Meacham et al. performed a genome-scale shRNA screen for modulators of B-cell leukemia progression in vivo and revealed dramatic distinctions between the relative effects of shRNAs on the growth of tumor cells in culture versus in their native microenvironment. They identified “context-specific” regulators of leukemia development, including the gene encoding the zinc finger protein Phf6. While inactivating mutations in Phf6 are commonly observed in human myeloid and T-cell malignancies, they found that Phf6 suppression in B-cell malignancies impairs tumor progression.
Keywords: Phf6, context dependency, in vivo screen, leukemia
Abstract
We performed a genome-scale shRNA screen for modulators of B-cell leukemia progression in vivo. Results from this work revealed dramatic distinctions between the relative effects of shRNAs on the growth of tumor cells in culture versus in their native microenvironment. Specifically, we identified many “context-specific” regulators of leukemia development. These included the gene encoding the zinc finger protein Phf6. While inactivating mutations in PHF6 are commonly observed in human myeloid and T-cell malignancies, we found that Phf6 suppression in B-cell malignancies impairs tumor progression. Thus, Phf6 is a “lineage-specific” cancer gene that plays opposing roles in developmentally distinct hematopoietic malignancies.
Retroviral insertional mutagenesis screens (Lund et al. 2002; Mikkers et al. 2002; Suzuki et al. 2002) and transposon-based screens (Collier et al. 2005; Dupuy et al. 2005) have been used as powerful tools for cancer gene discovery in hematopoietic and solid tumors. These screens have identified numerous previously uncharacterized genes that drive tumor initiation and progression. Using RNAi-based screening, it is possible to examine hypomorphic phenotypes in mammalian cells (Berns et al. 2004; Paddison et al. 2004; Ngo et al. 2006; Schlabach et al. 2008; Silva et al. 2008). Recently, pool-based shRNA screening approaches have been adapted to in vivo applications in transplantable tumors. shRNA pools were used to screen for tumor suppressors in mouse models of liver cancer (Zender et al. 2008), B-cell lymphoma (Bric et al. 2009), and skin carcinomas (Beronja et al. 2013). Here, we adapted a pool-based shRNA screening strategy for use in a transplantable mouse model of B-cell acute lymphoblastic leukemia (ALL) (Williams et al. 2006). Specifically, we introduced libraries of retrovirally expressed shRNAs into leukemia cells and transplanted transduced cells into recipient animals to screen for genes that impact leukemia cell growth. In this ALL model, the transplantation of murine p19Arf−/− pre-B cells that express the p185 BCR-ABL fusion protein (Arf-null, p185+) into immune-competent syngeneic mice produces an aggressive, disseminated polyclonal B-cell leukemia within ∼2 wk (Williams et al. 2006, 2007). Importantly, since leukemias are established within wild-type syngeneic recipient animals, tumor cell growth occurs in the context of a normal microenvironment and in the presence of a functional immune system.
Results and Discussion
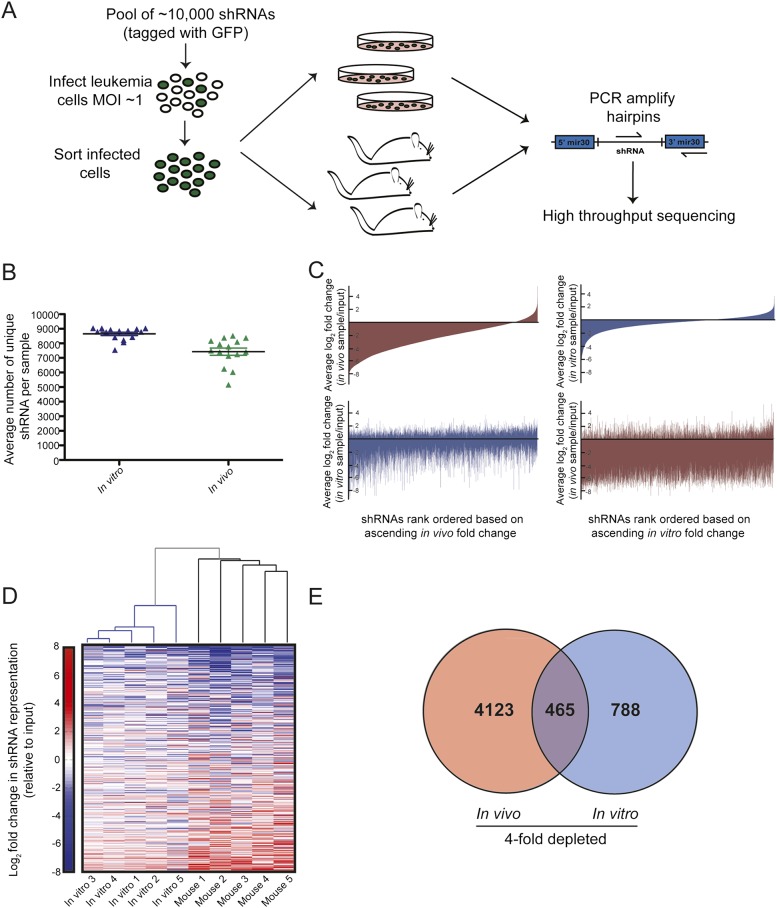
To determine whether large, unbiased shRNA sets could be introduced into disseminated leukemias in vivo, we made use of a genome-scale shRNA library composed of five pools of shRNAs containing ∼10,000 hairpins each. Leukemia cells were transduced with single pools of 10,000 shRNAs, and infected cells were injected into recipient mice or, in parallel, placed in culture. Growth in both contexts allowed us to compare shRNAs that impact leukemia cell proliferation in vitro and in vivo. In control experiments, we observed rapid homing of leukemia cells to the bone marrow and spleen following transplantation (Supplemental Fig. 1A,B), indicating that leukemia cell proliferation occurs within these physiological environments in vivo. Once overt disease developed, leukemia cells were harvested from the blood of the recipient animals and from cultured samples, and shRNA representation was deconvoluted via high-throughput sequencing (Fig. 1A). Here, we could detect, on average, between 7000 and 8000 unique hairpins in the peripheral leukemia burden derived from individual mice (Fig. 1B; Supplemental Fig. 2A). To assess the upper limit of the number of hairpins that can be detected in the leukemia burden from individual mice, we combined all five pools of shRNAs and introduced this set of 50,000 hairpins into tumor cells prior to transplantation. Here, we could identify as many as 30,000 unique shRNAs in tumors from individual animals at the onset of terminal disease (Supplemental Fig. 2B,C). Thus, given the high engraftment efficiency in this model of B-ALL, we can represent genome-scale shRNA libraries in vivo.
Figure 1.
Genome-scale in vivo RNAi screening in a transplantable model of ALL. (A) A schematic representation of the in vivo screening strategy. Leukemia cells were transduced with pools of shRNAs and then grown in culture or injected into recipient mice. shRNA representation at disease presentation was assessed by high-throughput sequencing. (B) A scatter plot showing the average number of hairpins detected across all in vitro and in vivo samples. Each data point represents an individual sample, and error bars show standard error of the mean (SEM). (C) Waterfall plots depicting the average log2 fold change of each shRNA in vivo (red) or in vitro (blue) relative to the input population. shRNAs are arranged in rank ascending order based on their behavior in vivo (left panel) or in vitro (right panel). (D) Unsupervised hierarchical clustering of samples based on the enrichment or depletion of shRNAs. Data are shown from an experiment in which a 50,000 shRNA set was infected into leukemia cells. Only hairpins represented by 100 or more sequencing reads in the input sample are shown. (E) A Venn diagram showing the overlap between the number of shRNAs that depleted an average of fourfold in the in vivo and in vitro settings.
As an initial survey of the data set produced from the 50,000 shRNA pool, the shRNA representation from cultured cells or mice was subjected to hierarchical clustering based on the enrichment and depletion of shRNAs within each sample. Here, the in vivo and in vitro samples clustered as distinct groups (Fig. 1D). In fact, phenotypes conferred by hairpins in vitro were poorly correlated with the phenotypes conferred by the same hairpins in vivo (Fig. 1C,E; Supplemental Fig. 3A), with limited mutual information provided when comparing rank-order lists of enriched and depleted shRNAs in cell culture and mice. For validation studies, we focused on shRNAs that depleted specifically in the in vivo setting, as these genes represent a putative new class of leukemia drug targets. To prioritize shRNAs for validation studies, hairpins that depleted an average of fourfold in vivo and did not deplete >25% in vitro were considered candidates for validation. Using these criteria, we identified ∼1700 scoring hairpins that targeted annotated genes, predicted genes, and predicted proteins (Supplemental Tables 1, 2).
Relative to the in vitro data set, the in vivo data set showed high mouse-to-mouse variability, (Fig. 1D; Supplemental Fig. 3), suggesting that transplantation and engraftment as well as the complexity of the in vivo microenvironment can substantially influence the measured effect of a relatively neutral shRNA. However, further inspection of this variation showed that a subset of shRNAs had lower coefficients of variation (CVs) between mice. When we examined the CVs between mice for our putative “scoring shRNAs,” the average CVs for these hairpins was significantly lower than the average CV in vivo (Supplemental Fig. 3B–D), and the CVs of candidate depletion hits were of a size comparable with those produced by shRNAs that exerted consistent biological effects in previous small-scale screens. In contrast, randomly selected shRNAs, validated for neutrality, showed greater variability and reduced fold change in our data set (Supplemental Fig. 3B). Thus, the set of low CV shRNAs from our screen is enriched for validating shRNAs.
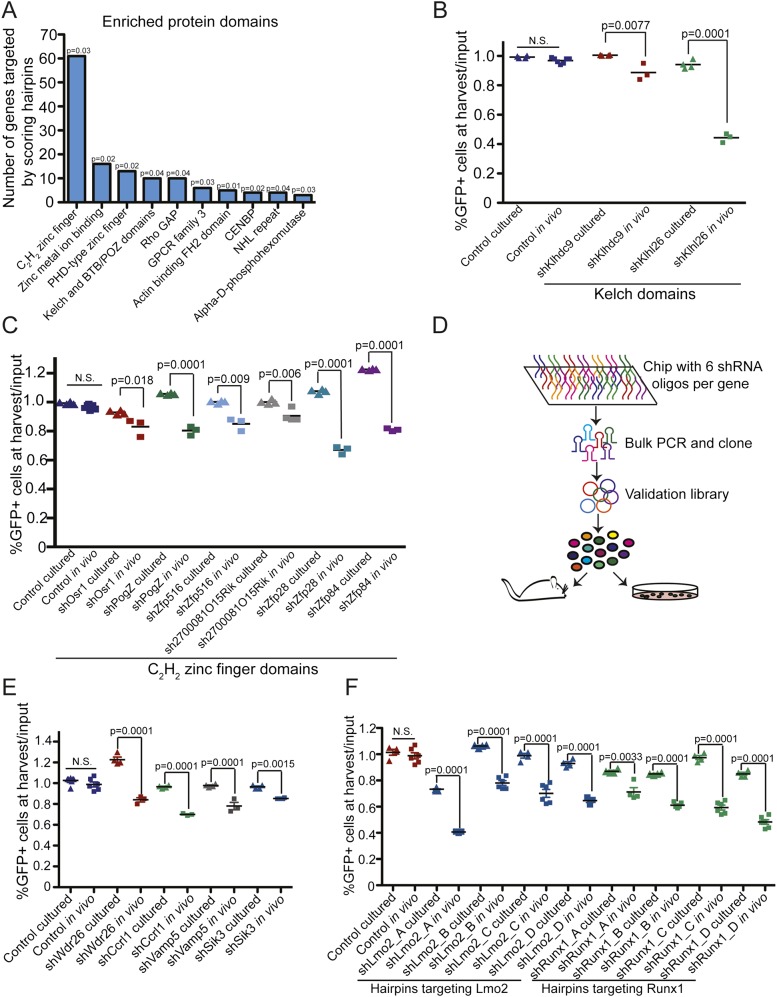
Functional categorization of the genes targeted by the 1700 scoring shRNAs (Huang et al. 2009a,b) based on structural features of proteins encoded by target genes revealed that these genes were enriched for a number of common domains (Fig. 2A; Supplemental Table 2B). These domains included C2H2-type zinc finger protein domains, which primarily regulate gene expression (Iuchi 2001); Kelch and BTB/POZ domains, which are involved in protein–protein interactions and regulate diverse cellular processes (Collins et al. 2001); and plant homeodomain (PHD) fingers, which recognize specific histone modifications and are part of multimeric complexes that can activate or repress gene transcription (Musselman and Kutateladze 2011; Sanchez and Zhou 2011). To assess the impact of a given hairpin on tumor cell growth, leukemia cells were partially transduced with single shRNAs coexpressing GFP and injected into recipient mice or maintained in culture. After a period of in vitro or in vivo proliferation, the percentage of GFP-positive (shRNA-expressing) cells was assayed by flow cytometry (Supplemental Fig. 4A). In total, out of the 18 shRNAs tested targeting genes containing Kelch and C2H2 zinc finger domains, nine depleted as single constructs. Importantly, eight of these nine shRNAs differentially impacted leukemia cell growth in vivo and in vitro (Fig. 2B,C). The deleterious effect of shRNA-mediated gene suppression in vivo was confirmed with second hairpins for these targets (Supplemental Fig. 4B). While these hairpins were neutral or, in some instances, provided a growth advantage in the in vitro context, they depleted during tumor expansion in mice. Thus, by performing screens in mice, we identified a number of genes that represent putative in vivo “context-specific” cancer dependencies that would not be identified in cell culture-based screens.
Figure 2.
The tumor microenvironment introduces distinct tumor growth dependencies relative to growth in culture. (A) A bar graph showing protein domains and motifs that were enriched in the set of hairpins that depleted specifically in vivo. (B,C) Scatter plots showing results from in vivo and in vitro validation assays for shRNAs targeting genes with Kelch (B) and C2H2 zinc finger (C) domains. Each data point represents the ratio of the percentage of hairpin-infected GFP-positive cells harvested from the blood of mice (in vivo samples) or from cells maintained in culture (in vitro samples) to the percentage of GFP-positive cells in the input population. P-values were determined using a Student’s t-test. (D) A schematic representation of the generation of the validation library and the secondary validation screen. (E) A scatter plot showing in vitro and in vivo validation assays of individual shRNAs. P-values were calculated using a Student’s t-test. (F) A scatter plot showing the effects of Runx1 or Lmo2 suppression in vitro and in vivo. shLmo2_A and shRunx1_A were the hairpins identified in the screen. P-values were calculated using a Student’s t-test.
To further prioritize shRNAs for validation, we generated transcriptional data from leukemia cells grown in vitro and in vivo (Supplemental Fig. 5A). Specifically, we compared the genes that were transcriptionally up-regulated in vivo with those that were targeted by hairpins that depleted specifically in vivo. Notably, only a small proportion of genes overlapped between the transcriptional and screening data sets (Supplemental Fig. 5B,C; Supplemental Table 3), a phenomenon that has also been observed when comparing functional and transcriptional data in other settings (Yeger-Lotem et al. 2009). To more extensively probe this overlapping gene set, we built a high-coverage validation library that contained six independent shRNAs directed against each of the 133 genes in this set (Fig. 2D). Only genes that had multiple independent shRNAs deplete specifically in vivo, but not in cell culture, in this secondary screen (Supplemental Table 4) were considered candidates for follow-up studies. By this criteria, this secondary screen validated 77 of the starting 133 genes (58%) as context-specific positive regulators of tumor growth, and a subset of these was further validated as single shRNA constructs (Fig. 2E; Supplemental Fig. 5C). Consistent with our previous observations, shRNAs targeting these 77 genes produced lower CVs than the nonscoring shRNAs (Supplemental Fig. 3D).
Two of the shRNAs that depleted specifically in the in vivo setting targeted Lmo2 and Runx1 (Fig. 2F), genes with established roles in hematopoietic malignancies. LMO2 translocations, resulting in overexpression of the full-length Lmo2 protein, are found in T-cell malignancies and have been suggested, at least in part, to promote tumorigenesis by blocking T-cell development (Curtis and McCormack 2010; McCormack et al. 2010). Thus far, LMO2 does not have a well-established role in the development of B-cell malignancies. Whereas inactivating mutations and translocations resulting in the truncation of RUNX1 are observed in myeloid and T-cell malignancies (Cameron and Neil 2004), full-length RUNX1 is expressed on translocated alleles in B-cell malignancies (Golub et al. 1995; Romana et al. 1995). Here, both Runx1 and Lmo2 are transcriptionally up-regulated in the in vivo setting (Supplemental Fig. 6A), and multiple hairpins targeting these genes confer a growth disadvantage specifically in vivo (Fig. 2F; Supplemental Fig. 6B,C). These data indicate that, in vivo, BCR-ABL-driven B-cell leukemia progression is dependent on Runx and Lmo2 function.
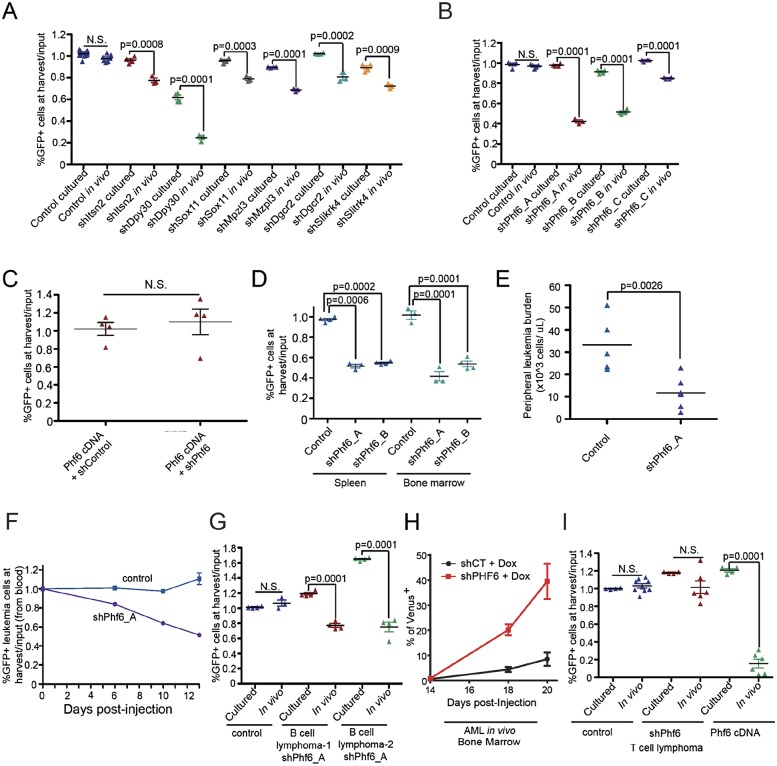
As a final means of examining our shRNA screening data using existing biological data sets, we compared genes targeted by shRNAs that depleted in vivo with genes contained within amplicons found in human ALL patients (Kuiper et al. 2007; Mullighan et al. 2008; Zhang et al. 2012). We mapped the genomic locations of the human orthologs of genes targeted by scoring shRNAs and looked for genes that fell within the amplified regions. In validation experiments, a number of shRNAs targeting genes contained within ALL amplicons conferred minor negative growth effects specifically in vivo (Fig. 3A). In contrast, hairpins targeting the PHD finger protein Phf6, located in the XQ26 amplicon, yielded a more pronounced negative growth effect in validation experiments, and selection against Phf6 suppression was observed in B-ALLs proliferating in vivo but not in vitro (Fig. 3B). Inactivating mutations in PHF6 are the cause of Börjeson-Forssman-Lehman syndrome (BFLS), an X-linked intellectual disability (XLID) disorder (Lower et al. 2002). Recently, inactivating PHF6 mutations have been identified in ∼25%–30% of human T-cell ALLs (Van Vlierberghe et al. 2010; Zhang et al. 2012) and in 2%–3% of acute myeloid leukemias (AMLs) (Supplemental Table 5; Van Vlierberghe et al. 2011; Patel et al. 2012; Yoo et al. 2012). While the mutational status of PHF6 has been examined in >100 human B-lineage ALLs (Van Vlierberghe et al. 2010; Zhang et al. 2012), no PHF6 mutations have been observed in these tumors (Supplemental Table 5), suggesting that inactivating mutations do not promote malignant growth in B-ALL. Strikingly, our data indicate that suppression of Phf6 actually impairs disease progression in B-cell ALLs, specifically in the in vivo setting.
Figure 3.
Phf6 is a developmentally specified, in vivo-specific regulator of tumor cell growth. (A) Scatter plot showing the behavior of single shRNAs in in vitro and in vivo validation assays. All hairpins target genes found within common amplicons in human ALL. P-values were calculated using a Student’s t-test. (B) A scatter plot showing the effect of hairpin-mediated Phf6 suppression on leukemia cell representation in vitro and in vivo. (C) A graph showing suppression of the in vivo effect of a Phf6 shRNA in a population of leukemia cells via expression of a nontargetable Phf6 cDNA. (D) Scatter plots showing the effect of Phf6 suppression in leukemia cells harvested from the spleen and bone marrow of tumor-bearing animals. (E) Peripheral leukemia cell counts 9 d following tumor cell transplantation. (F) Longitudinal monitoring of the percentage of vector control or shPhf6-infected leukemia cells in partially transduced tumor cell populations. (G) A scatter plot showing the effect of Phf6 suppression in distinct Eµ-myc transplanted B-cell lymphomas. (H) A graph showing the relative impact of Phf6 suppression in a transplanted AML. The Phf6 and control shRNAs were induced with doxycycline at day 14, and the relative percentage of infected cells over time is shown. (I) Scatter plots showing the impact of Phf6 suppression and overexpression in a transplanted T-cell lymphoma.
The strength of the phenotype conferred by Phf6 suppression correlated with the degree of hairpin-mediated gene knockdown (Supplemental Fig. 7A), and expression of a nontargetable Phf6 cDNA suppressed the deleterious effect of a Phf6 shRNA on tumor cell growth (Fig. 3C). Phf6 suppression negatively impacted B-cell tumor growth in all hematopoietic organs tested (Fig. 3D), and we observed a reduction in peripheral leukemia burden in sorted, transplanted populations of leukemia cells transduced with an shRNA targeting Phf6 relative to a vector control (Fig. 3E). This effect could be due to either impaired tumor growth or impaired tumor engraftment. To differentiate between these possibilities, we monitored the effect of Phf6 suppression on leukemia growth in vivo at sequential time points during disease progression and found that the proportion of cells suppressing Phf6 decreased progressively during disease progression (Fig. 3F). Thus, Phf6 suppression impairs leukemia cell growth or survival rather than impairing engraftment following transplantation.
To determine whether Phf6 is required for the growth of B-cell malignancies driven by other oncogenic lesions, we also tested the effect of Phf6 suppression in B-cell lymphomas derived from Eμ-Myc mice, which are driven by expression of high levels of c-Myc. Here, suppression of Phf6 was also specifically selected against in the in vivo context in transplanted B-cell lymphomas (Fig. 3G). Additionally, Phf6 suppression had no effect on nontransformed pre-B cells (Supplemental Fig. 7B). Together, these data suggest that Phf6 may be required for the maintenance of the transformed B-cell malignant state in B-cell diseases driven by distinct initiating oncogenes. We then tested the effect of Phf6 suppression in transplantable mouse models of acute leukemia and lymphoma and found that hairpin-mediated suppression of Phf6 had a neutral effect in T-cell lymphomas but promoted significant myeloid tumor cell (AML) growth in vivo (Fig. 3H,I). Together, these findings suggest differential requirements for Phf6 gene function in B-cell, T-cell, and myeloid tumors.
We next tested the effect of cDNA-mediated Phf6 overexpression in vivo in these transplantable murine tumor models and found that Phf6 overexpression was potently selected against in T-cell lymphomas in vivo (Fig. 3I), whereas it was neutral in B-cell ALLs in vivo (Supplemental Fig. 7C). These results support a lineage-specific role for Phf6 in hematopoietic malignancies such that overexpression of Phf6 impairs tumor cell growth in T-cell malignancies, and suppression of Phf6 hinders disease progression in B-cell malignancies. These findings are consistent with human mutational data, where inactivating mutations in PHF6 are observed in T-cell and myeloid malignancies but have never been identified in B-cell malignancies. These findings are similar to our observations describing a B-ALL dependency on Runx expression and to published findings showing RUNX inactivation in myeloid or T-cell malignancies. Such contrasting roles for an established cancer pathway have been documented for deregulated Notch signaling (Klinakis et al. 2011). However, unlike aberrant Notch activity, the effects of Phf6 and Runx suppression were observed only in vivo, suggesting that the tumor microenvironment can strongly influence these “lineage-specific” genetic dependencies.
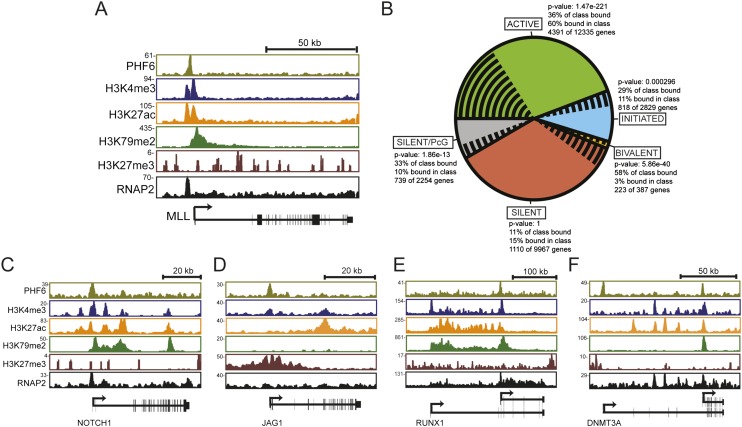
To gain insight into the molecular function of PHF6 in leukemia, we performed whole-genome chromatin immunoprecipitation (ChIP) coupled with high-throughput sequencing (ChIP-seq) using an antibody directed against Phf6 (Supplemental Fig. 8A,B). Using Jurkat cells, a system in which chromatin regulators and histone modifications have been extensively characterized, we found that Phf6 was bound to the promoter regions and within the gene body of many annotated genes (Supplemental Fig. 8C). Phf6 most frequently occupied the promoter region of active genes, as evidenced by co-occupancy with RNA polymerase II (RNAP2) and the histone modification H3K4me3 (Fig. 4A,B). Interestingly, when we ranked the genes bound by Phf6 based on ChIP-seq signal (Fig. 4C–F; Supplemental Table 6), we identified a set of target genes that have established roles in hematopoietic malignancies. This gene set included RUNX1, DMNT3A, NOTCH1, and JAG1 (a ligand of Notch). These data suggest that PHF6 may be involved in a gene regulatory network that influences disease progression through the modulation of other leukemia-associated genes. Consistent with this idea, Phf6 suppression resulted in a significant decrease in Runx1 and Notch levels in B-ALL cells (Supplemental Fig. 9A–C).
Figure 4.
(A) A gene track showing the binding of Phf6, Rnap2, and histone modifications (H3K4me3, H3K79me2, H3K27ac, and H3K27me3) in a T-ALL cell line (Jurkat) at the MLL1 locus. The X-axis indicates the linear sequence of genomic DNA, and the Y-axis represents the total number of mapped reads. The black horizontal bar indicates the genomic scale in kilobases. (B) A pie chart showing Phf6 binding to genes in the genome classified as active (green), initiated (blue), bivalent (orange), polycomb silent (gray), and silent (red) in T-ALL (Jurkat). The proportion of genes in each class bound by Phf6 is shown by black radial lines. P-values indicate significance of the difference within each class. (C–F) Phf6 occupies the promoters of genes/pathways that are frequently altered in hematopoietic malignancies. Gene tracks represent binding of Phf6, Rnap2, and histone modifications (H3K4me3, H3K79me2, H3K27ac, and H3K27me3) in the Jurkat T-ALL cell line at the NOTCH1 (C), JAG1 (D), RUNX1 (E), and DMNT3A (F) loci. The X-axis indicates the linear sequence of genomic DNA, and the Y-axis shows the total number of mapped reads.
The tractable characteristics of this murine pre-B-cell leukemia permit the introduction of very large, diverse libraries of shRNAs into individual animals. Specifically, malignant cells in this model show very high engraftment efficiency into syngeneic recipient mice. This efficiency allows for the simultaneous assessment of the impact of thousands of loss-of-function events in individual tumors. However, the ultimate relevance of this approach lies in the number of genes and gene classes that specifically impact leukemia cell growth in vivo. Here, we identified a large set of “context-specific” dependencies or acquired genetic dependency genes that become important for tumor progression only following transplantation into the tumor’s native microenvironment. These data also highlight the relevance of tumor type to cancer therapy. While many current therapeutic strategies focus on targeting cancer-specific alterations, our data suggest that the effects of targeting the same cancer-relevant pathways may yield dramatically distinct—and possibly opposite—outcomes depending on tumor cell identity.
Materials and methods
Cell culture
Bcr-Abl-driven mouse acute B-cell lymphoblastic leukemia cells (Williams et al. 2006) were cultured in RPMI supplemented with 10% FBS, 4 mM L-glutamine, and 5 µM β-mercaptoethanol. T-cell lymphomas were derived from p53-deficent animals and maintained in IMDM with 10% FBS and 5 μM β-mercaptoethanol. MLL-AF9-expressing AML cells were maintained in RPMI with 10% FBS, 4 mM L-glutamine, and 500 μM β-mercaptoethanol.
RNAi screening and validation assays
The shRNAs were expressed in a mir30 context, and the precise vector construction has been detailed in the referenced mir30 shRNA study (Dickins et al. 2005). Detailed screening and validation library information is provided in the Supplemental Material. PCR primer sequences are provided in Supplemental Table 7. Quantitative PCR (qPCR) reactions were performed on cDNA using the primer sequences in Supplemental Table 8. Knockdown data not shown in the figures are shown in Supplemental Table 9.
Microarray analysis
GFP-expressing leukemia cells (1 × 106) were injected into nonirradiated C57BL/6J syngeneic recipient animals or propagated in culture. After 12 d of growth in each condition, 2 × 106 leukemia cells were sorted from the peripheral blood of tumor-bearing animals or from cultured samples and flash-frozen. RNA was extracted from leukemia cells with an RNeasy kit (Qiagen), and microarrays were performed by the Massachusetts Institute of Technology BioMicro Center using Affymetrix Mouse Genome 430 2.0 arrays.
Western blotting
For Western blots, ALL cells were infected with individual shRNAs coexpressing GFP to a final infection percentage of 40%–60% and sorted based on GFP expression, and total protein was isolated from this sorted cell population. Proteins were detected with the following antibodies: Phf6 (1:1000 in TBS-T with 5% BSA; Novus Biologicals, NB100-68262), Runx1 (1:1000 in TBS-T with 5% BSA; Abcam, ab23980), and HSP60 (1:1000 in TBS-T with 5% BSA; Santa Cruz Biotechnology, SC1722). ImageJ was used to perform protein densitometry.
ChIP-seq
The antibody used for ChIP-seq was specific for PHF6 (Bethyl Laboratories, A301-451A). All protocols for Illumina sample preparation, sequencing, and quality control were provided by Illumina (http://www.illumina.com). Each gene in the human genome was classified into active, initiated, bivalent, or PcG-bound or silent groups based on the presence of co-occupancy of H3K4me3, H3K79me2, and H3K27me3. Using a hypergeometric test, the P-value for enrichment of PHF6 binding to the genes in each of these classes was determined.
Supplementary Material
Acknowledgments
We thank the Swanson Biotechnology Center for excellent technical assistance, and, particularly, A.J. Bhutkar for bioinformatics support. We also thank Francisco Sanchez-Rivera for supplying murine T-cell malignancies, Luke Gilbert and Eleanor Cameron for their comments on this manuscript, and members of the Hemann laboratory for helpful advice and discussions. C.E.M. was supported by the Massachusetts Institute of Technology Department of Biology training grant. M.T.H. is the Chang and Eisen Associate Professor of Biology. Funding was provided by Integrative Cancer Biology Program number U54-CA112967-06 and National Institutes of Health RO1-CA128803-05 (to M.T.H.). C.E.M. and M.T.H. conceived the idea for the research and designed experiments. J.Z., R.T.W., and R.A.Y. provided critical reagents and advice. C.E.M. and N.F. performed experiments. L.N.L. performed ChIP-seq experiments. Y.M.S.-F. performed qPCR and Western analysis of putative Phf6 targets. J.R.P. performed bioinformatics analysis of the screening data sets. B.A.J and T.E. constructed the shRNA design site. C.E.M. and M.T.H. wrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.254151.114.
References
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. . 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428: 431–437. [DOI] [PubMed] [Google Scholar]
- Beronja S, Janki P, Heller E, Lien WH, Keyes BE, Oshimori N, Fuchs E. 2013. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 501: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, Xuan Z, Zuber J, Wigler M, Hicks J, et al. . 2009. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell 16: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron ER, Neil JC. 2004. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene 23: 4308–4314. [DOI] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. 2005. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436: 272–276. [DOI] [PubMed] [Google Scholar]
- Collins T, Stone JR, Williams AJ. 2001. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol 21: 3609–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis DJ, McCormack MP. 2010. The molecular basis of Lmo2-induced T-cell acute lymphoblastic leukemia. Clin Cancer Res 16: 5618–5623. [DOI] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 37: 1289–1295. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. 2005. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436: 221–226. [DOI] [PubMed] [Google Scholar]
- Golub TR, Barker GF, Bohlander SK, Hiebert SW, Ward DC, Bray-Ward P, Morgan E, Raimondi SC, Rowley JD, Gilliland DG. 1995. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci 92: 4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S. 2001. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci 58: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E, et al. . 2011. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature 473: 230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, Hoogerbrugge PM. 2007. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia 21: 1258–1266. [DOI] [PubMed] [Google Scholar]
- Lower KM, Turner G, Kerr BA, Mathews KD, Shaw MA, Gedeon AK, Schelley S, Hoyme HE, White SM, Delatycki MB, et al. . 2002. Mutations in PHF6 are associated with Borjeson-Forssman-Lehmann syndrome. Nat Genet 32: 661–665. [DOI] [PubMed] [Google Scholar]
- Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D, Russell R, DePinho RA, Lenz J, van Lohuizen M. 2002. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet 32: 160–165. [DOI] [PubMed] [Google Scholar]
- McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, Jane SM, Curtis DJ. 2010. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 327: 879–883. [DOI] [PubMed] [Google Scholar]
- Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, Berns A. 2002. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet 32: 153–159. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. . 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453: 110–114. [DOI] [PubMed] [Google Scholar]
- Musselman CA, Kutateladze TG. 2011. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res 39: 9061–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, et al. . 2006. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441: 106–110. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O’Shaughnessy A, Gnoj L, Scobie K, et al. . 2004. A resource for large-scale RNA-interference-based screens in mammals. Nature 428: 427–431. [DOI] [PubMed] [Google Scholar]
- Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. . 2012. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romana SP, Poirel H, Leconiat M, Flexor MA, Mauchauffe M, Jonveaux P, Macintyre EA, Berger R, Bernard OA. 1995. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood 86: 4263–4269. [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. 2011. The PHD finger: a versatile epigenome reader. Trends Biochem Sci 36: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. . 2008. Cancer proliferation gene discovery through functional genomics. Science 319: 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. 2008. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319: 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG. 2002. New genes involved in cancer identified by retroviral tagging. Nat Genet 32: 166–174. [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, De Moerloose B, Philippe J, Gonzalez-Garcia S, Toribio ML, et al. . 2010. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet 42: 338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, Nicolas C, Payer AR, Fernandez HF, Tallman MS, et al. . 2011. PHF6 mutations in adult acute myeloid leukemia. Leukemia 25: 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RT, Roussel MF, Sherr CJ. 2006. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci 103: 6688–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RT, den Besten W, Sherr CJ. 2007. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev 21: 2283–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. . 2009. Bridging high-throughput genetic and transcriptional data reveals cellular responses to α-synuclein toxicity. Nat Genet 41: 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo NJ, Kim YR, Lee SH. 2012. Somatic mutation of PHF6 gene in T-cell acute lymphoblatic leukemia, acute myelogenous leukemia and hepatocellular carcinoma. Acta Oncol 51: 107–111. [DOI] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. . 2008. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135: 852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. . 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.