involved in RNA editing (20), and further RNA-binding proteins may also play a role (21). The MORF proteins form a small family of nine full-length proteins encoded in the nuclear genome of A. thaliana (12, 18, 22). In other flowering plant species, the number of MORF family genes can differ (23). MORF proteins are required for all RNA editing events in plastids and for many, possibly also all, sites in mitochondria of A. thaliana. In plastids, the two MORFs targeted exclusively to this organelle, MORF2 and MORF9, are both required for editing at several sites (18, 22). Other plastid sites depend on both proteins for efficient editing, either of the two MORFs providing only a low level of nucleotide conversion. In addition, the presence or absence of MORF8 (RIP1) influences editing at sites targeted by MORF2 and/or MORF9 (18, 22). Similarly in mitochondria, MORF1 and MORF3 are required at some editing sites, whereas at other sites, only MORF8 and MORF1 together sustain full editing levels (18, 19, 22). These observations suggest that different MORFs act in concert at some RNA editing events and that therefore MORFs are likely to contact each other. To investigate potential physical contacts between MORF proteins, we have now analyzed all possible MORF to MORF connections and report the homomer and heteromer MORF-MORF interactome.
EXPERIMENTAL PROCEDURES
MORF1 and MORF2 Pulldown Assays from A. thaliana Tissue Culture Cells and Affinity Purification-Mass Spectrometry Detection of Interacting Proteins
Identification of proteins interacting in vivo with MORF1 and MORF2, respectively, by affinity purification-mass spectrometry analysis was done with PSB-D A. thaliana cell suspension cultures essentially as described by Van Leene et al. (24). Open reading frames of MORF1 and MORF2 were cloned into pENTR1A (Life Technologies) and transferred into the pKCTAP vector with 35S promoter sequences in pEN-L4-2-R1 by the multisite Gateway LR reaction (Life Technologies) for constitutive expression of fusion proteins with a C-terminal tag combining the immunoglobulin G (IgG)-binding domains of protein G followed by the streptavidin-binding peptide (GS-tag). Transformation of PSB-D suspension culture cells was achieved as described (24). Protein complexes of MORF1 and MORF2 were tandem affinity-purified by IgG Sepharose and streptavidin Sepharose (GE Healthcare, Little Chalfont, UK) from 15 g of cells. Total protein in the purified fraction was concentrated by trichloroacetic acid precipitation and subjected to SDS-PAGE on 10 or 12% gels. Gels were stained by colloidal Coomassie Blue staining (25), and gel pieces containing proteins with various molecular masses were excised. In-gel tryptic digestion and LC-MS/MS protein identification were performed as described by Obata et al. (26). Database search was with Mascot 2.2 (Matrix Science Ltd., London, UK) setting criteria as described (26). The hits with probability-based Mowse scores above 50 were considered. Heat shock proteins were omitted from the analysis because they are supposed to interact with misfolded proteins caused by overexpression under the control of 35S promoter.
Plant Material and Preparation of Nucleic Acids
Growth of the A. thaliana plants and preparation of chloroplasts and DNA or RNA from leaves were as described (27, 28). Nicotiana benthamiana plants were grown on soil at 21 °C, 65% humidity, and long day illumination. Leaves were harvested after 5–6 weeks of growth.
Analysis of RNA Editing Sites
Specific cDNA fragments were generated by RT-PCR amplification following established protocols with respectively designed primers (27, 28). The cDNA sequences were compared for C to T differences resulting from RNA editing. RNA editing levels were estimated by the relative area under the respective nucleotide peaks in the sequence analyses.
Intracellular Location Analysis of MORF Proteins
The MORF genes were cloned in the pCF203 vector fused at their C terminus to the YFP open reading frame under expression control by the 35S promoter. The mitochondrial marker RFP was cloned as an in-frame fusion to the AOX1 target signal sequence (mtRFP1). Constructs were infiltrated into N. benthamiana leaf cells as described (29–32). Plants were grown on soil for 2 days, and epidermal cell layers were analyzed. Chloroplasts of the transiently transfected N. benthamiana leaf cells were identified by chlorophyll autofluorescence. Mitochondria were identified by the signal from the RFP marker. The fluorescent signals and the bright field images were obtained with the respective wavelength filters with a Leica TCS SP5 confocal microscope (Leica Microsystems, Heidelberg, Germany).
Bimolecular Fluorescence Complementation (BiFC) Analysis
YFP open reading frame N- and C-terminal fragments (amino acids 1–155 and 156–238, respectively) were fused to the individual MORF protein reading frames in the vector pMDC123 (33), containing the multiple cloning site from pET41 (Merck Millipore, Novagen), and in the YFP/YFP-N/YFP-C cassettes, respectively. Transformation into N. benthamiana leaves and expression analysis was as detailed above. Images were adjusted for brightness and contrast to improve quality and clarity.
Yeast Two-hybrid Analysis
The coding sequences of MORFs with their native termination codons and the vectors were amplified with respective primers to add 15-nucleotide adaptors for cloning via the In-Fusion HD cloning system (Clontech Laboratories). Open reading frames were integrated into the bait (pGBKT7) and prey (pGADT7) vectors of the Matchmaker GAL4 Two-Hybrid System 3 (Clontech Laboratories) for expression in yeast cells (PJ69-4A) according to the protocol and other publications (34–36). Yeast cells were generally grown on medium lacking leucine and tryptophan for selective pressure to maintain the respective plasmids. To obtain clear signals and to lower the rate of false positives, biosynthesis of histidine and biosynthesis of adenine were measured together as reporters on the respective selective medium. Drop assays were done as described in the next paragraph.
MORF1 Deletion Clones for the Yeast Two-hybrid Analysis
The coding sequence of MORF1 in the pGBKT7 vector was successively shortened by PCR, and the respective fragments were cloned in pGBKT7 via In-Fusion. Growth was assayed on synthetic dropout medium lacking adenine, histidine, leucine, and tryptophan and for rigorous probing also containing 2.5 mm 3-amino-1,2,4-triazole. In each assay, 5 μl of an overnight liquid culture adjusted to A600 nm of 0.3 was dropped to the agar medium plate.
Pulldown Assays
MORF proteins were expressed as fusion proteins with maltose-binding protein (MBP) label at their N termini on the one hand and N-terminal GST and His tags and GFP labels at the C termini on the other hand. GST-His-GFP-tagged proteins were purified from Escherichia coli via nickel-nitrilotriacetic acid-agarose (Qiagen) followed by a purification step with glutathione agarose (Sigma). MBP-tagged MORFs were bound to an amylose resin (New England Biolabs). To test for interactions between different MORF proteins, 10 μl of amylose resin saturated with one of the MORF proteins was incubated for 1 h at 4 °C with 10 μg of the respective other GFP-tagged MORF protein. Resins were boiled for 5 min in SDS-loading dye after five washing steps, and proteins were separated by SDS-PAGE on two gels in parallel. After blotting, proteins bound to the resin were detected by an anti-MBP antibody (New England Biolabs), and the respective pulled down GFP-labeled MORF proteins were identified with an anti-GFP antibody (Roche Applied Science).
RESULTS
In Vivo Interactions between MORF Proteins
Our previous in vitro co-immunoprecipitation and yeast two-hybrid (Y2H) analyses suggested the possible formation of homo- and heteromers of MORF proteins (18). To analyze whether MORF-MORF interactions indeed occur in vivo, we initiated tandem affinity purification studies in A. thaliana tissue culture cells. The MORF1 and MORF2 proteins, respectively, were introduced as GS-tagged constructs into the genomes of tissue culture cells (24–26). These bait proteins were immobilized on beads, and the attached proteins were analyzed by tandem mass spectrometry. Both the mitochondrially located MORF1 and the plastid MORF2 proteins pulled down the dual-targeted MORF8 protein among the prominent co-purifying proteins (Table 1). This finding suggests that MORF proteins can interact with each other in both plant organelles, mitochondria and plastids. To fathom the scale and specificity of MORF-MORF protein interactions, we now investigated all possible combinations, including the formation of homomers by yeast-2-hybrid assays, by pulldown assays, and by bimolecular fluorescence complementation in plant cells.
TABLE 1.
Identification of MORF8 co-purified with MORF1 and MORF2
Either mitochondrial MORF1 or chloroplast-located MORF2 proteins were expressed as GS-tagged proteins in A. thaliana tissue culture cells. MORF1 and MORF2 were enriched, and the co-purifying proteins were analyzed. The dual-targeted MORF8 protein is connected to both MORF1 and MORF2. Reps, repetitions.
| Bait |
Co-purified proteins |
Description | Founda | Best scoreb | Protein mass | Peptide matches |
|---|---|---|---|---|---|---|
| Description | Accession number | |||||
| MORF1 | AT3G15000.1 | MORF8 | 2 | 174 | 43014 | 7 |
| MORF2 | AT3G15000.1 | MORF8 | 1 | 89 | 43014 | 2 |
a Times of detection in two experimental repetitions.
b The highest probability-based Mowse score calculated by MASCOT in the two experimental repeats.
Intracellular Localization of the MORF Proteins
To be able to interpret the functional relevance of MORF protein interactions in the plant cell, we first analyzed their intracellular location with YFP fused to the respective MORF open reading frame in transiently transfected tobacco cells. Three MORF proteins had been located previously by different experimental approaches (37, 38); these were considered as controls to evaluate our results. The chloroplast localization of the MORF2 protein had been shown by import studies (37), and the MORF2 and MORF9 proteins had been detected in plastid proteome analyses (38). Our YFP fusion analyses confirm the exclusive plastid localization of both MORF2 and MORF9 proteins (Fig. 1).
FIGURE 1.
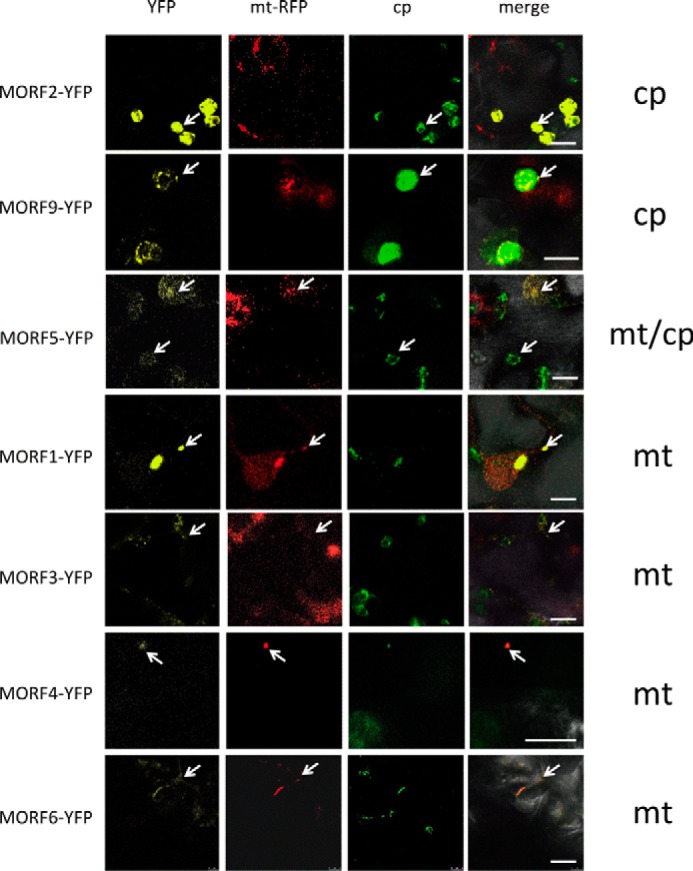
Intracellular localization of the MORF proteins. Open reading frames of the MORF proteins from A. thaliana were fused to YFP coding regions at their C termini to investigate the native N-terminal MORF protein sorting signals in transiently transfected N. benthamiana leaf cells. MORF2 and MORF9 proteins are located in plastids (upper panels). MORF5 proteins are seen in both plastids and mitochondria (center panels). MORF1 is seen in mitochondria. Signals of MORF3, MORF4, and MORF6 proteins suggest a mitochondrial location (lower panels). MORF7 and MORF8 did not yield clear signals. Respective left frames show the YFP signal (yellow), and central frames depict the mitochondrial marker RFP with the AOX1 target signal (mt-RFP) and the chlorophyll autofluorescence (cp), respectively. Right-hand frames (merge) show the overlap of all channels with the bright field image to assign the signals. Arrows point at samples for better orientation. Bars represent 10 μm.
The MORF8 protein (also termed RIP1) had been identified in a plastid protein fraction and affects RNA editing in mitochondria and plastids (18, 19, 22). In our analysis, MORF8 (and MORF7) did not yield a clear result. However, in the homo- and heteromer analyses, we find that the MORF8 protein is dually located and interacts with other MORF proteins in both organelles as detailed below. The MORF5 protein is similarly detected in both mitochondria and chloroplasts (Fig. 1), whereas the MORF1 protein is present in mitochondria. The data for MORF3, MORF4, and MORF6 are not clear; their localizations can be inferred from the homo- and heteromer locations presented below.
The Plastid-located MORF2 and MORF9 Proteins Form Homomers
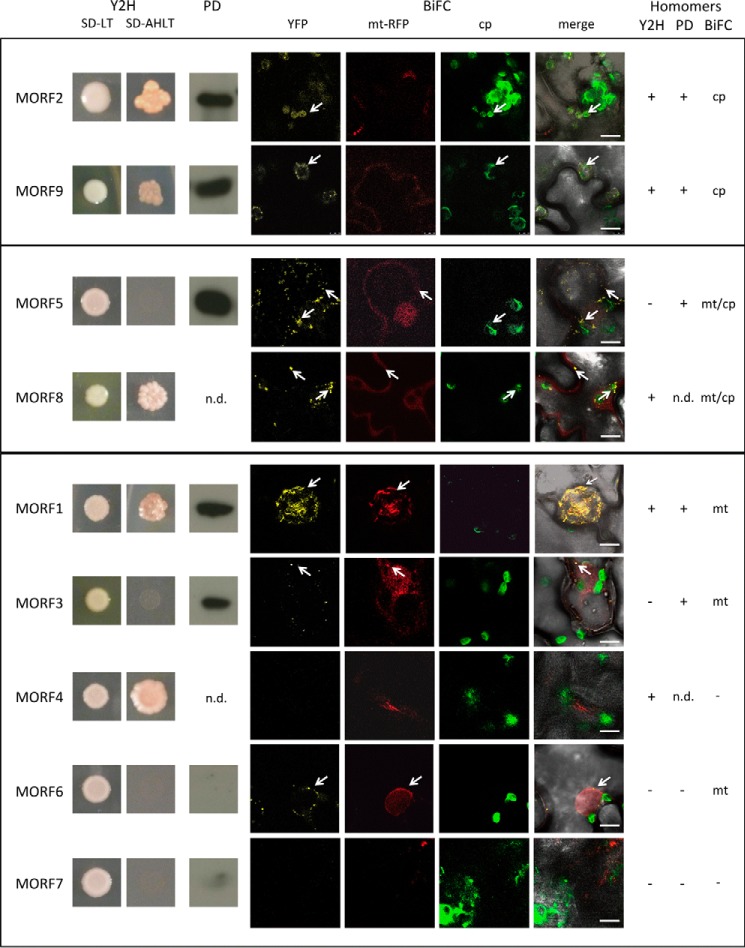
To investigate the competence of the MORF proteins to assemble homomers, we pursued three experimental approaches. Firstly, Y2H assays were performed, and secondly, reconstitution of fluorescent YFP was assayed in planta by transient transfections of tobacco leaves as BiFC. Because either method yields false negatives more often than false positives, (29, 30, 34, 35), we used pulldown experiments as an additional third method to probe for robust interactions.
The homomer assays of MORF2 and MORF9, respectively, document interactions in all three assay systems, Y2H, pulldown, and BiFC analyses, the latter observed in chloroplasts (Fig. 2). The consistent results suggest that homodimers or higher order homomers can form in vivo.
FIGURE 2.
Most MORF proteins can connect to homomers. The plastid-located MORF2 and MORF9 proteins form homomers in Y2H assays, in pulldown probings, and in BiFC experiments. The results of the Y2H analyses are shown in the left panels (Y2H), with SD-LT showing colony growth and SD-AHLT showing growth only upon MORF-MORF interaction on the selective medium lacking adenine and histidine. Pulldown probing results (PD) show the respective chemiluminescence signal after detection with an anti-GFP antibody (n.d. = not determined). The BiFC assays (BiFC) are displayed with YFP showing the interaction detecting fluorescence, mt-RFP showing the mitochondrial fluorescences, and cp showing the chloroplast fluorescences. These are laid over the bright field image in the merge panels. The dual-targeted MORF8 protein shows homomer interaction signals in the Y2H assays as well as in mitochondria and chloroplasts in the BiFC experiments. In addition, the dual-targeted MORF5 protein does not activate the yeast promoter through homomer interaction, but connects in the pulldown assays and yields a reconstituted BiFC fluorescence in mitochondria and chloroplasts. Of the mitochondrial MORF proteins, MORF1 shows strong homomer interaction in the Y2H as well as in the pulldown assays and BiFC experiments. MORF3 shows homomer interactions in the pulldown and in the BiFC assays. Of the other mitochondrial MORF proteins, only MORF4 undergoes a homomer interaction in the Y2H system, which is not seen in the BiFC experiments. The MORF6 protein yields a weak YFP signal in mitochondria, and MORF7 does not show homomer formation in any assay. Results of the three assay systems are summarized on the right. n.d. = not determined. Arrows point at samples for better orientation.
The Dual-targeted MORF5 and MORF8 Proteins Form Respective Homomers
Homomer interactions of the MORF5 protein are detected by BiFC in mitochondria and in chloroplasts, suggesting a dual localization of this MORF in both organelles (Fig. 2). The pulldown experiments confirm the formation of MORF5 homomers. Only in the Y2H assays is no MORF5-MORF5 interaction observed. MORF8-MORF8 interaction is detected in the Y2H assays, indicating that the MORF8 proteins can likewise form homomers (Fig. 2). In the BiFC experiments, homomer connections of the MORF8 protein are observed in mitochondria as well as in chloroplasts (Fig. 2).
Homomer Formations of MORF Proteins in Mitochondria
Of the mitochondrially located MORF proteins, MORF1 and MORF4, respectively, activate transcription of the reporter gene through homomer interactions in the Y2H interaction assays (Fig. 2). In the pulldown assays, MORF1 and MORF3, respectively, connect to homomers; unfortunately, MORF4 did not express sufficiently for analysis. Corresponding BiFC experiments confirm the homomer interaction of MORF1 and MORF3 in mitochondria, whereas homomer connections of MORF4 are not detected (Fig. 2). Homomer formation of MORF6 is indicated by BiFC signals in the mitochondria, whereas activating MORF6 homomers are not seen in the Y2H assays or in the pulldown experiments.
The MORF2 and MORF9 Proteins Connect to Respective Heteromers in Chloroplasts
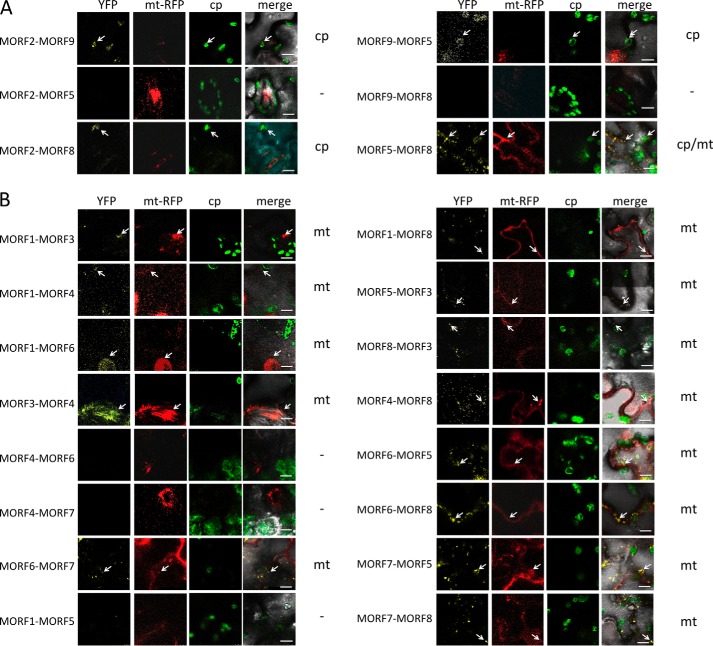
The Y2H analysis of MORF2-MORF9 and MORF9-MORF2 interactions yields positive signals in both directions, with either protein attached to the activation or binding domain (Fig. 3). The pulldown experiments likewise show interactions between these two MORFs in both directions (Fig. 4). This connection is further confirmed in planta with both N-terminal and C-terminal split YFP constructs yielding YFP signals in chloroplasts (Figs. 5 and 6).
FIGURE 3.
Yeast colony growth of all possible MORF-MORF protein combinations in both orientations in the Y2H system. Some MORF proteins such as MORF1, MORF8, and MORF9 show active interactions with most other MORF proteins, whereas others such as MORF5 or MORF7 connect to only a few MORF proteins. Left, growth of yeast colonies in the drop-assays on control medium (SD-LT). Right, selection medium without His and Ade (SD-AHLT), allowing growth only with the respective MORF-MORF interaction. Further details are discussed under “Results” and in the legend for Fig. 6.
FIGURE 4.
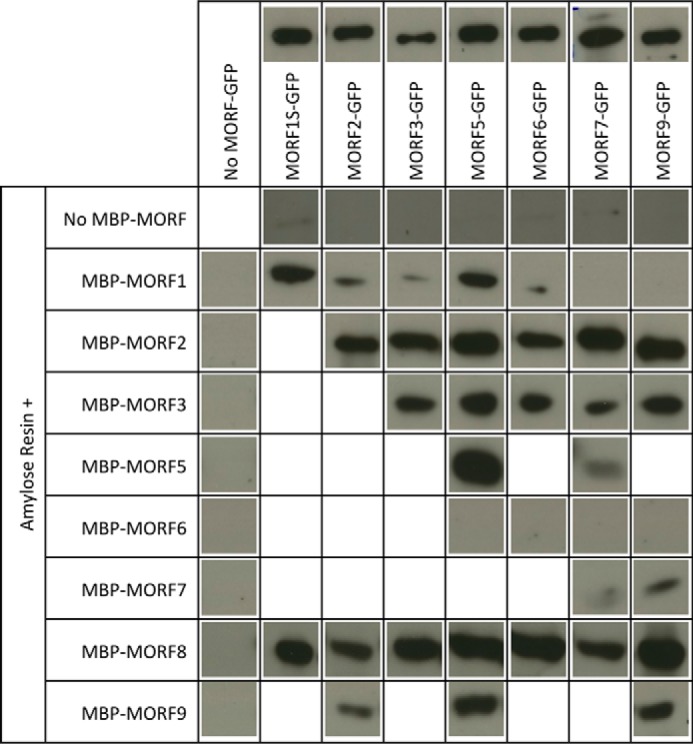
Pulldown probings of MORF-MORF protein interactions. Shown are the respective MORF-GFP GFP antibody chemiluminescence signals in the respective gel lanes pulled down with the respective MORF-MBP proteins bound to an amylose resin. MORF8 interacts with most other MORF proteins, whereas e.g. MORF7 binds only a few other MORF proteins. Without any MBP-MORF protein, no GFP-labeled MORF is retained by the amylose resin, and without a GFP-tagged MORF, no GFP antibody signal is detected. Above the respective MORF-GFP lanes, loading controls of 1 μg of the respective protein preparation are shown. Protein concentrations were determined by Bradford assays, and differences in the actual amounts of the respective GFP protein are due to variations of the impurities carried over from the overexpression in E. coli. For the pulldown assays, 10 μg of the respective GFP protein preparation was used. MORF1S-GFP is shortened in the C-terminal region outside of the MORF domain. Details are discussed under “Results” and in the legend for Fig. 6.
FIGURE 5.
MORF-MORF protein heteromer formation is observed in BiFC complementation assays. A, interactions of plastid-located MORF proteins. MORF2 and MORF9 proteins connect to heteromers, as indicated by the reconstituted YFP fluorescence from the N- and C-terminal YFP parts fused to the respective MORF reading frame. The MORF protein given first is fused to YFP-N, and the second is fused to YFP-C, respectively. MORF2 and MORF9 proteins are located in the plastid, and MORF5 and MORF8 proteins are dual-targeted to plastids and to mitochondria. MORF5 yields no detectable bimolecular interaction with MORF2, but connects with MORF8 in both organelles and with MORF9 in the plastid. The MORF8 protein contacts the MORF2 protein, but yields no signal with MORF9. Locations of the YFP signals are indicated on the right; the dash indicates that no YFP signal is detected. Arrows point out exemplary signals. B, interactions of mitochondrially located MORF proteins. Details of the various MORF-MORF protein heteromer connections are discussed under “Results” and in Figs. 6 and 7.
FIGURE 6.
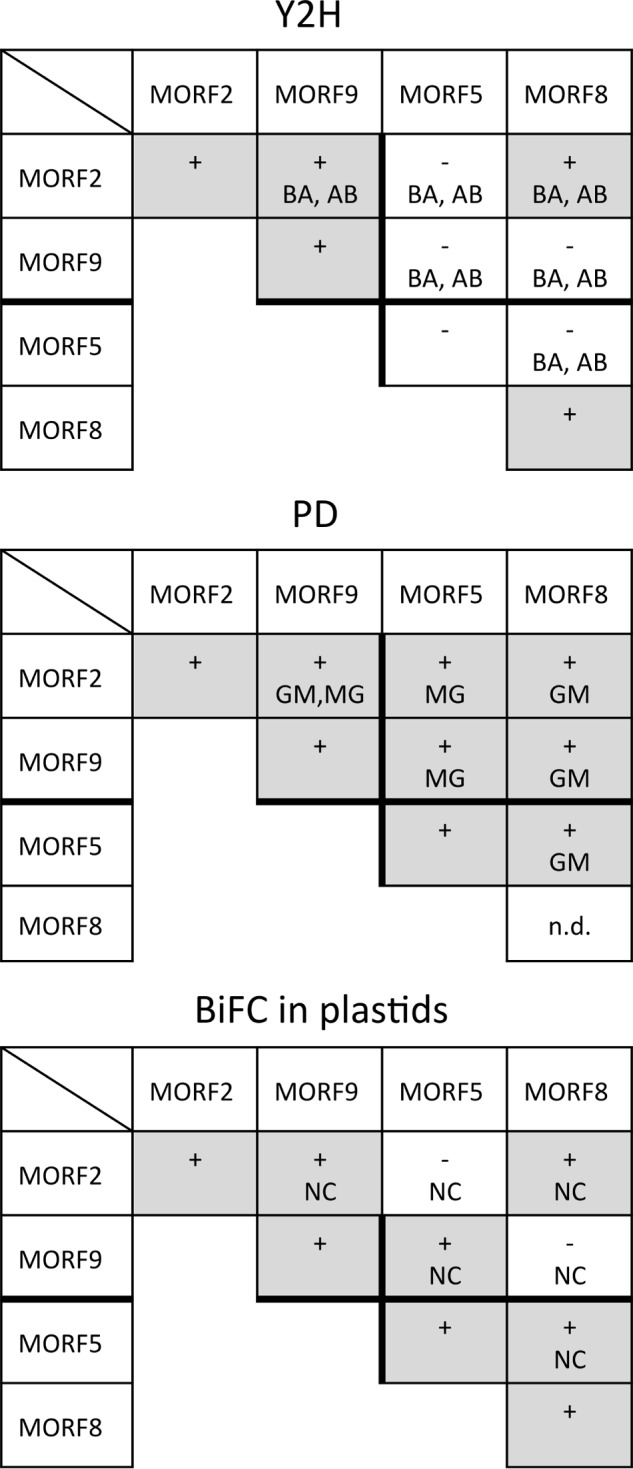
The plastid-located MORF2 and MORF9 proteins form homo- and heteromers and also interact with the dual-targeted MORF5 and MORF8 proteins. The comparative summary of the results from the three assay systems, the Y2H experiments (Fig. 3), the pulldown assays (PD; Fig. 4), and the BiFC complementations (Fig. 5), shows that the plastid-located MORF proteins connect to homo- and heteromers. Orientations of the respective assays are indicated with letters, positive signals are represented by a plus sign and shaded, and a minus sign indicates that no signal is seen. All interactions are detected in at least two of the three assays, and only MORF2-MORF5 and MORF8-MORF9 are seen in only one assay system. n.d. = not determined due to problems encountered with expression of the respective recombinant proteins.
The Dual-targeted MORF8 and MORF5 Proteins Interact with Several Mitochondrial and Plastid MORFs
MORF8 can attach to most plastid and mitochondrial MORFs in the Y2H interaction assays, in the pulldown experiments as well as in the BiFC analyses (Figs. 3–7). MORF8 connections are detected by Y2H assays with MORF1, MORF2, MORF3, MORF4, and MORF9 in both directions as activation domain (AD) and binding domain (BD) fusions and with MORF7 when attached to the BD. In the pulldown probings, MORF8 binds to MORF1, MORF2, MORF3, MORF5, MORF6, MORF7, and MORF9 in the MORF8-MBP direction for which sufficient MORF8 protein could be expressed. The interactions of the dual-targeted MORF8 with mitochondrial MORF1 and MORF3 and with MORF7 are seen in mitochondria, and its interactions with the plastid-located MORF2 proteins are appropriately detected in plastids in planta in the BiFC assays. Some BiFC interactions such as MORF2-MORF5 or MORF9-MORF8 do not yield interpretable signals and are therefore not considered.
FIGURE 7.
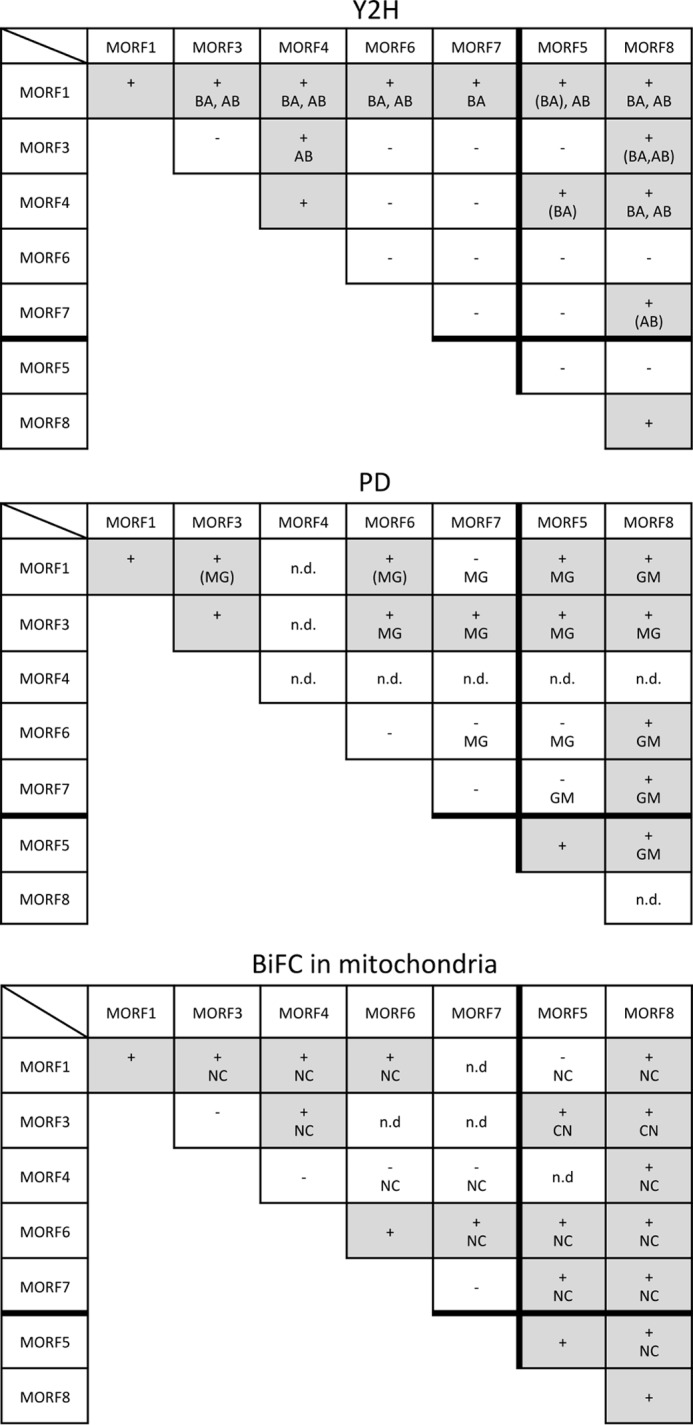
The interaction network between the mitochondrial MORF proteins. Interactions between the mitochondrially located MORF proteins 1, 3, 4, 6, and 7 and the dual-targeted MORF5 and MORF8 proteins were probed in Y2H (Fig. 3), pulldown (PD; Fig. 4), and BiFC analyses (Fig. 5). The mitochondrial MORF1 protein and the dual-targeted MORF8 protein contact all other MORFs in at least two of the three assay systems. MORF3, which is (like MORF1 and MORF8) involved in numerous RNA editing events, also contacts most other MORFs and forms heterodimers with MORF1, MORF4, MORF5, and MORF8 and in the pulldown assays, also with MORF6 and MORF7. The proteins involved in fewer editing events, MORF4, MORF6, and MORF7, form selected heterodimers mostly with MORF1 and a few other MORFs. n.d. = not determined due to problems encountered with expression of the respective recombinant proteins.
The other dual-targeted protein, MORF5, shows a Y2H interaction only with mitochondrial MORF1. This connection is not seen in BiFC assays (Fig. 5). In the BiFC experiments, however, MORF5 connects in plastids to MORF9 and MORF8, but not MORF2 (Fig. 5A), and in mitochondria, it connects to MORF3, MORF6, and MORF7, but not to MORF1 (Fig. 5B). The locations of the YFP signals in the correct organellar compartments in these instances argue in favor of these being bona fide connections. The pulldown assays reveal promiscuous interactions of MORF5 with most other MORFs, including the plastid MORF2 and MORF9 and the dual-targeted MORF8. For mitochondria, interactions with MORF1 and MORF3 are confirmed (Figs. 4, 6, and 7).
The Mitochondrial MORF1 Protein Interacts with Most MORFs
The mitochondrially located MORF1 protein is one of the most promiscuous MORFs in the Y2H interaction assays (Figs. 3 and 7). MORF1 interacts with the likewise mitochondrial MORF3, MORF4, MORF5, MORF6, and MORF8 in both directions as AD and BD fusions and in one direction with MORF7. Furthermore, interactions are seen with both plastid-located MORF2 and MORF9, which should not be relevant in planta due to their different organellar locations. In the BiFC assays, these two interactions are indeed not detected. The pulldown approaches show the MORF1 connections with MORF3, MORF5, MORF6, and MORF8, but do not detect MORF7. Plastid-located MORF2 is pulled down, but not MORF9 (Fig. 4). The BiFC assays confirm the MORF1-MORF3, MORF1-MORF4, MORF1-MORF6, and MORF1-MORF8 connections (Figs. 5 and 7).
Other Mitochondrial MORFs Form Selective Heteromers
In the Y2H assays, MORF3-MORF4 interactions are seen in one orientation (Figs. 3 and 7). The BiFC MORF3-MORF4 interaction is detected in mitochondria, suggesting that this is a genuine connection (Fig. 5 and data not shown). The BiFC assays of MORF3 with MORF5 and MORF8 yield signals in the mitochondria (Fig. 5B). For MORF3 with MORF6 or MORF7, no reconstituted YFP is detected, which may be partially due to intracellular mislocation seen in stressed or aging leaf cells (29–31). In the Y2H assays, the MORF3-MORF8 heteromer is seen as weak interactions in both orientations of the AD and BD domains. Pulldown experiments with MORF3 detect all of the interactions above, suggesting that MORF3 can promiscuously bind each of the other MORF proteins present in mitochondria.
MORF4 interacts in the Y2H system with MORF1, MORF3, and MORF8, and these connections to MORF1 are supported by the BiFC assays. For MORF4-MORF5, weak Y2H interaction is seen in one orientation, and BiFC YFP signals are not detected (Figs. 3, 5, and 7).
In the Y2H tests, MORF6 shows strong interaction with MORF1 in both directions, and in the BiFC assays, it shows strong interaction with MORF1 in mitochondria. The pulldown assays confirm the connection with MORF1. In addition, in the pulldown probings, MORF6-MORF3 and MORF6-MORF8 heteromers are detected, along with the MORF6 interaction with the plastid MORF2. MORF6 heteromers with MORF5, MORF7, and MORF8 are observed in the BiFC assays in mitochondria (Fig. 5B).
In Y2H assays, MORF7 connects with mitochondrial MORF1 in one orientation, whereas this is not seen in the BiFC or the pulldown assays. In the BiFC tests, MORF7 associates with MORF5 correctly localized in mitochondria; this connection is also detected in the pulldown assays, but not in Y2H assays. The MORF7-MORF8 combination is seen in one Y2H orientation, in the pulldown assays and it is seen as a YFP signal in mitochondria. MORF7 also interacts with MORF3 in the pulldown assays (Figs. 4 and 7).
MORF1-MORF Protein Interactions Require the C Terminus of the MORF Box
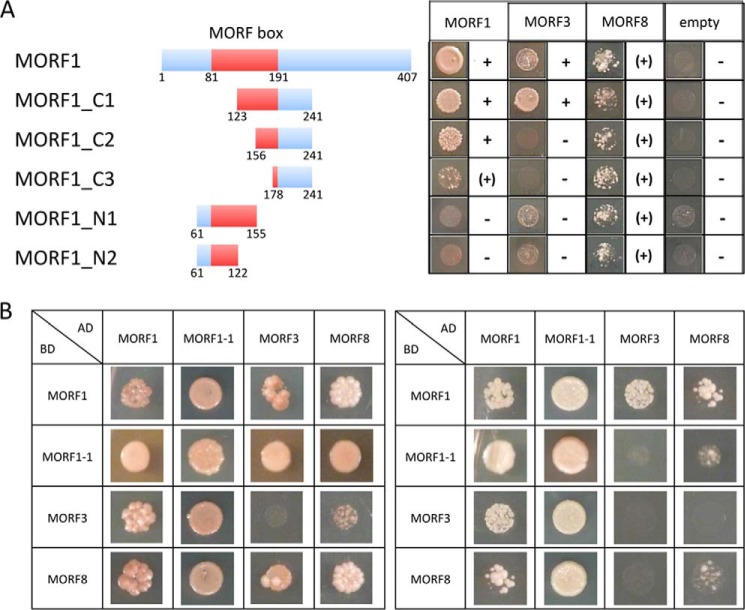
To investigate which parts of the MORF proteins are involved in homomer assembly and in heteromeric contacts with other MORF proteins, we selected the mitochondrial MORF1 protein, which readily forms homomers and interacts with several other MORF proteins. To probe heteromeric interactions with respective deletion clones of MORF1, we chose mitochondrial MORF3, which interacts with MORF1 in all assay systems, and the dual-targeted MORF8, which likewise interacts consistently with MORF1. For the Y2H assays, successively deleted fragments of MORF1 were fused to the BD, and full-length MORF1, MORF3, and MORF8, respectively, were fused with the AD (Fig. 8A).
FIGURE 8.
The C-terminal part of the MORF box domain mediates MORF-MORF interactions. A, yeast two-hybrid interaction assays with progressive truncations of the MORF1 protein reveal that the C-terminal part of the conserved central MORF box is required for the homomer MORF1-MORF1 protein interaction. In the morf1-1 mutant, a single amino acid alteration at position 165 severely compromises RNA editing at many mitochondrial sites (18). The C-terminal regions in deletion clones MORF1-C1 and MORF1-C2 contain this amino acid, whereas it is not present in clones MORF1-C3, MORF1-N1, and MORF1-N2. Fragments MORF1-C1 and MORF1-C2 show strong homomer interactions, indicated by +, whereas MORF1-C3 interacts less intensively, indicated by (+). MORF1-N1 and MORF1-N2 with MORF1 show weaker growth, but emerging cells are tinted red due to the lack of adenine, indicating the absence of interaction. In the heteromer assays with MORF3, the large C-terminal fragment of the MORF box contained in clone MORF1-C1 is necessary for interaction. The MORF8 protein displays a general, albeit weaker affinity to all fragments of MORF1. Respective fragments of MORF1 were fused to the BD of the GAL4 transcriptional reporter; small numbers indicate the border amino acid positions. Full-length MORF1, MORF3, and MORF8, respectively, were fused with the AD. B, interactions of the morf1-1 mutant with itself, MORF1, MORF3, and MORF8 are compared with the respective interactions of the wild type MORF1 in Y2H assays. The left panel shows drop assays on SD medium lacking adenine and the amino acids Trp-Leu-His. The right panel displays these combinations probed on medium containing in addition 3-amino-1,2,4-triazole for more stringent selection. Fused to the AD, the MORF1-1 mutant protein appears to connect similarly if not stronger than MORF1 to MORF3 and MORF8, but fused to the BD, the mutant MORF1-1 binds less tightly than the wild type MORF1 protein to MORF3 and MORF8, suggesting that the mutated amino acid position 165 in the C-terminal region of the conserved MORF box has an influence on heterologous MORF-MORF interactions.
Homomers of the MORF1 protein formed with truncated proteins containing the C-terminal region of the central conserved MORF box. Decreasing intensity of the interaction is observed upon shortening of the C-terminal fragments, whereas no interaction is seen with the N-terminal region. This suggests that the C terminus of the MORF box (and/or a few amino acids farther downstream) is involved in forming the homomer. The heteromer connection between the MORF1 and MORF3 proteins requires a larger part of the central MORF box; the shorter C-terminal fragments sufficient for MORF1-MORF1 affinity are not able to support detectable MORF1-MORF3 heteromers (Fig. 8A). The MORF8 protein also connects with the MORF1 conserved region. No decrease in the heteromeric MORF8-MORF1 interaction is seen with the progressively shorter fragments of MORF1, suggesting that MORF8 possibly connects with any part of the MORF box.
Analysis of the mutant protein encoded by the EMS (ethyl-methanesulfonate) mutant allele morf1-1 for its ability to sustain MORF-MORF interaction yielded affinities that appear stronger than those observed with the wild type MORF1 protein in Y2H assays (Fig. 8B). In the mutant, a single amino acid exchange at position 165 in the C-terminal region of the MORF box reduces RNA editing at more than 50 sites in mitochondria (18). This amino acid position is present in deletion clones MORF1-C1 and MORF1-C2, which form strong homomers, and is absent in MORF1-C3, MORF1-N1, and MORF1-N2 clones, which sustain only weak if any homomer interaction (Fig. 8).
DISCUSSION
MORF Proteins Interact in Vivo
The affinity purification assays in A. thaliana tissue culture cells now confirm in vivo the initial observations of interactions between MORF proteins in mitochondria and plastids by yeast two-hybrid assays (18). The mitochondrial MORF1 as well as the plastid MORF2 proteins co-purify the dual-targeted MORF8 protein. Firstly, this connection shows that these pairs of MORF proteins, MORF1-MORF8 and MORF2-MORF8, can form in vivo. Secondly, these pairs serve as positive controls in the evaluation of the MORF-MORF protein interactions we investigate here in all possible combinations, including the contacts in homomers, by yeast two-hybrid assays, by pulldown experiments, and by bimolecular fluorescence complementation. Both MORF1-MORF8 and MORF2-MORF8 are detected by all three methods, suggesting that the combination of these three procedures will detect a bona fide interaction with one, two, or all three systems.
For evaluation and discussion, it is important to note that the yeast two-hybrid system is more prone to not detect a bona fide interaction, i.e. to record false negatives, than to show an interaction artifact as various interactome analyses have documented (34, 35). Analogously, the BiFC assays are also more prone to false negatives than false positives (29–31). In addition, indirect interaction or requirement of a cofactor present in vivo in the plant but not in vitro or in the yeast cells may support an interaction only in BiFC. Also, interactions are often observed in only some combinations, which is why we probed various combinations of reporter domains and genes queried (39) and in addition employed as a third method pulldown experiments.
Plastid-located MORF Proteins Form Homo- and Heteromers
Two proteins of the MORF family, MORF2 and MORF9, are located in the plastid organelle, and two proteins, MORF5 and MORF8, are dual-targeted to plastids and mitochondria (Fig. 1). MORF2 and MORF9 form homomers in Y2H, in pulldown experiments, and in BiFC assays (Figs. 2 and 6). MORF8 readily connects with other MORF8 molecules in all assays, and MORF5 homomers are seen by pulldown experiments and in the BiFC assays in mitochondria.
The two dedicated chloroplast MORFs, MORF2 and MORF9, strongly interact with each other in heteromers in all assays, confirming the previous initial Y2H analysis (18). In all probings, MORF2 also interacts with the MORF8 protein, while MORF9 binds MORF8 in pulldown experiments and in both Y2H orientations (Fig. 6). The physical connection between MORF2 and MORF9 is likely to be a functional heteromer because both are required for common RNA editing events in the plastid (18, 22). MORF2 and MORF9 also connect with the MORF8 (RIP1) protein, for which 14 plastid RNA editing target sites have been identified in mutant analyses (18, 22). Editing at 11 of these 14 sites is also severely reduced in MORF2 and MORF9 mutants, and editing at three of these sites is increased in a rip1 (morf8) mutant and is detectably affected in MORF2 and MORF9 mutants, suggesting that at least two if not all three MORFs are required to act in concert for optimal RNA editing at these sites.
In conclusion, all MORFs present in the plastid interact with each other and also form homomers (Figs. 2 and 6). This finding suggests that several distinct “editosomes” assemble in plastids with at least two (different or identical) MORF proteins for efficient editing.
The Similar MORF5 and MORF6 Proteins Undergo Selective Interactions
The MORF5 and MORF6 proteins assemble homomers in the BiFC assays, and MORF5 homomers are also detected in the pulldown experiments (Figs. 2 and 4). Here we find MORF5 to be dual-targeted to plastids and mitochondria, whereas a recent analysis found only two mitochondrial sites to be influenced in a mutant (22), the mitochondrial location also being confirmed by proteomic data. We analyzed all plastid sites in the same mutant and, similar to Bentolila et al. (22), did not see any consistent influence of the MORF5 (RIP5) absence. Bentolila et al. (22) had also generated silenced plants that likewise did not show any defects in plastid editing. On the other hand, MORF5 can assemble in BiFC assays in heteromer formations with the plastid MORF9 and with the dual-targeted MORF8 in plastids. Furthermore, MORF5 binds to both plastid MORF2 and MORF9 proteins in the pulldown assays, which together suggests a functional connection most likely for RNA editing. In mitochondria, MORF5 interacts with MORF1 in the Y2H probings and with MORF6, MORF7, and MORF8 in one BiFC orientation (Fig. 7). The pulldown experiments detect MORF5 connections with MORF1, MORF3, and MORF8, either or all of these connections being possibly involved in editing at the two target sites identified for MORF5 (22). MORF5 connects weakly to MORF4 in Y2H assays. MORF6 interacts in all assays with MORF1, and in the pulldown experiments, it also interacts with MORF3 and MORF8 in one direction. For MORF6, one target site had been previously identified in mitochondria (18), whereas no significant effect was seen in the cDNA sequence analysis of the same mutant (22).
The issue of the function of MORF5 and MORF6 may be complicated by their high similarity. A recent evolutionary analysis revealed MORF5 and MORF6 homologs only in Arabidopsis relatives, whereas proteins similar to MORF1, MORF2, MORF3, MORF8, and MORF9 are encoded in all flowering plant species analyzed (23). In the sequence alignment, MORF5 and MORF6 branch from the plastid-located MORF2 proteins. The similarity between the MORF5 and MORF6 proteins is reflected by their analogous behavior; both form homomers and MORF5-MORF6 heteromers in the BiFC (but not the Y2H or pulldown assays), whereas strong connections to MORF1 are seen in the Y2H and pulldown assays, and between MORF1 and MORF6 also in the BiFC assays. MORF5 and MORF6 both connect to MORF3 and MORF8 in the pulldown experiments. These observations suggest that MORF5 and MORF6 can potentially substitute for each other in RNA editing in mitochondria and in other, unrelated functions.
The Dual-targeted MORF8 Protein Connects to Mitochondrial and Plastid MORFs
Consistent with its intracellular location in mitochondria and plastids, the MORF8 protein (also called RIP1 (19, 22)) interacts with MORFs in both organelles (Figs. 6 and 7). In mitochondria, connections are seen with MORF1 and MORF4, and rather weak Y2H assemblies are seen with MORF3 and MORF7. Apart from the MORF8-MORF4 combination, these interactions are also seen in the BiFC analysis and in the pulldown experiments, which in addition establish connections to MORF6 and MORF7. In plastids, strong interactions with MORF8 are observed with MORF2 and MORF9, and these results are confirmed in the pulldown experiments. The dual-targeted MORF5 and MORF8 proteins interact in both organelles in BiFC assays and in the pulldowns. In both Y2H and BiFC probings, MORF8 proteins form homomers.
The interactions of MORF8 with most other MORF proteins reflect the large number of editing sites in which MORF8 is involved in both mitochondria and plastids. The recent extensive RNA editing study of MORF8 (RIP1) mutants found that more than 50% of the ∼500 mitochondrial editing sites and 22% of the 36 plastid sites were affected (22).
In Mitochondria, MORF1 Forms Homomers and Interacts with All Other MORF Proteins
The interactions of MORF1 in homomers and with all other MORFs in mitochondria suggest that the MORF1 protein projects a sticky surface connecting to MORF proteins. The promiscuous interactions of MORF1 reflect the important role MORF1 (RIP8) plays in RNA editing at a large number of editing sites (18, 22).
Mitochondrial MORF3 and MORF4 Proteins Show Selective Interactions
The MORF3 protein interacts with all other MORFs similarly to the MORF1 protein at least in the pulldown assays. The MORF4 protein shows fewer interactions with other MORF proteins than MORF1, MORF3, or MORF8, but is still integrated in the mitochondrial MORF network. Homomers are detected for MORF4 as well as for MORF3. Both MORF3 and MORF4 can interact with one another, and both form heteromers with the MORF1 and MORF8 proteins (Fig. 7). The different heteromer connections made by MORF3 and MORF4 reflect the very different number of editing sites affected in respective mutants. MORF4 is involved in 6–19 sites, whereas MORF3 is required for more than 100 RNA editing sites in mitochondria (22). None of the MORF4 RNA editing targets overlap with those of other MORF proteins, and the MORF4 protein may act in a homomer configuration. MORF3 is, like MORF1 and MORF8 (RIP1), a major RNA editing cofactor, and almost 90% of the MORF3 target sites are also addressed by either or both of these other mitochondrial MORF proteins.
The Conserved MORF Box Mediates the MORF-MORF Protein Interactions
The involvement of the MORF box in homomer and heteromer MORF protein-protein interactions (Fig. 8) now explains the functional constraint on this region in evolution. The functional importance of the MORF-MORF interaction mediated by the MORF box is supported by the location of the morf1-1 mutant, with a single amino acid exchange at position 165 in the C-terminal region of the MORF box (18). This amino acid position is present in deletion clones C1 and C2, which form strong homomers, and is absent in C3, N1, and N2 clones, which sustain only weak if any homomer interaction (Fig. 8). However, in the direct interaction assays, there is little difference between this mutant and the wild type protein (Fig. 8B); the mutant actually seems to interact better with all MORFs. This observation suggests that the effect of the mutation is not simply a loss of MORF-MORF interactions, but something more complex. If it indeed binds better to other MORFs, it may less readily dissociate for further editing events and thus slow down the processivity of editing. In any case, this different quality of interactions of the morf1-1 mutant with a single amino acid alteration relative to the wild type MORF1 protein confirms that the C-terminal region of the conserved MORF box mediates MORF-MORF interactions.
Interestingly, the MORF1-MORF1 homomer formation requires a smaller protein-protein interface than the heteromer interactions between the MORF1 and MORF3 proteins. The observation that the surface of MORF8 can nonspecifically contact MORF1 fragments reflects the promiscuous interactions of MORF8 with many other MORFs. This property of MORF8 is possibly connected to the large number of sites it affects, many of which are also targets of other MORFs, MORF1 and MORF3 in mitochondria and MORF2 and MORF9 in plastids.
The recently identified ORRM proteins contain only the N-terminal region of the conserved MORF box (20). The loss of the C-terminal part suggests that these proteins cannot undergo the homo- or heteromer connections observed here with the MORF1 protein. The N-terminal MORF box fragment is duplicated in the ORRM proteins, but is not required to complement the RNA editing defects seen in an ORRM mutant (20).
Hetero- and Homomers of MORF Proteins Form Functional Units for RNA Editing in Plant Organelles
In mitochondria as well as in plastids, a given editing site requires the presence of different MORFs. This conclusion is based on two observations. Firstly, knock-out or knockdown mutants of two or three different MORF proteins can be compromised in editing at the same site, and secondly, a given site is often only reduced in a knock-out of one MORF protein (18, 22). The overlapping requirements for different MORFs suggest that these may act synergistically.
Our results reported here show that MORF proteins interact and connect in a complex network. The frequent homomer formations suggest that these may be functional in RNA editing. The homomers may be able to sustain a reduced level of editing at a given target site by working less efficiently than the heteromers, e.g. in mutants of one MORF protein where another MORF can perform, but cannot substitute as efficiently as the heteromer. It will now be interesting to determine the scale and specificity of the interactions between the various MORF, PPR, and other proteins (40) in plant organellar RNA editing.
Acknowledgments
We thank Dagmar Pruchner, Angelika Müller, and Bianca Wolf for excellent experimental help. We thank Christian Throm, Dorothea Kreuder, and Claudia Oecking at the Center for Plant Molecular Biology (ZMBP) in Tübingen for kindly introducing us to the yeast two-hybrid system and for providing material and support. We also thank Michael Heunemann and Klaus Harter at the ZMBP in Tübingen for kindly supporting us with the BiFC methodology. We are very grateful to Stefan Britsch and to the Department of Molecular Neurobiology at the Universität Ulm for kindly giving us access to microscopic facilities. We thank Alisdair Fernie and Lothar Willmitzer at the Max-Planck-Institute Golm for kindly giving us access to their facilities.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (to M. T. and A. B.).
- PPR
- pentatricopeptide repeat
- MORF
- multiple organellar RNA editing factor
- BiFC
- bimolecular fluorescence complementation
- Y2H
- yeast two-hybrid
- SD
- synthetic defined
- MBP
- maltose-binding protein
- AD
- activation domain
- BD
- binding domain
- ORRM
- organelle RNA recognition motif.
REFERENCES
- 1. Giegé P., Brennicke A. (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. U.S.A. 96, 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chateigner-Boutin A. L., Small I. (2010) Plant RNA editing. RNA Biol. 7, 213–219 [DOI] [PubMed] [Google Scholar]
- 3. Grewe F., Herres S., Viehöver P., Polsakiewicz M., Weisshaar B., Knoop V. (2011) A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 39, 2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kotera E., Tasaka M., Shikanai T. (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330 [DOI] [PubMed] [Google Scholar]
- 5. Zehrmann A., Verbitskiy D., van der Merwe J. A., Brennicke A., Takenaka M. (2009) A DYW domain containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21, 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hammani K., Okuda K., Tanz S. K., Chateigner-Boutin A. L., Shikanai T., Small I. (2009) A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21, 3686–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takenaka M., Verbitskiy D., Zehrmann A., Brennicke A. (2010) Reverse genetic screening identifies five E-class PPR proteins involved in RNA editing in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 285, 27122–27129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barkan A., Rojas M., Fujii S., Yap A., Chong Y. S., Bond C. S., Small I. (2012) A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PloS Genet. 8, e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B., Lecharny A., Le Ret M., Martin-Magniette M.-L., Mireau H., Peeters N., Renou J.-P., Szurek B., Taconnat L., Small I. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmitz-Linneweber C., Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13, 663–670 [DOI] [PubMed] [Google Scholar]
- 11. Takenaka M., Zehrmann A., Brennicke A., Graichen K. (2013) Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS ONE 8, e65343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takenaka M., Zehrmann A., Verbitskiy D., Härtel B., Brennicke A. (2013) RNA-editing in plants and its evolution. Annu. Rev. Genet. 47, 335–352 [DOI] [PubMed] [Google Scholar]
- 13. Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T. (2013) Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 8, e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H.-W., Wang J., Zhu J.-K., Shi Y., Yan N. (2013) Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171 [DOI] [PubMed] [Google Scholar]
- 15. Ke J., Chen R.-Z., Ban T., Zhou X. E., Gu X., Tan M. H. E., Chen C., Kang Y., Brunzelle J. S., Zhu J.-K., Melcher K., Xu H. E. (2013) Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 20, 1377–1382 [DOI] [PubMed] [Google Scholar]
- 16. Salone V., Rüdinger M., Polsakiewicz M., Hoffmann B., Groth-Malonek M., Szurek B., Small I., Knoop V., Lurin C. (2007) A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 581, 4132–4138 [DOI] [PubMed] [Google Scholar]
- 17. Boussardon C., Salone V., Avon A., Berthomé R., Hammani K., Okuda K., Shikanai T., Small I., Lurin C. (2012) Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell 24, 3684–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takenaka M., Zehrmann A., Verbitskiy D., Kugelmann M., Härtel B., Brennicke A. (2012) MORF family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bentolila S., Heller W. P., Sun T., Babina A. M., Friso G., van Wijk K. J., Hanson M. R. (2012) RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. U.S.A. 109, E1453-E1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun T., Germain A., Giloteaux L., Hammani K., Barkan A., Hanson M. R., Bentolila S. (2013) An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc. Natl. Acad. Sci. U.S.A. 110, E1169–E1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tillich M., Hardel S. L., Kupsch C., Armbruster U., Delannoy E., Gualberto J. M., Lehwark P., Leister D., Small I. D., Schmitz-Linneweber C. (2009) Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. U.S.A. 106, 6002–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bentolila S., Oh J., Hanson M. R., Bukowski R. (2013) Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9, e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takenaka M. (2014) How complex are the editosomes in plant organelles? Mol. Plant 7, 582–585 [DOI] [PubMed] [Google Scholar]
- 24. Van Leene J., Eeckhout D., Persiau G., Van De Slijke E., Geerinck J., Van Isterdael G., Witters E., De Jaeger G. (2011) Isolation of transcription factor complexes from Arabidopsis cell suspension cultures by tandem affinity purification. Methods Mol. Biol. 754, 195–218 [DOI] [PubMed] [Google Scholar]
- 25. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 26. Obata T., Matthes A., Koszior S., Lehmann M., Araújo W. L., Bock R., Sweetlove L. J., Fernie A. R. (2011) Alteration of mitochondrial protein complexes in relation to metabolic regulation under short-term oxidative stress in Arabidopsis seedlings. Phytochemistry 72, 1081–1091 [DOI] [PubMed] [Google Scholar]
- 27. Takenaka M., Brennicke A. (2007) RNA editing in plant mitochondria: Assays and biochemical approaches. Methods Enzymol. 424, 439–458 [DOI] [PubMed] [Google Scholar]
- 28. Takenaka M., Zehrmann A. (2011) Complementation of mutants in plant mitochondrial RNA editing by protoplast transfection. in RNA and DNA Editing (Aphasizhev R., ed), pp. 163–169, Humana Press, Springer, Heidelberg: [DOI] [PubMed] [Google Scholar]
- 29. Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 [DOI] [PubMed] [Google Scholar]
- 30. Schütze K., Harter K., Chaban C. (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol. Biol. 479, 189–202 [DOI] [PubMed] [Google Scholar]
- 31. Sparkes I. A., Runions J., Kearns A., Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025 [DOI] [PubMed] [Google Scholar]
- 32. Tanz S. K., Castleden I., Small I. D., Millar A. H. (2013) Fluorescent protein tagging as a tool to define the subcellular distribution of proteins in plants. Front. Plant Sci. 4, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Curtis M. D., Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Braun P. (2012) Interactome mapping for analysis of complex phenotypes: insights from benchmarking binary interaction assays. Proteomics 12, 1499–1518 [DOI] [PubMed] [Google Scholar]
- 35. Braun P., Aubourg S., Van Leene J., De Jaeger G., Lurin C. (2013) Plant protein interactomes. Annu. Rev. Plant Biol. 64, 161–187 [DOI] [PubMed] [Google Scholar]
- 36. James P., Halladay J., Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bisanz C., Bégot L., Carol P., Perez P., Bligny M., Pesey H., Gallois J.-L., Lerbs-Mache S., Mache R. (2003) The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol. Biol. 51, 651–663 [DOI] [PubMed] [Google Scholar]
- 38. Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K. J. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Horstman A., Tonaco I. A. N., Boutilier K., Immink R. G. H. (2014) A cautionary note on the use of split-YFP/BiFC in plant protein-protein interaction studies. Int. J. Mol. Sci. 15, 9628–9643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang F., Tang W., Hedtke B., Zhong L., Liu L., Peng L., Lu C., Grimm B., Lin R. (2014) Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. U.S.A. 111, 2023–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]