Background: miR-22 has a tumor-suppressive effect, but its regulation remains to be characterized.
Results: miR-22 is regulated by bile acid-activated FXR, and CCNA2 is a miR-22 target.
Conclusion: FXR-induced miR-22 in inhibiting CCNA2 is a novel pathway for FXR to exert its protective effect in the gastrointestinal tract.
Significance: The FXR-miR-22-CCNA2 axis is a novel mechanism for FXR-mediated anti-proliferative effect.
Keywords: Bile Acid, Colon Cancer, Liver Cancer, MicroRNA (miRNA), Nuclear Receptor
Abstract
Because of the significant tumor-suppressive role of microRNA-22 (miR-22), the current study was designed to understand the regulation of miR-22 and to identify additional downstream miR-22 targets in liver and colon cells. The data showed that miR-22 was transcriptionally regulated by bile acid receptor farnesoid X receptor (FXR) through direct binding to an invert repeat 1 motif located at −1012 to −1025 bp upstream from miR-22. Among the studied primary and secondary bile acids, chenodeoxycholic acid, which has the highest binding affinity to FXR, induced miR-22 level in both Huh7 liver and HCT116 colon cells in a dose- and time-dependent manner. In addition, cyclin A2 (CCNA2) was identified as a miR-22 novel target in liver and colon cancer cells. The sequence of miR-22, which is conserved in mice, rats, humans, and other mammalians, aligns with the sequence of 3′-UTR of CCNA2. Chenodeoxycholic acid treatment and miR-22 mimics reduced CCNA2 protein and increased the number of G0/G1 Huh7 and HCT116 cells. In FXR KO mice, reduction of miR-22 was accompanied by elevated hepatic and ileal CCNA2 protein, as well as an increased number of hepatic and colonic Ki-67-positive cells. In humans, the expression levels of miR-22 and CCNA2 are inversely correlated in liver and colon cancers. Taken together, our data showed that bile acid-activated FXR stimulates miR-22-silenced CCNA2, a novel pathway for FXR to exert its protective effect in the gastrointestinal tract.
Introduction
The tumor-suppressive effect of miR-222 has been demonstrated in different models. In breast cancer, miR-22 has an impact on cell proliferation by directly targeting estrogen receptor α (1). In lung cancer, the tumor-suppressive mechanism of miR-22 is associated with post-transcriptional regulation of ErbB3 (2). In colon cancer, miR-22 has an anti-angiogenic effect by inhibiting the HIF-1α and VEGF (3). In addition, miR-22 has a tumor-suppressive effect in p53-mutated colon cancer cells and can improve paclitaxel-induced chemoresistance (4). miR-22-induced apoptosis in p53 WT colon cancer cells is via P21 repression (5). In liver cancer, miR-22-targeted HDAC4 is implicated in anti-proliferative effects on hepatocellular carcinoma cells (6). Moreover, miR-22 repressed MYCBP, which in turn led to the reduction of oncogenic C-MYC activity in human cancer cell lines (7). Because of the significant tumor-suppressive role of miR-22, it is important to understand the mechanism by which miR-22 expression is regulated and to identify its additional downstream targets.
Cholesterol and bile acid homeostasis is regulated by the bile acid receptor FXR (8–10). FXR, which is abundantly expressed in the liver and intestines, plays a pivotal role in maintaining the health of the gastrointestinal tract. Lack of FXR causes spontaneous liver cancer and has an increased incidence of intestinal cancer in mice (11–15). FXR KO mice are predisposed to gallstone disease, but activation of FXR ameliorates this condition by restoring the bile acid to cholesterol ratio (16). In humans, FXR expression is markedly reduced in patients with severe cirrhosis and liver cancer (14, 15, 17). Together, FXR regulatory processes are involved in pathologies ranging from hyperlipidemia to diabetes, from cholestasis to enterohepatic tumors.
As a transcriptional factor, FXR can exert its biological effects by regulating microRNAs. Activation of FXR increases hepatic levels of miR-144, which in turn lowers hepatic ABCA1 and plasma HDL levels (18). FXR protects hepatocytes from injury by repressing miR-199a-3p and thereby increasing levels of LKB1 (17). Activation of FXR also increases the expression of miR-29a in hepatic stellate cells and inhibits extracellular matrix production (19). Moreover, the FXR/SHP pathway increases miR-34a levels in obese mice and contributes to decreased SIRT1 levels (20). Thus, FXR-regulated miRNA pathways are implicated in lipid homeostasis, fibrosis, and aging. Whether FXR-regulated miRNA has a role in enterohepatic tumorigenesis warrants further investigation.
In the current study, we report that miR-22 is transcriptionally regulated by FXR through direct binding to the invert repeat 1 (IR1) motif located at −1012 to −1025 bp upstream from miR-22. Chenodeoxycholic acid (CDCA) is effective in activating FXR and inducing miR-22 in both liver and colon cells. In addition, cyclin A2 (CCNA2) is a miR-22 novel target in liver and colon cancer cells. In FXR KO mice, the reduction of miR-22 is accompanied by increased hepatic and ileal CCNA2 protein levels, as well as cell proliferation. In human specimens, the expressions of miR-22 and CCNA2 are inversely correlated in liver and colon cancers. Taken together, our data showed that FXR-induced miR-22 in the suppression of CCNA2 is a potential novel pathway for FXR to inhibit cell proliferation in the gastrointestinal tract.
EXPERIMENTAL PROCEDURES
Clinical Samples
Twelve human hepatocellular carcinomas (HCCs) and nine normal liver specimens were included in the study. Among them, six tumors and adjacent normal tissues were paired and derived from six patients. Colon specimens were obtained from four pairs of colon rectal carcinomas and adjacent normal tissues. All the samples were obtained from the Translational Pathology Core Laboratory at the University of California, Los Angeles.
Mice
C57BL/6 WT mice (3–4 months) were purchased from the Jackson Laboratory (Sacramento, CA). FXR KO mice were provided by Dr. Frank Gonzalez (National Institutes of Health, Bethesda, MD) (21). Hepatocyte RXRα KO mice were generated as described in published papers (22, 23). Livers and ileums were frozen in liquid nitrogen immediately after collection and stored in −80 °C freezer for further assays. Animal protocols and procedures were approved by the Institutional Animal Care and Use Committee at the University of California, Davis.
Cell Culture
The reagents used for cell culture work were from Invitrogen unless otherwise noted. Huh7 (Japanese Collection of Research Bioresources) and HCT116 cell lines (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium and McCoy's medium supplemented with 10% fetal bovine serum, respectively. Cells were plated (1 × 106 cells per 60-mm dish, 2 × 105 cells per 6-well plates and 5 × 104 cells per 24-well plates) overnight prior to treatment or transfection. DMSO, chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), cholic acid (CA), lithocholic acid (LCA), hyodeoxycholic acid, rifampicin, TCPOBOP, WY14,643, vitamin D3, and all-trans-retinoic acid were used to treat the cell lines.
Quantification of RNA
Total RNA was extracted using TRIzol reagent (Invitrogen), and reverse transcribed into cDNA with a high capacity RNA-to-cDNA kit (Applied Biosystems, CA). miR-22 and mRNA expression levels were quantified by real time PCR on an ABI 7900HT system (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems). Primers were designed using Primer3 Input software, and the primers sequences are available upon request. U6 or U74 and GAPDH were used as internal controls to normalize the levels of miR-22 and mRNA, respectively.
Plasmid Construction and Luciferase Reporter Assay
PGL3-IR1 was constructed by cloning the IR1 motif (AGAGGGTCAGTGCCTG) into the XhoI and HindIII sites in the PGL3 vector (Promega, Madison, WI). The IR1 motif was located upstream from miR-22 (−1012 to −1025, Genome Browser, mouse database, 2007). Additional motifs that include DR1 (TTTGGCCTGTCACCCTG), ER6 (TGGACAGAGAGAAGGTCA), and ER5 (GGGTCAGGGCCAGTTCA), which are in proximity to IR1, were also studied by cloning into the PGL3 vector. Sequence (GCTGTCATGGTGCCAGAGAGTTGATGGAGCAGCTGGT) located 4 bp away from the IR1 motif was also cloned and served as a negative control (PGL3-Neg). The 3′-UTR of the CCNA2 gene that contains the putative binding sites for miR-22 was cloned into the psiCHECK2 vector (Promega) using the NotI and XhoI cloning sites. Restriction enzymes were purchased from the New England Biolabs.
For the luciferase assay, cells were transfected with PGL3-IR1 or PGL3-Neg using Lipofectamine 2000 (Invitrogen) for 6 h. Then the medium was replenished with fresh medium containing CDCA (100 μm) or DMSO for 24 h. For the 3′-UTR luciferase assay, cells were co-transfected with miR-22 mimics (50 nm) or scrambled control (50 nm) and psiCHECK2-CCNA2 using Lipofectamine 2000 (Invitrogen) for 24 h. After transfection, cells were collected to measure firefly and Renilla luciferase activity with the dual-luciferase reporter system (Promega). Renilla luciferase activity was standardized to the firefly luciferase activities.
Western Blot
Cells were lysed with M-PER mammalian protein extraction reagent (Thermo Scientific, Rockford, IL). Proteins (50 μg) were electrophoresed by 10% SDS-PAGE and transferred onto a PVDF membrane (Bio-Rad). The membrane was blocked with 5% nonfat milk in TBST (10 mm Tris, pH 7.5, 100 mm NaCl, 0.1% Tween 20) for 2 h at room temperature, followed by an overnight incubation at 4 °C with an anti-CCNA2 antibody (Abcam, Cambridge, MA). Membranes were then incubated with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). The signal was detected using the ECL system SuperSignal West Pico Chemiluminescent Substrates (Pierce). Protein levels were normalized to β-actin levels (Santa Cruz Biotechnology).
Cell Cycle Assay
For each treatment group, 1 × 106 cells were collected in PBS and fixed overnight in 70% ethanol at −20 °C. Then cells were resuspended in 300 μl of propidium iodide staining buffer and incubated for 30 min at room temperature. DNA content analyses were carried out using a FACScan flow cytometry (Becton Dickinson).
Ki-67 Immunostaining
Ki-67 immunostaining was performed with primary Ki-67 antibody (NeoMarkers, Fremont, CA) to monitor cell proliferation in livers and colons of wild type and FXR KO mice. The number of Ki-67-labeled nuclei was determined by counting the Ki-67-positive cells in at least six low magnification (20×) microscope fields for each section.
ChIP-Quantitative PCR Assay
ChIP assay was performed based on our previous publication (24–26). Briefly, chromatin lysates were cleared before incubation with a ChIP quality anti-FXR antibody (Santa Cruz Biotechnology). Antibodies to IgG (Santa Cruz Biotechnology) and RNA polymerase II (Millipore, Billerica, MA) were used as negative and positive controls, respectively. Samples were incubated with Dynase beads at 4 °C overnight followed by de-cross-linking and purification. DNA fragments generated (n = 3) served as templates for real time PCR using Power SYBR Green PCR Master Mix.
RESULTS
The Effect of Hepatic RXRα on miR-22 Expression
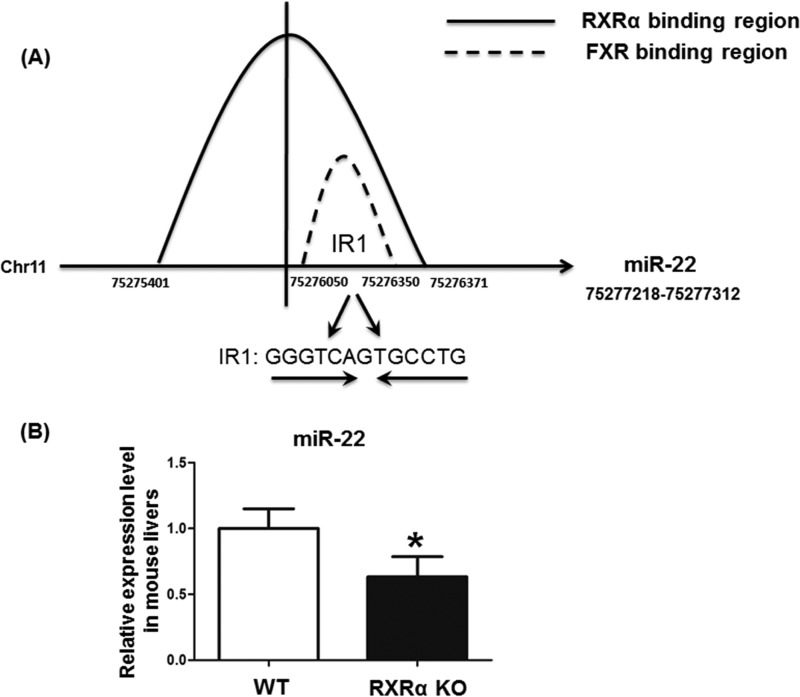
ChIP-seq data generated in our previous studies (24–26) showed that RXRα could bind to the region immediate upstream from miR-22 (located between −1817 to −367) (Fig. 1A). In addition, the level of miR-22 was reduced in 3-month-old male hepatocyte RXRα KO mice (Fig. 1B). Thus, RXRα and its heterodimeric partner could be involved in regulating miR-22 expression. By searching the UCSC Genome Browser, FXR binding sites were noted in the upstream sequence of miR-22 (−1268 to −868) (27). Thus, the FXR binding site overlaps with that of RXRα (Fig. 1A). Moreover, an IR1 motif, which could be a putative FXR binding site, was identified within the region (Fig. 1A).
FIGURE 1.
The interaction between nuclear receptors and miR-22. FXR and RXRα bind to the same region immediate upstream from miR-22. ChIP-seq data demonstrated that RXRα bound to a region immediate upstream of miR-22 (Chr11: 75275401–75276371) in the mouse liver. The UCSC Genome Browser database shows that FXR binds to a region located upstream of miR-22 (Chr11: 75276060–75276350). A, there is a putative IR1 motif (GGGTCAGTGCCTG) in the RXRα and FXR binding region. B, the hepatic miR-22 level was decreased in 3-month-old hepatocyte RXRα KO mice in comparison with that in WT mouse livers (B). The data are presented as the means ± S.D. *, p < 0.05.
Bile Acids Induce the Expression of miR-22
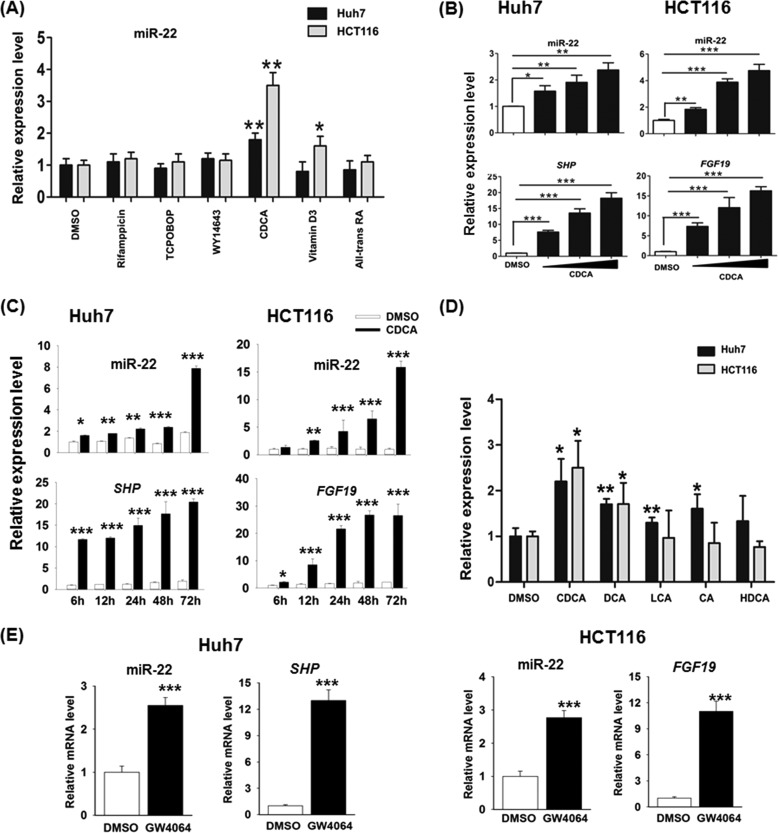
To study the potential effect of nuclear receptors in regulating miR-22, ligands for nuclear receptors were used to treat Huh7 and HCT116 cell lines for 24 h (Fig. 2A). The miR-22 level was induced significantly by CDCA treatment in both cell lines (Fig. 2A). Vitamin D3 also induced miR-22 in HCT116 cells, but not in Huh7 cells. The effect of CDCA in regulating miR-22 was further studied by dose- and time-responsive experiments. The tested doses ranged from 50 to 150 μm, and the studied times were 6, 12, 24, 48, and 72 h. Fig. 2 (B and C) shows that the induction of miR-22 was dose- and time-dependent in both liver and colon cancer cells. The up-regulation of SHP mRNA in human liver Huh7 cells and FGF19 mRNA in human colon HCT116 cells suggested that CDCA activated FXR in both cell lines (Fig. 2C).
FIGURE 2.
The effect of nuclear receptor ligands on miR-22 expression. A, ligands for nuclear receptors including rifampicin (10 μm), TCPOBOP (250 nm), WY14643 (100 μm), CDCA (100 μm), vitamin D3 (0.1 μm), and all-trans-retinoic acid (10 μm) were used to treat Huh7 and HCT116 cell lines for 24 h. B, expression of miR-22 along with SHP and FGF19 were quantified after 24 h of CDCA treatment (50, 100, and 150 μm). C, expression levels of miR-22 along with SHP and FGF19 RNA were quantified at the indicated times in CDCA-treated cells (150 μm). D, expression of miR-22 was quantified after 24 h treatment of LCA (20 μm), CDCA (50 μm), DCA (50 μm), CA (50 μm), and hyodeoxycholic acid (HDCA, 50 μm). E, expression levels of miR-22 along with SHP and FGF19 mRNA were quantified after 24 h of GW4064 treatment (5 μm). The data are presented as the means ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The effect of other bile acids in inducing miR-22 was also studied. DCA, LCA, CA, and hyodeoxycholic acid were used to treat Huh7 and HCT116 cells. Among those tested bile acids, CDCA was the strongest natural ligand for activation FXR when 10–100 μm of bile acids were used in the CAT assay (28). Other studies showed CDCA activated FXR with an EC50 in the 10–50 μm range, depending on the species (29). DCA at concentrations higher than 100 μm could induce apoptosis of colon cancer cells (30), and LCA is more toxic than DCA (31). Thus, 20 μm of LCA and 50 μm of other bile acids were used to compare their effectiveness in inducing miR-22. The data showed that CDCA was most effective inducing miR-22 in both cell lines (Fig. 2D). DCA, which has a lower binding affinity to FXR in comparison with CDCA, also induced miR-22 in both cell lines, whereas LCA and CA, which have the lowest binding affinity to FXR, induced miR-22 in Huh7 cells, but not in HCT116 cells. HDAC had no effect on the expression of miR-22. In addition, GW4064, a FXR agonist, also induced the level of miR-22 in Huh7 and HCT116 cells (Fig. 2E).
FXR and CDCA Regulates the Expression of miR-22
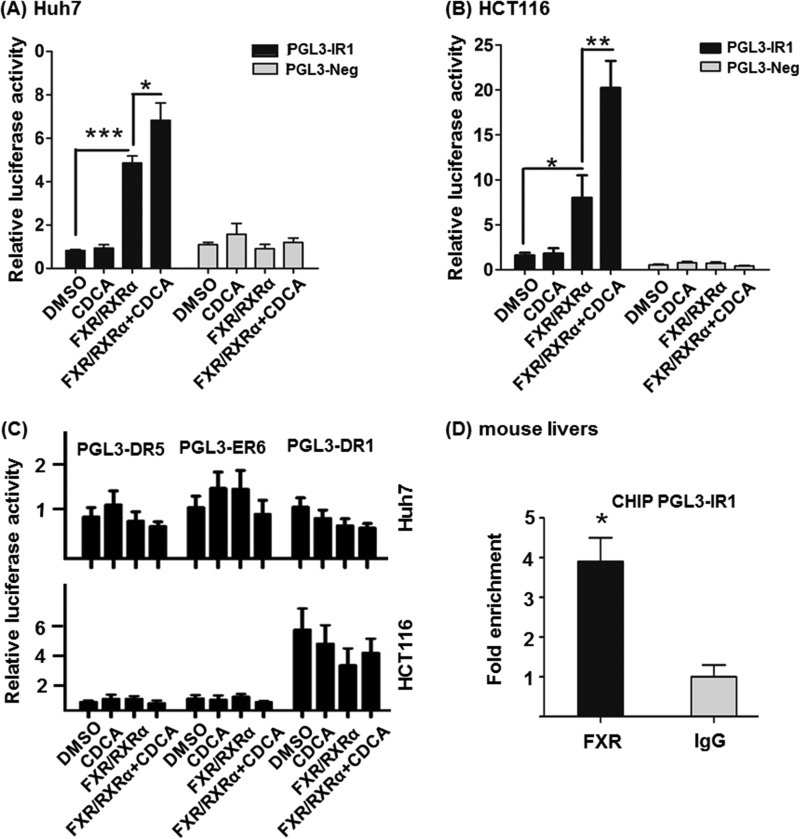
Based on the binding and ligand treatment data, FXR can be a potential transcriptional regulator that controls the expression of miR-22. To test whether the identified IR1 motif could respond to FXR, PGL3-IR1 was constructed and used for transient transfection assays in Huh7 and HCT116 cells. PGL3-Neg was used as a negative control. There was a 5-fold increase of luciferase activity after co-transfection with FXR and RXRα, and a 7-fold induction when CDCA was included in Huh7 cell line (Fig. 3A). Similarly, co-transfection of PGL3-IR1 along with FXR and RXRα in HCT116 cells increased the luciferase activity by 8-fold, and CDCA treatment increased the activity by 20-fold (Fig. 3B). In contrast, PGL3-Neg responded neither to FXR and RXRα overexpression nor to CDCA treatment in both cell lines (Fig. 3, A and B). Additional motifs that include putative DR1, ER6, and ER5, which are in proximity to IR1, were also studied (Fig. 3C). The data showed that neither FXR/RXRα nor CDCA was able to induce the expression of those cloned motifs. Because the sequence of miR-22 and the IR1 motif is conserved in human and mouse, we studied the binding of FXR to the IR1 motif using mouse livers. ChIP-quantitative PCR data revealed that FXR binding was enriched in the IR1 motif of the mouse hepatic genome (Fig. 3D). Taken together, FXR binds an IR1 motif located at −1025 to −1012 bp upstream from miR-22 and regulates its expression.
FIGURE 3.
FXR and CDCA regulate miR-22 expression. A and B, a putative IR1 motif was cloned into the PGL3 vector. FXR and RXRα were co-transfected with PGL3-IR1 or PGL3-Neg (control) plasmids for 6 h in Huh7 (A) and HCT116 cell lines (B). 6 h post-transfection, cells were treated with CDCA (100 μm) or DMSO for 24 h. Putative DR5, ER6, and DR1 motifs were cloned into a PGL3 vector and co-transfected with FXR and RXRα plasmids for 6 h in Huh7 and HCT116 cells. C, the transfected cells were treated with CDCA (100 μm) or DMSO for additional 24 h. D, fold enrichment of IR1 binding using anti-FXR antibody in comparison with IgG in mouse livers was measured by ChIP-quantitative PCR. The data are presented as the means ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
miR-22 Targets CCNA2
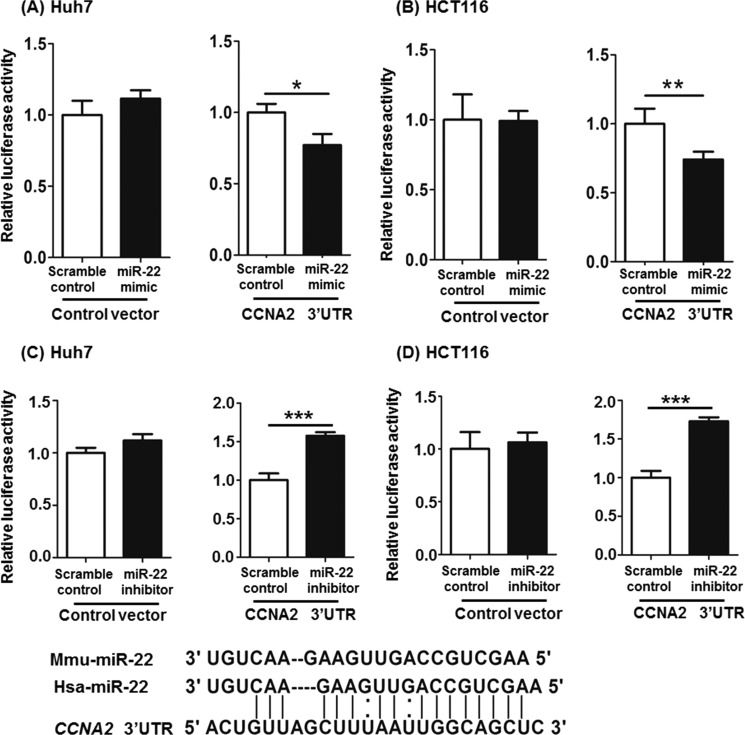
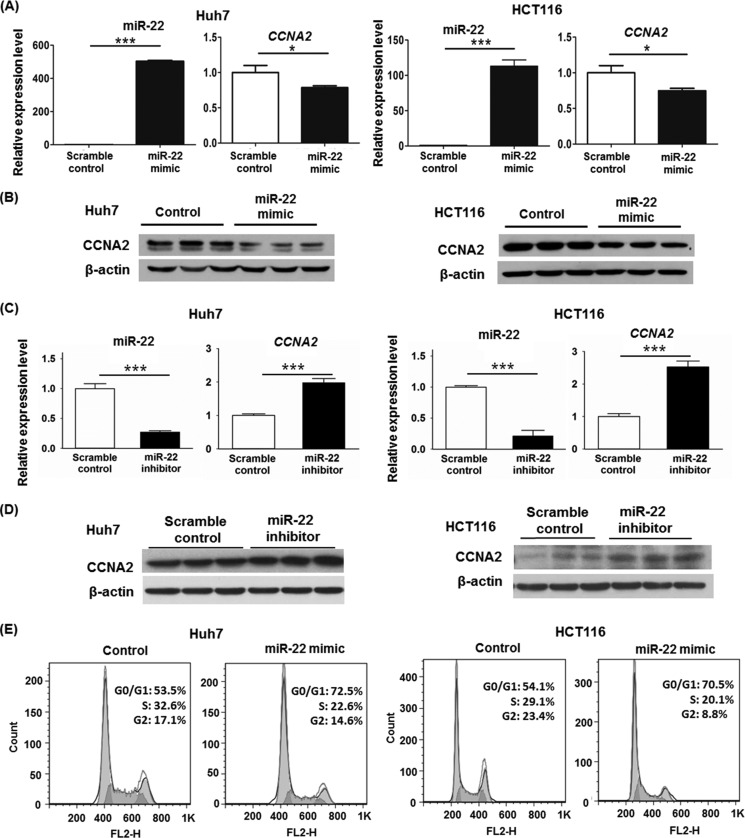
Based on miRBase database, CCNA2 is a potential target for miR-22. To establish the inhibitory role of miR-22 on CCNA2, in vitro functional assays were conducted using either miR-22 mimics or miR-22 inhibitors. Based on the sequence, miR-22 can partially pair with the 3′-UTR of the CCNA2. psiCHECK2-CCNA2, which contains the 3′-UTR of CCNA2, was constructed and used for transfection with either miR-22 mimics or miR-22 inhibitors into Huh7 and HCT116 cell lines. The scramble mimics or inhibitors were included as controls. In both cell lines, miR-22 mimics inhibited luciferase activity of psiCHECK2-CCNA2 by ∼20% (Fig. 4, A and B). However, miR-22 inhibitors increased luciferase activity of psiCHECK2-CCNA2 (Fig. 4, C and D). miR-22 mimic transfection significantly increased miR-22 levels in Huh7 and HCT116 cell lines. Accordingly, the endogenous CCNA2 mRNA levels were reduced (Fig. 5A). In addition, the protein levels of CCNA2 were also reduced by 65 and 50% in Huh7 and HCT116 cell lines, respectively (Fig. 5B). Conversely, miR-22 inhibitors reduced CDCA-induced miR-22 level, which in turn increased the expression of CCNA2 at both the mRNA and protein levels (Fig. 5, C and D). In addition, miR-22 mimics increased the percentage of Huh7 and HCT116 cells in the G0/G1 phase and reduced the percentage of cells in the S and G2 phases (Fig. 5E).
FIGURE 4.
miR-22 can partially pair with 3′-UTR of the CCNA2. The sequence of miR-22 is conserved in humans and mice. A and B, psiCHECK2-CCNA2, which contains the 3′-UTR of CCNA2, was constructed and used for transfection with either miR-22 mimics or scramble controls into Huh7 (A) and HCT116 (B) cells. C and D, psiCHECK2-CCNA2 was transfected with either miR-22 inhibitors or scramble controls into Huh7 (C) and HCT116 (D) cells. The data are presented as the means ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 5.
Repression of CCNA2 by miR-22 at the mRNA and protein level. A, expression of miR-22 and CCNA2 were quantified in Huh7 and HCT116 cells 48 h after transfection with either miR-22 mimics or scramble controls. B, protein level of CCNA2 was determined by Western blot. C, expression of miR-22 and CCNA2 were quantified in CDCA-treated Huh7 and HCT116 cells 48 h after transfection of miR-22 inhibitors or scramble controls. D, protein level of CCNA2 was determined by Western blot. E, cell cycle distribution of miR-22 mimic transfected-Huh7 or HCT116 cells was measured by flow cytometry after 72 h transfection. The data are presented as the means ± S.D. *, p < 0.05; ***, p < 0.001.
miR-22 and CCNA2 mRNA Levels Are Inversely Correlated in Human Hepatocellular Carcinoma and Colorectal Carcinoma Specimens
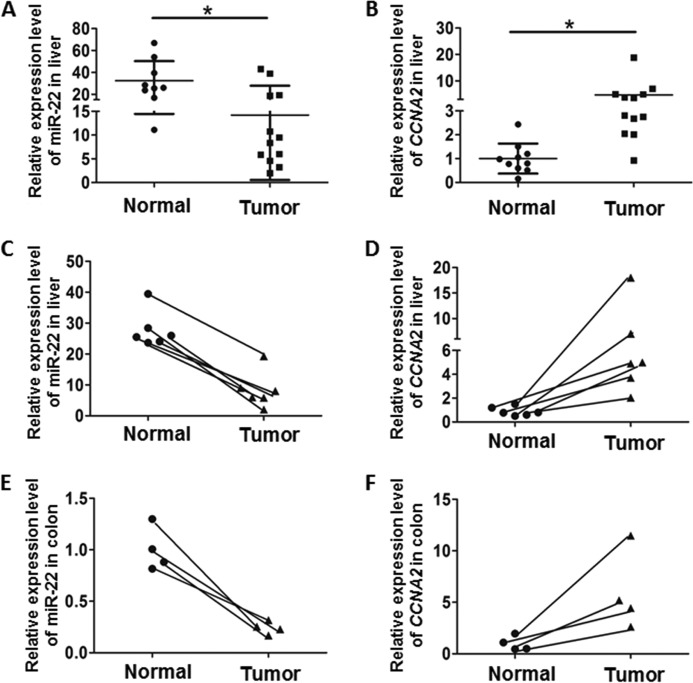
To demonstrate the clinical relevance of the findings, miR-22 and CCNA2 mRNA were studied in 12 HCC and 9 normal human liver specimens. The level of miR-22 was significantly lower in HCC than normal livers (Fig. 6A). In addition, the level of CCNA2 was higher in HCC than normal controls (Fig. 6B). Among the studied liver specimens, six pairs were derived from tumors and adjacent normal tissues from six patients. All six pairs had inversed expression levels of miR-22 and CCNA2 (Fig. 6, C and D). A similar trend and correlated expression pattern was also observed in four paired colon rectal carcinoma tissues and normal adjacent specimens (Fig. 6, E and F). Taken together, miR-22 could have an effect on inhibiting tumorigenesis by targeting CCNA2 in human liver and colon cancer.
FIGURE 6.
Correlation between miR-22 and CCNA2 expression level in liver and colon cancer specimens. A and B, relative expression of miR-22 (A) and CCNA2 (B) was quantified in HCC compared with normal livers. C and D, relative expression of miR-22 (C) and CCNA2 (D) in liver was quantified. Among the studied liver specimens, six pairs were derived from tumors and adjacent healthy tissues of the same individuals. E and F, expression of miR-22 (E) and CCNA2 (F) was also studied in four paired colon cancer tissues and normal adjacent specimens. *, p < 0.05.
Reversed Expression Pattern of FXR and CCNA2 in Cell Cultures and Mice
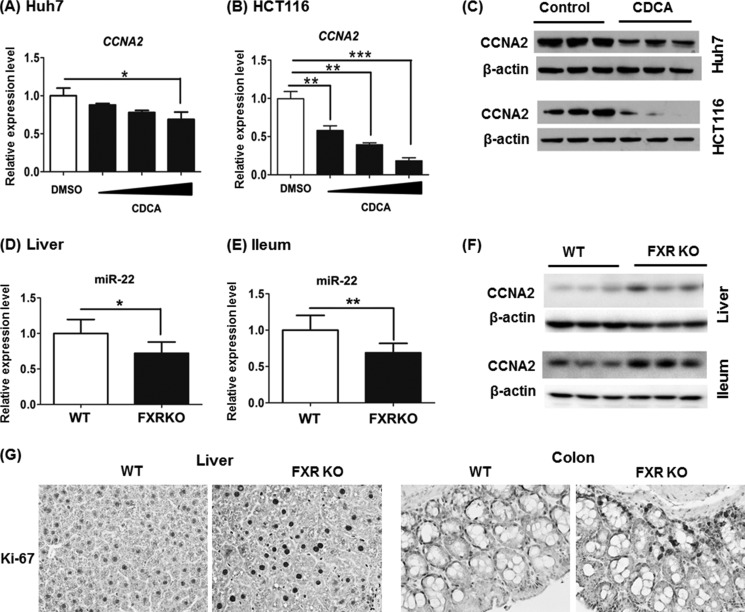
To further establish the relationship between bile acids, FXR, and CCNA2, CDCA was used to treat Huh7 and HCT116 cells. CDCA significantly decreased the mRNA and protein level of CCNA2 in both cell lines (Fig. 7, A–C). In addition, the miR-22 level was reduced, and CCNA2 protein level was increased in the livers and ileums of FXR KO mice (Fig. 7, D–F). Moreover, such changes were accompanied by increased Ki-67-positive cells in livers (12-fold) and colons (8-fold) of FXR KO mice (Fig. 7G). Taken together, activation of FXR is associated with increased miR-22 and reduced CCNA2. Not only does FXR maintain the basal expression level of miR-22, but the ligand for FXR induces miR-22 expression in cells that are of hepatic and colonic origin.
FIGURE 7.
Reversed expression pattern of FXR and CCNA2 in cell lines and mice. A and B, expression of CCNA2 was quantified in Huh7 (A) and HCT116 (B) cells after 24 h CDCA treatment (50, 100, and 150 μm). C, protein level of CCNA2 was determined by Western blot after 48 h treatment of 100 μm CDCA. D–F, the mRNA level of miR-22 and protein level of CCNA2 were determined in the livers and ileums of 3-month-old FXR KO mice. G, Ki-67 staining of liver and colon sections obtained from WT and FXR KO mice (magnification, 40×). The data are presented as the means ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
This is the first study to show that miR-22, a tumor suppressor, is regulated by bile acid-activated FXR. The regulation is mediated via direct binding of FXR to an IR1 motif located upstream of the miR-22 sequence, and this occurs in both human cells and mouse livers. The regulation is RXRα, FXR, and ligand-dependent. Among the studied bile acids, CDCA, which has the highest binding affinity to FXR, is the most effective in inducing miR-22 in cells that are of hepatic and colonic origin. The sequence of miR-22, conserved in mice, rats, humans, and other mammalians (miRBase database), aligns with the sequence of 3′-UTR of CCNA2. Our data also uncovered that CCNA2 is a novel miR-22 target, which could potentially explain the tumor-suppressive role of miR-22. In addition to bile acids, vitamin D3 also induced miR-22 expression in HCT116 cells, which is consistent with published findings demonstrated in colon cancer cell lines (32). It is of interest to note that vitamin D3 via its receptor, vitamin D receptor, can regulate bile acid detoxification and thus is also important for maintaining the health of the gastrointestinal tract (33). It would be important to uncover additional miR-22 targets to understand the common role of FXR and vitamin D receptor in the intestines.
The literature shows that most identified miR-22 targets are implicated in cancer, including estrogen receptor α (1), HDAC4 (6), and MYCBP (7). The hepatoprotective effect of FXR has been established using the FXR KO mouse model (11–15). In HepG2 cells, lentivirus-mediated overexpression of FXR can repress cell proliferation and tumor growth in nude mice (34). In mice, FXR KO mice develop spontaneous liver cancer when they are 12–15 months old (11, 12, 14, 15). FXR protects the intestinal epithelium by preventing bacterial overgrowth and subsequent mucosa deterioration (35). Azoxymethane treatment of FXR KO mice resulted in colon carcinogenesis (13). Therefore, FXR plays a vital role in liver and colon cancer development. In human, FXR expression is markedly reduced in patients with severe fibrosis (Ishak score > 5) and liver cancer (1, 17). The mechanism by which a lack of FXR promotes carcinogenesis has been investigated. Because cholestyramine treatment can prevent spontaneous liver carcinogenesis in FXR KO mice, elevated bile acid levels account for such tumor promoting effects (12). Other reported mechanisms include induction IL-1β by bile acids, which is an inflammatory signal that stimulates cell proliferation (11). In addition, the Wnt/β-catenin pathway plays an important role in liver carcinogenesis in FXR KO mice (14). It is also possible that overexpressed CCNA2 caused by repressed miR-22 plays a role in the carcinogenesis process when FXR is not expressed.
The induction of CCNA2 in human liver and intestinal cancer specimens may not be surprising because the induction of cell cycle genes is expected in the tumors. However, such induction was closely associated with reduced miR-22 in both types of tumors. Moreover, 3-month-old FXR KO mice that have not yet developed cancer also have elevated CCNA2, reduced miR-22, and increased Ki-67-positive cells in both the liver and intestines. Thus, there is a close linkage between FXR deficiency and miR-22 reduction, as well as CCNA2 induction, and the newly discovered FXR/miR-22/CCNA2 pathway could potentially be a novel mechanism that explains the FXR-mediated anti-proliferative effect. A time course experiment to determine whether miR-22 mimics can prevent liver and intestinal carcinogenesis in FXR KO is needed to validate the hypothesis. Taken together, using cell lines as well as human and mouse models, the current study establishes a novel FXR-miR-22-CCNA2 axis in the gastrointestinal tract, which potentially could be used for cancer prevention and treatment.
Acknowledgment
We thank Lisa Teixeira for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK092100 and CA53596 (to Y.-J. Y. W.).
- miR-22
- microRNA-22
- TCPOBOP
- 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene
- CDCA
- chenodeoxycholic acid
- CA
- cholic acid
- CCNA2
- cyclin A2
- DCA
- deoxycholic acid
- FXR
- farnesoid X receptor
- HCC
- hepatocellular carcinoma
- IR1
- invert repeat 1
- LCA
- lithocholic acid
- RXRα
- retinoid X receptor α.
REFERENCES
- 1. Xiong J., Yu D., Wei N., Fu H., Cai T., Huang Y., Wu C., Zheng X., Du Q., Lin D., Liang Z. (2010) An estrogen receptor α suppressor, microRNA-22, is downregulated in estrogen receptor α-positive human breast cancer cell lines and clinical samples. FEBS J. 277, 1684–1694 [DOI] [PubMed] [Google Scholar]
- 2. Ling B., Wang G. X., Long G., Qiu J. H., Hu Z. L. (2012) Tumor suppressor miR-22 suppresses lung cancer cell progression through post-transcriptional regulation of ErbB3. J. Cancer Res. Clin. Oncol. 138, 1355–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamakuchi M., Yagi S., Ito T., Lowenstein C. J. (2011) MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS One 6, e20291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J., Zhang Y., Zhao J., Kong F., Chen Y. (2011) Overexpression of miR-22 reverses paclitaxel-induced chemoresistance through activation of PTEN signaling in p53-mutated colon cancer cells. Mol. Cell Biochem. 357, 31–38 [DOI] [PubMed] [Google Scholar]
- 5. Tsuchiya N., Izumiya M., Ogata-Kawata H., Okamoto K., Fujiwara Y., Nakai M., Okabe A., Schetter A. J., Bowman E. D., Midorikawa Y., Sugiyama Y., Aburatani H., Harris C. C., Nakagama H. (2011) Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21. Cancer Res. 71, 4628–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang J., Yang Y., Yang T., Liu Y., Li A., Fu S., Wu M., Pan Z., Zhou W. (2010) microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br. J. Cancer 103, 1215–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiong J., Du Q., Liang Z. (2010) Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene 29, 4980–4988 [DOI] [PubMed] [Google Scholar]
- 8. Lambert G., Amar M. J., Guo G., Brewer H. B., Jr., Gonzalez F. J., Sinal C. J. (2003) The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J. Biol. Chem. 278, 2563–2570 [DOI] [PubMed] [Google Scholar]
- 9. Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. (1999) Identification of a nuclear receptor for bile acids. Science 284, 1362–1365 [DOI] [PubMed] [Google Scholar]
- 10. Chiang J. Y. (2009) Bile acids: regulation of synthesis. J. Lipid Res. 50, 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim I., Morimura K., Shah Y., Yang Q., Ward J. M., Gonzalez F. J. (2007) Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 28, 940–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang F., Huang X., Yi T., Yen Y., Moore D. D., Huang W. (2007) Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 67, 863–867 [DOI] [PubMed] [Google Scholar]
- 13. Maran R. R., Thomas A., Roth M., Sheng Z., Esterly N., Pinson D., Gao X., Zhang Y., Ganapathy V., Gonzalez F. J., Guo G. L. (2009) Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharmacol. Exp. Ther. 328, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolfe A., Thomas A., Edwards G., Jaseja R., Guo G. L., Apte U. (2011) Increased activation of the Wnt/β-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J. Pharmacol. Exp. Ther. 338, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu N., Meng Z., Lou G., Zhou W., Wang X., Zhang Y., Zhang L., Liu X., Yen Y., Lai L., Forman B. M., Xu Z., Xu R., Huang W. (2012) Hepatocarcinogenesis in FXR−/− mice mimics human HCC progression that operates through HNF1α regulation of FXR expression. Mol. Endocrinol. 26, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moschetta A., Bookout A. L., Mangelsdorf D. J. (2004) Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat. Med. 10, 1352–1358 [DOI] [PubMed] [Google Scholar]
- 17. Lee C. G., Kim Y. W., Kim E. H., Meng Z., Huang W., Hwang S. J., Kim S. G. (2012) Farnesoid X receptor protects hepatocytes from injury by repressing miR-199a-3p, which increases levels of LKB1. Gastroenterology 142, 1206–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Aguiar Vallim T. Q., Tarling E. J., Kim T., Civelek M., Baldán Á., Esau C., Edwards P. A. (2013) MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ. Res. 112, 1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J., Zhang Y., Kuruba R., Gao X., Gandhi C. R., Xie W., Li S. (2011) Roles of microRNA-29a in the antifibrotic effect of farnesoid X receptor in hepatic stellate cells. Mol. Pharmacol. 80, 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J., Padhye A., Sharma A., Song G., Miao J., Mo Y. Y., Wang L., Kemper J. K. (2010) A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 285, 12604–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sinal C. J., Tohkin M., Miyata M., Ward J. M., Lambert G., Gonzalez F. J. (2000) Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 [DOI] [PubMed] [Google Scholar]
- 22. Wan Y. J., Cai Y., Lungo W., Fu P., Locker J., French S., Sucov H. M. (2000) Peroxisome proliferator-activated receptor α-mediated pathways are altered in hepatocyte-specific retinoid X receptor α-deficient mice. J. Biol. Chem. 275, 28285–28290 [DOI] [PubMed] [Google Scholar]
- 23. Wan Y. J., An D., Cai Y., Repa J. J., Hung-Po Chen T., Flores M., Postic C., Magnuson M. A., Chen J., Chien K. R., French S., Mangelsdorf D. J., Sucov H. M. (2000) Hepatocyte-specific mutation establishes retinoid X receptor α as a heterodimeric integrator of multiple physiological processes in the liver. Mol. Cell. Biol. 20, 4436–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu Y., Liu H. X., He Y., Fang Y., Fang J., Wan Y. J. (2013) Transcriptome profiling and genome-wide DNA binding define the differential role of fenretinide and all-trans RA in regulating the death and survival of human hepatocellular carcinoma Huh7 cells. Biochem. Pharmacol. 85, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhan Q., Fang Y., He Y., Liu H. X., Fang J., Wan Y. J. (2012) Function annotation of hepatic retinoid x receptor α based on genome-wide DNA binding and transcriptome profiling. PLoS One 7, e50013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He Y., Gong L., Fang Y., Zhan Q., Liu H. X., Lu Y., Guo G. L., Lehman-McKeeman L., Fang J., Wan Y. J. (2013) The role of retinoic acid in hepatic lipid homeostasis defined by genomic binding and transcriptome profiling. BMC Genomics 14, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomas A. M., Hart S. N., Kong B., Fang J., Zhong X. B., Guo G. L. (2010) Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology 51, 1410–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., Lehmann J. M. (1999) Bile acids: natural ligands for an orphan nuclear receptor. Science 284, 1365–1368 [DOI] [PubMed] [Google Scholar]
- 29. Karpen S. J. (1999) Bile acids go nuclear! Hepatology 30, 1107–1109 [DOI] [PubMed] [Google Scholar]
- 30. Milovic V., Teller I. C., Faust D., Caspary W. F., Stein J. (2002) Effects of deoxycholate on human colon cancer cells: apoptosis or proliferation. Eur. J. Clin. Invest. 32, 29–34 [DOI] [PubMed] [Google Scholar]
- 31. Sharma R., Majer F., Peta V. K., Wang J., Keaveney R., Kelleher D., Long A., Gilmer J. F. (2010) Bile acid toxicity structure-activity relationships: correlations between cell viability and lipophilicity in a panel of new and known bile acids using an oesophageal cell line (HET-1A). Bioorg. Med. Chem. 18, 6886–6895 [DOI] [PubMed] [Google Scholar]
- 32. Alvarez-Díaz S., Valle N., Ferrer-Mayorga G., Lombardía L., Herrera M., Domínguez O., Segura M. F., Bonilla F., Hernando E., Muñoz A. (2012) MicroRNA-22 is induced by vitamin D and contributes to its antiproliferative, antimigratory and gene regulatory effects in colon cancer cells. Hum. Mol. Genet. 21, 2157–2165 [DOI] [PubMed] [Google Scholar]
- 33. Makishima M., Lu T. T., Xie W., Whitfield G. K., Domoto H., Evans R. M., Haussler M. R., Mangelsdorf D. J. (2002) Vitamin D receptor as an intestinal bile acid sensor. Science 296, 1313–1316 [DOI] [PubMed] [Google Scholar]
- 34. Su H. Y., Ma C., Liu J. F., Li N. B., Gao M. Q., Huang A. M., Wang X. C., Huang W. D., Huang X. F. (2012) Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am. J. Physiol. Gastr. L 303, G1245–G1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Inagaki T., Moschetta A., Lee Y. K., Peng L., Zhao G., Downes M., Yu R. T., Shelton J. M., Richardson J. A., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2006) Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 3920–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]