The transcriptional activator MyoD serves as a master controller of myogenesis. Singh et al. identify KAP1/TRIM28 as a key regulator of MyoD function. In myoblasts, KAP1 is present with MyoD and Mef2 at many muscle genes, where it acts as a scaffold to recruit not only coactivators such as p300 and LSD1 but also corepressors such as G9a and HDAC1, with promoter silencing as the net outcome. Upon differentiation, MSK1-mediated phosphorylation of KAP1 releases the corepressors from the scaffold, unleashing transcriptional activation by MyoD/Mef2 and their positive cofactors.
Keywords: KAP1, myogenesis, epigenetics, MyoD, MSK1 phosphorylation, G9a
Abstract
The transcriptional activator MyoD serves as a master controller of myogenesis. Often in partnership with Mef2 (myocyte enhancer factor 2), MyoD binds to the promoters of hundreds of muscle genes in proliferating myoblasts yet activates these targets only upon receiving cues that launch differentiation. What regulates this off/on switch of MyoD function has been incompletely understood, although it is known to reflect the action of chromatin modifiers. Here, we identify KAP1 (KRAB [Krüppel-like associated box]-associated protein 1)/TRIM28 (tripartite motif protein 28) as a key regulator of MyoD function. In myoblasts, KAP1 is present with MyoD and Mef2 at many muscle genes, where it acts as a scaffold to recruit not only coactivators such as p300 and LSD1 but also corepressors such as G9a and HDAC1 (histone deacetylase 1), with promoter silencing as the net outcome. Upon differentiation, MSK1-mediated phosphorylation of KAP1 releases the corepressors from the scaffold, unleashing transcriptional activation by MyoD/Mef2 and their positive cofactors. Thus, our results reveal KAP1 as a previously unappreciated interpreter of cell signaling, which modulates the ability of MyoD to drive myogenesis.
The discovery that fibroblasts could be induced to undergo myogenic differentiation by the forced expression of the DNA-binding basic helix–loop–helix (bHLH) MyoD protein was the first experimental evidence of genetically programmed transdifferentiation (Davis et al. 1987). MyoD was subsequently demonstrated to act as a master regulator of skeletal myogenesis driving myoblasts into a gene expression cascade that leads to their differentiation into multinucleated myotubes (Tapscott 2005; Aziz et al. 2010). It also became apparent that MyoD orchestrates this program by cooperating with a number of other modulators of gene expression, be they transcription factors or chromatin modifiers. How these multiple interactions are regulated and how MyoD-dependent gene targets are defined are still incompletely understood. In muscle precursor cells, MyoD indeed binds as a heterodimer with ubiquitously expressed E proteins to thousands of genomic sites containing a so-called E-box sequence (CAGSTG) (Cao et al. 2010; Soleimani et al. 2012). However, only a fraction of the genes situated nearby are activated during myotube differentiation (Cao et al. 2010). Interestingly, this subset of MyoD-recruiting genes is also enriched in Mef2 (myocyte enhancer factor 2) (Cao et al. 2010), and, more generally, MyoD- and Mef2-binding elements are overrepresented in cis-regulatory modules (CRMs) active during muscle differentiation (Kwon et al. 2011). MyoD and Mef2 also cooperate during muscle regeneration, where MyoD levels first increase in satellite cells, contributing to the expansion of these progenitors (Zhang et al. 2010; Singh and Dilworth 2013) before initiating with Mef2 the myogenic gene expression program that leads to cell cycle exit and formation of functional multinucleated myotubes (Penn et al. 2004; Liu et al. 2012; Singh and Dilworth 2013). This is consistent with the recruitment by the two factors of a combination of coregulatory molecules necessary for activating the muscle gene expression program (Molkentin et al. 1995; Rampalli et al. 2007). Supporting this model, MyoD interacts directly with components of the basal transcriptional machinery (Heller and Bengal 1998; Deato et al. 2008) and with the BAF60c subunit of the SWI/SNF complex (Forcales et al. 2012), while Mef2D recruits the Ash2L/MLL2–methyltransferase complex, which mediates trimethylation of histone H3 Lys4 (H3K4me3). A transcriptionally favorable epigenetic state is thus created, a prerequisite for high-level expression of muscle-specific genes (Rampalli et al. 2007).
However, in proliferating myoblasts, both MyoD and Mef2 are already bound to the CRMs of muscle differentiation genes yet are not expressed (Cao et al. 2010). Some of this paradox may stem from the methylation and hence inactivation of MyoD (Ling et al. 2012) and Mef2 (Choi et al. 2014) by the histone methyltransferase G9a in these cells. MyoD and Mef2 are indeed able to interact with either activators, such as the histone acetyltransferase p300 (Sartorelli et al. 1997) and the histone demethylase LSD1 (Choi et al. 2010), or repressors, such as G9a (Ling et al. 2012; Choi et al. 2014). Nevertheless, how the differential recruitment of these cofactors is regulated and hence how MyoD and Mef2D switch from repressors of muscle differentiation genes in proliferating myoblasts to activators of their expression in myotubes remain enigmatic.
Here, we identify KAP1 (KRAB [Krüppel-like associated box]-associated protein 1) as a crucial component of these events. KAP1, also known as TRIM28 (tripartite motif protein 28) or TIF1β (transcription intermediary factor 1β), is the universal cofactor of hundreds of KRAB domain-containing zinc finger proteins (KRAB-ZFPs), which are sequence-specific transcriptional repressors responsible for the early embryonic silencing of endogenous retroelements (EREs) (Wolf and Goff 2007; Rowe et al. 2010; Castro-Diaz et al. 2014; Turelli et al. 2014). Upon KRAB-ZFP-mediated tethering to DNA, KAP1 recruits the H3K9 methyltransferase SETDB1, the histone deacetylase (HDAC)-containing NuRD complex, heterochromatin protein 1 (HP1), and DNA methyltransferases, which results in heterochromatin formation and silencing of the targeted loci (Friedman et al. 1996; Schultz et al. 2001, 2002; Ivanov et al. 2007; Quenneville et al. 2012; Rowe et al. 2013a). KAP1 is also involved in DNA repair, where its phosphorylation at either one of two serine residues (473 and 824) transiently alters its heterochromatin-inducing activity (White et al. 2006, 2012; Iyengar and Farnham 2011). In the present study, we unveil KAP1 as an essential cofactor of MyoD-induced gene expression and MSK1-mediated KAP1 phosphorylation as a key regulator of this process.
Results
MyoD and Mef2D interact with KAP1 in myoblasts and myotubes
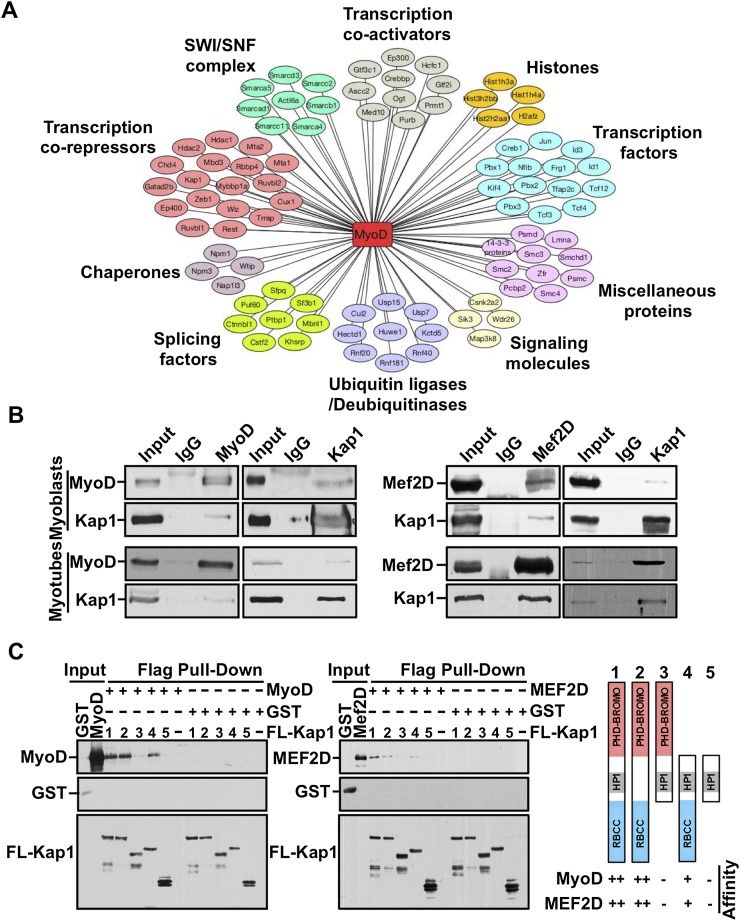
In proliferating myoblasts, the binding of MyoD to the promoter of muscle genes is not sufficient to induce their expression (Cao et al. 2010). To identify factors that might repress MyoD function in this setting, we performed proteomic analysis of MyoD-interacting proteins (Fig. 1A; Supplemental Fig. S1A). Mass spectrometry confirmed many known MyoD-interacting proteins (Supplemental Table S1), including its E-protein dimerization partners (TCF3, TCF4, and TCF12) (Lassar et al. 1991) and the transcriptional coactivator p300 (Sartorelli et al. 1997). Among several novel MyoD interactors detected through this analysis, our attention was attracted to KAP1 because of its known ability to recruit chromatin modifiers.
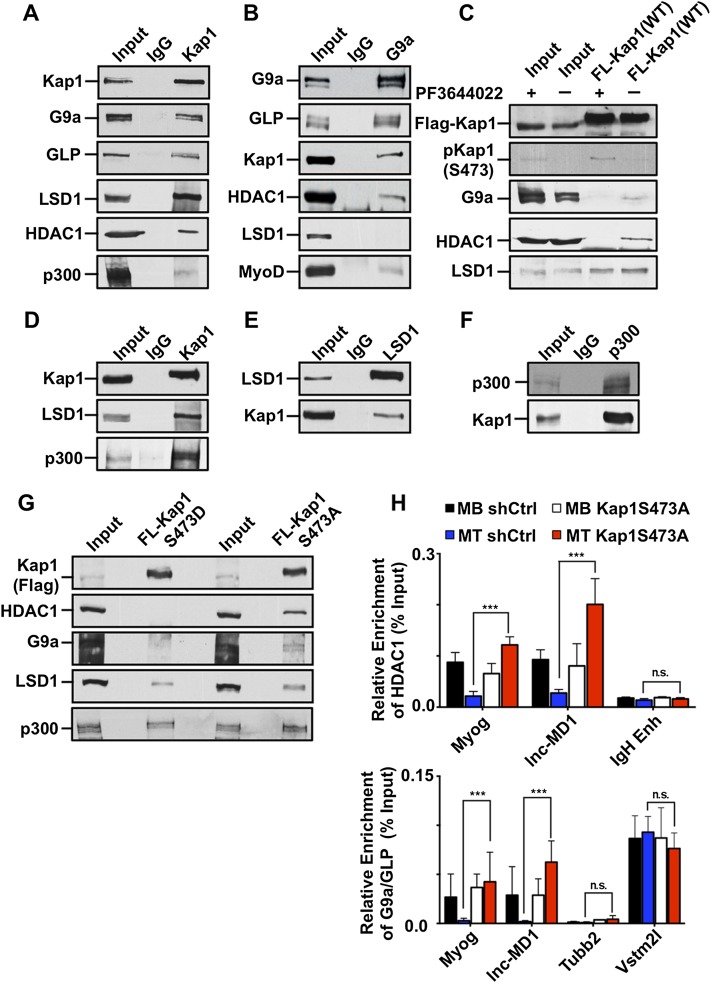
Figure 1.
KAP1 interacts with MyoD and Mef2D in muscle cells. (A) Nuclear extracts (NEs) prepared from C2 myoblasts expressing a doxycycline-inducible Flag-MyoD were subjected to immunoprecipitation using an anti-Flag antibody. Coimmunoprecipitated proteins were resolved on SDS-PAGE, visualized by silver staining, and identified by mass spectrometry. The interaction network was prepared using Cytoscape version 3.1.0 (see also Supplemental Fig. S1A). (B) Immunoprecipitations were performed using NEs prepared with either proliferating myoblasts or differentiating myotubes using antibodies against MyoD, Mef2D, or KAP1. Immunoprecipitated proteins were analyzed by Western blot using the indicated antibodies. (C) Exogenous Flag-KAP1 (full-length or its various domain deletion mutants) was purified from SF9 cell extracts using anti-Flag magnetic beads. Purified KAP1 proteins were left bound to the Flag beads and further incubated with purified MyoD and Mef2D for 3 h at room temperature. After extensive washing, Flag-KAP1-associated proteins were eluted and analyzed by Western blot using the indicated antibodies (see also Supplemental Fig. S1B).
Reciprocal coimmunoprecipitation experiments confirmed that MyoD and KAP1 form a complex in proliferating C2 myoblasts and, interestingly, also detected an association between these two proteins in differentiating myotubes (Fig. 1B). In vitro pull-down assays using purified MyoD and recombinant KAP1 demonstrated that their interaction is direct and requires the RBCC (RING B-box coiled-coil) domain of KAP1 (Fig. 1C), while immunoprecipitation of wild-type or deleted forms of HA-tagged MyoD indicated that it is dependent on the bHLH domain of this protein (Supplemental Fig. S1B). The ability to interact with KAP1 is a characteristic that is shared with the related muscle bHLH transcription factor Myf5 (Supplemental Fig. S2A). In addition, we found that KAP1 could also be immunoprecipitated with Mef2D in both myoblasts and myotubes (Fig. 1B), and in vitro pull-down experiments demonstrated that interaction between these two factors is direct and also mediated through the RBCC domain of KAP1 (Fig. 1C).
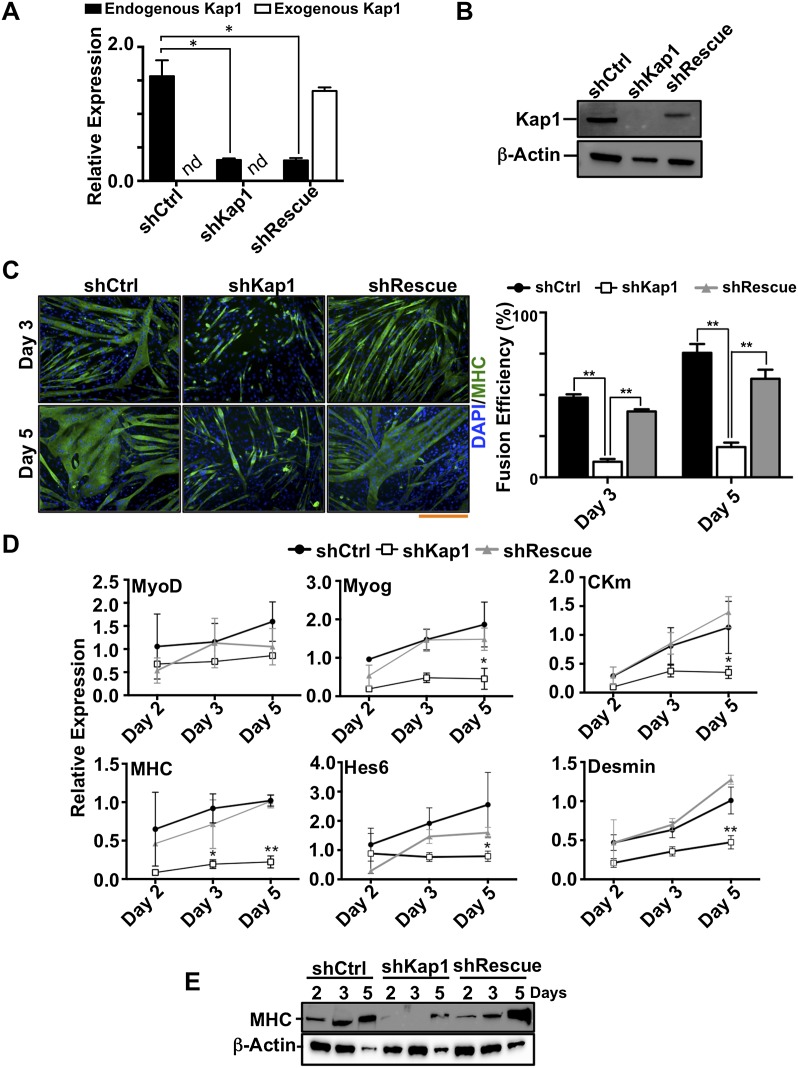
KAP1 is necessary for myoblast differentiation
To investigate the functional significance of the association of MyoD and Mef2D with KAP1, we depleted this factor in myoblasts by RNAi using lentiviral vectors expressing shRNAs against Kap1 with (shRescue) or without (shKap1) an shRNA-resistant codon-optimized Kap1 RNA for control complementation (Fig. 2A,B). KAP1 depletion did not result in detectable changes in the cell cycle progression of myoblasts (Supplemental Fig. S2B; data not shown) but blocked their differentiation into multinucleated myotubes (Fig. 2C). This correlated with decreased expression of the muscle-specific genes MyoD, Myogenin (Myog), myosin heavy chain (MHC), muscle creatine kinase (CKm), Hairy-Enhancer of Split homolog 6 (Hes6), and Desmin (Fig. 2D,E). Importantly, this differentiation block was directly attributable to the absence of KAP1, since the myogenic program could be restored by adding back this protein (Fig. 2C–E). Thus, KAP1 is required for the expression of key MyoD and Mef2 target genes such as Myog, a factor that mediates the cell cycle exit and terminal differentiation of myoblasts (Liu et al. 2012; Singh and Dilworth 2013).
Figure 2.
KAP1 is essential for myoblast differentiation. (A,B) KAP1 knockdown and recomplementation efficiency were assessed by RT-qPCR (A) and Western blot (B) in C2 myoblasts transduced with lentiviral vectors expressing either a control shRNA (shCtrl), a Kap1 targeting shRNA (shKAP1), or a Kap1 targeting shRNA together with a shRNA-resistant Kap1 allele (shRescue). In (A), values were calculated as the mean relative expression after normalization to three housekeeping genes ± SEM. n = 3; (*) P < 0.01; (nd) not determined. (C) Representative immunofluorescence images and myogenic fusion efficiency indicating the percentage of nuclei present within multinucleated cells, based on counting 10 unbiased fields per time point. Values were calculated as mean ± SEM. (**) P < 0.01. Bar, 200 μm. (D) RNA was isolated from infected C2 myoblasts that had been induced to differentiate. RT-qPCR analysis was performed using primers directed at specific genes, as indicated (see Supplemental Table S3 for primer sequences). Values were calculated as the mean relative expression after normalization to three housekeeping genes ± SEM. n = 3; (*) P < 0.05; (**) P < 0.01 shCtrl versus shKAP1. (E) Western blot analysis for MHC expression in shCtrl, shKAP1, and shRescue C2C12 myoblasts at different time points of differentiation. β-Actin was used as an internal loading control. One representative experiment out of three is presented.
KAP1 coregulates muscle gene expression
To explore fully the muscle gene expression program regulated by KAP1, we performed high-throughput sequencing of RNA (RNA-seq) isolated from myoblasts and differentiating myotubes in which KAP1 had been depleted by lentivector-mediated RNAi. Taking twofold as a cutoff, KAP1 knockdown altered the expression of 220 genes in proliferating myoblasts and 1363 genes in their differentiated counterparts, of which only a minority (71 and 418, respectively) was up-regulated (Supplemental Table S2). Gene set enrichment analysis (Subramanian et al. 2005) of the corresponding subset of deregulated genes was consistent with a role for KAP1 in regulating the activation of genes involved in muscle function (Supplemental Table S2).
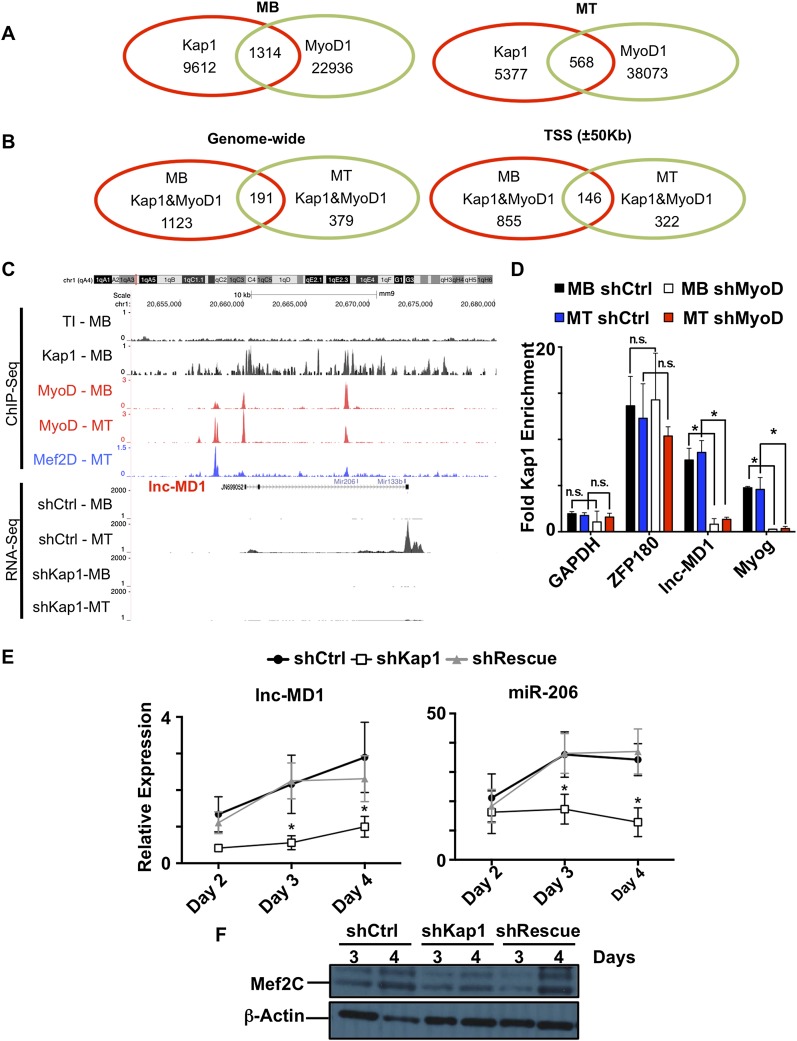
To identify the direct targets of KAP1 regulation among these genes, we proceeded to chromatin immunoprecipitation (ChIP) coupled to high-throughput sequencing (ChIP-seq). This analysis identified 19,549 and 6456 KAP1-enriched loci on the genome of proliferating myoblasts and differentiating myotubes, respectively. However, these KAP1 peaks were only rarely associated with the H3K9me3 mark (Supplemental Fig. S3A) typically found at KAP1-induced heterochromatin regions (Schultz et al. 2002). This is consistent with previous observations indicating that H3K9me3 does not significantly contribute to the regulation of muscle gene expression during myogenesis (Pauler et al. 2009; Asp et al. 2011). It further suggested that KAP1 might act in a nonprototypic fashion to regulate myogenesis. To probe this issue and ask more specifically how MyoD and KAP1 might coregulate muscle gene expression, we overlapped our KAP1 ChIP-seq data with those previously obtained for MyoD (Cao et al. 2010). This revealed that the two proteins colocalize at 1314 and 568 genomic loci in proliferating and differentiating myoblasts, respectively (Fig. 3A), which represent ∼10% of KAP1 genomic binding sites in each condition. Furthermore, a census of loci enriched for both KAP1 and MyoD in myotubes revealed that about a third of them already carried these two proteins in myoblasts (Fig. 3B).
Figure 3.
KAP1, MyoD, and Mef2D genomic corecruitment controls muscle genes. (A) Venn diagram showing the genome-wide overlap between the KAP1 and MyoD peaks identified by ChIP-seq in proliferating (MB) or differentiating (MT) C2 myoblasts. (B) Venn diagram illustrating the degree to which KAP1 and MyoD overlapping peaks are shared between myoblasts and myotubes either genome-wide or when limiting the analysis to gene regulatory regions (±50 kb TSS). (C) Sequencing reads were mapped across the lnc-MD1 locus for ChIP experiments performed using KAP1 (black), MyoD (red), and Mef2Dα2 (blue) antibodies and for RNA-seq experiments performed in shCtrl and shKAP1 myoblasts (MB) or myotubes (MT) (see also Supplemental Fig. S5B). (D) Knockdown of MyoD (shMyoD) or control (shCtrl) was induced in C2 myoblasts before subjecting myoblasts (MB) or differentiating myotubes (MT) to ChIP-qPCR with KAP1-specific antibodies. The well-characterized KAP1 target ZFP180 was used as a positive control, while GAPDH was used as a negative control. Values are represented as mean ± SEM and were calculated as relative enrichment over the Tubb2 promoter as a negative control, where P < 0.05 (*). (n.s.) Not significant. (E) RT-qPCR analysis for lnc-MD1 and miR-206 at various time points of differentiation in shCtrl, shKAP1, and shRescue myoblasts. Values were calculated as relative expression ± SEM and normalized over three housekeeping genes for lnc-MD1 and three housekeeping snoRNAs for miR-206 expression. n = 3; (*) P < 0.05 shCtrl versus shKAP1. (F) Western blot for the lnc-MD1-regulated transcriptional coactivator MEF2C at different time points of differentiation in shCtrl, shKap1, and shRescue myoblasts. β-Actin was used as an internal loading control. One representative experiment out of three is shown (see also Supplemental Fig. S6A).
MyoD characteristically binds to tissue-specific enhancers in muscle, whether at the myoblast or myotube stage (Blum et al. 2012). We thus asked whether KAP1 also associated with these elements. We examined sites of overlap between KAP1 and MyoD within 50 kb from a transcription start site (TSS) as a cutoff, as this has been reported as the median distance between an enhancer and its target promoter (Kim et al. 2010; Rada-Iglesias et al. 2011; Blum et al. 2012). We found that the majority of loci cobound by MyoD and KAP1 was within this interval in both myoblasts and myotubes (Fig. 3B) and that a significant proportion of these was at the gene promoters themselves, defined as a region ±2 kb from the TSS (Supplemental Fig. S5A). Consistent with MyoD and KAP1 associating at transcriptional enhancers, regions cobound by MyoD and KAP1 were significantly enriched for the enhancer-associated histone modifications H3K27ac and H3K4me1 but not the heterochromatin mark H3K9me3, which marks KAP1-induced heterochromatin (Supplemental Fig. S4). Taken together, these results strongly suggest that KAP1 and MyoD cooperate to control muscle gene expression both before and after myoblast differentiation.
To define further the importance of KAP1 in the regulation of muscle genes, we also examined the binding overlap between KAP1 and MyoD with published Mef2Dα2 ChIP-seq data of differentiating myotubes (Sebastian et al. 2013). This revealed that 134 of the 146 TSS-close (MyoD+KAP1)-bearing sites were also adorned with Mef2Dα2 (Supplemental Fig. S5B). Among genes potentially subjected to KAP1/MyoD/Mef2Dα2-mediated regulation in both myoblasts and myotubes, we identified the muscle-specific long noncoding RNA (lncRNA) lnc-MD1 (Fig. 3C; Cesana et al. 2011; Legnini et al. 2014), confirming the ChIP-seq data by quantitative PCR (qPCR) on independent chromatin immunoprecipitates (Fig. 3D). Focusing our analysis on this lncRNA and Myog as two key regulators of myogenic differentiation targeted by the MyoD/Mef2D/KAP1 complex, we could determine that the presence of KAP1 at the promoters of these genes was MyoD-dependent, since it was completely abrogated by MyoD depletion in both proliferating and differentiating myoblasts (Fig. 3D). In addition, lnc-MD1 levels were decreased upon Kap1 knockdown (Fig. 3E), as previously observed for Myog (Fig. 2D). Correspondingly, KAP1 depletion reduced the levels of lnc-MD1 target gene products, including miR-206, during myogenic differentiation (Fig. 3E,F; Supplemental Fig. S6A).
A KAP1 phosphorylation switch controls muscle differentiation
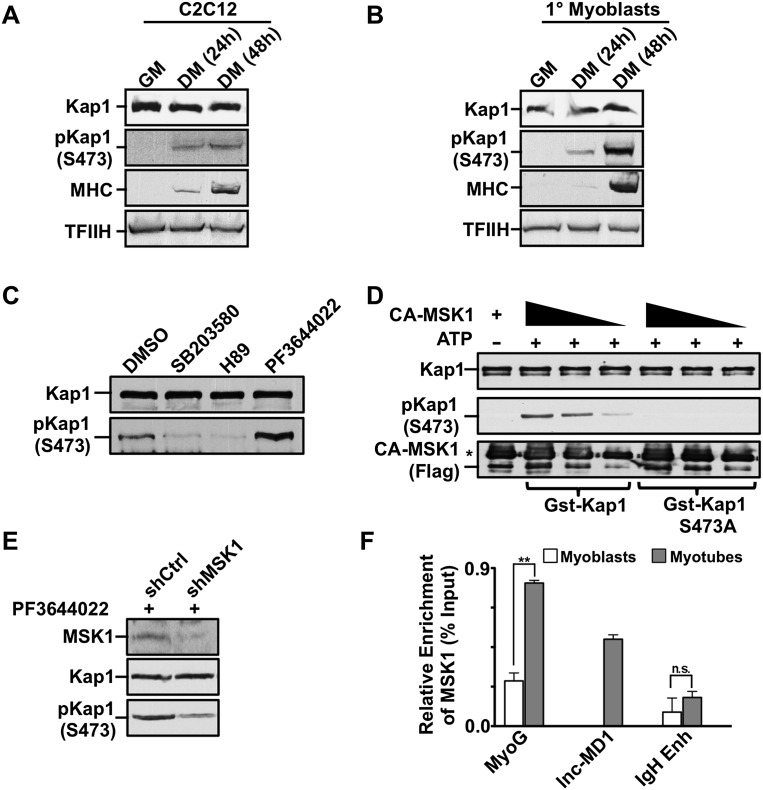
These results established KAP1 as a coactivator of MyoD/Mef2D-dependent gene expression, reminiscent of its previously reported stimulation of Nur77 transcriptional activating potential (Rambaud et al. 2009). However, the association of KAP1 with MyoD and Mef2D in both myoblasts, where their target genes are repressed, and myotubes, where they are induced, suggested that KAP1 action must be regulated. KAP1 was previously demonstrated to undergo post-translational modifications and have its corepressor activity reduced notably by phosphorylation on Ser473 (Chang et al. 2008; King 2013). Remarkably, while KAP1 protein levels remained relatively constant when myoblasts were induced to differentiate into myocytes, its S473pKAP1 isoform became detectable during this process (Fig. 4A). The transient nature of the KAP1 phosphorylation event and its occurrence at a limited number of genomic loci exclusively in differentiating myoblasts made it difficult to document strong phosphorylation of KAP1 in extracts prepared from regenerating adult mouse muscle (Supplemental Fig. S5C–E). However, we observed a dramatic augmentation of S473pKAP1 levels upon differentiation of primary mouse myoblasts isolated from the tibialis anterior muscle (Fig. 4B). We next set out to identify the kinase responsible for this process. Previous work suggested that S473KAP1 phosphorylation is mediated by the MK2 kinase in endothelial cells (King 2013). However, treatment of myoblasts with the MK2 inhibitor PF3644022 surprisingly led to increased levels of S473pKAP1 (Fig. 4C) and higher efficiency of myotube formation (Supplemental Fig. S7A). Instead, pharmacological inhibitors of two other kinases previously found to be active in muscle cells—SB20350 (active on p38 MAPK) and H89 (targeting MSK1)—prevented the accumulation of S473pKAP1 (Fig. 4C). As the S473 residue does not lie within a consensus p38 MAPK site, we probed further the potential role of MSK1 in S473KAP1 phosphorylation. Using purified GST-KAP1 protein and a constitutively active recombinant MSK1 protein, we found that the wild-type KAP1 protein, but not an S473 mutant (S473AKAP1), was efficiently phosphorylated by MSK1 in vitro (Fig. 4D). Furthermore, shRNA-mediated knockdown of MSK1 was sufficient to reduce PF3644022-induced S473KAP1 phosphorylation in proliferating myoblasts (Fig. 4E). Finally, we could document the recruitment of MSK1 at both the lnc-MD1 and Myog promoters in differentiating myotubes (Fig. 4F).
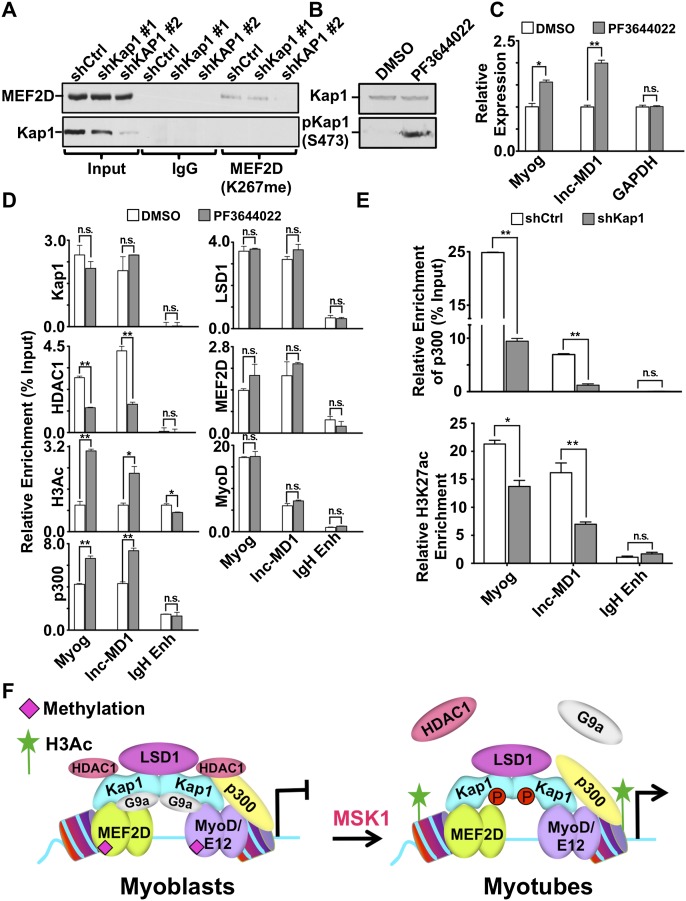
Figure 4.
MSK1 phosphorylates KAP1 during muscle differentiation. (A,B) Cell extracts were prepared from C2 myoblasts (A) or primary mouse myoblasts (B) at various stages of differentiation. Western blot was performed using antibodies as indicated. (C) Differentiating C2 myoblasts were treated with the pharmacological inhibitors SB203580 (p38 MAPK), H89 (MSK1), or PF3644022 (MK2) for 2 h prior to harvesting. Western blot analysis was performed for both KAP1 and S473pKAP1 (see also Supplemental Fig. S7A). (D) Phosphorylation of KAP1 by MSK1 was evaluated in vitro by incubating active Flag-MSK1 (CA-MSK1) with purified GST-KAP1 wild-type or S473A mutant proteins in the presence of ATP as outlined in the Supplemental Material. Kinase reactions were analyzed by Western blot using the indicated antibodies. An asterisk indicates a heavy chain of IgG. (E) C2 myoblasts were infected with lentivirus expressing either shMSK1 or shCtrl and incubated with the MK2 inhibitor PF3644022 for 2 h to induce KAP1 phosphorylation. Cell extracts were analyzed by Western blot using the indicated antibodies. (F) Chromatin was prepared from either proliferating myoblasts or differentiating myotubes and was subjected to ChIP-qPCR analysis using antibodies directed against MSK1. Immunoprecipitated DNA was quantitated relative to the input chromatin and is expressed as the mean ± SEM. (**) P < 0.01; (n.s.) not significant.
Consistent with an important role for S473KAP1 phosphorylation in MyoD-dependent myoblast differentiation, KAP1-depleted myoblasts complemented with the nonphosphorylatable S473AKAP1 mutant could neither morph into myotubes (Fig. 5A–C) nor efficiently induce the expression of MyoD/KAP1 target genes (Fig. 5D).
Figure 5.
KAP1 S473 phosphorylation is required for myoblast differentiation. (A–D) C2 myoblasts were infected with lentivirus expressing shCtrl, shKAP1, or KAP1 targeted shRNA together with an shRNA-resistant allele of either the wild type or a nonphosphorylatable S473 KAP1 mutant (S473AKAP1). (A) Representative immunofluorescence images. (B) Myogenic fusion efficiency showing the percentage of nuclei present within multinucleated cells taken from 10 unbiased fields per time point. Values were calculated as mean ± SEM. (**) P < 0.0001. Bar, 200 μm. (C) The efficiency of KAP1 knockdown and rescue with shRNA-resistant cDNAs was evaluated by Western blot analysis. (D) RNA was isolated from infected C2 myoblasts that had been induced to differentiate, and RT-qPCR analysis was performed using primers directed at specific genes as indicated. Values were calculated as the mean relative expression after normalization to three housekeeping genes ± SEM. n = 3; (*) P < 0.01 S473AKAP1 versus shCtrl.
KAP1 phosphorylation triggers the release of repressor chromatin modifiers
Having established KAP1 S473 phosphorylation as a molecular switch promoting myoblast differentiation, we sought to understand the mechanism of this phenomenon. KAP1 typically acts as a scaffold for the recruitment of chromatin-modifying enzymes and other transcription modulators (Friedman et al. 1996; Underhill et al. 2000; Schultz et al. 2001, 2002; Ivanov et al. 2007; Quenneville et al. 2012). Transcriptional repressors known to interact with both KAP1 and MyoD include HDAC1 and the protein methyltransferase G9a (Underhill et al. 2000; Schultz et al. 2001; Mal and Harter 2003; Fritsch et al. 2010; Ling et al. 2012). Accordingly, we could coimmunoprecipitate KAP1 with HDAC1 and G9a and its dimerization partner, GLP, in proliferating myoblasts (Fig. 6A,B). We also detected in these cells an interaction of KAP1 with the histone demethylase LSD1 and the histone acetyltransferases p300; that is, two enzymes with activities antagonistic to those of G9a/GLP and HDAC1, respectively (Fig. 6A). We next examined how KAP1 S473 phosphorylation influences these interactions. For this, we treated proliferating myoblasts with PF3644022, since we had found this MK2 inhibitor to induce KAP1 phosphorylation (Fig. 4C). This resulted in abrogating KAP1 association with G9a and HDAC1 but not with LSD1 (Fig. 6C). Consistent with this observation, we found that KAP1 interacts strongly with LSD1 and p300 in differentiating myotubes, where it exhibits significant rates of S473 phosphorylation (Fig. 6D–F). To strengthen the link between KAP1’s phosphorylation status and its association with these chromatin modifiers, we examined the binding partners of KAP1 point mutants that were either phosphomimetic (S473DKAP1) or phosphorylation-resistant (S473AKAP1) at position 473. We found that both G9a and HDAC1 interacted with S473AKAP1 but not with S473DKAP1 (Fig. 6G). In contrast, the transcriptional coactivators (LSD1 and p300) and transcription factors (MyoD and MEF2D) maintained their association with both mutant forms of the Kap1 (Fig. 6G; Supplemental Fig. S7B,C).
Figure 6.
KAP1 phosphorylation disrupts its association with corepressor complexes. (A) KAP1 and its associated proteins were immunoprecipitated from NEs prepared from proliferating C2 myoblasts and analyzed by Western blot. (B) G9a and its associated protein were identified in proliferating myoblasts as outlined in A. (C) Flag-KAP1(WT) was expressed in proliferating C2 myoblasts and subjected to treatment with 5 μM PF3644022 for 16 h before harvesting the cells. Flag-KAP1 was immunoprecipitated from cell extracts using an anti-Flag antibody and analyzed by Western blot using the indicated antibodies. (D) KAP1 and its associated proteins were immunoprecipitated from NEs prepared from differentiating C2 myotubes (48 h) and analyzed by Western blot. (E) LSD1 was immunoprecipitated from differentiating myotubes and examined for its ability to associate with KAP1 by Western blot. (F) Immunoprecipitation of p300 was performed in differentiating C2 myoblasts, and the interaction with KAP1 was determined by Western blot using antibodies as indicated. (G) C2 myoblasts expressing either the phosphomimic protein Flag-S473DKAP1 or the nonphosphorylatable mutant Flag-S473AKAP1 were induced to differentiate for 48 h. KAP1 proteins were immunopurified from NEs using an anti-Flag antibody, and the resulting eluates were analyzed by Western blot using the indicated antibodies. (H) C2 myoblasts expressing either the wild-type KAP1 or the nonphosphorylatable S473AKAP1 mutant were induced to differentiate for 48 h. Chromatin was immunoprecipitated using antibodies recognizing HDAC1 (top) and G9a/GLP (bottom) and quantitated by qPCR using primer sets specific for the indicated genomic regions. Values are represented as the mean ± SEM as a percentage of the input chromatin. n = 3; (**) P < 0.01; (n.s.) not significant.
We then performed ChIP-qPCR analyses in cells depleted for wild-type KAP1 and complemented with S473AKAP1 (Fig. 6H). The results revealed that, in the presence of this phosphorylation-resistant KAP1 mutant, both HDAC1 and G9a remained associated with the previously mapped MyoD/Mef2D-bound Myog and lnc-MD1 loci when myoblasts were placed in differentiation medium, consistent with a model in which it is KAP1 phosphorylation that normally triggers the release of these transcriptional corepressors from these sites.
In growing myoblasts, both MyoD and Mef2D are functionally inactivated by G9a-mediated methylation (Ling et al. 2012; Choi et al. 2014). Upon differentiation, these factors are demethylated, which allows them to activate muscle gene expression. For Mef2D, LSD1 was identified as the demethylase responsible for removing methyl residues deposited by G9a at Lys267 (Choi et al. 2014). Therefore, we examined the influence of KAP1 on this process (Fig. 7A). By comparing cells with different levels of KAP1 knockdown, we first observed that levels of methylated Mef2D (K267meMef2D) were proportional to residual KAP1 levels. We then measured the recruitment of coregulatory molecules to muscle genes in proliferating myoblasts exposed or not exposed to the KAP1 S473 phosphorylation-inducing compound PF3644022 (Fig. 7B). We found that PF3644022 treatment was sufficient to induce expression of both the Myog and lnc-MD1 genes in these cells (Fig. 7C). This transcriptional activation was not the result of increased recruitment of KAP1, MyoD, Mef2D, or LSD1 at either locus. Instead, it correlated with a drastic decrease in HDAC1 enrichment, mirrored by a significant increase in the presence of p300 and acetylated histone H3 (Fig. 7D).
Figure 7.
KAP1 phosphorylation induces a change in coregulatory molecules at muscle genes. (A) Cell extracts prepared from C2 myoblasts expressing either control (shCtrl) or shRNAs against KAP1 (shKAP1#1 or shKAP1#2) were subjected to immunoprecipitation using anti-methyl (K267) Mef2D antibody. Immunopurified proteins were analyzed by Western blot using the indicated antibodies. (B) C2 myoblasts were incubated with either 5 µM PF3644022 or DMSO vehicle for 16 h, and cell extracts were analyzed by Western blot using KAP1 or S473pKAP1 antibodies. (C) Gene expression was examined in C2 myoblasts after incubation with 5 µM PF3644022 (or DMSO vehicle) for 16 h. RT-qPCR was performed using primers for specific genes as indicated. Values are expressed as the mean ± SEM relative to the internal control GAPDH, where the expression was normalized to 1. (*) P < 0.05; (**) P < 0.01. (D) ChIP assays were performed for KAP1, HDAC1, H3Ac, p300, LSD1, Mef2D, or MyoD using chromatin prepared from C2 myoblasts that had been treated with 5 µM PF3644022 (or DMSO vehicle) for 16 h. Immunopurified DNA was quantified by qPCR using probes that recognize the indicated genomic regions and are represented as the mean ± SEM as a percentage of the input chromatin. (**) P < 0.01; (n.s.) not significant. (E) Chromatin was prepared from differentiating C2 myotubes (48 h) that expressed shKAP1 or shCtrl. Chromatin was then immunoprecipitated using antibodies recognizing p300 or H3K27ac and quantitated by qPCR using primer sets specific for the indicated genomic regions. Values are represented as mean ± SEM and were calculated as the percentage of input chromatin (p300) or relative enrichment over the ZFP180 3′ untranslated region (UTR) (H3K27ac). (*) P < 0.05; (**) P < 0.01. (See also Supplemental Fig. S6B,C.) (F) Model for the bimodal role of KAP1 in regulating MyoD transcriptional activity at muscle genes. In proliferating myoblasts, MyoD and Mef2D cooperate to allow efficient recruitment of KAP1. Promoter-tethered KAP1 then serves as a platform for the assembly of coregulatory complexes that include both corepressors (G9a and HDAC1) and coactivators (LSD1 and p300). The assembly of this complex results in the hypermethylation of Mef2D and hypoacetylation of histones, which establishes a transcriptionally poised state at the promoter. Upon differentiation, up-regulation of p38 MAPK signaling leads to activation of the downstream kinase MSK1 that phosphorylates KAP1 at S473. Phosphorylation of KAP1 at S473 results in the dissociation of corepressors (HDAC1 and G9a) but not coactivators (p300 and LSD1), leading to the formation of an open chromatin state that is permissive to high-level expression of muscle genes.
Finally, we could determine that the knockdown of KAP1 led to significant decreases in the recruitment of both p300 and LSD1 at the lnc-MD1 and Myog promoters in differentiating myoblasts, with a commensurate drop in levels of H3K27ac, a mark deposited by p300, but no change in H3K4me3 (Fig. 7E; Supplemental Fig. S6B,C).
Taken together, these results support a model in which KAP1 serves as a scaffold for the docking at MyoD/Mef2D targeted muscle gene CRMs of a set of chromatin modifiers, the composition of which is regulated by KAP1 phosphorylation, which acts as a molecular switch activating these genes when myoblasts are induced to differentiate into myotubes.
Discussion
We set out to understand how MyoD can bind the CRMs of hundreds of muscle genes in proliferating myoblasts and yet activate its targets only once these cells are induced to differentiate into myotubes. Here we conclusively demonstrate that the scaffold protein KAP1 acts as a key determinant of MyoD function during muscle differentiation. Indeed, KAP1 associates with the DNA-bound MyoD and Mef2D within the regulatory region of muscle genes in proliferating myoblasts, where it acts as a scaffolding for the recruitment of a combination of activating (p300 and LSD1) and repressive (G9a and HDAC1) chromatin modifiers, with transcriptional silencing as the net outcome. In response to differentiation cues, KAP1 becomes phosphorylated by MSK1 on Ser473, G9a and HDAC1 are released, and the transcriptional activation potential of the MyoD/Mef2D complex is unleashed, resulting in the expression of muscle genes (Fig. 7F).
KAP1 itself is not known to bind DNA directly but is recruited to genomic loci, notably through its RBCC-mediated interaction with KRAB-ZFPs that bind specific DNA sequences. Transcriptional silencing ensues at KRAB-ZFP-bound loci via the recruitment of SETDB1, NuRD-associated HDACs, and HP1 (Friedman et al. 1996). Here, we demonstrate that, in muscle cells, the RBCC domain also mediates the docking of KAP1 at MyoD/Mef2D-enriched cis-regulatory regions, but, in this case, the scaffold protein brings in a distinct set of chromatin modifiers, including G9a, HDAC1, LSD1, and p300. The bases for this difference are unknown. KRAB-ZFPs recruit KAP1 as a trimer (Peng et al. 2000). Whether similar high-order KAP1 complexes are formed at MyoD/Mef2D-bearing loci remains to be determined, but it would certainly allow for the assembly of a large platform spanning multiple transcription modulators at CRMs, consistent with our observations that an extensive group of corepressors and coactivators is associated with KAP1 at muscle genes. The specific domains and residues of KAP1 that are engaged in recruiting the various sets of chromatin modifiers identified in this and previous studies also need to be defined. Nevertheless, it is noteworthy that phosphorylation of KAP1 at Ser824 blocks its ability to recruit SETDB1 (Ivanov et al. 2007; Iyengar and Farnham 2011), whereas its phosphorylation at Ser473 was previously reported instead to impair its interaction with HP1 (Chang et al. 2008). Here, we demonstrate that targeting of Ser473 by MSK1 kinase releases G9a and HDAC1 from KAP1/MyoD/Mef2D complexes, confirming that motifs different from those involved in recruiting SETDB1 are engaged in binding these repressors.
A constitutively active recombinant MSK1 derivative could phosphorylate KAP1 in vitro, yet inhibitors of both this kinase and p38 MAPK could block this process in tissue culture. Consistent with previous findings that p38 MAPK phosphorylates MSK1 to activate its kinase activity (Pierrat et al. 1998), our results suggest that MSK1-mediated phosphorylation of KAP1 takes place upon the well-characterized activation of p38 MAPK signaling that occurs during differentiation (Zetser et al. 1999). Of note, MSK1-mediated KAP1 phosphorylation most likely happens only at specific gene promoters, ensuring that the repressive activity of KAP1 is not globally inactivated. This scenario is corroborated by the finding that MSK1 is targeted to at least a subset of muscle genes during muscle differentiation (Fig. 4A; Stojic et al. 2011) and is reminiscent of the ATM-mediated phosphorylation of KAP1 on Ser824 specifically at sites of DNA damage (White et al. 2006, 2012).
Our results indicate that KAP1 is essential to the efficient assembly of corepressor and coactivator complexes at muscle gene promoters. However, we note that previous studies have identified direct interactions between MyoD and G9a (Ling et al. 2012) or between MyoD and p300 (Sartorelli et al. 1997). Taken together, these results suggest that the MyoD/Mef2/KAP1 complex bound at muscle CRMs likely mediates multiple independent interactions with a set of coregulatory molecules, likely to maximize the specificity and efficiency of this recruitment at highly selected genomic loci. In this context, it is interesting to note that the corepressor G9a has previously been demonstrated to reduce the transcriptional activity of both MyoD (Ling et al. 2012) and Mef2D (Choi et al. 2014) through methylation of specific lysine residues within these transcription factors. Conversely, corresponding lysine residues are acetylated upon differentiation where MyoD and Mef2D serve as transcriptional activators. In the case of Mef2D, the conversion from the methylated to the acetylated state is facilitated by LSD1-mediated demethylation of K267meMef2D, promoting transcriptional activation of muscle genes (Choi et al. 2014). Our data suggest a regulatory mechanism by which G9a mediates methylation of Mef2D and MyoD to reinforce the transcriptionally repressive state at muscle genes in proliferating myoblasts. Upon differentiation, displacement of G9a following KAP1 phosphorylation allows LSD1-mediated demethylation of MyoD and Mef2D, which in turn ensures the assembly of the full complement of transcription coactivators required for high-level gene expression. Thus, we propose that the use of KAP1-associated coregulatory molecules to modify the transcriptional outcome of MyoD and Mef2D at muscle CRMs represents a self-reinforcing loop that helps ensure that transcription is repressed in proliferating myoblasts but can be rapidly activated in response to differentiation cues.
In conclusion, our results identify the KAP1/MyoD/Mef2D axis as a dynamic transcriptional regulatory system in which corepressor and coactivator complexes are superimposed in a combinatorial and signal-dependent fashion to modulate the efficient initiation of the muscle gene expression program. It remains to be determined whether the bivalent nature of KAP1 as a key regulator of the activity of transcription factor-bound CRMs is specific to MyoD/Mef2D and myogenesis or is used by additional combinations of proteins engaged in controlling cell fate determination in other lineages.
Materials and methods
Myoblast cultures
Mouse C2C12 myoblasts (Yaffe and Saxel 1977) were maintained in DMEM supplemented with 10% FCS. For differentiation studies, near confluent cells were switched to low-serum DMEM medium (1% [v/v] horse serum, 10 μg mL−1 insulin, 10 μg mL−1 transferrin). For analysis of kinase activity in C2C12 cells, either proliferating or differentiating cells were treated with the kinase inhibitor SB203580 (10 μM), PF3644022 (5 μM) Ly294002 (20 μM), or H89 (10 μM) for 2 h prior to harvesting (unless otherwise noted).
For knockdown and complementation studies, proliferating C2C12 cells were transduced at a multiplicity of infection (MOI) of 20 with lentiviral particles targeting KAP1, MyoD, MSK1, or a control (Luciferase). At 48 h after transduction, infected cells were selected in growth medium containing 2 μg mL−1 puromycin for an additional 72 h. Infected cells were maintained to near confluency and allowed to differentiate for 48 h.
Isogenic cell lines expressing C-terminal Flag-tagged cDNA for MyoD, KAP1(WT), S473AKAP1, and S473DKAP1 were generated as described earlier (Sebastian et al. 2013). Briefly, the mouse cDNAs encoding MyoD-Flag and KAP1(WT)-Flag were cloned into the multiple cloning site of a pCDNA5/FRT/TO plasmid. The plasmid pCDNA5/FRT/TO-S473AKAP1-Flag and pCDNA5/FRT/TO-S473DKAP1-Flag mutants were generated with QuickChange site-directed mutagenesis kit (Stratagene). Individual plasmids were transfected into C2iFRT cells (Sebastian et al. 2013) and selected for hygromycin B resistance as outlined in the Flp-In system protocol. These cell lines were termed C2iFRT-MyoD, C2iFRT-KAP1(WT), C2iFRT-S473AKAP1, and C2iFRT-S473DKAP1.
RNA-seq
Total RNA was isolated from proliferating or differentiating C2C12 cells using the RNeasy minikit (Qiagen) under standard conditions. Library preparation and 100-base-pair (bp) single-end RNA-seq were performed using standard Illumina procedures for the HiSeq 2000 platform. The reads were mapped to the mm9 genome using TopHat’s sensitive mode (Kim et al. 2013), not allowing for multiple hits, the discovery of novel junctions, or indels. After mapping, we normalized the counts and performed a likelihood ratio test as implemented in the Bioconductor package DEseq2. We tested the null hypothesis that log2 fold changes for multiple batches were equal to zero. Genes with a fold change bigger than two and a fold change <0.5 were considered to be differentially expressed.
ChIP and ChIP-seq
Histone modifications were analyzed using native ChIP as previously described (Brand et al. 2008). For cross-linked ChIP assay, chromatin was prepared from 107 C2C12 cells (proliferating or differentiating) as previously described (Rowe et al. 2013b). Fragmented chromatin was immunoprecipitated using the specified antibodies or control IgG. Immunoprecipitated DNA was quantified by Qubit (Life Technologies) and subjected to qPCR or high-throughput sequencing analysis. For ChIP-seq experiments, libraries for sequencing were prepared from total input chromatin and immunoprecipitated DNA as previously described (Santoni de Sio et al. 2012) using ∼10 ng of chromatin. Libraries were subjected to 100-bp single-end read analysis on an Illumina Genome Analyzer IIx. Reads were mapped to the murine genome assembly mm9 using Bowtie (Langmead and Salzberg 2012), allowing a maximum of three mismatches. In cases of multiple alignments, the one with best mapping quality was reported. In cases of multiple alignments with the same mapping quality, one of them was reported at random. The peaks were called using either the MACS program (Zhang et al. 2008) or the SICER algorithm for histone modification marks (Zang et al. 2009) and were normalized to the total input chromatin signal using the default software parameters. ChIP-seq data were aligned with available data in proliferating myoblasts and differentiating myotubes for MyoD (http://www.genome.ucsc.edu/ENCODE, generated by the Wold group at California Institute of Technology); H3K27ac, H3K4me3, and H3K4me1 (generated by the Brian Dynlacht group); and published Mef2Dα2 data sets (Sebastian et al. 2013). To assign co-occupancy, we ran the .BED files with the peaks coordinates generated by MACS or SICER using the intersectBed function of Bedtools software version 2.22.0 (Quinlan and Hall 2010). We asked for an overlap of at least one base to consider two peaks as overlapping. All ChIP-seq data reported in this study have been deposited in the Gene Expression Omnibus (GEO) database (GSE62664).
Statistical analyses
The statistical significance of the differences between relative expression was assessed with two-way ANOVA. The statistical significance of the differences between percentage values was assessed with a Kruskal-Wallis ranks test.
Supplementary Material
Acknowledgments
We thank Lawrence Puente for mass spectrometry analysis, Marjorie Brand for insightful discussions and critical reading of the manuscript, and Slimane Ait-Si-Ali, Christophe Rachez, and Bruce Conklin for providing plasmids. This work was funded by grants from the Canadian Institutes of Health Research (MOP-77778) to F.J.D. and from the Swiss National Science Foundation and the European Research Council to D.T. F.J.D. holds a Canada Research Chairs in the Epigenetic Regulation of Transcription. M.C., F.J.D., K.S., and D.T. conceived the study, interpreted the data, and wrote the manuscript. Other contributions were as follows: M.C. performed RNA-seq, ChIP-seq, and ChIP-qPCR analyses; knockdown/rescue experiments; and some bioinformatics and protein interactions analyses. K.S. performed proteomic analyses, identification of MSK1 as a mediator of KAP1 phosphorylation, and some protein interaction studies. S.S. performed some ChIP-qPCR experiments; E.P. performed bioinformatics analyses; S.M.J. performed in vitro protein interaction studies; G.S. helped with the characterization of KAP1 phosphorylation; H.F. characterized KAP1 in the regenerating mouse muscle; and J.C. and H.-D.Y. provided the methylated Mef2D antibody.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.254532.114.
References
- Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, Bowman C, Kluger Y, Dynlacht BD. 2011. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci 108: E149–E158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A, Liu QC, Dilworth FJ. 2010. Regulating a master regulator: establishing tissue-specific gene expression in skeletal muscle. Epigenetics 5: 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. 2012. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev 15: 2763–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. 2008. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation (N-ChIP) of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc 3: 398–409. [DOI] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. . 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. 2014. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28: 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Chou HY, Lin YS, Huang KH, Chang CJ, Hsu TC, Lee SC. 2008. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1β/KAP1. BMC Mol Biol 9: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Jang H, Kim H, Kim ST, Cho EJ, Youn HD. 2010. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem Biophys Res Commun 401: 327–332. [DOI] [PubMed] [Google Scholar]
- Choi J, Jang H, Kim H, Lee JH, Kim ST, Cho EJ, Youn HD. 2014. Modulation of lysine methylation in myocyte enhancer factor 2 during skeletal muscle cell differentiation. Nucleic Acids Res 42: 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000. [DOI] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. 2008. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell 32: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, Coutinho P, Saccone V, Consalvi S, Williams R, et al. . 2012. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. Embo J 31: 301–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ 3rd. 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev 10: 2067–2078. [DOI] [PubMed] [Google Scholar]
- Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S. 2010. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37: 46–56. [DOI] [PubMed] [Google Scholar]
- Heller H, Bengal E. 1998. TFIID (TBP) stabilizes the binding of MyoD to its DNA site at the promoter and MyoD facilitates the association of TFIIB with the preinitiation complex. Nucleic Acids Res 26: 2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. . 2007. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. 2011. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 286: 26267–26276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. . 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 13: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimental H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptiomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CA. 2013. Kaposi’s sarcoma-associated herpesvirus kaposin B induces unique monophosphorylation of STAT3 at serine 727 and MK2-mediated inactivation of the STAT3 transcriptional repressor TRIM28. J Virol 87: 8779–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon AT, Chou AY, Arenillas DJ, Wasserman WW. 2011. Validation of skeletal muscle cis-regulatory module predictions reveals nucleotide composition bias in functional enhancers. PLoS Comput Biol 7: e1002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. 1991. Functional activity of myogenic HLH proteins requires hetero- oligomerization with E12/E47-like proteins in vivo. Cell 66: 305–315. [DOI] [PubMed] [Google Scholar]
- Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. 2014. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell 53: 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling BM, Bharathy N, Chung TK, Kok WK, Li S, Tan YH, Rao VK, Gopinadhan S, Sartorelli V, Walsh MJ, et al. . 2012. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc Natl Acad Sci 109: 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QC, Zha XH, Faralli H, Yin H, Louis-Jeune C, Perdiguero E, Pranckeviciene E, Munoz-Canoves P, Rudnicki MA, Brand M, et al. . 2012. Comparative expression profiling identifies differential roles for Myogenin and p38α MAPK signaling in myogenesis. J Mol Cell Biol 4: 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A, Harter ML. 2003. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc Natl Acad Sci 100: 1735–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83: 1125–1136. [DOI] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. 2009. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res 19: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ 3rd. 2000. Reconstitution of the KRAB–KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol 295: 1139–1162. [DOI] [PubMed] [Google Scholar]
- Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ. 2004. A MyoD-generated feed forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev 18: 2348–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrat B, Correia JS, Mary JL, Tomas-Zuber M, Lesslauer W. 1998. RSK-B, a novel ribosomal S6 kinase family member, is a CREB kinase under dominant control of p38α mitogen-activated protein kinase (p38αMAPK). J Biol Chem 273: 29661–29671. [DOI] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. 2012. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Reports 2: 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 10: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaud J, Desroches J, Balsalobre A, Drouin J. 2009. TIF1β/KAP-1 is a coactivator of the orphan nuclear receptor NGFI-B/Nur77. J Biol Chem 284: 14147–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, Dilworth FJ. 2007. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol 14: 1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. . 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. 2013a. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140: 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. 2013b. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 23: 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio FR, Massacand J, Barde I, Offner S, Corsinotti A, Kapopoulou A, Bojkowska K, Dagklis A, Fernandez M, Ghia P, et al. . 2012. KAP1 regulates gene networks controlling mouse B-lymphoid cell differentiation and function. Blood 119: 4675–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Huang J, Hamamori Y, Kedes L. 1997. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol 17: 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ 3rd. 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev 15: 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & Dev 16: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Faralli H, Yao Z, Rakopoulos P, Palii C, Cao Y, Singh K, Liu QC, Chu A, Aziz A, et al. . 2013. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev 27: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Dilworth FJ. 2013. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J 280: 3991–4003. [DOI] [PubMed] [Google Scholar]
- Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CE, van Ijcken WF, Grosveld F, Rudnicki MA. 2012. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell 47: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic L, Jasencakova Z, Prezioso C, Stutzer A, Bodega B, Pasini D, Klingberg R, Mozzetta C, Margueron R, Puri PL, et al. . 2011. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)–Ezh2 and PRC2–Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. . 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132: 2685–2695. [DOI] [PubMed] [Google Scholar]
- Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. 2014. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res 24: 1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C, Qutob MS, Yee SP, Torchia J. 2000. A novel N-CoR complex contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem 275: 40463–40470. [DOI] [PubMed] [Google Scholar]
- White DE, Negorev D, Peng H, Ivanov AV, Maul GG, Rauscher FJ 3rd. 2006. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res 66: 11594–11599. [DOI] [PubMed] [Google Scholar]
- White D, Rafalska-Metcalf IU, Ivanov AV, Corsinotti A, Peng H, Lee SC, Trono D, Janicki SM, Rauscher FJ 3rd. 2012. The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Mol Cancer Res 10: 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57. [DOI] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270: 725–727. [DOI] [PubMed] [Google Scholar]
- Zang CZ, Schones DE, Zeng C, Cui K, Zhao K, Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics 25: 1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetser A, Gredinger E, Bengal E. 1999. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J Biol Chem 274: 5193–5200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. . 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Sha J, Harter ML. 2010. Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J Cell Biol 188: 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.