BRCA-associated cancers are sensitive to DNA-damaging agents such as cisplatin, but the efficacy of cisplatin is limited by the development of resistance. Guillemette et al. performed a genome-wide short hairpin (shRNA) screen and found that loss of the nucleosome remodeling factor CHD4 conferred cisplatin resistance. Cisplatin-resistant clones lacking genetic reversion of BRCA2 show de novo loss of CHD4 expression in vitro. BRCA2 mutant ovarian cancers with reduced CHD4 expression correlate with shorter progression-free survival and shorter overall survival.
Keywords: BRCA2, CHD4, DNA repair, hereditary cancer
Abstract
Hereditary cancers derive from gene defects that often compromise DNA repair. Thus, BRCA-associated cancers are sensitive to DNA-damaging agents such as cisplatin. The efficacy of cisplatin is limited, however, by the development of resistance. One cisplatin resistance mechanism is restoration of homologous recombination (HR), which can result from BRCA reversion mutations. However, in BRCA2 mutant cancers, cisplatin resistance can occur independently of restored HR by a mechanism that remains unknown. Here we performed a genome-wide shRNA screen and found that loss of the nucleosome remodeling factor CHD4 confers cisplatin resistance. Restoration of cisplatin resistance is independent of HR but correlates with restored cell cycle progression, reduced chromosomal aberrations, and enhanced DNA damage tolerance. Suggesting clinical relevance, cisplatin-resistant clones lacking genetic reversion of BRCA2 show de novo loss of CHD4 expression in vitro. Moreover, BRCA2 mutant ovarian cancers with reduced CHD4 expression significantly correlate with shorter progression-free survival and shorter overall survival. Collectively, our findings indicate that CHD4 modulates therapeutic response in BRCA2 mutant cancer cells.
BRCA1/2 mutant hereditary breast and ovarian cancers as well as Fanconi anemia (FA) derive from common genetic defects that compromise DNA repair by homologous recombination (HR). Thus, FA patient cells and BRCA-associated tumors are sensitive to DNA-damaging agents that require HR for repair processing, such as DNA breaks induced by the DNA cross-linking agent cisplatin. Loss of HR in BRCA1 and BRCA2 mutant cancer cells also elicits synthetic lethality with PARP1 inhibitors. As such, cisplatin and PARP1 inhibitors alone or in combination are actively being tested on a diverse set of tumors with suspected BRCA- or HR-associated pathway mutations (Lord et al. 2015).
The efficacy of these therapies is limited, however, by the development of resistance. Currently, the most well-described mechanism known to underlie resistance in BRCA cancers is re-established DNA repair. In both BRCA1 and BRCA2 mutant cancer cells, genetic rewiring events, including secondary or reversion mutations, can reinstate functional gene products and HR-based DNA repair (Edwards et al. 2008; Sakai et al. 2008, 2009). In BRCA1 mutant cells, HR can also be restored through loss of the DNA repair protein 53BP1, which restores DNA end resection required for HR (Bunting et al. 2010). Triple-negative breast cancers that express low levels of BRCA1 and 53BP1 significantly correlate with shorter progression-free survival (PFS) and shorter overall survival (OS) (Bouwman et al. 2010). In BRCA2 mutant cancer cells, 53BP1 depletion does not promote therapeutic resistance or improve HR. Aside from genetic reversion, which occurs in about half of BRCA2 mutant ovarian cancers (Norquist et al. 2011), the mechanisms that mediate resistance to therapy in the remaining half is unknown.
Results and Discussion
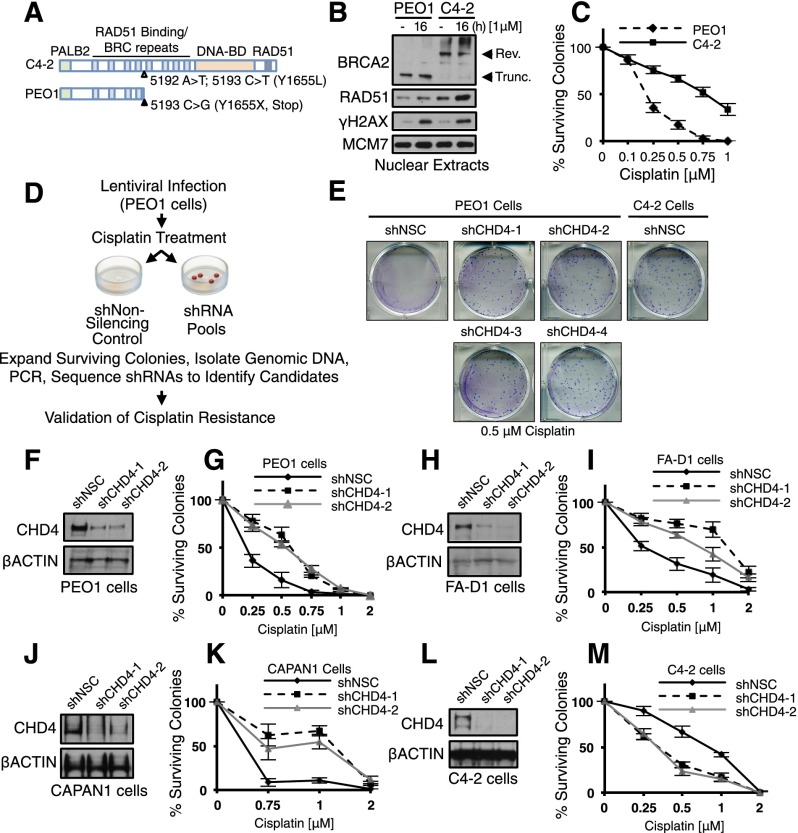
To identify factors that mediate resistance to cisplatin chemotherapy, we performed a survival-based, loss-of-function RNAi screen in the cisplatin-sensitive PEO1 ovarian cancer cell line. PEO1 cells carry a hemizygous nonsense mutation (5193C>G) that generates an N-terminal-truncated form of BRCA2, which cannot carry out HR (Fig. 1A; Sakai et al. 2009). Previous selection for cisplatin-resistant clones revealed BRCA2 reversion mutations that restore HR, as in the C4-2 clone (Fig. 1A–C), as well as clones that were resistant by an unknown mechanism (Sakai et al. 2009). Thus, the screen was performed in PEO1 cells because they reflect BRCA2 mutant ovarian cancers that develop both reversion and reversion-independent mechanisms of cisplatin resistance (Norquist et al. 2011).
Figure 1.
Genome-wide screen identifies CHD4 as a mediator of cisplatin response in BRCA2 mutant cells. (A) Schematic of BRCA2 species found in BRCA2 revertant C4-2 cells or PEO1 cells. (B) Nuclear extracts were analyzed by immunoblot with the indicated antibodies. Truncated BRCA2 in PEO1 cells migrates (∼220 kDa), while the revertant BRCA2 protein in C4-2 cells migrates (∼390 kDa). (C) Cells were left untreated or treated with cisplatin and analyzed for colony survival. (D) Schematic of viral infection with shRNA pools and screening conditions. (E) Colony survival of PEO1 cells containing four unique shRNA vectors targeting CHD4 or NSC as compared with BRCA2 revertant C4-2 cells (SF50 [in micromolar]: PEO1+shNSC, 0.21; shCHD4 (1–4), 0.55–0.56; and C4-2+shNSC, 0.7). PEO1 cells (F,G), FA-D1 cells (H,I), CAPAN1 cells (J,K), and BRCA2 revertant C4-2 cells (L,M) containing two unique pGIPZ shRNA vectors (one or two) targeting CHD4 or NSC were analyzed by immunoblot, left untreated or cisplatin-treated, and analyzed for colony survival. Where shown, error bars represent the standard deviation of the mean of three independent experiments.
Cells were infected with viral pools containing the pGIPZ library comprised of ∼81,000 shRNAs directed against 28,000 unique targets or a nonsilencing control (NSC), selected with puromycin, cisplatin-treated, and analyzed for colony formation (see the Materials and Methods) (Fig. 1D). Of the ∼89 gene targets identified in the initial screen (Supplemental Table S1), we elected to focus on CHD4, a member of the nucleosome remodeling (NuRD) complex (Denslow and Wade 2007; Ramirez and Hagman 2009), for several reasons. Two unique unrelated shRNAs targeting CHD4 were identified in the RNAi screen, and CHD4 was recently implicated as a tumor suppressor in several female malignancies (Le Gallo et al. 2012; Zhao et al. 2013). Moreover, PEO1 cells expressing any of four unrelated CHD4 shRNAs displayed substantially increased colony survival following cisplatin treatment, similar to the BRCA2 revertant line C4-2 (Fig. 1E–G; Supplemental Fig. S1A,B). Ectopic expression of CHD4 in cells containing an shRNA targeting the 3′ untranslated region (UTR) of CHD4 significantly restored cisplatin sensitivity, ruling out off-target effects (Supplemental Fig. S1C,D). In contrast, depletion of two other NuRD subunits, MBD2 and CHD3, had no effect on colony survival after treatment with cisplatin (Supplemental Fig. S1E–H). Furthermore, in two other cell lines that contain a truncated BRCA2 mutant (the FA EUFA423 cell line FA-D1 and the pancreatic cancer cell line CAPAN-1) (Howlett et al. 2002; Edwards et al. 2008), CHD4 depletion also increased colony survival following cisplatin treatment (Fig. 1H–K). In addition, CHD4 depletion had little effect on untreated cells but alleviated cisplatin-induced cell cycle progression defects such that the CHD4-depleted cells more closely resembled the revertant cell lines (Fig. 2A; Supplemental Fig. S2A,B). Finally, CHD4 has been shown to contribute to the DNA damage response (for reviews, see O’Shaughnessy and Hendrich 2013; Stanley et al. 2013).
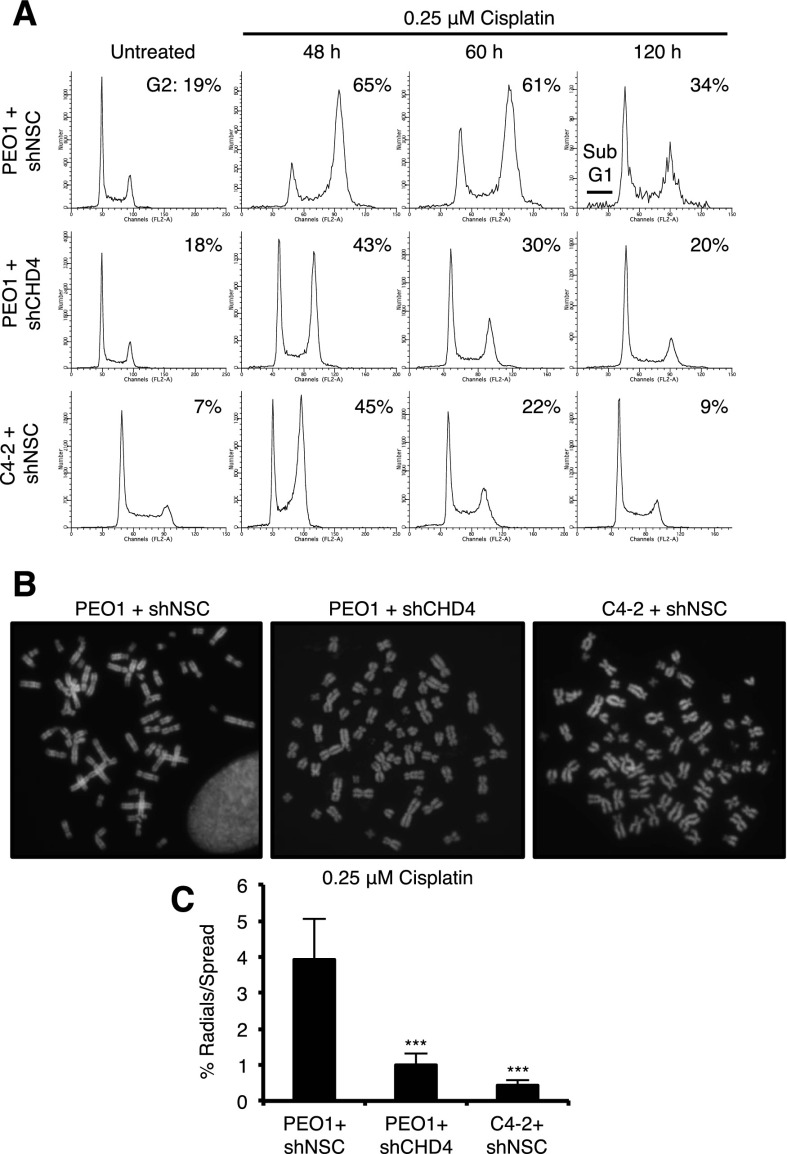
Figure 2.
CHD4 depletion corrects cisplatin-induced cell cycle progression defects and chromosomal aberrations in PEO1 cells. (A) Cells were left untreated or cisplatin-treated and analyzed for cell cycle with propidium iodide. (B,C) Cells were treated with cisplatin (B), and metaphase spreads were analyzed for percentage of radials per metaphase spread (C). At least 75 metaphase spreads were analyzed for each variable (25 per variable in triplicate). Where shown, error bars represent the standard deviation of the mean of three independent experiments. Asterisks denote significance from Student’s two-tailed, unpaired t-test: (***) P ≤ 0.005.
CHD4 loss enhances sensitivity to DNA-damaging agents (Larsen et al. 2010; Smeenk et al. 2010; Sims and Wade 2011; Pan et al. 2012). Thus, we considered that our findings were unique to the cisplatin response and/or BRCA2 mutant cells. Following depletion of CHD4, C4-2 cells, which have functional BRCA2, were sensitive to cisplatin (Fig. 1L,M), the PARP inhibitor Olaparib, the double-strand break-inducing agent zeocin, and the DNA polymerase inhibitor aphidicolin (Supplemental Fig. S3A–C). Moreover, in the absence of exogenous DNA-damaging agents, C4-2 cells displayed a significant induction of γ-H2AX and cleaved Caspase3 along with reduced colony-forming efficiency and growth upon depletion of CHD4 or the NuRD subunit MBD2 (Supplemental Fig. S4A,C,F–I). In contrast, CHD4 depletion in BRCA2 mutant PEO1 cells enhanced survival against these DNA-damaging agents (Supplemental Fig. S3D–F). In untreated PEO1 cells, γ-H2AX and cleaved Caspase3 were not induced, and colony-forming efficiency was not affected (Supplemental Fig. S4B,D,J). CHD4 depletion did not enhance resistance of PEO1 cells to all drugs examined, including 6-thioguanine (6-TG) or melphalan (Supplemental Fig. S3G,H). Thus, in response to diverse DNA-damaging agents, CHD4 depletion improves fitness in BRCA2 mutant, but not BRCA2-proficient, cells.
To test the possibility that CHD4 depletion uniquely enhances cisplatin resistance when DNA repair is defective, we analyzed other HR-defective cell lines. In the BRCA1 mutant HCC1937 breast cancer cell line, CHD4 depletion greatly enhanced γ-H2AX foci (Supplemental Fig. S4E). Examination of either the HCC1937 or the BRCA1 mutant SUM1315MO2 breast cancer cell line revealed that CHD4 depletion dramatically reduced proliferation such that cisplatin toxicity could not be evaluated (Supplemental Fig. S5A–D). Moreover, CHD4-depleted PEO1 cells were resensitized to cisplatin upon BRCA1 depletion (Supplemental Fig. S5E,F). In DNA repair-defective cell lines, including FANCJ mutant (FA-J) cells, FANCD2 mutant (PD20) cells, or XPF mutant (XP-F) cells, CHD4 depletion did not affect proliferation, and cisplatin resistance was not enhanced (Supplemental Fig. S5G–L). Taken together, CHD4 depletion mediates resistance to cisplatin specifically in a BRCA2 mutant background.
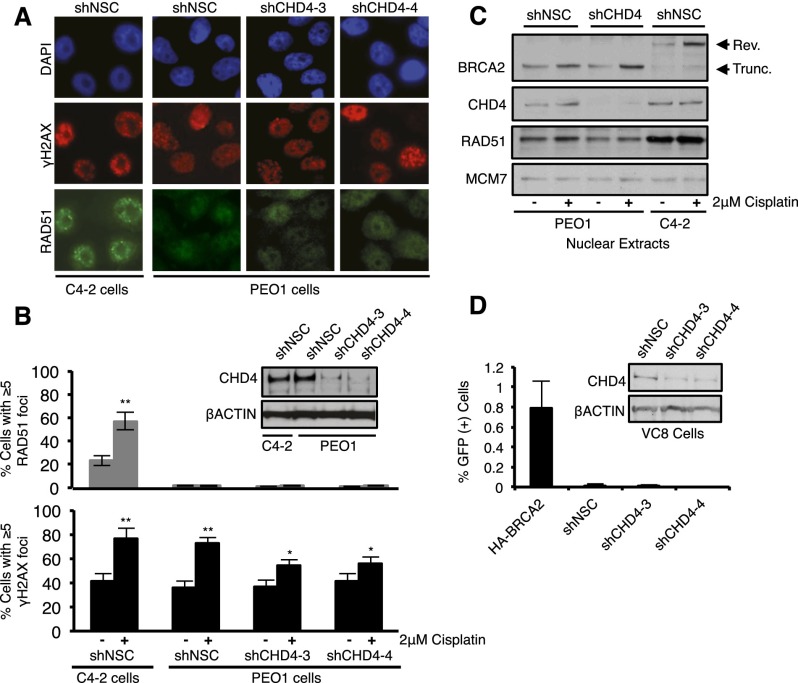
Due to HR defects, BRCA2 mutant cells display chromosomal aberrations following cisplatin treatment. The average number of radials induced by cisplatin per metaphase spread in PEO1 cells significantly decreased following CHD4 depletion and was comparable with that in C4-2 cells (Fig. 2B,C), suggesting that HR was restored. However, depletion of CHD4 in PEO1 cells did not increase RAD51 foci formation or its nuclear localization (Fig. 3A–C; Supplemental Fig. S6A). We also found that CHD4 depletion did not change the expression and size of BRCA2 in PEO1 cells (Fig. 3C). Consistent with the conclusion that HR is not restored, CHD4 depletion did not enhance break-induced HR in the BRCA2 mutant VC-8 hamster cell line that includes an integrated SCE-1 break-inducible DR-GFP reporter, (Pierce et al. 1999), but GFP-positive cells were recovered, as expected, by introduction of a functional BRCA2 (Fig. 3D; Siaud et al. 2011).
Figure 3.
CHD4 depletion in BRCA2 mutant cells does not restore HR. (A) Cells were left untreated or treated with cisplatin and coimmunostained with the indicated antibodies. A representative image of cisplatin-treated cells is shown. (B) Depletion of NSC verses CHD4 was confirmed by immunoblot with the indicated antibodies, and the percentage of cells with RAD51 foci or γH2AX foci was quantified. (C) Cells were left untreated or cisplatin-treated, and nuclear extracts were analyzed with the indicated antibodies. (D) VC8 hamster cells were analyzed as in B and quantified for the percentage of GFP-positive cells by flow cytometry using the DR-GFP HR reporter assay.
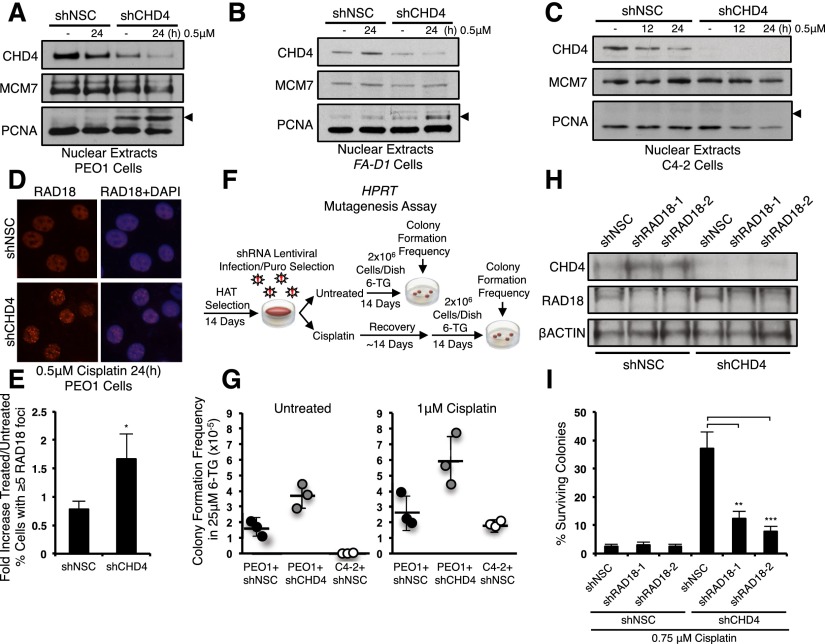
Cisplatin resistance can also result from DNA damage tolerance mechanisms that include error-prone translesion synthesis (TLS) polymerases that bypass DNA lesions (Lehmann et al. 2007; Wang et al. 2009; Doles et al. 2010; Xie et al. 2010). TLS is initiated by RAD6/RAD18-dependent ubiquitination of the proliferating cell nuclear antigen (PCNA) (Lehmann et al. 2007). Notably, CHD4 depletion in BRCA2 mutant PEO1 and FA-D1 cells induced a modification on PCNA that increased its size by ∼8.5 kDa, and this modification was increased by treatment with cisplatin. This modification was not present in PEO1 cells expressing a control shRNA, BRCA2-proficient C4-2 cells, or MBD2-depleted PEO1 cells (Fig. 4A–C; Supplemental Fig. S6D). The PCNA modification also stained positively with antibodies to ubiquitin, suggesting that the modification represents a monoubiquitinated form of PCNA (Supplemental Fig. S6B,C). Correspondingly, CHD4 depletion significantly increased the average number of PEO1 cells with RAD18 foci (Fig. 4D,E).
Figure 4.
CHD4 depletion in BRCA2 mutant cells enhances DNA damage tolerance. (A–C) BRCA2 mutant PEO1 cells (A), BRCA2 mutant FA-D1 cells (B), and BRCA2 revertant C4-2 cells (C) expressing pLKO.1 vectors targeting NSC or CHD4 were left untreated or cisplatin-treated, and nuclear extracts were analyzed with the indicated antibodies. Arrows indicate molecular weight of ∼37 kDa. (D) Cells were left untreated or cisplatin-treated and coimmunostained with the indicated antibodies. A representative image of cisplatin-treated cells is shown. (E) Quantification of the average fold increase in cells with RAD18 foci. (F) Schematic of HPRT mutagenesis assay. (G) Quantitation of HPRT mutagenesis assay. (H) PEO1 cells expressing pLKO.1 shRNAs targeting NSC or CHD4 were codepleted of RAD18 with two unique vectors versus NSC and analyzed by immunoblot blot with the indicated antibodies. (I) Cells were left untreated or cisplatin-treated and quantified for colony survival. Where shown, error bars represent the standard deviation of the mean of three independent experiments.
We also examined whether CHD4 depletion enhanced TLS by measuring the induction of inactivating mutations at the endogenous hypoxanthine phosphoribosyl-transferase (HPRT) locus (Fig. 4F; Chiu et al. 2006). Suggesting induction of a mutagenic pathway, CHD4 depletion increased the mutation frequency in both untreated and cisplatin-treated PEO1 cells as compared with PEO1 or C4-2 cells expressing control shRNAs, as evidenced by enhanced colony formation in 6-TG (Fig. 4G). We next sought to determine whether a RAD18-dependent pathway was required for cisplatin resistance. Compared with CHD4 depletion alone, colony formation after treatment with cisplatin was significantly reduced by codepletion of CHD4 and RAD18 (Fig. 4H,I). These findings suggest that in BRCA2 mutant cells, CHD4 depletion primes a RAD18-dependent mechanism that confers cisplatin resistance.
Finally, we evaluated the clinical relevance of our results. First, we examined and validated a series of previously derived PEO1 clones that lack BRCA2 reversion mutations and RAD51 foci formation but were cisplatin-resistant by an unknown mechanism (Fig. 5A–C; Sakai et al. 2009). Remarkably, in cisplatin-resistant clones C4-4, C4-11, and C4-13, CHD4 protein levels are reduced as compared with parental PEO1 cells or the BRCA2 revertant C4-2 (Fig. 5D). Moreover, ectopic expression of CHD4 increased apoptosis more dramatically in clones C4-4, C4-11, and C4-13 than in PEO1 cells (Fig. 5E,F). Suggesting that resistant clones with reduced CHD4 expression operate similarly to CHD4-depleted PEO1 cells that were reliant on DNA damage tolerance, RAD18 depletion also reduced cisplatin resistance in C4-4, C4-11, and C4-13 clones but not in PEO1 or C4-2 cells (Fig. 5G,H).
Figure 5.
In BRCA2 mutant cancer cells, reduced CHD4 expression correlates with cisplatin resistance and poor patient response to chemotherapy. (A) Nuclear extracts of C4-2 cells, PEO1 cells, and derived cisplatin-resistant clones C4-4, C4-11, and C4-13 were analyzed by immunoblot with the indicated antibodies. (B) Cells were treated with cisplatin and immunostained with the indicated antibodies. A representative image is shown at 16 h. (C) Cells were left untreated or treated with cisplatin and analyzed for colony survival. (D) Cells were analyzed by immunoblot with the indicated antibodies. (E) Empty vector or HA CHD4 were ectopically expressed and analyzed by immunoblot with the indicated antibodies. (F) Quantification of average fold increase in apoptotic cells from three experiments. (G) Cells expressing pGIPZ shRNAs targeting NSC or RAD18 were analyzed by immunoblot with the indicated antibodies. (H) Cells were left untreated or cisplatin-treated and quantified for colony survival. (I,J) Above median or below median CHD4 mRNA levels were correlated with PFS and OS in sporadic ovarian cancers (I) and BRCA2 mutant ovarian cancers (J). P-values were determined by two-tailed log rank tests. (K) Model depicting the response to cisplatin-induced cross-links in BRCA2 mutant cells expressing or depleted of CHD4.
We next asked whether there was a relationship between CHD4 expression and the response of ovarian cancer patients. CHD4 mRNA levels were analyzed in serous ovarian cancers for which OS (n = 315) and PFS (n = 260) were documented (The Cancer Genome Atlas Research Network 2011; Cerami et al. 2012; Gao et al. 2013). Median CHD4 expression was used as a threshold to classify cancers as CHD4-overexpressed (above median) or CHD4-underexpressed (below median), and PFS and OS were assessed. In sporadic cancers, CHD4 expression levels displayed no significant correlation with PFS or OS (Fig. 5I). Strikingly, analysis of CHD4 mRNA specifically in BRCA2 mutant ovarian cancers revealed that low CHD4 expression significantly correlated with shorter PFS (n = 29) and shorter OS (n = 32), (Fig. 5J). Low CHD4 expression also correlated with shorter OS in 22 BRCA2 mutant breast cancers (Supplemental Fig. S7A,B). These results suggest that CHD4 mRNA expression levels could serve as a valuable predictor of response to chemotherapy in ovarian and potentially other cancers carrying a BRCA2 mutation.
Here, through an unbiased RNAi screen, we identified the NuRD protein CHD4 as a factor that modulates response to cisplatin and other DNA-damaging agents in BRCA2 mutant cells. Suggesting that CHD4 also modulates chemotherapeutic responsiveness in the clinic, reduced levels of CHD4 mRNA significantly correlate with shorter PFS and OS in BRCA2 mutant ovarian cancers. Loss of CHD4 expression promotes resistance to cisplatin by a mechanism independent of HR but dependent on RAD18 (see model, Fig. 5K). TLS has been shown to mediate cisplatin resistance in cancer models (Wang et al. 2009; Doles et al. 2010; Xie et al. 2010), but aside from monoubiquitination of PCNA, limited biomarkers exist to determine whether TLS is up-regulated. TLS activation could also be a consequence of changes in chromatin conformation, as CHD4 has a defined role in nucleosome remodeling. Our study demonstrates that CHD4 depletion enhances PCNA monoubiquitination uniquely in BRCA2 mutant cells. However, CHD4 depletion also enhanced PCNA monoubiquitination in cells depleted for ZBTB1, a zinc finger transcription factor that promotes RAD18 accessibility during chromatin remodeling (Kim et al. 2014). Taken together, this suggests that CHD4 serves as a negative regulator of PCNA monoubiquitination and TLS. Our study did not reveal any correlation with CHD4 depletion and restored cisplatin resistance in BRCA1, FANCJ, FANCD2, or XPF mutant cells. In these repair-compromised cells, it remains to be determined whether PCNA monoubiquitination and/or TLS activation or processing is dependent on CHD4. Defining the genetic background for which CHD4 depletion enables TLS will be an important future direction to determine whether CHD4 mediates chemotherapeutic response beyond BRCA2 mutant ovarian or breast cancers.
Materials and methods
Cell lines
Ovarian cancer cell lines PEO1, C4-4, C4-11, C4-13, and C4-2 were grown in DMEM with 10% FBS and 1% Glutmax, and VC8, FA-D1, XP-F, PD20, and FA-J cells were grown in DMEM with 15% FBS. We thank Toshi Taniguchi for PEO1 cells series, Orlando Scharer for XP-F cells, and Maria Jasin for VC8 cells.
Immunofluorescence
Cells grown on coverslips were fixed for 10 min with 3% paraformaldehyde/2% sucrose in phosphate-buffered saline (PBS). For RAD18 foci, 3.7% formaldehyde in PBS was used as fixative. Cells were then permeabilized for 5 min with 0.5% Triton X-100 in PBS. Coverslips were rinsed three times in PBS prior to each step. For primary and secondary staining, cells were incubated for 40 min each in a humid chamber, face down on a 100-μL meniscus of antibodies diluted in 3% BSA in PBS. Primary antibodies used were anti-RAD51 poly (1:500; Santa Cruz Biotechnology), anti-γ H2AX mono (1:500; Millipore), and anti-RAD18 mono (1:400; Abcam). Secondary antibodies included Rhodamine Red-X-conjugated AffiniPure goat anti-rabbit IgG and fluorescein (FITC)-conjugated AffiniPure goat anti-mouse IgG (Jackson ImmunoResearch Laboratories. Inc.). Coverslips were mounted on slides using VectaShield mounting medium with DAPI (Vector Laboratories, Inc.) and analyzed on a fluorescence microscope (Leica, DM 5500B) with a Qimaging Retiga 2000R fast 1394 camera. For flow cytometry experiments examining apoptosis, live cells were collected and stained using the Annexin V-FITC and propidium iodide apoptosis detection kit (Abcam).
RNAi
The packaging cell line 293TL was used to produce lentiviral particles containing pGIPZ or pLKO.1 vectors. Cells were transfected with 1:1:2 μg of DNA packaging versus insert using Effectene transfection reagent (Qiagen) 48 h prior to harvesting supernatants. Supernatants were filtered and added to recipient cell lines with 1 μg/mL polybrene. Cells infected with shRNA vectors were selected with puromycin (pGIPZ and pLKO.1). For shRNA-mediated silencing, the mature sense was used for pGIPZ, CHD4-1 (5′-ATTCATAGGATGTCAGCAG-3′) and CHD4-2 (5′-TTAGTTCTGTCTTGGAGGG-3′); pLKO.1, CHD4-3 (5′-GCTGCTGACATCCTATGAATT-3′) and CHD4-4 (5′-GCTGACACAGTTATTATCTAT-3′); pLKO.1, CHD4-(UTR)3′ (5′-GCCTGTTACACACAAACTGT-3′); pLKO.1, CHD3-1 (5′-CCTCCCACACTGCCAAGTATA-3′); pLKO.1, CHD3-2 (5′-GCCATTATCAACGAGCCATTT-3′); pLKO.1, MBD2-1 (5′-GCCTAGTAAATTACAGAAGAA-3′) and MBD2-2 (5′-GTACGCAAGAAATTGGAAGAA-3′); pGIPZ, RAD18-1 (5′-TTTGGTCTTTGCAGCAGGGCTC-3′); and pLKO.1, RAD18-2 (5′-CCCTCGACATCCACTTTGAAA-3′).
shRNA screen
The human shRNAmir pGIPZ library (release 6, Open Biosystems) was obtained through the University of Massachusetts Medical School RNAi core facility. Lentiviral pools were generated and used to transduce PEO1 cells as described at a multiplicity of infection (MOI) of 0.2 (Gazin et al. 2007). Cells that bypassed two cycles of 2 μM cisplatin treatment (enough to induce ∼100% lethality in the shNSC pool) underwent cellular proliferation to form colonies. Colonies were pooled and expanded, and the shRNAs were identified by sequence analysis as previously described (Gazin et al. 2007). Individual knockdown cell lines were generated by stable transduction of 6 × 105 cells with a single shRNA.
Immunoblot
Cells were lysed in 150 mM NETN lysis buffer (20 mM Tris at pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail) for 30 min on ice. Cell extracts were clarified by centrifugation at 14,000 rpm, protein was quantified by Bradford assay, and lysates were boiled in SDS loading buffer. Nuclear extracts were prepared with a nuclear extraction kit (ThermoFisher) with 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail, and 20 mM N-ethylmaleimide, and proteins were separated by SDS-PAGE on 4%–12% bis Tris or 3%–8% Tris acetate gels (Novex, Life Technologies) and electrotransferred onto nitrocellulose membranes. Membranes were blocked in 5% milk diluted in PBS. Antibodies used for immunoblot analysis included anti-BRCA2 poly (1:1000; Abcam), anti-CHD4 mono (1:1000; Abcam), anti-RAD51 poly (1:500; Santa Cruz Biotechnology), anti-RAD51 mono (1:1000; Millipore), anti-MCM7 mono (1:1000; Santa Cruz Biotechnology), anti-MBD2 mono (1:1000; Santa Cruz Biotechnology), anti-CHD3 mono (1:1000; Abcam), anti-γ H2AX mono (1:500; Millipore), anti-PCNA mono (1:1000; Abcam), anti-ubiquitin mono (1:1000; Abcam), anti-RAD18 mono (1:500; Abcam), anti-HA mono (1:2000; Cell Signaling), anti-βACTIN (1:5000; Abcam), and Caspase3 (1:1000; Cell Signaling). Membranes were washed and incubated with horseradish peroxidase-linked secondary antibodies (1:5000; Amersham) and detected by chemiluminescence (Ambersham).
Metaphase spreads
Cells were treated with 0.25 μM cisplatin for 60 h and incubated with 150 μL of 10 μg/mL colecemid (Life Technologies) for 2–3 h, and medium and cells were collected. Cells were lysed with 75 mM prewarmed KCl (hypotonic buffer) in a 37°C water bath, and 100 μL of 3:1 methanol/acetic acid was added dropwise to stop the reaction. Cells were then collected and resuspended in 3:1 methanol/acetic acid fixative solution for 20 min on ice, which was repeated three times. Cells were dropped from a bench top onto sides and mounted with VectaShield mounting medium with DAPI (Vector Laboratories, Inc.). Spreads were viewed under high magnification with oil to observe radials.
Survival analysis
The serous ovarian cancer Affymetrix U133 raw gene expression data set or breast cancer data set was retrieved from the Cancer Genome Atlas (TCGA), and survival analysis was performed using BRB-ArrayTools version 4.2 (Biometrics Research Branch, National Cancer Institute). Median CHD4 mRNA expression was used as a threshold to classify cancers as CHD4-overexpressed (above median) or CHD4-underexpressed (below median).
Supplementary Material
Acknowledgments
We thank T. Taniguchi, E. Swisher, and S. Elledge for comments on the manuscript; A.Virbasius for help with lentiviral preparation; and the J. Mao, T. Fazzio, S. Elledge, and S. Ganesan laboratories for sharing reagents. The GFP-HA-CHD4 constructs were generous gifts from S. Jackson, and the HA-BRCA2 construct was a generous gift from R. Scully. We thank A. Venkatesh, B. Morehouse, and S. Nayak for technical assistance. This work was supported by the Department of Defense (W81XWH-10-1-0585 to P.A.K.) and the National Institutes of Health (RO1 11150917 to S.B.C.) as well as charitable contributions from the Lipp Family Foundation and Mr. and Mrs. Edward T. Vitone, Jr. M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.256214.114.
References
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17: 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. 2006. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet 2: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. 2007. The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438. [DOI] [PubMed] [Google Scholar]
- Doles J, Oliver TG, Cameron ER, Hsu G, Jacks T, Walker GC, Hemann MT. 2010. Suppression of Rev3, the catalytic subunit of Polζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci 107: 20786–20791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. 2008. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. 2007. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature 449: 1073–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609. [DOI] [PubMed] [Google Scholar]
- Kim H, Dejsuphong D, Adelmant G, Ceccaldi R, Yang K, Marto JA, D’Andrea AD. 2014. Transcriptional repressor ZBTB1 promotes chromatin remodeling and translesion DNA synthesis. Mol Cell 54: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, et al. 2010. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 190: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et al. 2012. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet 44: 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. 2007. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 6: 891–899. [DOI] [PubMed] [Google Scholar]
- Lord CJ, Tutt AN, Ashworth A. 2015. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med 66: 455–470. [DOI] [PubMed] [Google Scholar]
- Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W, Karlan BY, Taniguchi T, Swisher EM. 2011. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol 29: 3008–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy A, Hendrich B. 2013. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem Soc Trans 41: 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M-R, Hsieh H-J, Dai H, Hung W-C, Li K, Peng G, Lin S-Y. 2012. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J Biol Chem 287: 6764–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J, Hagman J. 2009. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics 4: 532–536. [DOI] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, et al. 2008. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451: 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Jacquemont C, Chandramohan KV, Couch FJ, Langdon SP, Wurz K, Higgins J, Villegas E, Taniguchi T. 2009. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res 69: 6381–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaud N, Barbera MA, Egashira A, Lam I, Christ N, Schlacher K, Xia B, Jasin M. 2011. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet 7: e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, Wade PA. 2011. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol Biol Cell 22: 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. 2010. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 190: 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley FKT, Moore S, Goodarzi AA. 2013. CHD chromatin remodelling enzymes and the DNA damage response. Mutat Res 750: 31–44. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang S-Y, Wang S, Lu J, Wu W, Weng L, Chen D, Zhang Y, Lu Z, Yang J, et al. 2009. REV3L confers chemoresistance to cisplatin in human gliomas: the potential of its RNAi for synergistic therapy. Neuro-oncol 11: 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Doles J, Hemann MT, Walker GC. 2010. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci 107: 20792–20797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Choi M, Overton JD, Bellone S, Roque DM, Cocco E, Guzzo F, English DP, Varughese J, Gasparrini S, et al. 2013. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci 110: 2916–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.