Background: In biofilms, bacteria communicate via quorum-sensing (QS) molecules.
Results: The specific binding affinity of QS molecules to a functional amyloid is determined.
Conclusion: Functional amyloids can transiently bind and retain QS molecules.
Significance: Functional amyloids are important for cell signaling within biofilms.
Keywords: Amyloid, Biofilm, Pseudomonas, Pseudomonas aeruginosa (P. aeruginosa), Quorum Sensing, Functional Amyloid
Abstract
The mechanism by which extracellular metabolites, including redox mediators and quorum-sensing signaling molecules, traffic through the extracellular matrix of biofilms is poorly explored. We hypothesize that functional amyloids, abundant in natural biofilms and possessing hydrophobic domains, retain these metabolites. Using surface plasmon resonance, we demonstrate that the quorum-sensing (QS) molecules, 2-heptyl-3-hydroxy-4(1H)-quinolone and N-(3-oxododecanoyl)-l-homoserine lactone, and the redox mediator pyocyanin bind with transient affinity to functional amyloids from Pseudomonas (Fap). Their high hydrophobicity predisposes them to signal-amyloid interactions, but specific interactions also play a role. Transient interactions allow for rapid association and dissociation kinetics, which make the QS molecules bioavailable and at the same time secure within the extracellular matrix as a consequence of serial bindings. Retention of the QS molecules was confirmed using Pseudomonas aeruginosa PAO1-based 2-heptyl-3-hydroxy-4(1H)-quinolone and N-(3-oxododecanoyl)-l-homoserine lactone reporter assays, showing that Fap fibrils pretreated with the QS molecules activate the reporters even after sequential washes. Pyocyanin retention was validated by electrochemical analysis of pyocyanin-pretreated Fap fibrils subjected to the same washing process. Results suggest that QS molecule-amyloid interactions are probably important in the turbulent environments commonly encountered in natural habitats.
Introduction
Amyloids are insoluble fibers formed by either regulated or unregulated protein folding. Although they are more commonly associated with human diseases, amyloids fulfill important microbial functions, including as structural components in biofilms and in the cell wall of bacterial spores, adhesins mediating specific binding to and internalization into host cells, and as a means to attenuate the cytotoxicity of bacteriocins (1–4). Furthermore, their assembly is regulated (5), suggesting that such functions are not chance occurrences.
Bacterial amyloids have been detected in natural biofilms from a range of aquatic systems, including water reservoirs, seawater, and wastewater treatment-activated sludges (6, 7). Given their apparent abundance in natural biofilms and the tantalizing prospect of using them to better understand the formation of disease-related amyloids (4), there is a surprising paucity of information on the structure-function dependence of amyloids and the means by which this is impacted by interactions with other molecules. The curli fibers of Escherichia coli and Salmonella enterica, Fap fibrils of Pseudomonas spp., and TasA of Bacillus subtilis have each received attention (2, 8, 9). Their roles in biofilms have been described variously as those of adhesins, stabilizers (e.g. B. subtilis) (8), and promoters of the initial biofilm formation (9). They have also been suggested to be virulence determinants, as with the curli fimbriae of enterohemorrhagic, enterotoxigenic, and sepsis-derived strains of E. coli (10). However, with some notable exceptions (11), as with functional analyses of other extracellular polymeric substances (EPS),3 work with amyloids has generally been observational rather than mechanistic.
Pseudomonas aeruginosa is an opportunistic bacterial pathogen (12). Its virulence is associated with the ability to form biofilms (13), which are the dominant mode of microbial life in many engineered, medical, and natural settings (14). Furthermore, the extracellular domain of P. aeruginosa biofilms is well understood chemically in terms of the EPS composition and metabolites produced. Thus, P. aeruginosa is often used in biofilm studies, including those addressing the effects of its amyloids on biofilm formation. Dueholm et al. (15) discovered a six-gene operon encoding the expression of functional amyloids (fapA–F) in P. aeruginosa, Pseudomonas fluorescens, and Pseudomonas putida. Overexpression of Fap fibrils, which are composed mainly of FapC, has since been shown to increase biofilm formation (9).
Bacteria collectively coordinate their behavior and communicate with each other using quorum-sensing (QS) systems, whereby small molecules are secreted and elicit changes in gene expression in those cells responding to them (16). Four QS systems have been identified for P. aeruginosa, including the Las and Rhl systems, which employ the autoinducers N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) and N-butyryl-l-homoserine lactone, respectively (17, 18); the cis-2-decenoic acid signaling system (19); and the 4-quinolone signal system, unique to the members of the genus Pseudomonas, which involves 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS) and the transcriptional regulator PqsR (20). PQS is a putative iron chelator, which positively regulates the Rhl QS system (21). The Rhl system in turn modifies elastase (LasB) and lectin (LecA) production. Furthermore, the PQS system utilizes the redox shuttle pyocyanin as its terminal signaling factor (16). Pyocyanin is a heterocyclic compound that belongs to the phenazine group, which when produced by P. aeruginosa enhances bacterial virulence (e.g. in chronic lung infections) and may also provide the bacterium with a competitive advantage as a result of its antimicrobial activity (22). QS molecules in P. aeruginosa are chemically distinct with different targets and a cascade of integrated and secondary effects that confound our understanding of their phenotypic consequences. However, they are all transmitted extracellularly, and they are all hydrophobic (23).
Although both the P. aeruginosa QS systems and its extracellular matrix have been studied extensively, the importance of possible interactions between the matrix components and QS molecules has not attracted the same level of interest. QS chemicals are transmitted through the extracellular domain. Mashburn and Whiteley (24) addressed the question of how transmission of hydrophobic QS molecules might be managed extracellularly by the packaging of a QS molecule into membrane vesicles before export. However, this was specific to PQS. Interactions between extracellular polymers and signaling chemicals are also probably crucial to their movement through biofilms. For example, it is believed that the immediate precursor of PQS, 4-hydroxy-2-heptylquinoline (HHQ), acts as a “messenger” molecule because it is secreted extracellularly and is then converted to PQS by other responding cells (21). Biofilms can form in flowing, often highly turbulent aquatic environments like rivers or wastewater treatment plants (25, 26), where the loss of small molecules by high diffusion rates is a significant challenge for bacteria. These habitats also mostly consist of complex, mixed microbial communities, raising interesting challenges around signal cross-talk and uptake by non-self organisms (14). Dilution or interception and potential interference or corruption of chemical signals represent a serious challenge to the bacteria. Although it is assumed that the EPS matrix allows biofilms to retain such molecules under non-quiescent conditions (27), the ability of a single EPS to facilitate this by a reversible binding process has never been demonstrated.
There are currently no high-resolution protein structures of biofilm-associated functional amyloids. However, amyloid fibers of the HET-s(218–289) prion and the β-amyloid peptide (Aβ(1–40)) associated with Alzheimer disease arise from β-solenoids that are stabilized by a hydrophobic triangular core (28, 29). The current structural model of the Fap fibrils suggests that their amyloids also contain similar hydrophobic domains (9). Such domains represent potential binding sites for hydrophobic signaling molecules. In the present study, amyloids were identified as an EPS component that may play a role in binding signaling metabolites. We investigated such a functional role by characterizing the binding affinities of QS molecules and other small hydrophobic and non-hydrophobic metabolites to amyloid polymers using surface plasmon resonance (SPR) biosensor technology. We present two important physiological consequences of this binding. Under non-quiescent conditions, amyloids can act as storage reservoirs to enhance information retention and electroconductivity, thus providing the first direct evidence for a specific role for an EPS component of a bacterial biofilm in signal molecule binding.
EXPERIMENTAL PROCEDURES
Strains
P. aeruginosa PAO1 was obtained from the Genetic Stock Center (strain PAO0001). The PQS reporter strain was constructed by introducing the promoter-probe plasmid pAC37, containing a PpqsA::gfp(ASV) transcriptional fusion, into P. aeruginosa PAO1 ΔpqsC, as described by Yang et al. (30). PQS positively regulates the pqsABCDE operon via PqsR (21). Thus, pqsABCDE expression indicates interaction of PQS with the cell (31). The P. aeruginosa PAO1 ΔpqsC mutant, which is unable to synthesize the PQS precursor HHQ, was obtained from a transposon mutagenesis library screen (31). The 3-oxo-C12-HSL reporter strain was obtained by introducing a PlasB::gfp(ASV) transcriptional fusion and an extra copy of lasR under the influence of the lac promoter, which is constitutively expressed in P. aeruginosa into P. aeruginosa PAO1 ΔlasIΔrhlI via a mini-Tn5 insert, as described by Yang et al. (32). 3-Oxo-C12-HSL is thus sensed through the LasR reporter, which in turn regulates the lasB promoter (32). Construction of the isogenic P. aeruginosa PAO1 ΔlasIΔrhlI strain, which is unable to synthesize 3-oxo-C12-HSL, is described elsewhere (33).
Metabolites
PQS and HHQ were synthesized as described previously (21), and 25 mm stock solutions were prepared by dissolution in DMSO (25 mm). A stock solution of pyocyanin from P. aeruginosa (from Sigma-Aldrich) (480 μm) was prepared in DMSO.
Expression and Isolation of Monomeric FapC
A synthetic gene corresponding to the mature FapC protein from P. aeruginosa PAO1 flanked by XbaI and XhoI restriction sites was designed using CLC Main Workbench version 6.0 and synthesized by GenScript (Germany). The synthetic gene was cloned into the pET32b(+) vector (Novagen) using the XbaI and XhoI restriction sites. This added a C-terminal His tag to the construct. The plasmid was transformed into E. coli BL21(DE3) (Invitrogen). Protein expression was carried out in 500 ml of LB medium (37 °C, 200 rpm). Induction was done with 1 mm isopropyl 1-thio-β-d-galactopyranoside at A600 nm = 0.7–1.2, and cells were harvested 3 h after induction by centrifugation (10,000 × g, 30 min). The cell pellet was resuspended in 25 ml of extraction buffer (6 m guanidinium chloride, 20 mm potassium phosphate, 500 mm NaCl, pH 7.4) and sonicated three times for 1 min each. The sample was placed on ice for 1 min between sonications. The lysate was then incubated overnight at 4 °C with gentle shaking. Insoluble material was pelleted by centrifugation (20,000 × g, 30 min). The His-tagged FapC proteins were isolated from the supernatant using a 5-ml HisTrap HP column (GE Healthcare) and an elution buffer corresponding to the extraction buffer containing 500 mm imidazole.
Preparation of Natively Seeded FapC Fibrils
Purified monomers in elution buffer were desalted using a PD-10 desalting column (GE Healthcare) equilibrated with deionized water. Protein concentration was estimated by UV absorbance of the peptide bond using the following extinction coefficient for the recombinant protein: ϵ280 nm = 0.701 cm−1·(g/liter)−1. The protein was diluted in deionized water to 1 mg/ml and mixed with an equal amount of buffer (100 mm Tris-HCl, pH 7.4) supplemented with 50 μg/ml sonicated native Fap fibrils, which acted as a template to ensure the assembly of FapC fibrils with a native structure. The native Fap fibrils were purified from P. aeruginosa PAO1 overexpressing the fap operon as described by Dueholm et al. (9). 200-μl samples were loaded in a 96-well plate. Thioflavin T was added to a final concentration of 40 μm in a subset of the wells to visualize the fibrillation process. Immediately afterward, the plate was transferred to a Tecan GENios Pro plate reader, and ThT fluorescence was measured using excitation at 448 nm, emission at 485 nm, and a gain of 60. Measurements were obtained by bottom reads every 2 min, and 30 s of shaking (orbital, amplitude 2.5 mm) was applied between the reads. Reads were integrated for 40 μs. Three fibrillation curves were averaged to reduce the signal/noise ratio. The samples without ThT were collected, sonicated, and used to seed a new generation of recombinant FapC fibrils as described above. This step was repeated twice to ensure a uniform fibril batch without non-protein contaminants originating from the native fibrils.
Expression, Isolation, and Fibrillation of α-Synuclein
Recombinant α-synuclein was purified and fibrillated in vitro by shaking-induced fibrillation as described elsewhere (34).
Transmission Electron Microscopy
Amyloid fibrils were mounted on 400-mesh carbon-coated, glow-discharged nickel grids for 30 s. Grids were washed with one drop of double-distilled water and stained with three drops of 10 μg/ml phosphotungstic acid at pH 7.2. Samples were inspected in a transmission electron microscope (JEOL, 1010) at 60 kV. Images were obtained using an electron-sensitive CCD camera (Olympus, KeenView). For size determination, a cross-line carbon replica grid (2160 lines/mm) was used.
FTIR
FTIR was carried out using a Tensor 27 (Bruker) FTIR spectrophotometer equipped with a DTGS mid-infrared detector and a Golden Gate single reflection diamond attenuated total reflectance cell (Specac). Fibrils were dried on the attenuated total reflectance crystal using dry nitrogen. Attenuated total reflectance spectra were recorded from 4000 to 1000 cm−1 using a nominal resolution of 2 cm−1 and 128 accumulations. Resulting spectra were baseline-corrected, and interfering signals from H2O and CO2 were removed using the atmospheric compensation filter in the OPUS version 5.5 system (Bruker).
Circular Dichroism
Circular dichroism (CD) spectra from 250 to 190 nm were collected on a Jasco J-810 spectropolarimeter using 0.2-nm steps, a scan speed of 50 nm/min, a bandwidth of 1 nm, and a response time of 1 s. A 1-mm quartz cuvette (Hellma) was used, and the temperature was kept constant at 20 °C with a thermostatically controlled cell holder (Jasco PTC 423S). The protein concentration was diluted to 0.1 mg/ml with double-distilled H2O, and all spectra were baseline-corrected with respect to double-distilled H2O. To improve the signal/noise ratio, five scans were averaged on each sample. Data points with a high tension voltage higher than 600 V were removed. The results were expressed as mean residue ellipticity.
SPR
FapC fibrils washed in double-distilled H2O were resupended in HBS-EP (10 mm HEPES, 150 mm NaCl, 3 mm EDTA, and 50 μm P20 (GE Healthcare), pH 7.4). Amyloid fibers were fragmented by sonication (80 Hz, 10 s) and centrifuged (14,000 × g, 30 s) to remove any large particles. 40 μl of centrate was mixed with 120 μl of 10 mm sodium acetate, pH 5.5, for the immobilization.
SPR experiments were performed at 25 °C using Biacore T-200 biosensor with research grade carboxymethylated (CM5S) sensor chips (Biacore, Uppsala, Sweden) with the exception of the binding of Congo Red, which was done in a Biacore 3000 instrument. Carboxymethylated CM5S chips were activated using 70 μl of 0.2 m N-ethyl-N′-(dimethylaminopropyl)carbodimide and 0.05 m N-hydroxysuccinimide in a 1:1 ratio. 100 μl of amyloids in sodium acetate solution (10 mm, pH 4.5) were injected over the activated surface, and an immobilization level of 8800 response units was reached. To confirm immobilization of the amyloids on the chip, solutions of 100 μm Congo Red (Sigma-Aldrich) or ThT (Sigma-Aldrich) in HBS-EP were injected over the immobilized amyloids for 45 s at a flow rate of 20 μl/min. Dissociation of the bound analytes was monitored for 1 min after the end of the injection. 20 μm stock solutions of analytes were prepared in HBS-EP with 5% DMSO. Serial 2-fold dilutions were performed in the same buffer to eight concentrations ranging from 0.16 to 20 μm. The chip surface was regenerated with HBS-EP for 100 s. All assays were performed in duplicate, and the averages are reported.
QS Reporter Assays
FapC fibrils and α-synuclein were suspended separately in buffer (10 mm HEPES, pH 7.4) to 4–5 μg/ml. 400 μl of each was then transferred to new 2.0-ml Eppendorf tubes, and QS molecules were added to a final concentration of 200 μm from a 25 mm stock in DMSO. The fibrils were incubated with the QS molecules overnight (22 °C, 200 rpm) and centrifuged (5 min, 1000 × g). The centrate was discarded, and the retentate was resuspended in 1 ml of buffer. This was repeated three times. Following the fourth centrifugation, the retentate was resuspended in 400 μl of buffer. This was performed for PQS and 3-oxo-C12-HSL.
P. aeruginosa PAO1 ΔpqsC PpqsA::gfp(ASV) and ΔlasIΔrhlI PlasB::gfp(ASV) were grown overnight (37 °C, 200 rpm) in LB medium containing carbenicillin (100 μg/ml) and gentamicin (30 μg/ml), respectively. Overnight cultures of the reporter strains were mixed in ratios of 1:100 (v/v) with ABTGC medium (AB basal salts + 0.1% (w/v) thiamine + 0.2% (w/v) glucose + 0.5% (w/v) casamino acids). 180-μl aliquots of the diluted cultures were dispensed into the wells of a clear-bottom 96-well cell culture plate (Thermo Scientific). The volume in each well was then made up to 200 μl with buffer or the amyloid stocks before and after exposure to QS molecules diluted to 100, 50, 21, 7, and 4% of the original concentration. Plates were placed into a Tecan Infinite M200Pro (Switzerland) and incubated at 37 °C with shaking for 10 s prior to measurement, and fluorescence was detected every 20 min. At least three replicates were performed per assay.
Electrical Characterization
Electrical conductivity of the amyloid fibrils was characterized through a two-terminal electrode measurement. The fibrils were first dissolved in double-distilled H2O and drop-casted onto a 200-nm silicon nitride surface for uniform dispersion across the channel length. Silicon nitride was deposited through the plasma-enhanced chemical vapor method, and chromium was employed as the adhesion layer for the interdigitated gold electrodes (channel width, 1000 μm; channel length, 30 μm). This layer was fabricated through a series of lithography and electron beam deposition procedures. The substrates went through a standard cleaning process: acetone sonication (5 min), isopropyl alcohol sonication (5 min), deionized water sonication (5 min), and ultraviolet air plasma (2 min). 100 μl of dissolved fibrils were then drop-casted onto the cleaned substrate and allowed to dry overnight in the dark in ambient conditions to achieve a thin film through complete solvent evaporation. Current-voltage profiles were measured by a Keithley 4200 semiconductor characterization system interfaced to a Desert cryogenic probe station.
RESULTS
Preparation and Immobilization of Amyloid Fibers onto SPR Chip
Native Fap fibrils purified from Fap-overexpressing P. aeruginosa strains usually contain considerable amounts of non-protein contaminants, such as extracellular polysaccharides. These contaminants hinder determination of the specific binding affinity between metabolites and the Fap fibrils. In vitro formed fibrils derived from recombinant FapC are clean of contaminants, but their structure may not be the same as the native amyloid structure due to the polymorphism associated with non-constrained fibril assembly (35).
In order to obtain clean native folded PAO1 Fap fibrils for binding studies, we took advantage of the autocatalytic nature of amyloid fibrils (36). Fibrillation kinetics of recombinant PAO1 FapC were greatly enhanced in the presence of 5% sonicated native PAO1 Fap fibrils, ensuring transmission of the native structure to the in vitro formed fibrils (Fig. 1A). The formed fibrils were then sonicated and used as seeds for another in vitro fibrillation in order to dilute contaminants originating from the native fibril seeds. This process was repeated to produce third generation PAO1 FapC fibrils.
FIGURE 1.
A, fibrillation kinetics of FapC unseeded or in the presence of 5% fibrils seeds visualized by ThT fluorescence. B, FTIR spectrum of third generation PAO1 FapC fibrils showing the amyloid characteristic peak between 1620 and 1630 cm−1. C, circular dichroism spectrum of third generation PAO1 FapC fibrils showing a clear β-sheet signature with a single minimum at 217 nm. D, transmission electron microscopy images of third generation PAO1 FapC fibrils.
The amyloid structure of the produced fibrils was confirmed by FTIR by the presence of the major amyloid characteristic peak between 1615 and 1630 cm−1 (37) (Fig. 1B). The amyloid structure was furthermore supported by the circular dichroism (CD) spectra of the same fibrils, which showed a clear β-sheet signature (38) (Fig. 1C). Transmission electron microscopy confirmed the presence of clean, homogeneous fibrils similar to the native fibrils described previously for P. aeruginosa PAO1 (9) (Fig. 1D). SDS-PAGE of the produced fibrils confirmed that all monomers were converted into amyloids (data not shown).
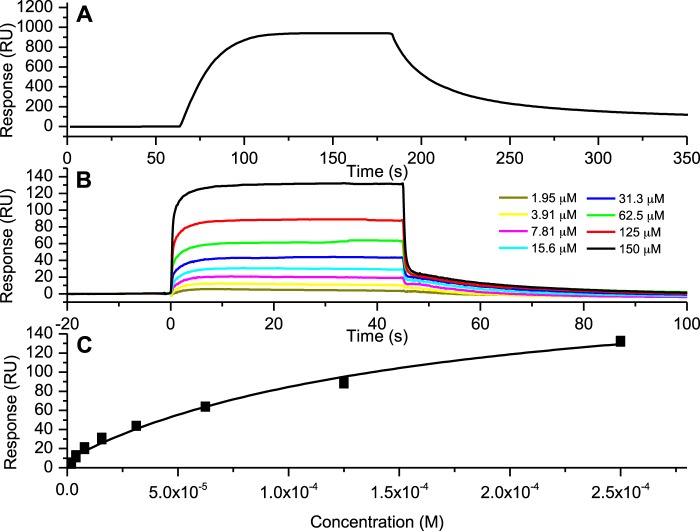
The third generation FapC fibrils were fragmented by sonication and immobilized onto CM5S sensor chips for binding studies using amine-coupling chemistry. Congo Red and ThT bind to amyloids and can be used as diagnostic indicators of amyloidosis (39). Therefore, to confirm FapC fibril immobilization on the chip surface, the binding responses of Congo Red and ThT were followed. The sensorgram for Congo Red (Fig. 2A) clearly shows an association and a dissociation response. For ThT, on the other hand (Fig. 2B), a box-shaped response was observed, which is typical for weak or transient interactions. A concentration dependence of the response unit value following ThT injection confirmed that the amyloids had bound ThT (Fig. 2C). The equilibrium dissociation constant (KD) for this ThT binding to amyloids was close to 100 μm, which is consistent with ThT acting as a transient affinity binding compound, as noted by Kawatake et al. (40). The binding responses of both Congo Red and ThT to the Pseudomonas amyloids were similar to those reported for Congo Red and ThT to amyloid prion proteins (40). These results indicate that amyloids had been immobilized successfully on the CM5S chip.
FIGURE 2.
A and B, sensorgrams showing the interaction of Congo Red at 10 μm (A) and ThT at concentrations of 1.95–150 μm (B) with FapC fibrils. C, dose-response curve for the interaction between ThT and FapC fibrils. RU, response units.
QS Molecules Display Transient Binding to FapC Fibrils
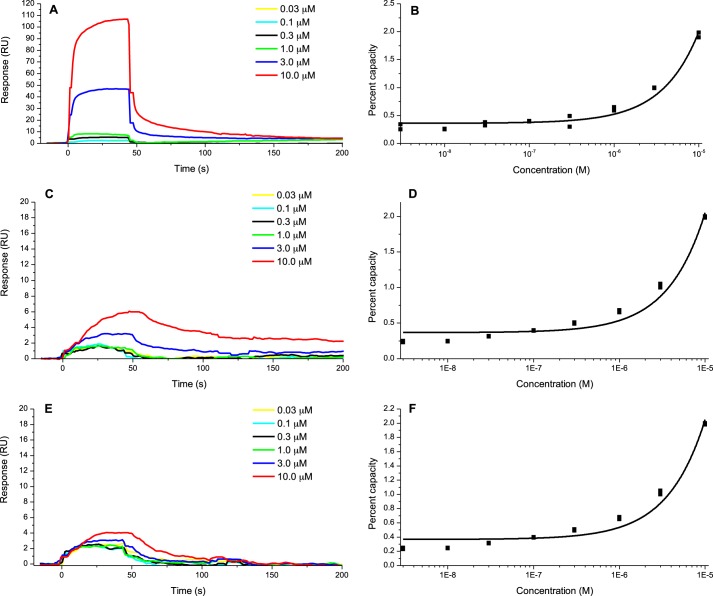
The binding affinities of the QS molecules pyocyanin, PQS, and 3-oxo-C12-HSL to the FapC fibrils were measured at concentrations ranging from 0.03 to 10 μm (Fig. 3, A, C, and E). Due to the limited solubility of each of these molecules in SPR running buffers, it was not possible to exceed this concentration, although these are within the range of biological relevance (41, 42). As with the binding response for ThT, a concentration-dependent box-shaped sensorgram was observed for the interactions of pyocyanin with the FapC fibrils, displaying clear association following the onset of injection and then dissociation at cessation of injection. For PQS and 3-oxo-C12-HSL the response was less pronounced, and the association and dissociation responses were more protracted. The magnitude of change in RI depends on the thickness and refractive layer of the molecular layer (43). The greater absolute change in RI elicited by pyocyanin interactions with FapC fibrils could reflect only that colored molecules like pyocyanin alter RI by a greater amount. It is therefore important to look at the concentration dependence of the responses. Here there is clearly concentration dependence, demonstrating binding between all of these metabolites and the FapC fibrils.
FIGURE 3.
A, C, and E, sensorgrams showing the interaction of pyocyanin (A), PQS (C), and 3-oxo-C12-HSL (E) with FapC fibrils at concentrations ranging from 0.03 to 10 μm. B, D, and F, dose-response curves for the interaction between pyocyanin (B), PQS (D), and 3-oxo-C12-HSL (F) and FapC fibrils. Data are fit to a non-linear dose-response curve (i.e. sigmoidal), with 100% site occupation determined from extrapolation. RU, response units.
The average maximum binding responses (in response units) were plotted against pyocyanin, PQS, and 3-oxo-C12-HSL concentrations, and the data were fitted to the steady state model of the evaluation software of Biacore T-200 (44) (Fig. 3, B, D, and F). KD values between the FapC fibrils and pyocyanin, PQS, and 3-oxo-C12-HSL were 400, 250, and 100 μm, respectively. These values and the box-shaped interaction kinetics indicate that they have transient binding to the fibrils (45). Metabolite binding was therefore similar to that of ThT, which is commonly used as a diagnostic tool for amyloids because of its altered spectral properties upon amyloid binding (46).
Hydrophobicity Predisposes Metabolites to FapC Fibril Binding
The three QS molecules screened are structurally different from each other and the positive control ThT (Fig. 4). To investigate whether features specific to P. aeruginosa metabolites predispose them to Fap fibril binding, binding responses to FapC fibrils of a range of biological molecules with different hydrophobicities and structures were determined (Figs. 5 and 6).
FIGURE 4.
Representative chemical structures of molecules belonging to three classes of P. aeruginosa quorum-sensing molecules, pyocyanin (A), PQS (B), and 3-oxo-C12-HSL (C), and thioflavin T (D).
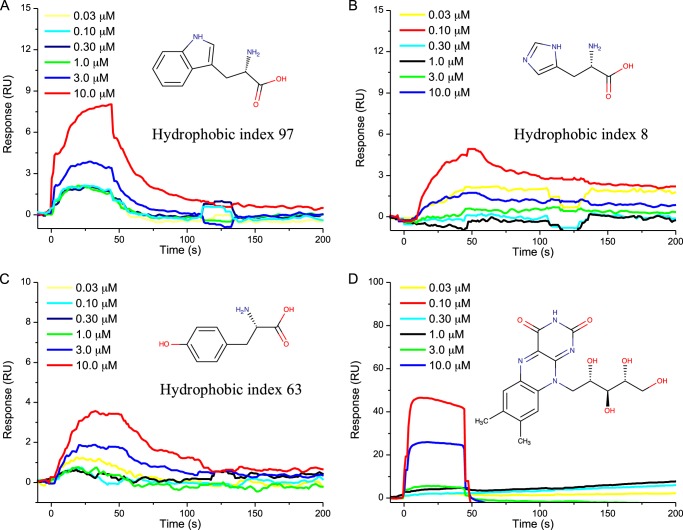
FIGURE 5.
Sensorgrams showing the interaction of l-tryptophan (A), l-histidine (B), l-tyrosine (C), and riboflavin (D) with FapC fibrils at concentrations ranging from 0.03 to 10 μm. Chemical structures and hydrophobicity indices (for amino acids only) are presented in the insets. RU, response units.
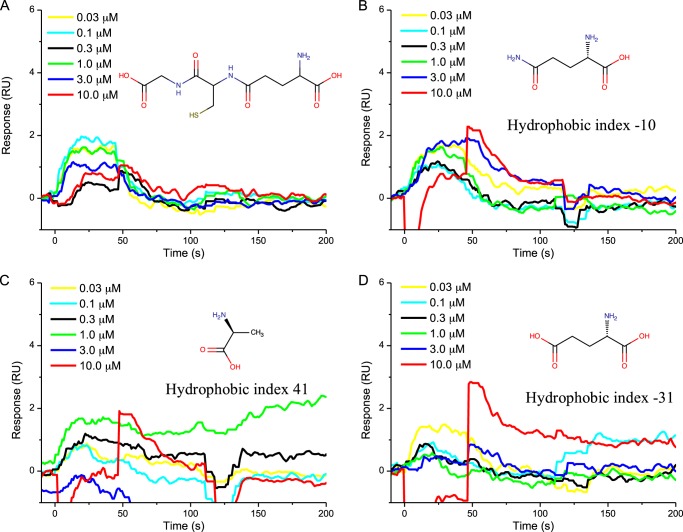
FIGURE 6.
Sensorgrams showing the non-interaction of l-glutathione (A), l-glutamine (B), l-alanine (C), and l-glutamate (D) with FapC fibrils at concentrations ranging from 30 nm to 10 μm. Chemical structures and hydrophobicity indices (for amino acids only) are presented in the insets. RU, response units.
For the amino acids, the highest binding response was observed for l-tryptophan, which also has the highest hydrophobic index of all amino acids screened, as illustrated by the sensorgram (Fig. 5A) displaying gradual association and dissociation responses. Furthermore, average maximum binding response for l-tryptophan to FapC fibrils increased with molecular concentration (KD = 3 mm). Weaker binding responses were detected for the next most hydrophobic molecules, l-tyrosine (Fig. 5C) and l-histidine (Fig. 5B), with KD = 100 mm for both. No measurable binding response was observed for glutathione, l-glutamine, l-alanine, or l-glutamate (Fig. 6). They all have lower hydrophobic indices.
To investigate whether molecules functionally and structurally analogous to P. aeruginosa QS molecules, expressed by other organisms, bind with similar affinities to FapC fibrils, the binding response of riboflavin to FapC fibrils was screened. Like pyocyanin, riboflavin is hydrophobic with a central annulated pyrazine (Fig. 4D). As a soluble redox mediator, it performs a similar role for Shewanella oneidensis MR-1 as pyocyanin for P. aeruginosa (47). A binding response analogous to the one obtained for pyocyanin was observed for riboflavin (Fig. 5D), with strong response signal, box-shaped sensorgram, clear association and dissociation responses, and KD = 500 μm.
Binding therefore appears not to be specific for P. aeruginosa metabolites. Hydrophobicity, however, appears to predispose molecules to Fap fibril binding, as indicated by the fact that only the most hydrophobic amino acid tested achieved significant binding affinities to the amyloid fibrils. This has implications not only for binding of P. aeruginosa QS molecules to amyloids but also retention of hydrophobic QS molecules and redox mediators expressed by Gram-negative bacteria more generally (48).
Binding Responses of QS Molecules Are Characteristic of Fap Fibrils
The specificity of QS molecule binding behavior for Fap fibrils versus non-amyloid proteins was tested using bovine serum albumin (BSA) and tobacco etch virus protease (i.e. His-tagged). Negligible binding to BSA and tobacco etch virus protease was recorded for pyocyanin, PQS, and 3-oxo-C12-HSL (data not shown).
Binding responses were screened for QS molecules to FapC monomers to determine whether fibrillation of FapC is a requirement for QS molecule binding. FapC monomers were immobilized onto the CM5S sensor within 30 min of desalting. In the absence of a fibril seed and within this short time period, the non-fibril state of the FapC monomers was probably preserved during immobilization (Fig. 1A). No measurable binding response was detected for any of the QS molecules screened against the FapC monomers (data not shown).
The binding responses of QS molecules to amyloid fibrils of the mammalian protein α-synuclein were then screened to determine whether their binding responses reflect a general feature of all amyloids and not just FapC fibrils from Pseudomonas (Fig. 7). Although pyocyanin was the only metabolite examined that bound to α-synuclein, this metabolite displayed a 10-fold lower affinity for the α-synuclein compared with that of binding to the FapC fibrils (KD = 4 mm). This shows that metabolite binding observed in this study is not a generic feature of all amyloid fibrils.
FIGURE 7.
A, transmission electron microscopy image of α-synuclein fibrils; B, dose-response curves for the interaction between quorum-sensing molecules and control amino acids with α-synuclein fibrils. RU, response units.
Fap fibrils can act as a reservoir for quorum-sensing compounds under non-quiescent conditions. The transient binding affinity of QS molecules to the FapC fibrils suggests that the fibrils are able to retain QS compounds under non-quiescent conditions and then act as a reservoir for these compounds. To provide evidence for this model, we employed P. aeruginosa reporter assays for PQS and 3-oxo-C12-HSL. The pqsC gene is required for the synthesis of the PQS precursor HHQ, and the lasI and RhlI genes are required for synthesis of 3-oxo-C12-HSL. The accompanying deletion strains are therefore unable to synthesize the two QS molecules on their own, and the fusion of the GFP gene to the pqsABCDE and lasB promoters allows PQS and 3-oxo-C12-HSL, respectively, to be monitored in terms of GFP fluorescence intensity (30, 32).
FapC fibrils were incubated separately with PQS and 3-oxo-C12-HSL for 24 h and then washed four times in buffer to remove unbound QS molecules, thus simulating non-quiescent conditions. The reporter strains were then challenged with various concentrations of the washed FapC fibrils, and the accessible QS molecules were detected directly by GFP fluorescence (Fig. 8).
FIGURE 8.
A and C, transcription of pqsA in a P. aeruginosa pqsA::gfp(ASV) reporter strain unable to synthesize PQS upon the addition of various concentrations of pure PQS (A) or PAO1 FapC fibrils (C) preincubated with PQS. B and D, transcription of lasB in a P. aeruginosa lasB::gfp(ASV) reporter strain unable to synthesize 3-oxo-C12-HSL upon the addition of various concentrations of pure 3-oxo-C12-HSL (B) or PAO1 FapC fibrils preincubated with 3-oxo-C12-HSL (C). E and F, transcription of pqsA in a P. aeruginosa pqsA::gfp(ASV) reporter strain upon the addition of various concentrations of FapC fibrils without PQS preincubation (E) and α-synuclein fibrils preincubated with PQS (F). All fibrils were washed four times with buffer to remove unbound QS molecules.
When buffer was added as a negative control, a minimal increase in the GFP signal was recorded for both reporter strains, confirming their inability to synthesize the QS molecules. A clear increase was observed when standard solutions of PQS and 3-oxo-C12-HSL were added at the onset of the growth experiment, thus demonstrating the sensitivity of the reporter strain to the QS molecules (Fig. 8, A and B). The intensity of the GFP signal generated by both reporter strains normalized by cell density increased with concentration of QS-exposed FapC fibrils added even after four cycles of washing (Fig. 8, C and D). FapC fibrils not preincubated with QS molecules did not affect the GFP signal of the reporter strains (Fig. 8E), suggesting that the increase is due to the retention of QS molecules and not the fibrils themselves. The presence of FapC fibrils therefore prevented the loss of these QS molecules by dilution, which is consistent with the observation by SPR that they bind to FapC fibrils.
On the other hand, no binding of either PQS or 3-oxo-C12-HSL to α-synuclein was observed by SPR. To validate SPR as a predictor of QS molecule retention, the serial dilution assays were repeated for both QS molecules with α-synuclein (Fig. 8F). α-Synuclein preincubated with PQS and subjected to sequential washing failed to increase the GFP signal intensity of the reporter strains. The same was observed for α-synuclein preincubated with 3-oxo-C12-HSL (data not shown). Unlike FapC fibrils, the mammalian amyloid protein α-synuclein therefore could not prevent the loss of QS molecules by dilution, also consistent with the predictions by SPR.
Pyocyanin Binding Can Enhance Electrical Conductivity of Fap Fibrils under Non-quiescent Conditions
It has previously been shown that amyloid fibrils, such as those composed of the elastin-related polypeptide poly(Val-Gly-Gly-Leu-Gly), can conduct electrical signals (49). This conductivity was attributed to partial delocalization of peptide electrons caused by the high density of hydrogen bonding in β-sheet structures.
As a redox mediator, pyocyanin can enhance conductivity in biofilms (50). Binding of pyocyanin may therefore improve the conductive properties for Fap fibrils. To determine whether this is the case, FapC fibrils were incubated with or without pyocyanin and then washed four times in distilled water to remove unbound PQS and deposited by drop-casting onto interdigitated gold. Without exposure to pyocyanin, average current values of ∼1 nA were recorded for the FapC fibrils, similar to that determined for elastin-related amyloid fibrils (49) but considerably lower than that measured for α-synuclein (∼17 nA) (Fig. 9). Drop-casted pyocyanin onto the electrodes produced a maximum current of ∼600 nA (Fig. 9). Preincubation of FapC fibrils and α-synuclein with pyocyanin elevated their conductivities to ∼200 and 170 nA, respectively (Fig. 9), despite four cycles of washing with distilled water. This confirms the robust nature of the pyocyanin binding onto the amyloid fibrils. Additionally, current generation in the fibrils remained relatively stable following each scan.
FIGURE 9.
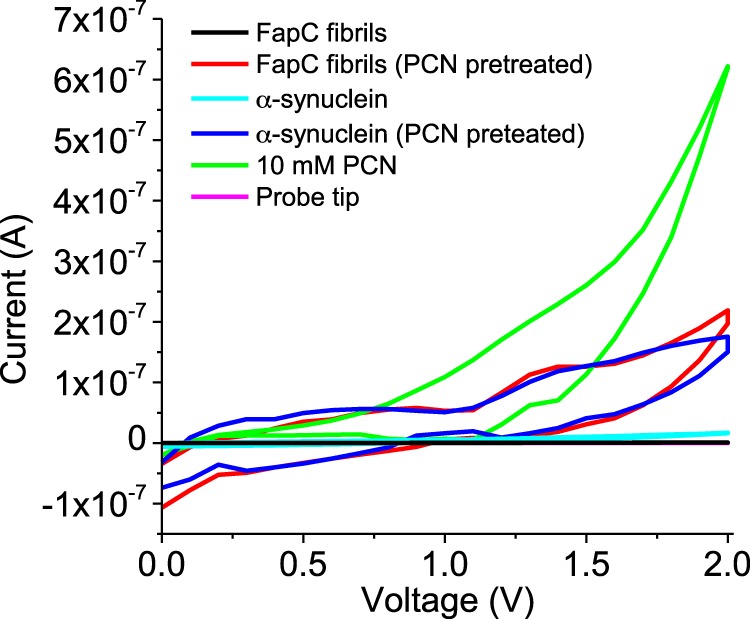
Current-voltage (I-V) characteristics of PAO1 FapC and α-synuclein fibrils with and without preincubation with pyocyanin (PCN). A 10 mm PCN solution following deposition and evaporation and the probe tip alone were used as positive and negative controls, respectively. All fibrils were washed four times with buffer to remove unbound QS molecules.
DISCUSSION
Direct and quantitative evidence is provided here that strongly suggests a function for Fap fibrils in binding extracellular metabolites, such as quorum-sensing molecules and redox mediators, with pyocyanin, PQS, and 3-oxo-C12-HSL used in this study to illustrate this. Pyocyanin is a direct determinant of P. aeruginosa virulence (51, 52) and enables the cells to maintain redox balance under anaerobic conditions (53). PQS protects against oxidative stress (54) and induces lasB and rhlI expression to disrupt host immune responses (55, 56). Interactions between such clinically important agents and a constituent of the extracellular matrix of P. aeruginosa biofilms (9) are implicitly significant and show that Fap fibrils can facilitate the retention and release of extracellular metabolites when required. Thus, molecular retention needs to be added to the properties assigned to bacterial amyloids (i.e. their ability to facilitate interactions with host proteins, through curli (2, 57), and to contribute to biofilm formation and adhesion, as is the case for TasA and Fap) (8, 58).
There appears to be growing evidence that matrix components have multiple functions, as demonstrated by extracellular DNA, which are involved in the chelation of cations and antibiotic resistance (59), horizontal gene transfer (60), and matrix self-organization (61). Multiple biological roles are also seen for the Psl exopolysaccharide of P. aeruginosa, which, besides being a structural component of biofilms (62), also displays regulatory functions (63). Whether such effects are primary or secondary effects of the EPS synthesis remain to be established. For example, the ability of amyloids to capture signaling molecules suggests an indirect role in mediating cell function. Nonetheless, as with amyloid fibrils, synthesis of these additional matrix components has a range of phenotypic consequences, and indeed products and traits with multiple functions of individual components or products are commonplace adaptive features of bacteria and may be necessary for resilience.
The task of assigning motive for EPS synthesis and understanding whether secondary effects are deliberate strategies of the bacteria or unintended consequences is therefore complicated. Performing direct and quantitative functional assignment on a matrix constituent does not address the question of motive. It does, however, provide key information in the causal chain between an EPS and phenotype. Similar to observations made for the exopolysaccharide Psl, a regulatory role was recently also demonstrated for Fap fibrils on the basis that they elicit major changes to the proteome of P. aeruginosa (64). Fap proteins are not expected to have any direct regulatory capacities, and the proteomic changes were postulated as being secondary effects resulting from interference by Fap fibrils of quorum-sensing signaling (64). By means of single molecular functional analysis on FapC fibrils, we can therefore hone in specifically on these interactions between matrix constituents and QS molecules and provide further support to the hypothesis that such interactions have important consequences. Such QS molecules are highly concentrated in biofilms (65), and we demonstrate in this work that as long as these QS molecules are expressed in a biofilm containing FapC fibrils, the fibrils will serve to prevent their loss from dilution under non-quiescent conditions.
Additionally, demonstrating an interdependence between matrix constituents and QS molecules highlights a shortcoming of a popular approach toward allocating a functional assignment of EPS in biofilms, which is to generate EPS knockdown mutants and then indirectly assign functions by observing any phenotypic differences (12, 66). As demonstrated here, changing the EPS balance will set off a cascade of secondary effects by, for example, affecting local concentrations of metabolites in the cell environment. It is thus important to employ a method for direct functional assignment for individual EPS.
Transient interactions are common and often advantageous in biological systems. In dense biofilm matrices, there are many binding sites within close proximity to each other. Rapid association and dissociation kinetics, as demonstrated here, allow for greater bioavailability (45). Rapidly dissociated QS molecules will quickly rebind to the nearest site with parallel and serial bindings that can augment binding strength (67). This is the basis of improving efficacy of many drugs and small inhibitors (68). FapC fibrils are therefore likely to be important in turbulent and competitive environments commonly encountered in non-laboratory environments (e.g. wastewater treatment plants and acute respiratory infections). The sorptive properties of amyloids ensure that they would minimize the loss or interception by other bacterial cells of molecules required to transfer information between individual producing cells. Synthesis of amyloids could therefore minimize the loss of metabolites to other bacteria as well as enable P. aeruginosa to capture and store metabolites produced by other bacteria. Amyloid binding may also be significant for siderophores and a range of other public good molecules.
Unlike the dissimilatory metal reducers Geobacter and Shewanella, conductivity is not considered an essential property of P. aeruginosa biofilms any more than it is considered a necessary function of pyocyanin (53, 69). However, this study illustrates quantitatively how an EPS might contribute to extracellular electron transfer. Researchers are still divided on how cells might achieve extracellular electron conduction, with metallic-like conductivity (70) and superexchange, in which electrons are conducted by a succession of electron transfer reactions (71), identified as two possible mechanisms. Enhanced conductivity due to reversible binding of redox-active agents along the length of the EPS is therefore consistent with the superexchange explanation.
Furthermore, although extracellular polymeric substances are thought to serve an important function in retaining molecules and proteins, both endogenous and exogenous, in biofilms (27), to the authors' best knowledge, direct binding of a metabolite to any individual EPS component has hitherto not been quantified. Das et al. (72) demonstrated pyocyanin binding to extracellular DNA, based on the displacement of ethidium bromide from DNA following pyocyanin addition. SPR was used here not only to demonstrate binding between clinically relevant metabolites and an EPS component but furthermore to quantify this interaction and describe binding specificity. Quantification of binding affinities is essential in establishing and assessing the significance of metabolite-EPS interactions.
Establishing that bacterial amyloids bind QS molecules in an orderly manner suggests a function for them as reservoirs for information transfer. These results unveil a mechanism by which biofilm communities can manage information transfer in non-quiescent and competitive environments. Amyloid expression may thus confer a competitive advantage on some populations under such conditions. It remains to be seen whether amyloids are matrix components of general benefit in biofilms by their ability to retain all signaling molecules or only those expressed by certain strains. A better understanding of binding specificity could lead to biofilm mitigation or management strategies that target metabolite binding sites on matrix components like amyloid polymers.
The Singapore Centre on Environmental Life Sciences Engineering is supported by Singapore's National Research Foundation, Ministry of Education, Nanyang Technological University (NTU), and National University of Singapore (NUS), and hosted by NTU in partnership with NUS. Electrical characterization was performed at the Energy Research Institute at NTU.
- EPS
- extracellular polymeric substance(s)
- ThT
- thioflavin T
- QS
- quorum-sensing
- 3-oxo-C12-HSL
- N-(3-oxododecanoyl)-l-homoserine lactone
- PQS
- 2-heptyl-3-hydroxy-4(1H)-quinolone
- HHQ
- 4-hydroxy-2-heptylquinoline
- SPR
- surface plasmon resonance
- FAP
- functional amyloid(s) from Pseudomonas.
REFERENCES
- 1. Bieler S., Estrada L., Lagos R., Baeza M., Castilla J., Soto C. (2005) Amyloid formation modulates the biological activity of a bacterial protein. J. Biol. Chem. 280, 26880–26885 [DOI] [PubMed] [Google Scholar]
- 2. Barnhart M. M., Chapman M. R. (2006) Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shewmaker F., McGlinchey R. P., Wickner R. B. (2011) Structural insights into functional and pathological amyloid. J. Biol. Chem. 286, 16533–16540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dueholm M. S., Nielsen P. H., Chapman M., Otzen D. (2013) in Amyloid Fibrils and Prefibrillar Aggregates (Otzen D. E., ed) pp. 411–438, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany [Google Scholar]
- 5. Blanco L. P., Evans M. L., Smith D. R., Badtke M. P., Chapman M. R. (2012) Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20, 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Larsen P., Nielsen J. L., Dueholm M. S., Wetzel R., Otzen D., Nielsen P. H. (2007) Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9, 3077–3090 [DOI] [PubMed] [Google Scholar]
- 7. Larsen P., Nielsen J. L., Otzen D., Nielsen P. H. (2008) Amyloid-like adhesins produced by floc-forming and filamentous bacteria in activated sludge. Appl. Environ. Microbiol. 74, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romero D., Aguilar C., Losick R., Kolter R. (2010) Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 2230–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dueholm M. S., Søndergaard M. T., Nilsson M., Christiansen G., Stensballe A., Overgaard M. T., Givskov M., Tolker-Nielsen T., Otzen D. E., Nielsen P. H. (2013) Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen 2, 365–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olsen A., Herwald H., Wikstrom M., Persson K., Mattsson E., Björck L. (2002) Identification of two protein-binding and functional regions of curli, a surface organelle and virulence determinant of Escherichia coli. J. Biol. Chem. 277, 34568–34572 [DOI] [PubMed] [Google Scholar]
- 11. Seviour T., Malde A. K., Kjelleberg S., Yuan Z., Mark A. E. (2012) Molecular dynamics unlocks atomic level self-assembly of the exopolysaccharide matrix of water-treatment granular biofilms. Biomacromolecules 13, 1965–1972 [DOI] [PubMed] [Google Scholar]
- 12. Ghafoor A., Hay I. D., Rehm B. H. A. (2011) Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77, 5238–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mathee K., Ciofu O., Sternberg C., Lindum P. W., Campbell J. I. A., Jensen P., Johnsen A. H., Givskov M., Ohman D. E., Molin S., Høiby N., Kharazmi A. (1999) Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 145, 1349–1357 [DOI] [PubMed] [Google Scholar]
- 14. Hall-Stoodley L., Costerton J. W., Stoodley P. (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 [DOI] [PubMed] [Google Scholar]
- 15. Dueholm M. S., Petersen S. V., Sønderkaer M., Larsen P., Christiansen G., Hein K. L., Enghild J. J., Nielsen J. L., Nielsen K. L., Nielsen P. H., Otzen D. E. (2010) Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020 [DOI] [PubMed] [Google Scholar]
- 16. Dietrich L. E. P., Price-Whelan A., Petersen A., Whiteley M., Newman D. K. (2006) The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61, 1308–1321 [DOI] [PubMed] [Google Scholar]
- 17. Pearson J. P., Gray K. M., Passador L., Tucker K. D., Eberhard A., Iglewski B. H., Greenberg E. P. (1994) Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U.S.A. 91, 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Passador L., Tucker K. D., Guertin K. R., Journet M. P., Kende A. S., Iglewski B. H. (1996) Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J. Bacteriol. 178, 5995–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davies D. G., Marques C. N. H. (2009) A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pesci E. C., Milbank J. B. J., Pearson J. P., McKnight S., Kende A. S., Greenberg E. P., Iglewski B. H. (1999) Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96, 11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diggle S. P., Matthijs S., Wright V. J., Fletcher M. P., Chhabra S. R., Lamont I. L., Kong X., Hider R. C., Cornelis P., Cámara M., Williams P. (2007) The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 14, 87–96 [DOI] [PubMed] [Google Scholar]
- 22. Hunter R. C., Klepac-Ceraj V., Lorenzi M. M., Grotzinger H., Martin T. R., Newman D. K. (2012) Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am. J. Respir. Cell Mol. Biol. 47, 738–745 [DOI] [PubMed] [Google Scholar]
- 23. Mashburn-Warren L., Howe J., Garidel P., Richter W., Steiniger F., Roessle M., Brandenburg K., Whiteley M. (2008) Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol. Microbiol. 69, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mashburn L. M., Whiteley M. (2005) Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425 [DOI] [PubMed] [Google Scholar]
- 25. Purevdorj B., Costerton J. W., Stoodley P. (2002) Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 68, 4457–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seviour T., Yuan Z., van Loosdrecht M. C. M., Lin Y. (2012) Aerobic sludge granulation: a tale of two polysaccharides? Water Res. 46, 4803–4813 [DOI] [PubMed] [Google Scholar]
- 27. Flemming H.-C., Wingender J. (2010) The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 [DOI] [PubMed] [Google Scholar]
- 28. Paravastu A. K., Leapman R. D., Yau W.-M., Tycko R. (2008) Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 105, 18349–18354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wasmer C., Lange A., Van Melckebeke H., Siemer A. B., Riek R., Meier B. H. (2008) Amyloid fibrils of the HET-s(218–289) prion form a β solenoid with a triangular hydrophobic core. Science 319, 1523–1526 [DOI] [PubMed] [Google Scholar]
- 30. Yang L., Barken K. B., Skindersoe M. E., Christensen A. B., Givskov M., Tolker-Nielsen T. (2007) Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153, 1318–1328 [DOI] [PubMed] [Google Scholar]
- 31. Wang V. B., Chua S.-L., Cao B., Seviour T., Nesatyy V. J., Marsili E., Kjelleberg S., Givskov M., Tolker-Nielsen T., Song H., Loo J. S. C., Yang L. (2013) Engineering PQS biosynthesis pathway for enhancement of bioelectricity production in Pseudomonas aeruginosa microbial fuel cells. PLoS One 8, e63129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang L., Rybtke M. T., Jakobsen T. H., Hentzer M., Bjarnsholt T., Givskov M., Tolker-Nielsen T. (2009) Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 53, 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hentzer M., Wu H., Andersen J. B., Riedel K., Rasmussen T. B., Bagge N., Kumar N., Schembri M. A., Song Z., Kristoffersen P., Manefield M., Costerton J. W., Molin S., Eberl L., Steinberg P., Kjelleberg S., Høiby N., Givskov M. (2003) Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22, 3803–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lorenzen N., Lemminger L., Pedersen J. N., Nielsen S. B., Otzen D. E. (2014) The N-terminus of α-synuclein is essential for both monomeric and oligomeric interactions with membranes. FEBS Lett. 588, 497–502 [DOI] [PubMed] [Google Scholar]
- 35. Pedersen J. S., Dikov D., Flink J. L., Hjuler H. A., Christiansen G., Otzen D. E. (2006) The changing face of glucagon fibrillation: structural polymorphism and conformational imprinting. J. Mol. Biol. 355, 501–523 [DOI] [PubMed] [Google Scholar]
- 36. Dueholm M. S., Nielsen S. B., Hein K. L., Nissen P., Chapman M., Christiansen G., Nielsen P. H., Otzen D. E. (2011) Fibrillation of the major curli subunit CsgA under a wide range of conditions implies a robust design of aggregation. Biochemistry 50, 8281–8290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zandomeneghi G., Krebs M. R., McCammon M. G., Fändrich M. (2004) FTIR reveals structural differences between native β-sheet proteins and amyloid fibrils. Protein Sci. 13, 3314–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Herva M. E., Zibaee S., Fraser G., Barker R. A., Goedert M., Spillantini M. G. (2014) Anti-amyloid compounds inhibit α-synuclein aggregation induced by protein misfolding cyclic amplification (PMCA). J. Biol. Chem. 289, 11897–11905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Groenning M. (2010) Binding mode of Thioflavin T and other molecular probes in the context of amyloid fibrils: current status. J. Chem. Biol. 3, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kawatake S., Nishimura Y., Sakaguchi S., Iwaki T., Doh-ura K. (2006) Surface plasmon resonance analysis for the screening of anti-prion compounds. Biol. Pharm. Bull. 29, 927–932 [DOI] [PubMed] [Google Scholar]
- 41. Usher L. R., Lawson R. A., Geary I., Taylor C. J., Bingle C. D., Taylor G. W., Whyte M. K. B. (2002) Induction of neutrophil apoptosis by the Pseudomonas aeruginosa exotoxin pyocyanin: a potential mechanism of persistent infection. J. Immunol. 168, 1861–1868 [DOI] [PubMed] [Google Scholar]
- 42. Zhang L., Gao Q., Chen W., Qin H., Hengzhuang W., Chen Y., Yang L., Zhang G. (2013) Regulation of pqs quorum sensing via catabolite repression control in Pseudomonas aeruginosa. Microbiology 159, 1931–1936 [DOI] [PubMed] [Google Scholar]
- 43. Stenberg E., Persson B., Roos H., Urbaniczky C. (1991) Quantitative determination of surface concentration of protein with surface plasmon resonance using radiolabeled proteins. J. Colloid Interface Sci. 143, 513–526 [Google Scholar]
- 44. Roden L. D., Myszka D. G. (1996) Global analysis of a macromolecular interaction measured on BIAcore. Biochem. Biophys. Res. Commun. 225, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 45. Ohlson S. (2008) Designing transient binding drugs: a new concept for drug discovery. Drug Discovery Today 13, 433–439 [DOI] [PubMed] [Google Scholar]
- 46. LeVine H. (1993) Thioflavine T interaction with synthetic Alzheimer's disease β-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 2, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marsili E., Baron D. B., Shikhare I. D., Coursolle D., Gralnick J. A., Bond D. R. (2008) Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 3968–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Williams P., Winzer K., Chan W. C., Cámara M. (2007) Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 1119–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Del Mercato L. L., Pompa P. P., Maruccio G., Della Torre A., Sabella S., Tamburro A. M., Cingolani R., Rinaldi R. (2007) Charge transport and intrinsic fluorescence in amyloid-like fibrils. Proc. Natl. Acad. Sci. U.S.A. 104, 18019–18024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rabaey K., Boon N., Höfte M., Verstraete W. (2005) Microbial phenazine production enhances electron transfer in biofuel cells. Environ. Sci. Technol. 39, 3401–3408 [DOI] [PubMed] [Google Scholar]
- 51. Jimenez P. N., Koch G., Thompson J. A., Xavier K. B., Cool R. H., Quax W. J. (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Britigan B. E., Railsback M. A., Cox C. D. (1999) The Pseudomonas aeruginosa secretory product pyocyanin inactivates α1 protease inhibitor: implications for the pathogenesis of cystic fibrosis lung disease. Infect. Immun. 67, 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Price-Whelan A., Dietrich L. E. P., Newman D. K. (2007) Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 6372–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Häussler S., Becker T. (2008) The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 4, e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Calfee M. W., Coleman J. P., Pesci E. C. (2001) Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 98, 11633–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Diggle S. P., Winzer K., Chhabra S. R., Worrall K. E., Cámara M., Williams P. (2003) The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50, 29–43 [DOI] [PubMed] [Google Scholar]
- 57. Dueholm M. S., Albertsen M., Otzen D., Nielsen P. H. (2012) Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One 7, e51274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dueholm M. S., Otzen D., Nielsen P. H. (2013) Evolutionary insight into the functional amyloids of the pseudomonads. PLoS One 8, e76630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mulcahy H., Charron-Mazenod L., Lewenza S. (2008) Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4, e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Harmsen M., Lappann M., Knøchel S., Molin S. (2010) Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 76, 2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gloag E. S., Turnbull L., Huang A., Vallotton P., Wang H., Nolan L. M., Mililli L., Hunt C., Lu J., Osvath S. R., Monahan L. G., Cavaliere R., Charles I. G., Wand M. P., Gee M. L., Prabhakar R., Whitchurch C. B. (2013) Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Natl. Acad. Sci. U.S.A. 110, 11541–11546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Colvin K. M., Irie Y., Tart C. S., Urbano R., Whitney J. C., Ryder C., Howell P. L., Wozniak D. J., Parsek M. R. (2012) The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Irie Y., Borlee B. R., O'Connor J. R., Hill P. J., Harwood C. S., Wozniak D. J., Parsek M. R. (2012) Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 109, 20632–20636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Herbst F.-A., Søndergaard M. T., Kjeldal H., Stensballe A., Nielsen P. H., Dueholm M. S. (2014) Major proteomic changes associated with amyloid-induced biofilm formation in Pseudomonas aeruginosa PAO1. J. Proteome Res. 10.1021/pr500938x [DOI] [PubMed] [Google Scholar]
- 65. Ramos I., Dietrich L. E. P., Price-Whelan A., Newman D. K. (2010) Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res. Microbiol. 161, 187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Byrd M. S., Sadovskaya I., Vinogradov E., Lu H., Sprinkle A. B., Richardson S. H., Ma L., Ralston B., Parsek M. R., Anderson E. M., Lam J. S., Wozniak D. J. (2009) Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73, 622–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mammen M., Choi S.-K., Whitesides G. M. (1998) Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. [DOI] [PubMed] [Google Scholar]
- 68. Myszka D. G. (2004) Analysis of small-molecule interactions using Biacore S51 technology. Anal. Biochem. 329, 316–323 [DOI] [PubMed] [Google Scholar]
- 69. Britigan B. E., Roeder T. L., Rasmussen G. T., Shasby D. M., McCormick M. L., Cox C. D. (1992) Interaction of the Pseudomonas aeruginosa secretory products pyocyanin and pyochelin generates hydroxyl radical and causes synergistic damage to endothelial cells: implications for Pseudomonas-associated tissue injury. J. Clin. Invest. 90, 2187–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Malvankar N. S., Vargas M., Nevin K. P., Franks A. E., Leang C., Kim B.-C., Inoue K., Mester T., Covalla S. F., Johnson J. P., Rotello V. M., Tuominen M. T., Lovley D. R. (2011) Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 6, 573–579 [DOI] [PubMed] [Google Scholar]
- 71. Strycharz-Glaven S. M., Snider R. M., Guiseppi-Elie A., Tender L. M. (2011) On the electrical conductivity of microbial nanowires and biofilms. Energy Environ. Sci. 10.1039/C1EE01753E [DOI] [Google Scholar]
- 72. Das T., Kutty S. K., Kumar N., Manefield M. (2013) Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PLoS One 8, e58299. [DOI] [PMC free article] [PubMed] [Google Scholar]