Background: The PHF6 gene is mutated in patients with Börjeson-Forssman-Lehmann syndrome, T-cell acute lymphoblastic leukemia, and acute myeloid leukemia. The PHF6 protein is a newly identified interactor with the NuRD complex.
Results: The complex structure of the NoLS region of PHF6 bound to RBBP4 was solved.
Conclusion: By interacting with the RBBP4 component, PHF6 associates with the NuRD complex.
Significance: Association with the NuRD complex implicates a role for PHF6 in chromatin structure modulation and gene regulation.
Keywords: Chromatin Regulation, Gene Regulation, Protein Crystallization, Protein-Protein Interaction, Structural Biology, BFLS, NuRD Complex, PHF6, RBBP4/RbAp48, T-ALL
Abstract
The NuRD complex is a conserved transcriptional coregulator that contains both chromatin-remodeling and histone deacetylase activities. Mutations of PHF6 are found in patients with Börjeson-Forssman-Lehmann syndrome, T-cell acute lymphoblastic leukemia, or acute myeloid leukemia. Recently, PHF6 was identified to interact with the NuRD complex, and this interaction is mediated by the RBBP4 component. However, little is known about the molecular basis for the interaction. Here, we present the crystal structure of the complex of the NuRD subunit RBBP4 bound to the PHF6 peptide (residues 162–170). The PHF6 peptide binds to the top surface of the RBBP4 β-propeller. A pair of positively charged residues of the PHF6 peptide insert into the negatively charged pocket of RBBP4, which is critical for the interaction between PHF6 and RBBP4. Corresponding PHF6 mutants impair this interaction in vitro and in vivo. Structural comparison shows that the PHF6-binding pocket overlaps with FOG1 and histone H3 on RBBP4/Nurf55, but it is distinct from the pocket recognizing histone H4, Su(z)12, and MTA1. We further show that the middle disordered region (residues 145–207, containing the RBBP4-binding motif) is sufficient for the transcriptional repression mediated by PHF6 on the GAL4 reporter, and knockdown of RBBP4 diminished the PHF6-mediated repression. Our RBBP4-PHF6 complex structure provides insights into the molecular basis of PHF6-NuRD complex interaction and implicates a role for PHF6 in chromatin structure modulation and gene regulation.
Introduction
Regulation of gene transcription relies on many multiprotein complexes that are coordinately recruited and assembled onto the specific chromatin region, acting by changing histone modification marks or by induction of conformational changes in chromatin to fine-tune the transcriptional control of specific sets of genes. The Mi-2/NuRD (nucleosome remodeling and deacetylase) complex is one of the four major types of ATP-dependent chromatin-remodeling complexes (1–3). The Mi-2/NuRD core complex consists of ATPases of CHD3 (chromodomain helicase DNA-binding protein 3; also known as Mi-2α) and CHD4 (also known as Mi-2β); HDAC1 (histone deacetylase 1) and HDAC2; MBD2 (methyl-CpG-binding domain 2) and MBD3; MTA1 (metastasis-associated gene 1), MTA2, and MTA3; RBBP4 (retinoblastoma binding protein 4; also known as RbAp48) and RBBP7 (also known as RbAp46); and p66α and p66β (1–5). The NuRD complex is widely conserved in animal and plant species and has been shown to play important roles in gene expression regulation and development (6, 7). Additionally, multiple lines of evidence show that the NuRD complex functions primarily in gene transcriptional repression through its association with diverse transcription factors (6, 8). However, the mechanism by which transcription-associated factors recruit the NuRD complex to regulate specific gene expressions is still not well understood.
PHF6, located on the X chromosome, was first identified as the gene associated with human Börjeson-Forssman-Lehmann syndrome (OMIM 301900) (9, 10), an X-linked mental retardation disorder characterized by moderate-to-severe mental disability, epilepsy, hypogonadism, and obesity (11, 12). PHF6 was identified as a new X-linked tumor suppressor gene (13). Inactivating mutations, including missense, truncation, frameshift, and deletion mutations, in the PHF6 gene have been detected in T-cell acute lymphoblastic leukemia and acute myeloid leukemia patients (13, 14). Recently, de novo aberrations in PHF6 were identified in female patients with Coffin-Siris syndrome (OMIM 135900) and with a phenotype not identical to Börjeson-Forssman-Lehmann syndrome (15, 16). These loss-of-function mutations and disruptions of the PHF6 gene may selectively impair the tumor suppressor function or other unknown functions of the protein. The PHF6 gene is highly conserved among orthologs in other species and appears to be absent from non-vertebrate genomes (17). The human PHF6 gene is highly expressed in the developing brain as well as in other embryonic tissues (10, 13, 17). The PHF6 protein contains two extended plant homeodomain zinc fingers and four putative nuclear localization sequences (NLSs)3 for nuclear and nucleolar localization, indicating the roles of PHF6 in transcriptional regulation and ribosome biogenesis (10, 18).
PHF6 has been found to be a novel interactor with the NuRD complex in the nucleoplasm, which confirms a role for PHF6 in modifying chromatin structure and gene regulation (19). Furthermore, in our previous work, we showed that the PHF6 protein can directly interact with the NuRD complex component RBBP4 and that the interaction is mediated by the nucleolar localization sequence (NoLS) composed of NLS3 and NLS4 of PHF6 (20). To study the molecular basis of this interaction, we subsequently determined the crystal structure of RBBP4 in complex with a PHF6 peptide (residues 157–171). The complex structure reveals that the PHF6 peptide binds to the top surface of the WD40 domain of RBBP4. The interaction between RBBP4 and PHF6 is highly specific, and mutations or deletions of the NoLS region of PHF6 reduced or abolished the interaction in vitro and in vivo. The RBBP4-binding motif of PHF6 is highly conserved in terrestrial vertebrates. In addition, we demonstrated that the middle disordered region of PHF6 (residues 145–207, containing the RBBP4-binding motif) is sufficient for transcriptional repression and that the repression is moderately dependent on RBBP4. RBBP4 knockdown by shRNA reduced the PHF6-mediated transcriptional repression.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Full-length human RBBP4 (residues 1–425) was cloned into a pFastBac HT-B vector encoding an N-terminal His6 tag and a tobacco etch virus protease cleavage site and expressed in Tn5 cells infected with baculovirus using the Bac-to-Bac expression methodology (Invitrogen) according to the manufacturer's instructions. Infected cells were resuspended on ice in lysis buffer (20 mm Tris (pH 8.0) and 500 mm NaCl), lysed by sonication, and clarified by centrifugation. The supernatant was incubated with nickel-nitrilotriacetic acid beads (GE Healthcare) for 2 h at 8 °C. The beads were collected and washed with lysis buffer containing 30 mm imidazole. RBBP4 was eluted with 0.5 m imidazole, and the N-terminal His6 tag was removed with tobacco etch virus protease. RBBP4 was further purified with 16/60 Superdex 200 size exclusion columns (GE Healthcare) and concentrated up to 10 mg/ml in 20 mm Tris (pH 7.4) and 150 mm NaCl.
Peptide Synthesis
Wild-type PHF6 (residues 157–171) and mutant peptides used in the study were chemically synthesized by GL Biochem (Shanghai, China).
Isothermal Titration Calorimetry Experiments
Isothermal titration calorimetry (ITC) experiments were performed using a MicroCal iTC200 titration calorimeter (GE Healthcare). PHF6 peptides were injected into a sample cell containing 50 μm RBBP4 in 20 mm Tris (pH 7.4) and 150 mm NaCl at 24 °C. The titration data were analyzed using Origin 7 (OriginLab Corp.), provided with the iTC200 calorimeter.
Protein Crystallization, Data Collection, and Structure Determination
The RBBP4-PHF6 peptide (residues 157–171) complex was prepared by incubating 10 mg/ml protein with peptide at a 1:1.2 molar ratio overnight at 4 °C and crystallized by mixing equal volumes of protein and crystallization solution using the hanging-drop vapor-diffusion method. Single crystals were obtained in 0.1 m potassium thiocyanate and 30% PEG MME 2000 at 16 °C, and the crystals were harvested, soaked in the mother liquor containing 25% glycerol, and flash-frozen in liquid nitrogen. Data sets were collected on beamline 17U at the Shanghai Synchrotron Radiation Facility. The initial data were indexed and integrated with iMosflm (21), and scaled with SCALA (22) from the CCP4 program suite (23). The structure of the complex was solved by molecular replacement using the program MOLREP (24) and free-state RBBP4 (Protein Data Bank ID 3GFC) as a search model (25). The model was further built and refined using Coot and Refmac (26, 27), respectively. Crystal diffraction data and refinement statistics are displayed in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses are for highest resolution shell. r.m.s.d., root mean square deviation.
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 50.63, 87.55, 95.84 |
| α, β, γ | 90°, 90°, 90° |
| Wavelength (Å) | 0.979 |
| Resolution (Å) | 44.77-1.85 (1.95-1.85) |
| Rmerge (%) | 10.4 (57.1) |
| I/σI | 12.5 (3.6) |
| Completeness (%) | 96.6 (91.9) |
| Redundancy | 6.3 (6.4) |
| Refinement | |
| Resolution (Å) | 43.83-1.85 |
| No. of reflections | 33,938 |
| Rwork/Rfree (%) | 17.64/20.69 |
| No. of atoms | |
| Protein | 2902 |
| Peptide | 69 |
| Water | 267 |
| B-factors (Å2) | |
| Protein | 18.47 |
| Peptide | 27.06 |
| Water | 26.51 |
| r.m.s.d. | |
| Bond length (Å) | 0.0065 |
| Bond angle | 1.178° |
| Ramachandran values (%) | |
| Most favored | 98.1 |
| Additionally allowed | 1.6 |
| Outliers | 0.3 |
Cell Culture, Transient Transfections, and Reporter Assays
The HEK293T cell line was cultivated in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% FBS and antibiotics (Gibco) at 37 °C under an atmosphere of 5% CO2 in air. Cells were transfected using Lipofectamine Plus (Invitrogen) with the following plasmids: pGAL-TK-Luc (0.3 μg), pRK5 (GAL4 DNA-binding domain) or pRK5-PHF6 (wild-type or mutation or deletion mutants; GAL4 DNA-binding domain-PHF6 fusion; 0.4 μg), or the Renilla reporter vector pRL-luciferase (25 ng) as a normalization control for transfection efficiency. The cells were harvested 24 h after transfection, and luciferase activity was measured using the luciferase assay system (Promega).
Gene Knockdown by shRNA
A lentivirus-mediated knockdown system (pLKO.1, vesicular stomatitis virus G, Gag, and Rev) was used to knock down the RBBP4 gene. The RBBP4 knockdown vectors and control vector were present in the pLKO.1-puro plasmids and were purchased from Sigma-Aldrich. The RBBP4 shRNA sequence was 5′-CCGGGCCTTTCTTTCAATCCTTATACTCGAGTATAAGGATTGAAAGAAAGGCTTTTTG-3′. HEK293T cells were grown in the presence of 1 μg/ml puromycin for selecting the positive cells.
Co-immunoprecipitation and Antibodies
Cells were lysed by incubation in TBST buffer (50 mm Tris (pH 7.4), 150 mm NaCl, 1% Triton X-100, and protease inhibitor mixture) for 15–30 min on a shaker, and the extract was cleared by centrifugation. The supernatant was incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) overnight at 4 °C. The resin was washed three times with TBST buffer and boiled in loading buffer. The following antibodies were used: anti-RBBP4 (ab79416, Abcam), anti-FLAG (DYKDDDDK tag antibody, 26368, Cell Signaling Technology), and anti-β-actin (sc-47778, Santa Cruz Biotechnology).
RESULTS
PHF6 Binds to RBBP4/RbAp48, Forming a Stable Complex
We previously reported that residues 152–171 of PHF6 can directly interact with RBBP4 (RbAp48) in GST pulldown assays in vitro (20). ITC was conducted using a synthetic optimized PHF6 peptide (residues 157–171) to titrate RBBP4 (Fig. 1A) and therefore determined the binding affinity of RBBP4 for PHF6. The ITC experimental data revealed that this peptide bound to RBBP4 with a stoichiometry of ∼1:1 and a dissociation constant (Kd) of 7.1 ± 0.4 μm (Fig. 1B).
FIGURE 1.
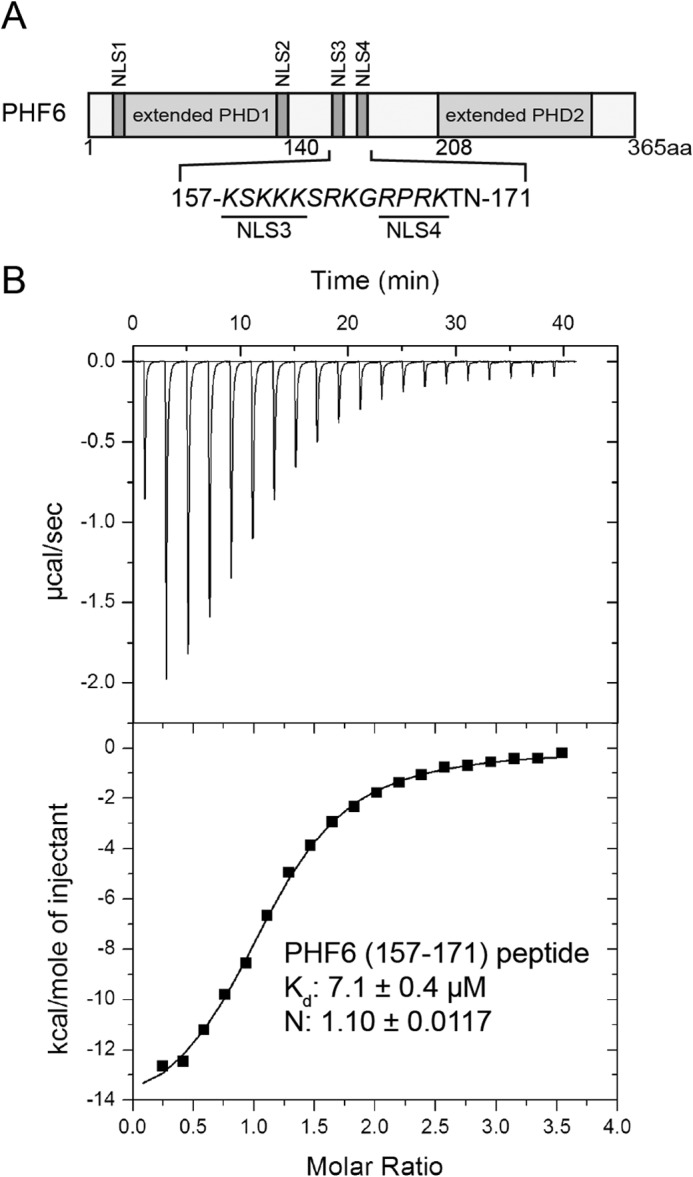
Interaction of the NoLS of PHF6 with RBBP4. A, schematic diagram of PHF6, including two extended plant homeodomain (PHD) zinc-finger domains and four NLSs (NSL1–NLS4). The sequence of the putative NoLS (composed of NLS3 and NLS4) is shown below in italic type, and the NLS3 and NLS4 sequences are underlined. aa, amino acids. B, determination of the affinity of the PHF6(157–171) peptide for the RBBP4 protein using ITC. Data were fitted to a one-site binding model using Origin 7, and calculated binding parameters were ΔH = −15,180 ± 225.2, and ΔS = −27.5.
Complex Structure of RBBP4/RbAp48 with the PHF6 Peptide
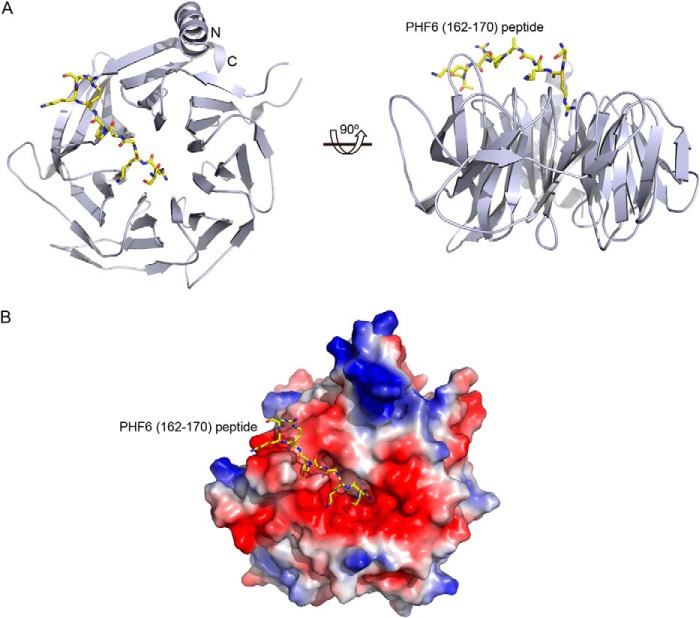
Full-length RBBP4 was co-crystallized with the PHF6(157–171) peptide to gain insight into how RBBP4 recognizes PHF6. The complex crystallized in space group P21, and the structure was solved at 1.85 Å by molecular replacement using the RBBP4 structure as an initial search model. Data collection and refinement statistics are outlined in Table 1. There was one molecule/asymmetric unit. Among the 15 amino acids in the PHF6 peptide, only 9 amino acids (residues 162–170) are visible in the final model (Fig. 2A). Furthermore, the structure of RBBP4 (residues 15–410) encompasses seven WD40 repeats forming a β-propeller structure with an additional α-helix at the N terminus (Fig. 2A). The N-terminal 14 residues; loop region residues 55–60, 89–113, and 354–359; and C-terminal residues 411–425 are disordered and invisible in the final electron density maps. The complex structure shows that the PHF6 peptide binds to RBBP4 in an extended conformation across the central channel on the smaller face of the β-propeller disc (Fig. 2B).
FIGURE 2.
Crystal structure of the PHF6 peptide bound to RBBP4. A, two orthogonal views of RBBP4 bound to the PHF6 peptide, with the N and C termini of RBBP4 labeled. The PHF6 peptide is shown in yellow. B, electrostatic surface of RBBP4 showing the PHF6 peptide-binding pocket. The PHF6 peptide is shown as in A.
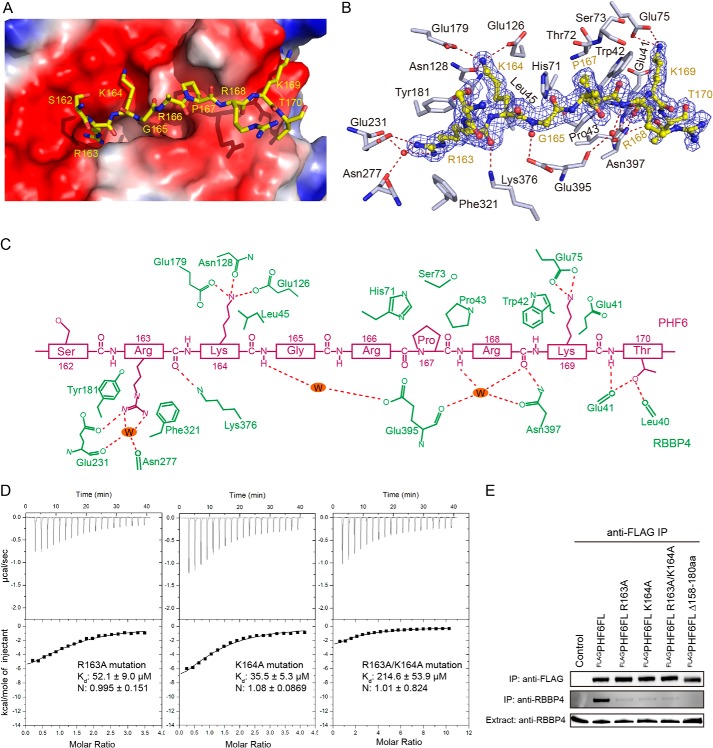
Interaction between RBBP4/RbAp48 and the PHF6 Peptide
An electrostatic potential surface representation of the PHF6 peptide-binding pocket of RBBP4 shows that a highly negatively charged surface with a hole at the top of RBBP4 is used to recognize the PHF6 peptide (Fig. 3A). A refined 2Fo − Fc map at 1σ reveals a clear electron density for residues 162–170 in the PHF6 peptide (Fig. 3B). The clear electron density for the PHF6 peptide in the map shows the orientation of the peptide in the complex and multiple hydrogen bond interactions with RBBP4 (Fig. 3B).
FIGURE 3.
Recognition of the PHF6 peptide by RBBP4. A, electrostatic surface potential representation of the binding pocket with the PHF6 peptide (shown in yellow stick model). B, a simulated annealing omit map (blue) contoured at 1.0σ shows the electron density for the PHF6 peptide bound to RBBP4. RBBP4 residues are shown in gray, and water molecules are shown as red dots. C, schematic representation of the interactions observed between RBBP4 and the PHF6 peptide. Residues in RBBP4 (green labels) that engage in van der Waals contacts and hydrogen bonds or salt bridge interactions with the PHF6 peptide (purple labels) are shown. Hydrogen bonds and salt bridge interactions are delineated by red dashed lines. D, ITC assays for determining the interaction between RBBP4 and PHF6 peptide mutants. E, co-immunoprecipitation (IP) assays with PHF6 and RBBP4. FL, full-length; aa, amino acids.
A network of hydrogen bonds and van der Waals contacts anchor the PHF6 peptide into its binding cleft at the top of RBBP4 (Fig. 3, B and C). The side chain of Arg-163 directly inserts into the acidic central channel, which traverses the core of RBBP4 (Fig. 3A). Furthermore, its guanidinium group is sandwiched between the aromatic side chains of Tyr-181 and Phe-321 in this cleft, forming cation-π interactions (Fig. 3, B and C). The guanidinium group of Arg-163 is further pinioned through hydrogen bonding to the side chain of Glu-231, water-mediated hydrogen bonding to the main chain carbonyl groups of Glu-231 and Asn-277, and its main chain carbonyl group interacting with the Lys-376 side chain in RBBP4 (Fig. 3, B and C). Lys-164 in the PHF6 peptide interacts with RBBP4 through its side chain, forming polar interactions with the side chains of Glu-179 and Glu-126 and a hydrogen bond with the side chain of Asn-128. In addition, van der Waals contacts occur between the side chain of Leu-45 in RBBP4 and the aliphatic part of Lys-164 (Fig. 3, B and C). The backbone amide of Gly-165 in the peptide forms a water-mediated hydrogen bond with the side chain of Glu-395 in RBBP4. Pro-167 in the peptide forms van der Waals contacts within a small pocket composed of His-71, Thr-72, Ser-73, and Pro-43 in the peptide-binding cleft. The backbone amide and carbonyl oxygen of Arg-168 participate in direct and water-mediated hydrogen bonds with the side chain of Asn-397 and the main chain carbonyl oxygen of Glu-395 in RBBP4 (Fig. 3, B and C). Lys-169 in the peptide forms polar interactions with the side chain of Glu-75, and further van der Waals contacts occur between its aliphatic part and the side chains of Trp-42 and Glu-41 in RBBP4 (Fig. 3, B and C). The final visible residue in the PHF6 peptide, Thr-170, interacts with RBBP4 through three hydrogen bonds between its backbone amide, the hydroxyl group, and the main chain carbonyl groups of Leu-40 and Glu-41 (Fig. 3, B and C).
Functional Analysis of PHF6 Recognition by RBBP4/RbAp48
To elucidate the determinants that are crucial for conferring binding specificity, the affinities of RBBP4 for PHF6 peptide mutants were measured by ITC. Affinities between the PHF6(157–171) peptide with mutations R163A, K164A, or R163A/K164A and RBBP4 were reduced by 7-, 5-, and 30-fold, respectively, compared with that of the wild-type PHF6 peptide (Fig. 3D).
To assess the interaction of PHF6 with RBBP4 in vivo, co-immunoprecipitation assays were performed using full-length PHF6. Lysates from HEK293T cells expressing FLAG-tagged full-length PHF6 or mutants were immunoprecipitated with anti-FLAG M2 affinity gel, followed by Western blot analysis with anti-RBBP4 antibody. Compared with FLAG-tagged wild-type full-length PHF6, the mutants led to a significant reduction or loss of binding to RBBP4 (Fig. 3E). These results support the structural basis for PHF6 recognition by RBBP4 in vivo and in vitro.
Structure Comparison of the RBBP4-PHF6 Complex with Other RbAp46/48 Complexes
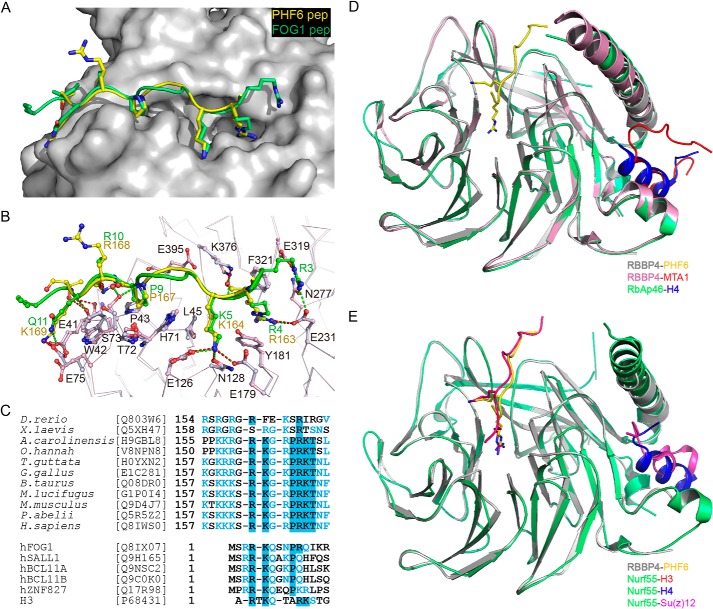
The PHF6-binding site on RBBP4 has previously been shown to be occupied by N-terminal residues 1–15 of FOG1 (friend of GATA1) (28). Comparison of the RBBP4-PHF6 peptide complex structure with the reported crystal structure of the RBBP4-FOG1 complex reveals that the PHF6 peptide binds to the same groove in an extended conformation as the FOG1(1–12) peptide on the top surface of the WD40 repeats of RBBP4 (28). Similar to the recognition of FOG1 by RBBP4, Arg-163 of the PHF6 peptide extends inside the same central cavity of RBBP4 as FOG1 Arg-4, and PHF6 Lys-164 and Pro-167 interact with the same residues of RBBP4 as FOG1 Lys-5 and Pro-9, respectively (Fig. 4, A and B). The detailed comparison of the RBBP4-PHF6 structure with that of FOG1 bound to RBBP4/RbAp48 strongly suggests that the binding of PHF6 and FOG1 is mutually exclusive (Fig. 4, A and B). Sequence alignment between PHF6 orthologs reveals that the RBBP4-binding motif is highly conserved in terrestrial vertebrates, but not in fishes and frogs (Fig. 4C). Interestingly, the structure revealed that Arg-163 and Lys-164 of PHF6 interact with RBBP4 in a manner similar to FOG1 with RBBP4/RbAp48 and histone H3 with Nurf55 (28, 29), suggesting that this pair of positive residues is important for recognition and binding (Fig. 4C). The structural and biochemical data provide clear evidence that RBBP4/RbAp48, RbAp46, and Nurf55 can interact with different partners using different sides of their surfaces. The FOG1, PHF6, or histone H3 peptide binds to the top surface of the WD40 domain of RBBP4 or Nurf55 (Fig. 4, D and E), whereas the histone H4, Su(z)12, or MTA1 peptide-binding site on RbAp46, RBBP4, or Nurf55 is located on the side of the propeller between the N-terminal helix and the PP-loop (Fig. 4, D and E) (29–31). The two recognition sites on the WD40 domain surface are distinct and separable, and the two binding interactions are independent.
FIGURE 4.
Comparative analysis of PHF6 peptide-binding properties of RBBP4 with other reported complex structures. A, superimposition of RBBP4-bound PHF6 (yellow) and FOG1 (green) peptides on RBBP4. B, detailed interaction analysis of PHF6 and FOG1 peptides with RBBP4. The RBBP4 residues from the RBBP4-FOG1 and RBBP4-PHF6 complexes are colored pink and gray, respectively. Hydrogen bonds and salt bridge interactions are depicted as green and red dashed lines, respectively. C, sequence alignment of PHF6 proteins. Only the NoLS region involved in the interaction with RBBP4 is shown. Identical residues are in blue boxes. Representative sequences of the corresponding FOG1, SALL1, BCL11A/B, ZNF827, and histone H3 motifs are aligned with PHF6. D, structural alignment of RBBP4 (gray and pink) and RbAp46 (green). The PHF6, histone H4, and MTA1 peptides are shown in yellow, blue, and red, respectively. E, structural alignment of RBBP4 (gray) and Nurf55 (green). The PHF6, histone H3, H4, and Su(z)12 peptides are shown in yellow, red, blue, and magenta, respectively.
Transcriptional Repression Is Mediated by the Middle Disordered Region of PHF6 and Is Moderately RBBP4-dependent
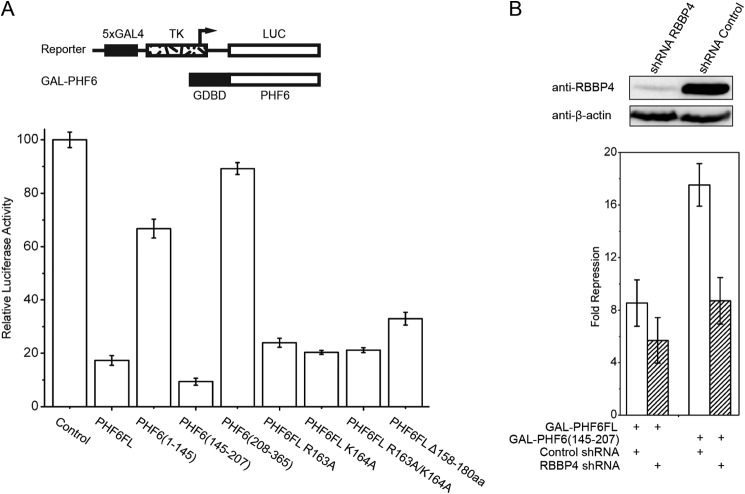
A reporter system in which PHF6 was expressed as a fusion with the GAL4 DNA-binding domain and coexpressed with reporters controlled by GAL4 response elements (GAL4-TK-luciferase) was used to investigate the function of the PHF6-NuRD complex. Our results demonstrate that targeting PHF6 to a promoter resulted in repression of transcription, and the middle disordered region of PHF6 (residues 145–207) was sufficient for transcriptional repression (Fig. 5A). However, mutating or deleting the NoLS region (residues 158–180) of full-length PHF6 could only moderately recover transcriptional repression (Fig. 5A), suggesting that more mechanisms are involved in PHF6-mediated transcriptional repression. Because the RBBP4-interacting motif is located in the middle disordered region of PHF6, we conducted reporter assays in RBBP4 knockdown cells to investigate whether the NuRD complex is involved in transcriptional repression mediated by PHF6, which we predicted would be inhibited. Indeed, RBBP4 knockdown significantly reduced PHF6-mediated transcriptional repression, demonstrating that RBBP4 is required for this function (Fig. 5B).
FIGURE 5.
Repression mediated by PHF6 is diminished as a result of knockdown of RBBP4. A, luciferase reporter assays of PHF6 constructs and mutants. A schematic representation of the reporter and expression vectors used in the transfection experiments is also shown. TK, thymidine kinase; LUC, luciferase; GDBD, GAL4 DNA-binding domain; FL, full-length; aa, amino acids. B, knockdown of RBBP4 reduces the repression mediated by PHF6. The quantifications represent means ± S.D. of three independent experiments.
DISCUSSION
The RBBP4-PHF6 Interaction Is Highly Conserved
Previous studies using co-immunoprecipitation and mass spectrometry identified PHF6 as a new interaction partner with the NuRD complex (19). In addition, our previous work showed that the NoLS region of PHF6 mediates the direct interaction between PHF6 and RBBP4, a component of NuRD complex (20). Here, our biochemical and crystallographic data reveal that a short motif located in the NoLS region of PHF6 is able to bind independently to RBBP4. Specifically, residues 162–171 of PHF6 adopt an extended conformation, binding to the top smaller surface of RBBP4. In particular, PHF6 Arg-163 and Lys-164 are important for the interaction because mutations or deletions of the motif (both in the PHF6 peptide and full-length PHF6) result in a reduction or loss of RBBP4 binding. Although PHF6 is highly conserved in vertebrates, a sequence alignment shows that the RBBP4-binding motif is conserved only in terrestrial vertebrates. This sequence is an evolutionary phenomenon that may be associated with some specific functions of PHF6 in higher species.
A similarly conserved N-terminal RRKQXXP motif is also found in several corepressors and transcription factors, including FOG1/2, SALL12/3/4, BCL11A/B, and ZNF827 (28, 32). In particular, Arg-4 and Lys-5 of FOG1 and SALL1 have been found to be important for recruitment of the NuRD complex; Arg-3 does not appear to be absolutely required (28, 33). Consistent with our ITC data, RBBP4 mutants (E126A/E179A or E231A/D248A/E275A/E219A) impaired the interactions with the FOG1(1–15) peptide (28), suggesting that they are likely to recruit the NuRD complex through a similar mechanism.
The Interactions of PHF6, FOG1, and Histone H3 with RBBP4 Are Mutually Exclusive
Our data clearly show that PHF6 occupies the same RBBP4 pocket that was previously shown to bind the N-terminal of FOG1, and histone H3 also shared the same surface on Nurf55 (28, 29). The structure of the RBBP4/Nurf55 β-propeller domain and its ability to bind histone H3 (Kd: 0.8 ± 0.1 μm), the FOG1 peptide (Kd: 0.6 ± 0.13 μm), and PHF6 NoLS motif (Kd: 7.1 ± 0.4 μm) are well documented, thus it is unlikely that PHF6, FOG1 and histone H3 could be interacting simultaneously with the same molecule of RBBP4. Similarly, there are overlapping binding sites at the propeller edge of RBBP4/RbAp46/Nurf55 for binding histone H4, Su(z)12 and MTA1 (29–31). These two separate binding interactions are compatible and independent. Previous studies have shown that the N-terminal residues 1–15 of FOG1 can interact with two different components of the NuRD complex, MTA1 and RBBP4 (28). Because of the significant sequence similarity between the NoLS region of PHF6 and the N-terminal residues 1–15 of FOG1, PHF6 may also be able to associate with MTA1 in a similar way. It is possible that RBBP4 and MTA1 can simultaneously interact with separate PHF6 molecules at target loci in vivo.
PHF6 Mutations Might Affect the Developmental Pathways Regulated by the NuRD Complex
The NuRD complex is a multifunctional regulator and has important roles in processes such as transcriptional regulation, cell cycle progression, and normal development (6, 7). For example, during hematopoiesis, transcriptional repression of multiple lineage-specific genes by the NuRD complex is mediated by FOG1, which binds MTA proteins and RBBP4/RbAp46 and recruits NuRD to GATA family transcription factors (34, 35). Additionally, other hematopoiesis lineage-specific transcription factors such as BCL11B are also associated with the NuRD complex (36). PHF6 was identified as a new key X-linked tumor suppressor gene, and tumorigenic mutations have been almost exclusively detected in male T-cell acute lymphoblastic leukemia or acute myeloid leukemia patients (13, 14). Any event impairing the association of PHF6 with the NuRD complex or with the targeted promoters may affect recruitment of the NuRD complex to the promoters and control of the target genes. The NuRD complex is the only RBBP4-containing complex known to interact with the PHF6 protein, but it is possible that PHF6 might associate with other RBBP4-containing complexes, such as the CAF-1 and PRC2 complexes, in a parallel manner. RBBP4/RbAp48 decline in the dentate gyrus is responsible for age-related memory loss (37), and the PHF6 gene is associated with human Börjeson-Forssman-Lehmann syndrome, an X-linked mental retardation disorder, and is also expressed in the dentate gyrus in the adult brain (17). However, it is unclear whether there is a relationship between RBBP4-PHF6 interaction and human mentality and memory.
In summary, we have demonstrated that PHF6 binds to RBBP4 via a mechanism involving a highly specific interaction of its NoLS region with the top surface of RBBP4. This RBBP4-binding motif of PHF6 is highly conserved, and a similar motif is also likely to exist in other transcription cofactors to recruit the NuRD complex in a similar mechanism. The middle region (containing the RBBP4-binding motif) of PHF6 is sufficient to mediate the transcriptional repression of the GAL4 reporter, and RBBP4 is required for this function. Our data provide the structural basis and functional role of PHF6 associated with the NuRD complex. Further identification and analysis of PHF6 target genes will provide more insight into the mechanisms of PHF6-mediated repression and regulation and an understanding of the roles of PHF6 during normal development and in the pathogenesis of Börjeson-Forssman-Lehmann syndrome, T-cell acute lymphoblastic leukemia, and acute myeloid leukemia.
Acknowledgments
We thank the staff at beamline 17U of the Shanghai Synchrotron Radiation Facility for assistance with x-ray data collection. We thank Prof. Yide Mei for providing the GAL4 response reporter system.
This work was supported by National Basic Research Program of China 973 Program Grants 2011CB966302, 2012CB917201, and 2011CB911104; Chinese National Natural Science Foundation Grants 31170693 and 31330018; and Chinese Academy of Sciences Grant KJZD-EW-L05 and Strategic Priority Research Program Grants XDB08010101 and XDB08030302.
The atomic coordinates and structure factors (code 4R7A) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- NLS
- nuclear localization sequence
- NoLS
- nucleolar localization sequence
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Zhang Y., LeRoy G., Seelig H. P., Lane W. S., Reinberg D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289 [DOI] [PubMed] [Google Scholar]
- 2. Tong J. K., Hassig C. A., Schnitzler G. R., Kingston R. E., Schreiber S. L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921 [DOI] [PubMed] [Google Scholar]
- 3. Xue Y., Wong J., Moreno G. T., Young M. K., Côté J., Wang W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861 [DOI] [PubMed] [Google Scholar]
- 4. Feng Q., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Zhang Y. (2002) Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol. Cell. Biol. 22, 536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brackertz M., Boeke J., Zhang R., Renkawitz R. (2002) Two highly related p66 proteins comprise a new family of potent transcriptional repressors interacting with MBD2 and MBD3. J. Biol. Chem. 277, 40958–40966 [DOI] [PubMed] [Google Scholar]
- 6. Denslow S. A., Wade P. A. (2007) The human Mi-2/NuRD complex and gene regulation. Oncogene 26, 5433–5438 [DOI] [PubMed] [Google Scholar]
- 7. Ramírez J., Hagman J. (2009) The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics 4, 532–536 [DOI] [PubMed] [Google Scholar]
- 8. Bowen N. J., Fujita N., Kajita M., Wade P. A. (2004) Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta 1677, 52–57 [DOI] [PubMed] [Google Scholar]
- 9. Börjeson M., Forssman H., Lehmann O. (1962) An X-linked, recessively inherited syndrome characterized by grave mental deficiency, epilepsy, and endocrine disorder. Acta Med. Scand. 171, 13–21 [DOI] [PubMed] [Google Scholar]
- 10. Lower K. M., Turner G., Kerr B. A., Mathews K. D., Shaw M. A., Gedeon A. K., Schelley S., Hoyme H. E., White S. M., Delatycki M. B., Lampe A. K., Clayton-Smith J., Stewart H., van Ravenswaay C. M., de Vries B. B., Cox B., Grompe M., Ross S., Thomas P., Mulley J. C., Gécz J. (2002) Mutations in PHF6 are associated with Börjeson-Forssman-Lehmann syndrome. Nat. Genet. 32, 661–665 [DOI] [PubMed] [Google Scholar]
- 11. Visootsak J., Rosner B., Dykens E., Schwartz C., Hahn K., White S. M., Szeftel R., Graham J. M. (2004) Clinical and behavioral features of patients with Börjeson-Forssman-Lehmann syndrome with mutations in PHF6. J. Pediatr. 145, 819–825 [DOI] [PubMed] [Google Scholar]
- 12. Turner G., Lower K. M., White S. M., Delatycki M., Lampe A. K., Wright M., Smith J. C., Kerr B., Schelley S., Hoyme H. E., De Vries B. B., Kleefstra T., Grompe M., Cox B., Gécz J., Partington M. (2004) The clinical picture of the Börjeson-Forssman-Lehmann syndrome in males and heterozygous females with PHF6 mutations. Clin. Genet. 65, 226–232 [DOI] [PubMed] [Google Scholar]
- 13. Van Vlierberghe P., Palomero T., Khiabanian H., Van der Meulen J., Castillo M., Van Roy N., De Moerloose B., Philippé J., González-García S., Toribio M. L., Taghon T., Zuurbier L., Cauwelier B., Harrison C. J., Schwab C., Pisecker M., Strehl S., Langerak A. W., Gécz J., Sonneveld E., Pieters R., Paietta E., Rowe J. M., Wiernik P. H., Benoit Y., Soulier J., Poppe B., Yao X., Cordon-Cardo C., Meijerink J., Rabadan R., Speleman F., Ferrando A. (2010) PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat. Genet. 42, 338–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Vlierberghe P., Patel J., Abdel-Wahab O., Lobry C., Hedvat C. V., Balbin M., Nicolas C., Payer A. R., Fernandez H. F., Tallman M. S., Paietta E., Melnick A., Vandenberghe P., Speleman F., Aifantis I., Cools J., Levine R., Ferrando A. (2011) PHF6 mutations in adult acute myeloid leukemia. Leukemia 25, 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zweier C., Rittinger O., Bader I., Berland S., Cole T., Degenhardt F., Di Donato N., Graul-Neumann L., Hoyer J., Lynch S. A., Vlasak I., Wieczorek D. (2014) Females with de novo aberrations in PHF6: clinical overlap of Börjeson-Forssman-Lehmann with Coffin-Siris syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 290–301 [DOI] [PubMed] [Google Scholar]
- 16. Kosho T., Miyake N., Carey J. C. (2014) Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: historical review and recent advances using next generation sequencing. Am. J. Med. Genet. C Semin. Med. Genet. 166C, 241–251 [DOI] [PubMed] [Google Scholar]
- 17. Voss A. K., Gamble R., Collin C., Shoubridge C., Corbett M., Gécz J., Thomas T. (2007) Protein and gene expression analysis of Phf6, the gene mutated in the Börjeson-Forssman-Lehmann syndrome of intellectual disability and obesity. Gene Expr. Patterns 7, 858–871 [DOI] [PubMed] [Google Scholar]
- 18. Wang J., Leung J. W., Gong Z., Feng L., Shi X., Chen J. (2013) PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J. Biol. Chem. 288, 3174–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Todd M. A., Picketts D. J. (2012) PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J. Proteome Res. 11, 4326–4337 [DOI] [PubMed] [Google Scholar]
- 20. Liu Z., Li F., Ruan K., Zhang J., Mei Y., Wu J., Shi Y. (2014) Structural and functional insights into the human Börjeson-Forssman-Lehmann syndrome-associated protein PHF6. J. Biol. Chem. 289, 10069–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 23. Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 25. Xu C., Min J. (2011) Structure and function of WD40 domain proteins. Protein Cell 2, 202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28. Lejon S., Thong S. Y., Murthy A., AlQarni S., Murzina N. V., Blobel G. A., Laue E. D., Mackay J. P. (2011) Insights into association of the NuRD complex with FOG-1 from the crystal structure of an RbAp48·FOG-1 complex. J. Biol. Chem. 286, 1196–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmitges F. W., Prusty A. B., Faty M., Stützer A., Lingaraju G. M., Aiwazian J., Sack R., Hess D., Li L., Zhou S., Bunker R. D., Wirth U., Bouwmeester T., Bauer A., Ly-Hartig N., Zhao K., Chan H., Gu J., Gut H., Fischle W., Müller J., Thomä N. H. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 [DOI] [PubMed] [Google Scholar]
- 30. Alqarni S. S., Murthy A., Zhang W., Przewloka M. R., Silva A. P., Watson A. A., Lejon S., Pei X. Y., Smits A. H., Kloet S. L., Wang H., Shepherd N. E., Stokes P. H., Blobel G. A., Vermeulen M., Glover D. M., Mackay J. P., Laue E. D. (2014) Insight into the architecture of the NuRD complex: structure of the RbAp48-MTA1 subcomplex. J. Biol. Chem. 289, 21844–21855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murzina N. V., Pei X. Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J. V., McLaughlin S. H., Ben-Shahar T. R., Verreault A., Luisi B. F., Laue E. D. (2008) Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Conomos D., Reddel R. R., Pickett H. A. (2014) NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol. 21, 760–770 [DOI] [PubMed] [Google Scholar]
- 33. Lauberth S. M., Rauchman M. (2006) A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. J. Biol. Chem. 281, 23922–23931 [DOI] [PubMed] [Google Scholar]
- 34. Hong W., Nakazawa M., Chen Y. Y., Kori R., Vakoc C. R., Rakowski C., Blobel G. A. (2005) FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 24, 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao Z., Huang Z., Olivey H. E., Gurbuxani S., Crispino J. D., Svensson E. C. (2010) FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis. EMBO J. 29, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cismasiu V. B., Adamo K., Gecewicz J., Duque J., Lin Q., Avram D. (2005) BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene 24, 6753–6764 [DOI] [PubMed] [Google Scholar]
- 37. Pavlopoulos E., Jones S., Kosmidis S., Close M., Kim C., Kovalerchik O., Small S. A., Kandel E. R. (2013) Molecular mechanism for age-related memory loss: the histone-binding protein RbAp48. Sci. Transl. Med. 5, 200ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]