Endogenous retroviruses (ERVs) are epigenetically silenced during development, yet the cellular factors recognizing ERVs in a sequence-specific manner remain elusive. Wolf et al. find that ZFP809 initiates the silencing of ERVs in a sequence-specific manner via recruitment of heterochromatin-inducing complexes. ERV reactivation is accompanied by an epigenetic shift from repressive to active histone modifications. ZFP809 is required to initiate ERV silencing during embryonic development but becomes largely dispensable in somatic tissues.
Keywords: endogenous retrovirus, epigenetic silencing, retrovirus restriction factors, KRAB zinc finger proteins
Abstract
Retroviruses have been invading mammalian germlines for millions of years, accumulating in the form of endogenous retroviruses (ERVs) that account for nearly one-tenth of the mouse and human genomes. ERVs are epigenetically silenced during development, yet the cellular factors recognizing ERVs in a sequence-specific manner remain elusive. Here we demonstrate that ZFP809, a member of the Krüppel-associated box zinc finger protein (KRAB-ZFP) family, initiates the silencing of ERVs in a sequence-specific manner via recruitment of heterochromatin-inducing complexes. ZFP809 knockout mice display highly elevated levels of ZFP809-targeted ERVs in somatic tissues. ERV reactivation is accompanied by an epigenetic shift from repressive to active histone modifications but only slight destabilization of DNA methylation. Importantly, using conditional alleles and rescue experiments, we demonstrate that ZFP809 is required to initiate ERV silencing during embryonic development but becomes largely dispensable in somatic tissues. Finally, we show that the DNA-binding specificity of ZFP809 is evolutionarily conserved in the Muroidea superfamily of rodents and predates the endogenization of retroviruses presently targeted by ZFP809 in Mus musculus. In sum, these data provide compelling evidence that ZFP809 evolved to recognize foreign DNA and establish histone modification-based epigenetic silencing of ERVs.
Endogenous retroviruses (ERVs) and long terminal repeat (LTR) transposons are remnants of ancient retroviral germline infections that have been identified in large numbers in all mammalian genomes investigated to date (Weiss 2006). Although rare cases of infectious ERVs have been reported (Baylac-Kalabokias et al. 1991; Le Tissier et al. 1997), the majority of ERVs have lost their ability to form infectious viral particles over time due to mutations (Weiss 2006). However, replication-defective ERVs may still impact their host by introducing retroviral promoters and enhancer elements that in some cases modulate the expression of nearby genes (Maksakova et al. 2006; Jern and Coffin 2008; Macfarlan et al. 2011, 2012; Gifford et al. 2013). Moreover, several human diseases have been linked to ERVs (Lamprecht et al. 2010; Nexo et al. 2011; Kassiotis 2014). To restrict infectious exogenous retroviruses and potentially hazardous ERVs, mammals have developed a wide range of defense mechanisms, including epigenetic transcriptional repression via covalent DNA methylation and histone modifications (Yoder et al. 1997; Wolf and Goff 2008; Rowe and Trono 2011; Leung and Lorincz 2012; Wolf et al. 2013). While DNA methylation represses ERVs in somatic tissues, embryonic stem cells (ESCs) silence ERVs predominantly by histone 3 Lys9 trimethylation (H3K9me3) (Mikkelsen et al. 2007; Matsui et al. 2010; Rowe et al. 2013a), which is associated with tightly packed and transcriptionally inactive chromatin (Grewal and Jia 2007; Kouzarides 2007). However, the factors that recruit silencing machinery to ERVs in a sequence-specific manner remain elusive.
Two lines of evidence support the hypothesis that Krüppel-associated box zinc finger proteins (KRAB-ZFPs), the largest single family of mammalian transcription factors (Urrutia 2003), function as DNA-binding recognition modules that facilitate sequence-specific silencing of ERVs. First, ZFP809, a member of the KRAB-ZFP family, was shown to repress de novo integrated retroviral vectors in embryonic carcinoma cells (ECCs), an ESC-like pluripotent teratocarcinoma cell line, via binding to the proline tRNA primer-binding site (PBS-pro), which is used by some retroviruses to prime reverse transcription (Wolf and Goff 2009). ZFP809 recruits the KRAB domain-binding corepressor KAP1 (TRIM28, TIF1β), which induces silencing via recruitment of histone deacetylases, heterochromatin protein 1 (HP1), and the histone methyltransferase SETDB1 (ESET, KMT1E) (Sripathy et al. 2006; Wolf and Goff 2007; Wolf et al. 2008). Second, several ERV families reactivate in KAP1 and SETDB1 knockout ESCs (Matsui et al. 2010; Rowe et al. 2010, 2013b; Turelli et al. 2014). Furthermore, KRAB-ZFP genes are abundant in mammalian genomes (Corsinotti et al. 2013), are correlated with the number of LTR transposons in vertebrates (Thomas and Schneider 2011), and have diversified in different species through gene duplications and positive selection (Emerson and Thomas 2009), possibly indicating adaptive evolution against invading ERVs. Nevertheless, the role that specific KRAB-ZFPs play in ERV repression has not been widely investigated to date. Although the function of ZFP809 in the repression of engineered retroviral vectors has been described in detail (Wolf and Goff 2009; Schlesinger and Goff 2013; Schlesinger et al. 2014), the endogenous targets of ZFP809 and its requirement in establishing ERV silencing during development have not been explored.
In this study, we performed a detailed functional analysis of ZFP809 using genome-wide binding assays, mouse knockouts, and evolutionary analyses. Our findings demonstrate that ZFP809 is required to establish epigenetic silencing of ancient ERVs during embryonic development and provide a detailed case study for the adaptive evolution of a KRAB-ZFP against genomic parasites.
Results
Genome-wide mapping of ZFP809-binding sites
ZFP809 was identified based on its ability to bind to the PBS-pro of murine leukemia virus (MuLV)-based retroviral vectors (Wolf and Goff 2009). However, since MuLV proviruses are absent in most wild Mus species (Stocking and Kozak 2008), we speculated that exogenous and endogenous MuLV are not the only targets of ZFP809. We thus attempted to explore the function of ZFP809 using genome-wide binding analysis and knockout mice. To date, few studies have successfully achieved genome-wide binding profiles of KRAB-ZFPs due to the unavailability of specific antibodies. We thus performed chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq) using an ECC line stably transfected with a transposon-based Flag-tagged ZFP809 expression vector. This vector integrates in multiple genomic copies per cell (Xue et al. 2009) and resulted in high levels of transgene expression (Supplemental Fig. 1A), which we found necessary to enrich enough DNA for library construction and accurate peak detection with a low false discovery rate (FDR). More than 9000 Flag-ZFP809 ChIP-seq peaks were called using this approach (Supplemental Fig. 1B).
To confirm ZFP809 binding to these sites in a more developmentally relevant cell type, we generated an ESC line containing a single copy of a Flag-ZFP809 expression construct inserted at the HPRT locus but driven by a doxycycline-inducible promoter. After doxycycline addition, Flag-ZFP809 protein was expressed approximately threefold greater than endogenous ZFP809 (Supplemental Fig. 1A). Furthermore, we generated a custom-made anti-ZFP809 polyclonal antibody (ZFP809_5763) to analyze binding of endogenous ZFP809 in ESCs. Although these latter strategies did not allow us to reliably identify ZFP809-binding sites due to a high FDR of the called peaks (Supplemental Fig. 1B), heat map analysis confirmed that genomic regions covered by the strongest Flag-ZFP809 peaks identified in ECCs were also bound by Flag-ZFP809 and endogenous ZFP809 in ESCs (Supplemental Fig. 1C). Therefore, we focused our further analysis on “strong” (>50-fold enrichment over input) peaks. In an independent ChIP-seq replicate with Flag-tagged ZFP809 in ECCs, 96% of these peaks were called again with high confidence (data not shown).
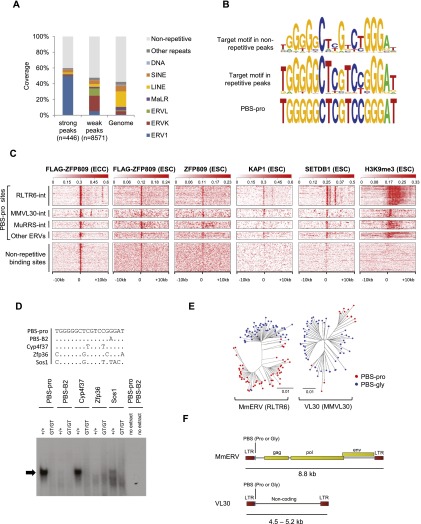
More than half of the 446 genomic regions identified as strong peaks were annotated as ERVs belonging to the ERV1 class (Fig. 1A; Supplemental Fig. 2A), and ∼90% of the ∼150 endogenous PBS-pro sequences in the mouse genome were found within these peaks (Supplemental Fig. 2B). However, ∼40% of the strong Flag-ZFP809 peaks were located in nonrepetitive genomic regions (Fig. 1A). The consensus ZFP809 target motif inferred from the 100 top-scored nonrepetitive peaks strikingly resembled the PBS-pro motif deduced from peaks in repetitive sequences (Fig. 1B). Unlike a large proportion of the inferred binding sites in repetitive peaks, none of the binding sites in nonrepetitive peaks were identical to the canonical PBS-pro sequence. Nevertheless, the majority of the >9000 Flag-ZFP809 peak regions contained a PBS-pro-like sequence (Supplemental Fig. 2B). Importantly, although Flag-ZFP809 bound ERV PBS-pro and imperfect nonrepetitive sites equally well when overexpressed in ECCs, endogenous ZFP809 showed a clear preference for the intact PBS-pro sequence (Fig. 1C; Supplemental Fig. 2C). Taken together, these results indicate that ERV1-associated PBS-pro loci are the preferred endogenous ZFP809-binding sites.
Figure 1.
Genome-wide mapping of ZFP809-binding sites. (A) Repeat content of strong (enrichment >50) and weak (enrichment <50) Flag-ZFP809 ChIP-seq peaks and the mouse genome as determined by genomic overlap with University of California at Santa Cruz (UCSC) RepeatMasker annotations. (B) Consensus Flag-ZFP809 target motif derived from the 100 top-scored nonrepetitive peaks (top) and the corresponding target consensus in strong repetitive peaks (middle). The PBS-pro sequence is shown at the bottom. (C) Heat maps of ChIP-seq data at ERV-associated PBS-pro sites and strong nonrepetitive Flag-ZFP809 ChIP-seq peak regions. Ten-kilobase regions are displayed with the PBS-pro sequence or nonrepetitive peaks located at the center. PBS-pro sites were grouped according to flanking ERV sequences and are orientated 5′ to 3′. (D) EMSA with nuclear extracts from wild-type and Zfp809 mutant (Zfp809GT/GT) ESCs. Sequences of dsDNA probes (+ strand, 5′ to 3′) are shown in the top panel. The repressor complex binding to the PBS-pro probe is indicated by an arrow. (E) Neighbor-joining phylogenetic trees of genomic MmERV and VL30 elements containing PBS-pro or PBS-gly. The pol gene or the noncoding internal region was used to create trees for MmERV and VL30 elements, respectively. (F) Schematic representation of MmERV and VL30 consensus sequences. Yellow rectangles indicate coding regions for retroviral proteins.
To explore whether ZFP809-binding sites overlap with known KRAB-ZFP corepressors in ESCs, we compared our ZFP809 ChIP-seq data with previously published ChIP-seq data sets for KAP1 (Rowe et al. 2013b), SETDB1 (Yuan et al. 2009), and H3K9me3 (Rowe et al. 2013b) in ESCs. Indeed, repeat-associated PBS-pro loci strongly correlated with KAP1- and SETDB1-binding sites and were marked by H3K9me3 (Fig. 1C). In contrast, only very weak SETDB1 binding was observed at strong nonrepetitive Flag-ZFP809 peaks, and these regions were devoid of H3K9me3 and KAP1 in ESCs (Fig. 1C). The lack of repressive H3K9me3 at nonrepetitive ZFP809-binding sites could be due to the lower binding affinity of endogenous ZFP809 to these sites and/or in cis acting flanking regions that prevent heterochromatin formation. We therefore extracted the putative PBS-pro target motifs in strong Flag-ZFP809 ChIP-seq peaks immediately upstream of four genes and tested the ability of these sequences to silence a retroviral reporter in cultured cells (Supplemental Fig. 3A,B). Of all tested motifs, only the canonical PBS-pro sequence was efficiently silenced (Supplemental Fig. 3B), indicating that ZFP809 fails to recruit silencing machinery to these imperfect target sites due to a lower binding affinity.
To further analyze binding of the KAP1/ZFP809 repressor complex to imperfect Flag-ZFP809 target sites near gene promoters in vitro, we performed electrophoretic mobility shift assays (EMSAs) (Yamauchi et al. 1995). In accordance with our previous results, the two tested nonrepetitive PBS-pro-like target sites at gene promoters that were devoid of H3K9me3 in ESCs (Zfp36 and Sos1) did not show ZFP809 repressor complex binding (Fig. 1D). In contrast, a Flag-ZFP809 target sequence that was found at the promoter region of Cyp4f37 was bound by the repressor complex at levels comparable with the canonical PBS-pro (Fig. 1D). Notably, we observed high levels of H3K9me3 at the Cyp4f37 promoter region in ESCs, and this target was not characterized as a nonrepetitive binding site due to several flanking ERV1 fragments. Despite the lack of repressor binding to nonrepetitive Flag-ZFP809 targets, Flag-ZFP809 overexpression in ECCs resulted in increased KAP1 recruitment to these sites and, in some cases, also induction of H3K9me3 and H4K20me3 and reduced gene expression levels (Supplemental Fig. 4A,B). In sum, these results indicate that, at physiological concentrations, ZFP809 exhibits strict sequence specificity, achieving effective epigenetic silencing predominantly at the canonical PBS-pro motif.
To further investigate ZFP809 binding to ERVs, we performed an in silico screen of murine ERVs and their corresponding PBS sequences (Supplemental Table 1). Notably, two of the ZFP809-targeted ERV1 groups, Mus musculus ERV (MmERV, RLTR6) and virus-like 30 (VL30, MMVL30) elements, can be phylogenetically divided into two closely related subgroups that use either PBS-pro or a PBS matching to a glycine tRNA (PBS-gly) (Fig. 1E). MmERV elements are predominantly intact ERVs with coding regions for retroviral Gag, Pol, and Env proteins, whereas VL30 elements lack coding regions and are likely derived from MmERV elements that lost their coding region (Fig. 1F). Since the PBS-gly and the majority of PBS sequences used by murine ERVs are not able to initiate silencing in ESCs and ECCs (Supplemental Table 1 and references therein), we explored whether there was differential recruitment of KAP1/SETDB1 complexes to these ERV subgroups. Whereas only MmERV-pro and VL30-pro elements are bound by ZFP809, KAP1, and SETDB1 directly at the PBS, both MmERV and VL30 subgroups contain additional SETDB1-targeted sites outside of the PBS (Supplemental Fig. 5). These findings indicate that MmERV-gly and VL30-gly elements escape repression by ZFP809 but are targeted by other silencing factors that recruit SETDB1 and induce H3K9me3. Furthermore, this analysis demonstrates that the regulation of ERV family members must be considered on a case-by-case basis, since even closely related ERVs can be silenced by distinct mechanisms via subtle changes of their nucleotide content.
ZFP809 depletion leads to ERV reactivation in adult tissues
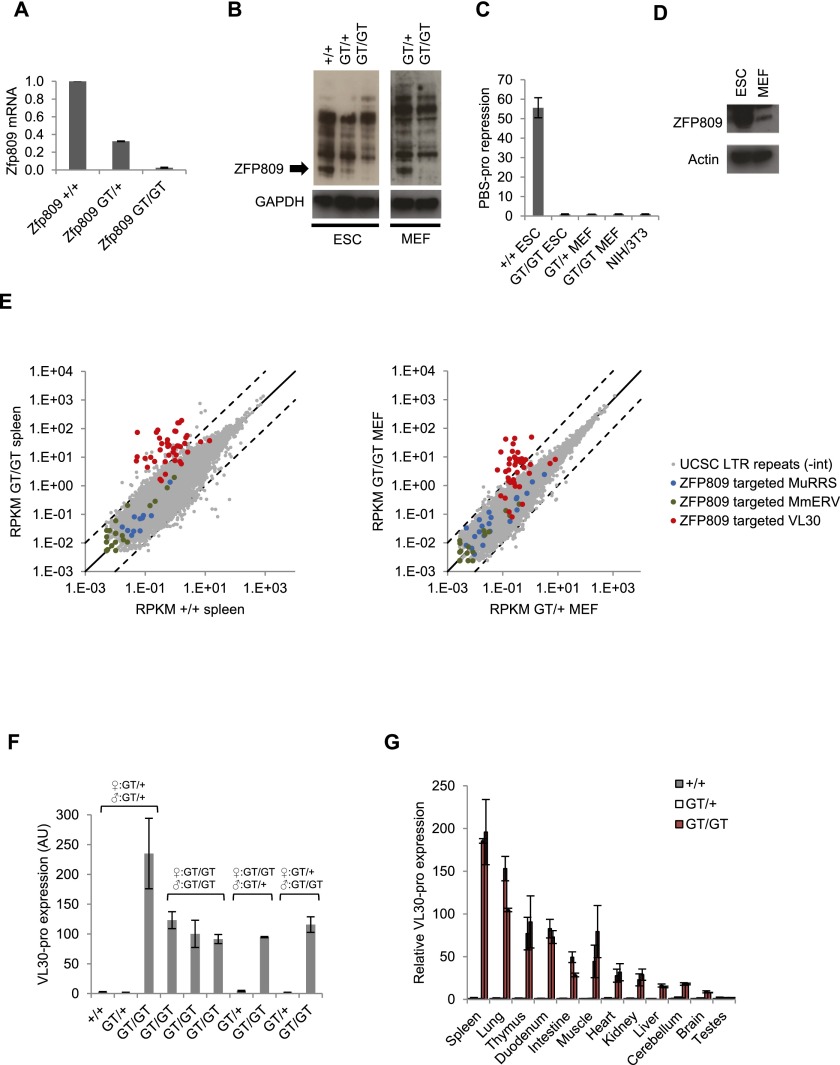
To investigate the functional requirement of ZFP809 in ERV silencing, we generated Zfp809 mutant mice carrying a gene trap (Zfp809GT) or a conditional Zfp809 knockout-first (Zfp809KO-first) allele (Supplemental Fig. 6). Zfp809GT/GT and Zfp809−/− mice were viable and fertile and showed no signs of physical distress or disease. Efficient ZFP809 knockout in Zfp809GT/GT mouse embryonic fibroblasts (MEFs) and ESCs was confirmed by RT-qPCR (Fig. 2A) and Western blots (Fig. 2B). In line with previous studies (Barklis et al. 1986; Wolf and Goff 2009), a PBS-pro-containing retroviral vector was efficiently silenced in wild-type ESCs but not in Zfp809GT/GT ESCs, whereas MEFs failed to repress the PBS-pro vector even in the presence of a Zfp809 wild-type allele (Fig. 2C). To test whether the lack of PBS-pro silencing in MEFs could be due to reduced ZFP809 levels, we performed RT-qPCR and Western blots on ESCs and MEFs. As previously reported (Wolf and Goff 2009), we found that Zfp809 mRNA levels were comparable in ESCs and MEFs (data not shown). However, Western blotting with our custom ZFP809 antibody demonstrated a >20-fold decrease in ZFP809 protein levels in MEFs compared with ESCs (Fig. 2D), indicating that a post-transcriptional control mechanism reduces ZFP809 protein levels in MEFs.
Figure 2.
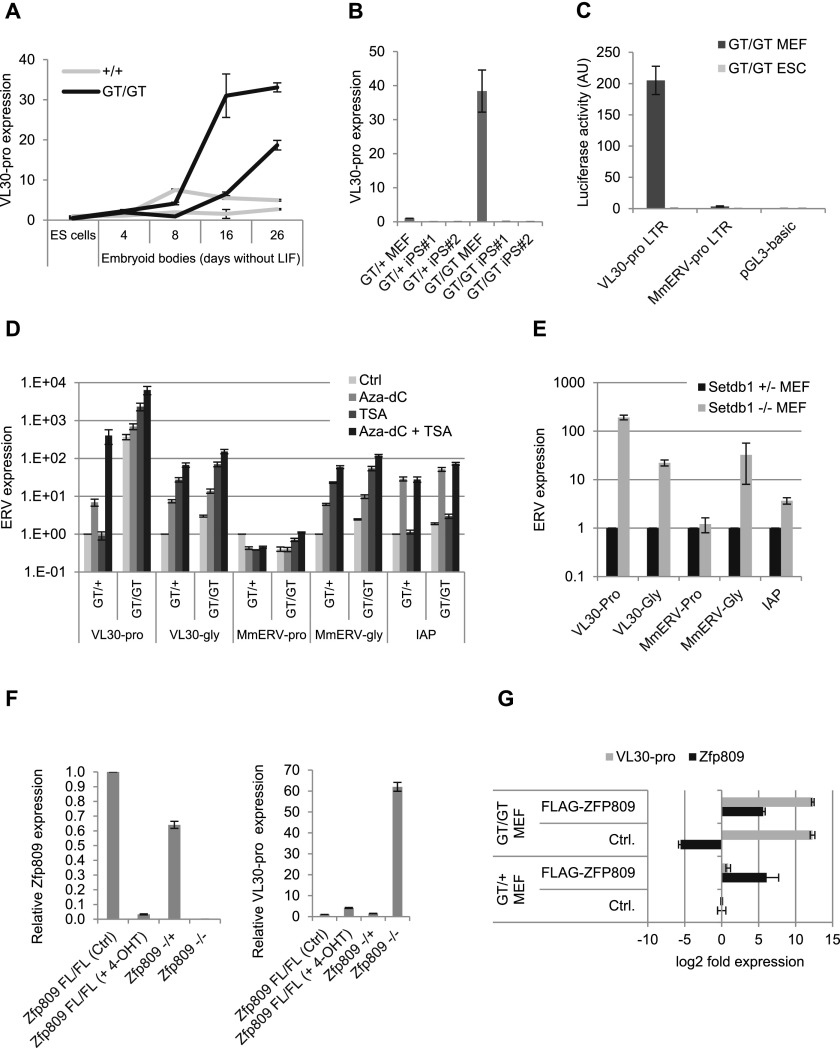
ZFP809 is required for transcriptional repression of VL30 elements. (A) RT-qPCR analysis of Zfp809 mRNA levels in Zfp809GT ESCs (normalized to ActB). Graphs show the mean fold expression (n = 3) relative to wild-type ESCs. Error bars represent SD. (B) Western blot with whole-cell lysates of wild-type and Zfp809GT ESCs and MEFs using antibodies against ZFP809 (5763) or GAPDH (significantly longer exposure times were used for MEFs to observe signal). (C) ESCs and MEFs were transduced with retroviral vectors containing PBS-pro or the repression escape mutant PBS-B2 upstream of a puro resistance gene and treated with puromycin for 10 d to determine viral titers (colony-forming units per milliliter). Graphs show the mean ratios of PBS-B2/PBS-pro transduction efficiencies (indicating PBS-pro repression activity) as determined in three replicates. Error bars represent SD. (D) Western blot with whole-cell lysates of wild-type ESCs and MEFs using antibodies against ZFP809 or Actin. (E) RNA-seq analysis of Zfp809GT MEFs (left) and spleens (right). Mapped RNA-seq reads overlapping with annotated internal RepeatMasker LTR repeats were counted and plotted as reads per kilobase of transcript per million mapped reads (RPKM). The three largest ZFP809-targeted ERV groups (repeats located within 10 kb of strong Flag-ZFP809 peaks) are highlighted. Dotted lines mark 10-fold up-regulation and down-regulation. (F) RT-qPCR analysis of VL30-pro expression in whole blood of adult female mice. The genotypes of the animals’ parents are indicated. Values represent mean expression (normalized to Hprt) in arbitrary units (AU) (n = 3 technical replicates). Error bars represent SD. (G) VL30-pro expression in somatic tissues and organs of male adult mice (9–12 wk old) measured by RT-qPCR (normalized to Hprt). Two animals of each genotype were analyzed. Values represent mean fold expression (n = 2 technical replicates) relative to wild-type mouse tissues. Error bars represent SD.
To investigate the functional consequences of ZFP809 loss of function, we performed gene expression studies on Zfp809 mutant cell lines and tissues. RNA sequencing (RNA-seq) analysis of Zfp809GT/GT MEFs and adult spleen samples revealed highly increased expression levels of VL30-pro elements, whereas other PBS-pro-containing ERVs remained unaffected (Fig. 2E). VL30-pro reactivation was also observed by RT-qPCR in whole blood (Fig. 2F) and all other investigated tissues and organs of Zfp809 mutant mice, with the sole exception of testis (Fig. 2G). Our data thus demonstrate that ZFP809 is required to silence a subset of PBS-pro-containing ERVs in mice.
ZFP809 recruits epigenetic silencing machinery to ERVs
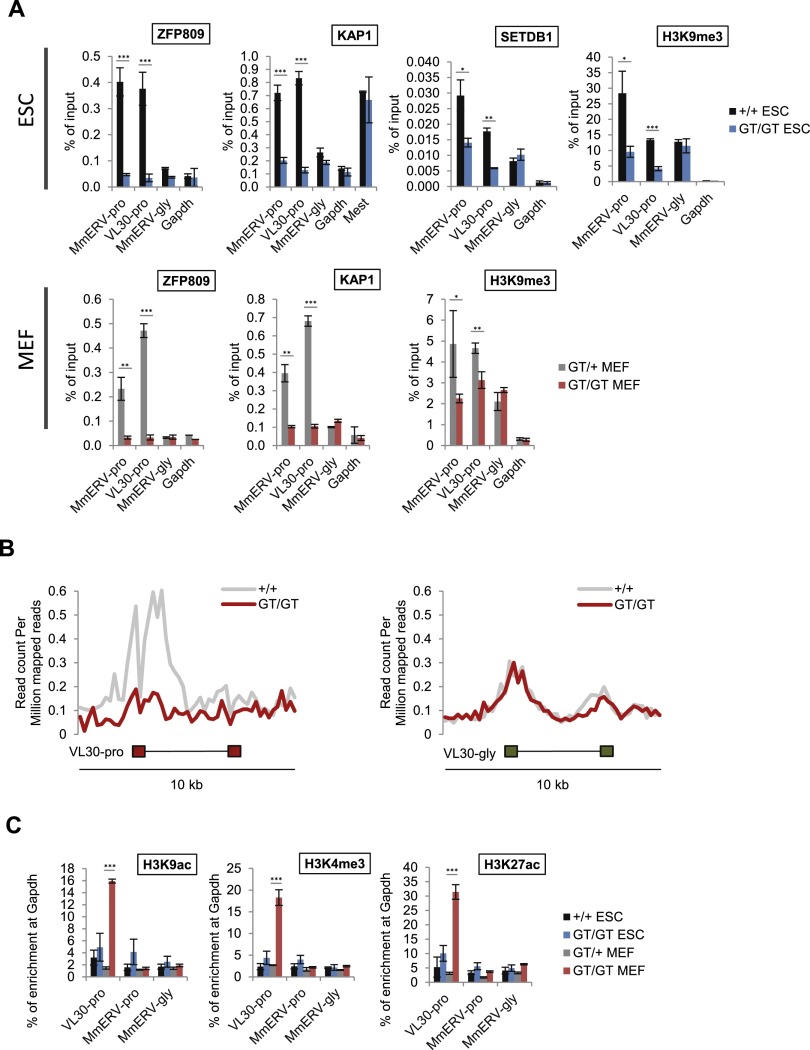
Our data to this point indicate that ZFP809 is required for the site-specific recruitment of heterochromatin machinery to PBS-pro-containing ERVs. To test this hypothesis further, we analyzed the chromatin status of ZFP809-targeted ERVs in Zfp809 mutants. As expected, we observed a decrease in ZFP809, KAP1, SETDB1, and H3K9me3 enrichment at PBS-pro-containing ERVs in Zfp809GT/GT ESCs (Fig. 3A). Furthermore, genome-wide ChIP-seq analysis confirmed a loss of H3K9me3 at VL30-pro elements in Zfp809GT/GT ESCs (Fig. 3B).
Figure 3.
ZFP809 is required to recruit KAP1/SETDB1 to ERVs and establish heterochromatin marks. (A) ChIP-qPCR (ZFP809, KAP1, and SETDB1) or native ChIP (NChIP)-qPCR (H3K9me3) analysis of ERVs in Zfp809GT ESCs (top panel) and MEFs (bottom panel). Enrichment is shown as mean percentage of input DNA (n = 3 technical replicates). Error bars represent SD. P-values (by unpaired t-tests) are indicated as follows: (***) P < 0.0005; (**) 0.0005<P < 0.005; (*) 0.005<P < 0.05. (B) NGS plots showing average H3K9me3 NChIP-seq read densities at 10-kb regions around genomic VL30-pro (n = 15) and VL30-gly (n = 65) elements in Zfp809+/+ and Zfp809GT/GT ESCs. (C) ChIP-qPCR analysis of markers of active chromatin in Zfp809GT MEFs and ESCs. Relative enrichment was normalized to input DNA and is shown as mean percentage (n = 3 technical replicates) of enrichment at the Gapdh promoter. Error bars represent SD. P-values (by unpaired t-tests) are indicated as described above.
Reduced levels of ZFP809, KAP1, and H3K9me3 at these ERVs were also found in Zfp809GT/GT MEFs (Fig. 3A), indicating that ZFP809 still binds to ERVs in MEFs and recruits KAP1 despite its significantly reduced levels in MEFs relative to ESCs (Fig. 2D). In addition to the reduction in repressive marks, we found a strong increase in histone modifications associated with active transcription, including H3K9ac, H3K4me3, and H3K27ac at VL30-pro elements in Zfp809GT/GT MEFs but not ESCs (Fig. 3C). Interestingly, VL30-pro elements in Zfp809GT/GT MEFs appeared to be enriched in both repressive H3K9me3 and active marks such as H3K4me3 (Fig. 3A,C). To test whether those marks colocalize on the same VL30-pro elements, we performed sequential ChIP (Re-ChIP). This analysis shows that H3K4me and H3K9me3 at VL30-pro elements are mutually exclusive (Supplemental Fig. 7A), indicating that only a subset of these elements becomes epigenetically derepressed in Zfp809GT/GT MEFs. Indeed, a more detailed RNA-seq analysis of all VL30-pro elements using only uniquely mappable RNA-seq reads confirmed that only a small number of these elements is strongly expressed upon ZFP809 depletion (Supplemental Table 2).
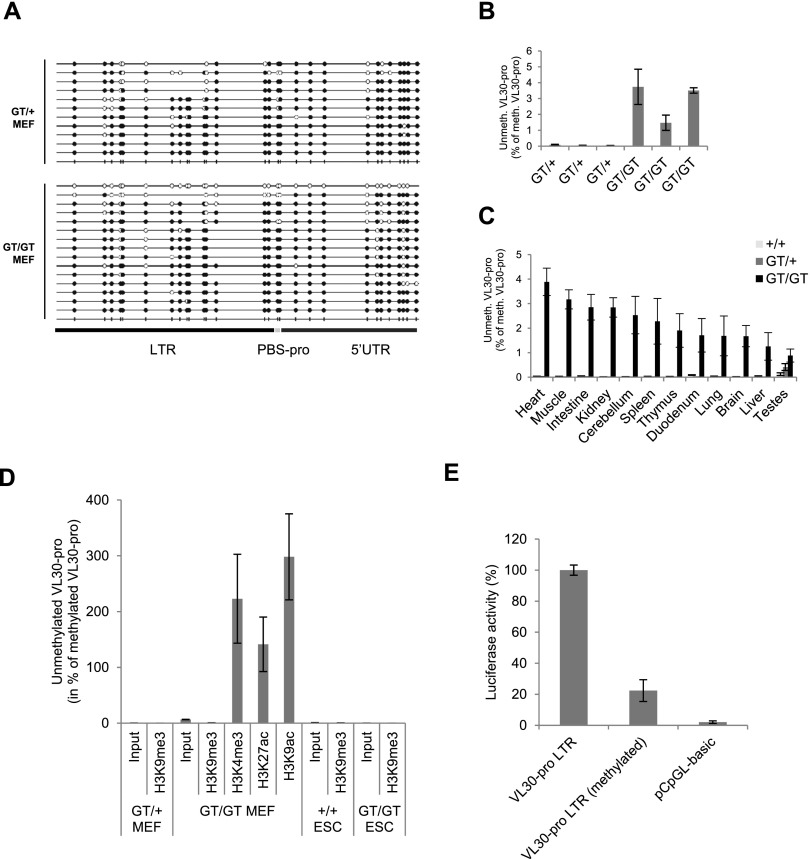
To address whether VL30-pro activation is associated with lower levels of DNA methylation, we performed bisulfite sequencing in Zfp809GT/+ and Zfp809GT/GT MEFs. In both cell lines, the majority of VL30-pro elements were highly methylated at CpG sites, indicating that bulk CpG methylation of VL30-pro ERVs is largely independent of ZFP809 (Fig. 4A). However, we found a single VL30-pro sequence derived from Zfp809GT/GT MEFs that was entirely devoid of CpG methylation (Fig. 4A). Quantitative methylation-specific PCR (qMSP) analysis of bisulfite-converted DNA with primers specific for methylated or unmethylated VL30-pro elements confirmed that only a small proportion of these elements had lost their methylation pattern in Zfp809GT/GT MEFs (Fig. 4B). Similarly, a partial loss of VL30-pro DNA methylation was also observed in adult Zfp809GT/GT tissues (Fig. 4C).
Figure 4.
VL30 reactivation is associated with partial loss of DNA methylation. (A) Bisulfite sequencing analysis of VL30-pro ERVs in Zfp809GT MEFs. Full and empty circles represent methylated and unmethylated CpG sites, respectively. (B) qMSP analysis of Zfp809GT MEFs. Graphs show the genomic occurrence of unmethylated VL30-pro elements as percentage of methylated VL30-pro elements (n = 3 technical replicates). Error bars represent SD. (C) qMSP analysis was performed as above but with genomic DNA isolated from tissues of adult wild-type and Zfp809GT/GT mice. (D) qMSP analysis of MEF and ESC DNA after ChIP with the indicated antibodies. Graphs show the occurrence of unmethylated VL30-pro elements as percentage of methylated VL30-pro elements (n = 3 technical replicates). Error bars represent SD. (E) Dual-luciferase assay of Zfp809GT/+ MEFs transfected with a single unmethylated or in vitro CpG methylated VL30-pro LTR (+PBS-pro) cloned into a CpG-free luciferase vector. LTR activity was normalized to CMV promoter activity of a cotransfected renilla luciferase plasmid and is displayed in arbitrary units (AU). Assay was performed in triplicates. Error bars represent SD.
To test a possible association between active histone marks and DNA methylation at VL30-pro elements, we performed qMSP on DNA that had been immunoprecipitated with antibodies against various histone modifications. Strikingly, VL30-pro elements marked by active histone marks in Zfp809GT/GT MEFs displayed low levels of DNA methylation, whereas elements marked by H3K9me3 were heavily methylated (Fig. 4D). Importantly, VL30-pro elements were also heavily methylated in both Zfp809 wild-type and Zfp809GT/GT ESCs (Fig. 4D). These data suggest that the demethylated state is strongly correlated with actively transcribed VL30-pro elements.
To further examine the effect of DNA methylation on VL30-pro transcription, we cloned a single VL30-pro LTR into a CpG-free luciferase expression vector and in vitro methylated these vectors with the CpG-specific methyltransferase M.SssI (Supplemental Fig. 7B). CpG methylation strongly decreased promoter activity of the VL30-pro LTR, yet some transcriptional activity remained (Fig. 4E), indicating that individual VL30-pro elements might become reactivated to some degree despite persistent DNA methylation. In sum, our data show that KAP1/SETDB1 complexes fail to be recruited to ERV-associated PBS-pro in the absence of ZFP809, resulting in reduced levels of heterochromatin marks (H3K9me3) and the presence of active histone modifications at a subset of ZFP809 targets that contain active LTR elements. However, the majority of CpG methylation as well as some H3K9me3 at ZFP809-targeted ERVs is independent of ZFP809.
ZFP809 is required to establish ERV silencing during development
To determine the timing of ERV reactivation phenotypes in Zfp809 mutants, we performed RT-qPCR analysis on several Zfp809GT/GT ESC and MEF lines. VL30-pro elements were up-regulated in Zfp809GT/GT MEFs but, surprisingly, not in ESCs (Supplemental Fig. 8A). Concordantly, we detected increased VL30-pro expression in Zfp809−/− embryonic day 9.5 (E9.5) embryos but not in E3.5 preimplantation embryos (Supplemental Fig. 8B,C). However, Zfp809GT/GT ESCs differentiated as embryoid bodies in the absence of leukemia inhibitory factor (LIF) increasingly expressed VL30-pro during the course of differentiation (Fig. 5A). Conversely, VL30-pro elements were resilenced in induced pluripotent stem cells (iPSCs) derived from Zfp809GT/GT MEFs (Fig. 5B). Notably, both KAP1 and ZFP809 protein levels were increased in iPSCs as compared with MEFs (Supplemental Fig. 8D). Altogether, these data indicate that VL30-pro elements remain silent in ESCs but become gradually activated during differentiation in Zfp809 mutants.
Figure 5.
ZFP809 is required for the initiation, but not the maintenance, of ERV silencing. (A) RT-qPCR analysis of VL30-pro expression in ESCs during differentiation (normalized to Hprt). Values represent mean fold expression (n = 3 technical replicates) relative to wild-type ESCs. Results from two independent ESC lines are shown for each genotype. Error bars represent SD. (B) VL30-pro expression in MEFs and iPSCs derived from these MEF lines (normalized to Gapdh). Two iPSC clones were analyzed by RT-qPCR for each MEF line. Values represent mean fold expression (n = 3 technical replicates) over Zfp809GT/+ MEFs. Error bars represent SD. (C) Dual-luciferase reporter assay in transiently transfected Zfp809GT/GT cells. LTR activity was normalized to CMV promoter activity on cotransfected renilla luciferase plasmids and is displayed in arbitrary units (AU). Assay was performed in triplicates; error bars represent SD. (D) ERV expression analysis of MEFs treated with Aza-dC and/or TSA by RT-qPCR (normalized to Hprt). Graphs show mean fold expression (n = 3 technical replicates) over Zfp809GT/+ untreated control. Error bars represent SD. (E) ERV expression in Setdb1−/+ and Setdb1−/− MEFs was measured by RT-qPCR (normalized to Hprt) and is shown as mean fold expression (n = 3 technical replicates) relative to Setdb1-/+ MEFs. Error bars represent SD. (F) RT-qPCR analysis of Zfp809 and VL30-pro expression in inducible Zfp809 knockout (Zfp809FL/FL; RosaCreERT2) and Zfp809−/− MEFs. Values represent mean fold expression (n = 3 technical replicates) relative to untreated Zfp809FL/FL MEFs (normalized to Hprt). Error bars represent SD. (G) Zfp809GT/+ and Zfp809GT/GT MEFs stably transfected with a transposon-based Flag-ZFP809 expression vector or an empty control vector were grown under puromycin selection for 2 wk before RT-qPCR analysis. Values represent mean fold expression (n = 3 technical replicates) relative to Zfp809GT/+ MEFs transfected with control vector (normalized to Hprt). Error bars represent SD.
The lack of VL30-pro derepression in Zfp809GT/GT ESCs and iPSCs, and of MmERV-pro derepression in general might be due to additional ZFP809-independent epigenetic repression mechanisms (as indicated by additional SETDB1-binding sites at these ERVs) (Supplemental Fig. 5) and/or differences in the transcriptional activity of their LTR elements. To assess the transcriptional potential of these LTRs separately from the effects of chromosomal context and flanking sequences, we cloned genomic VL30-pro and MmERV-pro LTR libraries as well as single LTRs into a luciferase reporter vector and determined their transcriptional activity in transiently transfected Zfp809GT/GT MEFs and ESCs. In accordance with our ERV expression data, VL30-pro LTRs were active in MEFs but not ESCs, whereas MmERV-pro LTRs were inactive in both MEFs and ESCs (Fig. 5C; Supplemental Figs. 8E, 9). Furthermore, global chromatin derepression by the histone deacetylase inhibitor trichostatin A (TSA) and the DNA demethylating agent 5-aza-2′-deoxycytidine (Aza-dC) or genetic depletion of SETDB1 did not activate MmERV-pro elements in MEFs, whereas VL30-pro, VL30-gly, and MmERV-gly elements were strongly activated under these conditions (Fig. 5D,E). These data point out that epigenetic derepression is not sufficient for the transcriptional activation of ERVs that lack active LTR promoters in a given cell type.
The observed up-regulation of endogenous VL30-pro in Zfp809GT/GT MEFs stands in contrast to the lack of ZFP809-dependent silencing of retroviral vectors in the same cell line (Fig. 2C). We thus speculated that ZFP809 is required to initiate stable epigenetic silencing during development but not to maintain silencing in somatic cells. To test this hypothesis, we generated conditional Zfp809 knockout MEFs by crossing Zfp809 floxed (FL) mice with mice carrying a CreERT transgene that can be induced with 4-hydroxytamoxifen (4-OHT). 4-OHT treatment resulted in efficient deletion of Zfp809 but only a marginal increase in VL30-pro expression levels relative to MEFs derived from knockout mice lacking ZFP809 throughout development (Fig. 5F). To test whether restoring ZFP809 expression can reverse the observed phenotypes in MEFs derived from ZFP809-depleted embryos, we stably transfected Zfp809GT/GT MEF with our transposon-based Flag-Zfp809 expression vector. Flag-ZFP809 protein was efficiently expressed (Supplemental Fig. 10A,B) and was capable of immunoprecipitating KAP1 (Supplemental Fig. 10C) and VL30-pro elements in ChIP assays (Supplemental Fig. 10D). Nevertheless, restoring ZFP809 expression in MEFs did not rescue silencing of VL30-pro elements (Fig. 5G) or repression of de novo integrated retroviral vectors (Supplemental Fig. 10E). Furthermore, unlike in ESCs (Supplemental Fig. 10F), Flag-ZFP809 overexpression did not restore H3K9me3 at endogenous VL30-pro in MEFs (Supplemental Fig. 10G). These data indicate that the failure of retroviral repression in MEFs is not exclusively due to reduced ZFP809 protein levels (Fig. 2D). Potentially, low expression levels of KAP1 (Supplemental Fig. 8D) and/or other cofactors required for retroviral repression or possibly post-translational modifications of one or several of these factors contribute to the lack of retroviral repression in MEFs. Altogether, our results indicate that the ERV reactivation phenotype in Zfp809 mutant somatic tissues is likely due to a failure to initiate silencing during embryo development and not to a persistent requirement of ZFP809 function in all somatic cells.
The ZFP809 DNA-binding domain is evolutionarily conserved in Muroidea
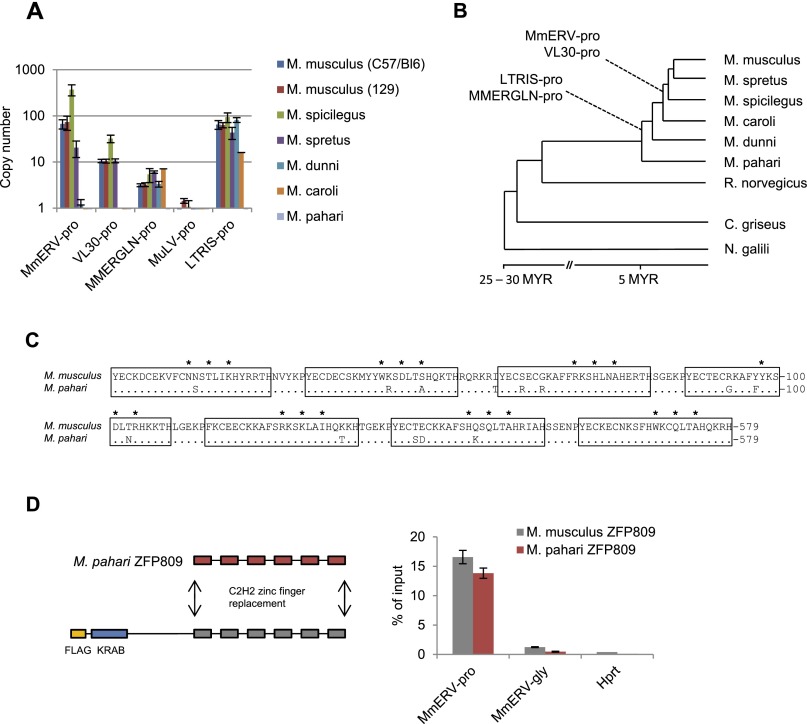
It has been speculated that KRAB-ZFPs evolve to restrict newly emerging ERVs that have colonized the host genome (Thomas and Schneider 2011). To test this hypothesis, we determined the approximate age of ZFP809-targeted ERVs and the evolutionary conservation of the ZFP809 DNA-binding domain. Both MmERV-pro and VL30-pro were exclusively present in Mus species closely related to M. musculus, but none of the five PBS-pro-containing ERV groups identified in M. musculus could be detected in the genome of Mus pahari, indicating that these ERVs integrated into the germline of M. musculus ancestors <5 million years ago, after the radiation of the Mus genus (Fig. 6A,B). However, conserved Zfp809 DNA-binding domains were found in all screened Mus species (Supplemental Fig. 11). The target specificity of KRAB-ZFPs is mainly determined by three amino acids per zinc finger that directly interact with DNA (Emerson and Thomas 2009). Within the Mus genus, only M. pahari ZFP809 was found to have a mutation in one of these amino acids as compared with M. musculus (Fig. 6C). However, this mutation did not decrease the binding affinity of ZFP809 to the PBS-pro target, as determined using ChIP assays with a recombinant Flag-ZFP809 protein carrying the M. pahari DNA-binding domain (Fig. 6D). Furthermore, we identified two potential Zfp809 orthologs with conserved DNA-binding domains in Cricetulus griseus (Chinese hamster) and Nannospalax galili (blind mole rat) by computational data mining (Supplemental Fig. 11). These species are believed to have shared a common ancestor with mice >25 million to 30 million years ago (Jansa and Weksler 2004; Steppan et al. 2004), indicating that ZFP809 evolved to restrict PBS-pro-containing retroviruses before the Muroidea radiation that gave rise to >1500 different species (Musser and Carleton 2005). However, we were unable to identify a Zfp809 ortholog by BLAST searches of the sequenced rat genome or by genomic PCR screening. Interestingly, a BLAST screen identified >500 PBS-pro sequences associated with >10 different ERV1 groups in the rat genome (Baylor 3.4/rn4 assembly), indicating that Zfp809 has been lost in some Muroidea species, possibly leaving them more susceptible to germline colonization by retroviruses.
Figure 6.
The ZFP809 DNA-binding specificity is conserved in Muroidea and predates germline colonization by ZFP809-targeted ERVs. (A) Genomic ERV copy numbers in wild Mus species were determined by genomic qPCR using plasmid dilution series as references and Zfp809 primers for normalization (n = 3 technical replicates). Error bars represent SD. (B) Approximate phylogenetic tree of Mus species and related Muroidea with the estimated historical appearance of PBS-pro-containing ERVs as determined by genomic qPCR. (C) Alignment of the ZFP809 DNA-binding domain in M. musculus and its sequenced ortholog in M. pahari. The seven C2H2 zinc fingers are underlined, and residues known to be directly involved in sequence-specific DNA binding are marked by asterisks. Identical amino acids in M. pahari are shown as dots. (D, left) Strategy for replacement of the M. musculus ZFP809 DNA-binding domain with the homologous region of M. pahari ZFP809. (Right) ChIP with Flag antibody in F9 ECCs stably transfected with M. musculus Flag-ZFP809 or recombinant M. pahari Flag-ZFP809 expression vectors. Enrichment at MmERV-pro and control regions was determined by qPCR and is shown as mean percentage (n = 3 technical replicates) of input DNA. Error bars represent SD.
ERV insertions induce gene silencing through ZFP809 recruitment
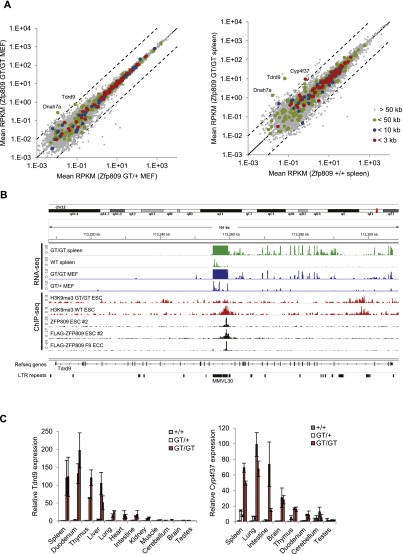
Our evolutionary analyses demonstrate the recurrent colonization of Muroidea genomes with PBS-pro-containing ERVs, and we thus speculated that novel integrations near genes could lead to ZFP809-dependent silencing of nonviral sequences due to ERV proximity. We therefore correlated the transcriptional start site (TSS) of RefSeq genes with Flag-ZFP809 ChIP-seq peaks and analyzed gene expression levels in Zfp809GT/GT MEFs and spleen samples by RNA-seq. Although expression of the vast majority of genes was unchanged in Zfp809 mutants, we identified at least three genes in proximity to ZFP809-binding sites that were reproducibly expressed at increased levels in Zfp809GT/GT MEFs and/or spleen: Tdrd9, Cyp4f37, and Dnah7a (Fig. 7A; Supplemental Table 3; Supplemental Fig. 12A). Closer inspection of the ZFP809-binding site within Tdrd9 revealed the presence of a VL30-pro element that has integrated in antisense orientation between exons 13 and 14 and is itself highly up-regulated in Zfp809 mutants (Fig. 7B). A VL30-pro element that is expressed at increased levels in Zfp809GT/GT MEFs and spleen was also identified near the TSS of Dnah7a (Supplemental Fig. 12B).
Figure 7.
ERVs repress host genes via ZFP809 recruitment. (A) Differential gene expression analysis of RefSeq genes in Zfp809GT MEFs (left panel) and spleens (right panel). RNA-seq RPKM values were determined for the exon proportion of RefSeq genes. Genes with TSSs that are located near strong Flag-ZFP809 ChIP-seq peaks are highlighted as indicated. Dotted lines mark 10-fold up-regulation and down-regulation. (B) RNA-seq and ChIP-seq read density at the Tdrd9 gene. (C) Tdrd9 (left) and Cyp4f37 (right) mRNA expression in somatic tissues and organs of male adult mice (9–12 wk old) measured by RT-qPCR (normalized to Hprt). For each genotype, two animals were analyzed. Values represent mean fold expression (n = 2 technical replicates) relative to wild-type mouse tissues.
In the case of Cyp4f37, no VL30-pro element was identified near the gene, and the ZFP809 ChIP-seq target sequence, which is located ∼300 base pairs (bp) upstream of the TSS in antisense orientation, differs from the PBS-pro by two nucleotides. However, in vitro binding assays determined that these two mismatches do not abrogate repressor complex binding (Fig. 1D), and retroviral reporter assays confirmed that the imperfect target sequence efficiently silences retroviral vectors in ECCs (data not shown). Notably, the motif is located next to several partial internal ERV1 fragments (Supplemental Fig. 12B), and both nucleotide mismatches are C-to-T mutations at the two CpG sites within the PBS-pro (Fig. 1D). Since CpG sites within repetitive elements in mammals are heavily methylated and therefore subject to an increased C-to-T transition rate, it seems likely that the ZFP809-binding site close to Cyp4f37 is a degenerated PBS-pro of an ancient ERV that has been inactivated by several deletions, insertions, and the loss of both LTRs. Notably, increased expression of spliced Tdrd9 and Cyp4f37 mRNAs was also detected in several other tested organs and tissues of Zfp809GT/GT mice (Fig. 7C). Altogether, these data demonstrate that a few ancient retroviral integrations altered gene expression patterns in mice through ZFP809 recruitment.
Discussion
Epigenetic silencing of ERVs and other transposable elements requires a highly specific targeting system that reliably detects and represses these genomic parasites without disturbing overall gene expression patterns. Here, we demonstrated that ZFP809 is able to bind a large number of genomic targets in pluripotent cells, yet heterochromatin formation occurs at only the highest-affinity binding sites found exclusively at ERV-associated PBS-pro sequences. These findings are in accordance with previously published in vitro binding assays (Wolf and Goff 2009) and highlight the accurate target specificity of KRAB-ZFPs, facilitated by their use of multiple zinc finger domains. Furthermore, we demonstrated that overexpression of a Flag-tagged KRAB-ZFP is a valid approach to identify KRAB-ZFP binding sites within ERVs. However, we also showed that unnaturally high transgene expression levels may lead to KRAB-ZFP binding at loci that are not bound by endogenous KRAB-ZFPs when expressed at physiological levels. Additional experimental analysis such as in vitro binding assays and confirmatory ChIP analysis with antibodies raised against the endogenous KRAB-ZFP are therefore required to differentiate between true binding sites and artifacts that result from KRAB-ZFP overexpression.
Several lines of evidence support the hypothesis that ZFP809 is essential for the establishment of ERV silencing at a critical window of time near implantation: (1) ZFP809 protein levels are significantly higher in ESCs (derived from blastocyst stage embryos) relative to differentiated cells such as MEFs, (2) ZFP809 is bound to target ERVs in ESCs, (3) ZFP809 depletion in ESCs leads to a loss of H3K9me3, and (4) ERV reactivation phenotypes are observed in all somatic tissues as well as ESCs differentiated in vitro. Furthermore, the majority of our own and previously published findings argue against an active role of ZFP809 in differentiated cell types such as MEFs: (1) ZFP809 is unable to silence exogenous retroviruses in MEFs, (2) ZFP809 and KAP1 protein levels are dramatically reduced in MEFs compared with ESCs, (3) conditional ZFP809 or KAP1 knockout in MEFs has little effect on ERV expression (Rowe et al. 2010), and (4) phenotypes in Zfp809 mutant MEFs cannot be rescued by re-expression of ZFP809.
However, it has been shown that repression of retroviral vectors containing the PBS-pro sequence is not restricted to ESCs and ECCs but is also evident in murine hematopoietic stem cells (Haas et al. 2003) and ZFP809 expressing human embryonic kidney cells (Wolf and Goff 2009). Also, conditional KAP1 knockout in neural progenitor cells has recently been shown to result in ERV reactivation (Fasching et al. 2015), indicating that ERV repression by KRAB-ZFPs and KAP1 plays a role in several somatic cell types. Furthermore, our data demonstrate that SETDB1 and H3K9me3 are required to maintain epigenetic repression of certain ERVs in MEFs, possibly independent of KAP1 and KRAB-ZFPs.
Whereas both KAP1 and SETDB1 knockout leads to early embryonic lethality (Matsui et al. 2010; Rowe et al. 2010), Zfp809 mutant mice develop normally and show no signs of disease. These mouse lines therefore represent valuable models in which ERV repression and reactivation can be investigated in any tissue of interest. VL30 derepression in Trim24 knockout hepatocytes has been previously shown to be accompanied by antiviral interferon responses and deregulation of neighboring genes via VL30 LTR activation (Herquel et al. 2013). Further studies will be required to assess whether the elevated expression of VL30-pro elements in Zfp809 knockout mice causes additional phenotypes under certain circumstances, possibly in response to stress or viral infection.
Interestingly, testis was the only adult organ in which no VL30-pro activation was observed in Zfp809 mutant mice. This might be due to the unique chromatin structure in sperm cells in which the majority of histones is replaced by protamines that are unlikely to be modified by the ZFP809/KAP1/SETDB1 complex. Previously, high-copy retrotransposons such as LINEs and IAP ERVs have been shown to be repressed by the piRNA pathway in testis (Aravin et al. 2007), suggesting that piRNAs and not KRAB-ZFPs are the dominant transposon repressors in sperm cells. However, piRNA-dependent VL30 repression has not been reported to date.
Previous reports have demonstrated that ERVs can exert repressive effects spreading over large distances due to recognition by KRAB/KAP1 complexes (Groner et al. 2010; Rowe et al. 2013b). Here we show that a handful of PBS-pro-containing ERVs have caused ZFP809-mediated transcriptional repression of nearby genes in mice. It is tempting to speculate that PBS-pro virus integration and the associated ZFP809-dependent silencing of such nonviral genes may have played an important role throughout Muroidea evolution. What are less in doubt are the recent findings that ERVs have continually rewired gene expression networks in mammals by providing platforms for DNA-binding transcriptional activators (Macfarlan et al. 2011, 2012; Chuong et al. 2013; Gifford et al. 2013; Rowe et al. 2013b). Our study now provides an example of a negative regulatory sequence carried by an ERV, the PBS-pro, which can directly repress nearby genes.
Despite the finding that KRAB/KAP1 recognition of newly integrated ERVs can lead to the emergence of new gene repression programs, our data also provide evidence that KRAB-ZFP-binding sites are unlikely to arise by chance. We found no evidence for silencing of PBS-pro-like motifs that were not associated with an ERV despite the fact that these sites were weakly bound by ZFP809. Altogether, our data suggest that ZFP809 has evolved to exclusively silence a subset of exogenous and endogenous retroviruses and by proxy a few genes closely associated with viral integrations, which is facilitated by highly specific tandem zinc fingers and fine-tuned protein expression levels that prevent recruitment of silencing machinery to imperfect target sites.
Our evolutionary analysis also revealed that the ZFP809 DNA-binding specificity is remarkably conserved considering the rate of germline colonizations by newly emerging ERV groups and rapidly evolving infectious exogenous retroviruses. It has been proposed that ERV-repressing KRAB-ZFPs evolve in response to invading ERVs, as indicated by a correlation between numbers of ERV LTRs and KRAB-ZFPs in mammalian species (Thomas and Schneider 2011). An ancient germline colonization event >25 million years ago, before or at the base of the Muroidea radiation, might have forced the evolution of the ZFP809 repressor. Since current ZFP809-targeted ERVs in M. musculus entered the mouse germline <5 million years ago, it seems likely that reoccurring waves of newly emerging infectious PBS-pro retroviruses prevented the loss of ZFP809 or a modification of its target specificity in most Muroidea species.
In conclusion, our findings add to a growing body of evidence that at least some KRAB-ZFPs, including ZFP809, Gm6871 (Castro-Diaz et al. 2014), ZNF91, ZNF93 (Jacobs et al. 2014), and Ssm1b (Ratnam et al. 2014) recognize and repress foreign DNA in mammals. Our approach of using epitope-tagged KRAB-ZFPs for ChIP-seq combined with genetic deletions of KRAB-ZFPs will be a valuable tool to identify additional ERV targeting KRAB-ZFPs and in general lead to important insights into the function of this large, rapidly evolving, and enigmatic protein family.
Materials and methods
Mouse lines
Zfp809GT mice were generated by injection of 129S2/SvPas ESCs (German Gene Trap Consortium, no. F065A07), which contain a Zfp809 allele with a gene trap vector (pT1ATG βGeo) inserted between exons 1 and 2, into B6D2F2 blastocysts. Chimeric mice were intercrossed to obtain Zfp809GT/+ and Zfp809GT/GT mice. Zfp809KO-first mice were generated by injection of C57/Bl6 ESCs harboring a conditional Zfp809 knockout allele (Knockout Mouse Project [KOMP] Repository, University of California at Davis) into C57/Bl6 blastocysts. Zfp809KO-first mice were crossed with Flp mice (Jackson Laboratory, stock no. 003946) to facilitate germline FLP-FRT recombination and the birth of Zfp809FL mice. Zfp809FL mice were crossed with mice harboring either constitutively active Cre (Jackson Laboratory, stock no. 003724) or a 4-OHT-inducible RosaCreERT2 gene (Jackson Laboratory, stock no. 004847) to generate Zfp809 knockout mice or inducible Zfp809FL/FL; RosaCreERT2 mice, respectively.
Cell line derivation and culture
Zfp809GT/+ and Zfp809GT/GT MEFs were prepared from E12.5 embryos harvested from Zfp809GT/+ mice that had been paired with Zfp809GT/GT males and cultivated in DMEM supplemented with 10% FBS. MEFs were analyzed before passage 5 or between passages 15 and 20, after MEFs had undergone growth crisis and showed stable cell growth again. Inducible Zfp809FL/FL; RosaCreERT MEFs were prepared from E12.5 embryos that were derived from Zfp809FL/+ × Zfp809FL/+; RosaCreERT2 crosses. Zfp809 knockout was induced by 4-OHT treatment (1 µM) for 72 h, and cells were analyzed after growing in normal culture medium for five more days. Zfp809+/+, Zfp809GT/+, and Zfp809GT/GT ESCs were generated from blastocysts derived from Zfp809GT/+ × Zfp809GT/GT and Zfp809GT/GT × Zfp809GT/GT pairings, respectively. These and Flag-Zfp809-expressing ESCs were grown on mitomycin C inactivated feeder cells in DMEM supplemented with 16% FBS, 104 U/mL LIF, HEPES, NEAA, L-glutamine, β-ME, and antibiotics. Embryoid bodies were derived from ESCs grown in mDiff medium (1:1 knockout DMEM:DMEM/F12 [Invitrogen], 5% KOSR [Invitrogen], 1× NEAA [Mediatech], 2 mM L-glutamine, 14.3 mM β-ME) in bacterial-grade plates under constant shaking. Retinoic acid (1 µM) was added 2 d after changing to mDiff medium. Medium was changed every other day. iPSCs were generated by lentiviral expression of OSKM using the Stem CCA system (Millipore). iPSC colonies were picked and expanded at day 14 and stained with alkaline phosphatase and OCT3/4 to confirm pluripotency. For chemical inhibition of DNA methylation and histone deacetylation, MEFs were treated with 1 µM Aza-dC for 24 h followed by 24 h of recovery in medium without Aza-dC. TSA was applied to MEFs at a concentration of 166 nM for 24 h. Double treatment was performed by growing cells in medium with Aza-dC for 24 h followed by treatment with TSA for additional 24 h in the absence of Aza-dC.
Library construction for ChIP-seq
N-terminal 3XFlag-tagged ZFP809 (1–353), a kind gift of Stephen Goff (Wolf and Goff 2009), was cloned into a Sleeping Beauty (SB) transposon-based vector (Dalsgaard et al. 2009) that was modified by removing the GFP cassette and replacing the RSV promoter of pSB/RGIP with a PGK promoter. From the resulting vector, pSB/Flag-ZFP809, Flag-tagged ZFP809, and puromycin resistance were expressed as a bicistronic mRNA. F9 ECCs were cotransfected with pSB/Flag-ZFP809 and pCMV-SB100X (a plasmid coding for the hyperactive SB100X transposase; a kind gift of Zoltán Ivics) (Xue et al. 2009) and selected with 1 µg/mL puromycin for 2 wk before analysis. Cross-linked chromatin from Flag-ZFP809-overexpressing F9 ECCs was prepared from ∼108 cells as described previously (O’Geen et al. 2010) and immunoprecipitated with 10 µg of monoclonal anti-Flag M2 antibody (Sigma-Aldrich).
Flag-ZFP809-expressing ESCs were produced using the inducible cassette exchange (ICE) system as described (Iacovino et al. 2011). Briefly, A2lox ESCs were transfected with a p2lox vector encoding for Flag-tagged Zfp809 (1–353) 24 h after treatment with 1 µg/mL doxycycline for 24 h. Cross-linked chromatin from an expanded individual G418-resistant clone was prepared from ∼108 cells and immunoprecipitated with 10 µg of monoclonal anti-Flag M2 antibody (Sigma-Aldrich) as described previously (Barish et al. 2010). Endogenous ZFP809 ChIP-seq was performed with ∼108 A2lox ESCs and 10 µg of polyclonal ZFP809 antibody (produced by injection of peptide amino acids 338–354 into rabbits). Immunoprecipitated and input control DNA was submitted for library construction using the ChIP-seq DNA sample preparation kit (Illumina) and sequenced (single-end) on an Illumina HiSeq system (O’Geen et al. 2010). A second set of ZFP809 and Flag-ZFP809 ESC ChIP-seq data (marked as #2 in Fig. 7B; Supplemental Fig. 12B) was generated with the same A2lox cell lines as described above but with a different ChIP protocol (O’Geen et al. 2010). H3K9me3 native ChIP (NChIP) was performed as descried previously (Karimi et al. 2011) using 2 × 106 Zfp809+/+ or Zfp809GT/GT ESCs and 6 µL of H3K9me3 antibody (Active Motif, no. 39161). DNA was further sonicated after ChIP to reduce fragment size to 150–500 bp, and libraries were constructed using the Library Builder fragment core kit for SOLiD 5500 and sequenced on a SOLiD sequencing system (paired-end).
Computational ChIP-seq analysis
For Illumina sequencing, ∼12 million to 20 million high-quality 50-bp single-end reads per ChIP-seq and input control were mapped to the mouse (mm9) genome using the Bowtie alignment tool (version 1.1.2) with preset parameters and Best mode turned on (-best). Peaks were called by model-based analysis of ChIP-seq (MACS) (version 1.0.1) using input control data and preset parameters. Repeat content of ChIP-seq peaks (base pairs in repeats per base pairs in peaks) was determined based on genomic overlaps with repeats annotated in the University of California at Santa Cruz (UCSC) RepeatMasker track. For further analysis, peaks were categorized into repetitive (>80% repeat content) and nonrepetitive (<20% repeat content). The Flag-ZFP809 consensus target motif was derived from the 100 top-scored nonrepetitive (<20% repeat content) ChIP-seq core peaks (200-bp region around the peak summit) using the CisFinder tool (http://lgsun.grc.nia.nih.gov/CisFinder). Remaining ChIP-seq core peaks were screened for motifs using CisFinder with settings allowing five wrong hits per 10 kb. The following previously published ChIP-seq data sets were mapped to the mm9 genome as described above and used to generate heat maps: KAP1 (Sequence Read Archive [SRA] accession: SRR611529), SETDB1 (SRA accession: SRR031683), and H3K9me3 (SRA accession: SRR611521). SOLiD ChIP-seq reads were aligned to the mm9 genome using the Novocraft Colorspace aligner using the following parameters: -r Random -i PE 200,99 -p 7,10 0.3,10.
EMSA
Nuclear extracts were prepared from ESCs as described previously (Yamauchi et al. 1995). Twenty-nine-base-pair-long double-stranded probes containing 18 bp of target sequence and an additional 3′ 11 bp, which were shown to improve repressor binding (Yamauchi et al. 1995), were end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (Fermentas) and purified using Bio-Gel P-6 purification columns (Bio-Rad), and activity was measured using a scintillation counter prior to normalization. EMSA binding reactions were carried out with nuclear extracts from ∼106 cells per reaction in modified Thornell binding buffer (25 mM HEPES at pH 7.9, 1 mM EDTA, 12.5% [v/v] glycerol, 5 mM DTT, 25 ng of poly(dI-dC) per microliter, 150 mM NaCl, 5 mM KCl, 3 mM MgCl2, 0.1 mM ZnCl2, 15% fetal calf serum, 1× protease inhibitor mix [Invitrogen]) for 30 min at 30°C and analyzed by electrophoresis on 4% native polyacrylamide gels.
RNA-seq
RNA was prepared from MEFs (p.4) and spleen samples (prepared from 8-wk-old male animals) using TRIzol reagent (Invitrogen) followed by purification and DNase treatment with RNeasy columns (Qiagen) and submitted for library construction by the TruSeq RNA sample preparation kit version 2 or TruSeq stranded mRNA sample preparation kit (Illumina), respectively, and sequencing at the BGI sequencing facility on a HiSeq instrument (Illumina). Approximately 25 million trimmed and filtered high-quality 100-bp paired-end reads per sample were mapped to the mouse genome (mm9) using the TopHat2 splice junction mapper (version 0.6) (Kim et al. 2013) with preset parameters. Repeat expression analysis was performed using BEDTools on the Galaxy platform (https://usegalaxy.org). Differential gene expression analysis was performed by counting reads overlapping with exons of RefSeq genes by BEDTools. For analysis of uniquely mappable reads, RNA-seq reads were mapped using the Bowtie aligner (version 1.1.2) under settings that suppress all alignments for which more than one reportable alignment exists (-m 1).
RT-qPCR
RNA was purified using TRizol (Invitrogen) or RNeasy columns (Qiagen), DNase-treated, and reverse-transcribed with random hexamer oligos using RevertAid H Minus first strand cDNA synthesis kit (Fermentas). RT-qPCR was performed using Platinum SYBR Green qPCR Supermix UDG (Invitrogen). The amplification efficiency of each primer pair was determined using dilution series of template cDNA. Primer sequences are shown in Supplemental Table 4.
ChIP and NChIP
ChIP with ZFP809 and KAP1 antibodies was performed using the MAGnify ChIP kit (Invitrogen) according to the manufacturer’s protocol. Chromatin from 2 × 105 cells was immunoprecipitated with 1 µg of ZFP809ab (custom-made), 1 µL of KAP1ab (Nielsen et al. 1999), and 5 µL of anti-SETDB1 serum. ChIP with SETDB1 antibody (a kind gift of F. Rauscher) was performed as described previously (O’Geen et al. 2010). ChIP with antibodies against markers for active transcription was performed using a modified protocol kindly provided by the Barish laboratory (Barish et al. 2010). Briefly, cross-linked chromatin from 1.4 × 106 cells was immunoprecipitated with 1 µg of H3K9ac (Abcam, ab4441), 1 µg of H3K4me3 (Abcam, ab8580), or 1 µg of H3K27ac (Abcam, ab4729) antibody. NChIP was performed according to a modified protocol kindly provided by M. Lorincz (Karimi et al. 2011). Briefly, ESC and MEF chromatin was digested for 9 or 12 min, respectively, to obtain predominantly mononucleosomal fragments using 150 U/mL micrococcal nuclease (Worthington Biochemicals). Per ChIP, chromatin purified from 1.25 × 106 cells was immunoprecipitated with 3 µL of anti-H3K9me3 antibody (Active Motif, no. 39161). Relative enrichment of target sequences in immunoprecipitated DNA was quantified using Platinum SYBR Green qPCR Supermix UDG (Invitrogen). For all ChIP assays, an unspecific IgG antibody was included to monitor unspecific binding and washing efficiency. Primer sequences are shown in Supplemental Table 4.
Dual-luciferase reporter assays
VL30-pro and MmERV-pro LTR sequences were amplified from genomic MEF DNA and cloned into the pGL3-basic vector by in-fusion cloning (Clontech). Primer sequences are shown in Supplemental Table 4. Plasmid DNA was prepared without prior subcloning to obtain a mixed population of genomic VL30-pro or MmERV-pro LTR elements. Reporter plasmids were cotransfected using Lipofectamine 2000 (Invitrogen) together with the Renilla luciferase-expressing pRL vector for internal normalization. Luciferase activity was measured 2 d after transfection using the dual-luciferase reporter assay system (Promega). For analysis of CpG methylation dependence of LTR activity, a single VL30-pro LTR was cloned into the CpG-free pCpGL-basic vector (a kind gift of Michael Rehli), methylated with the CpG methyltransferase M.SssI (New England Biolabs), and subsequently purified with a PCR purification kit (Qiagen). Unmethylated control plasmids were treated identically but in the absence of M.SssI.
Viral titer assays
Retroviral particles were produced in PlatE packaging cells (Morita et al. 2000) transiently transfected with Akv-MLV-derived retroviral vectors pPBS-pro and pPBS-B2 (Modin et al. 2000), of which the neomycin resistance cassette had been replaced with a puromycin gene. Target cells were transduced with dilution series of viral particles in the presence of 5 µg/mL polybrene and grown under 1 µg/mL puromycin selection until single resistant colonies became visible. Viral titers were calculated as colonies per milliliter of viral supernatant.
Bisulfite sequencing and qMSP
Genomic DNA was purified from MEFs and bisulfite converted using the Cells-to-CpG bisulfite conversion kit (Invitrogen). PCR products were amplified using TrueStart polymerase (Thermo Scientific) and subcloned into Topo plasmids for sequencing. CpG methylation was analyzed using the QUMA tool (http://quma.cdb.riken.jp). qMSP was performed using specific primer pairs (Supplemental Table 4) and PerfeCTa SYBR Green FastMix (Quanta Biosciences) qPCR reaction mix. A dilution series of plasmids containing sequences corresponding to fully methylated or unmethylated VL30-pro elements was used as a reference to determine the ratio of methylated to unmethylated sequences.
Genomic PCR screening of wild Mus species
Genomic M. musculus and Mus dunni DNA was extracted from NIH/3T3 cells or immortalized M dunni tail fibroblasts, respectively. The following DNA samples from wild Mus species were purchased from The Jackson Laboratory: Mus caroli/EiJ, M. pahari/Ei, PANCEVO/Ei (Mus spicilegus), and SPRET/Ei (Mus spretus). The Zfp809 DNA-binding domain was amplified with Taq-polymerase (Invitrogen) and sequenced without subcloning. Primer sequences are shown in Supplemental Table 4. Synonymous/nonsynonymous mutation rates were calculated using MEGA5 (Tamura et al. 2011). ERV copy numbers in the genomes of wild Mus species were determined by qPCR using DNA samples as described above. PCR products amplified from NIH/3T3 DNA were cloned, sequenced, and used to spike Peromyscus DNA (Jackson Laboratories) in equimolar dilution series as reference. Primers amplifying a conserved Zfp809 sequence were used for normalization.
Phylogenetic ERV analysis
MmERV and VL30 ERVs were identified by BLAST screening of the mouse genome for intact PBS-pro or PBS-gly sequences. Flanking regions of PBS sequences were screened for ERV sequences as in the UCSC RepeatMasker track in order to categorize elements. Full-length and nearly intact ERVs were then aligned with muscle aligner and used to create neighbor-joining trees using the p-distance model in MEGA5. Alignment regions with gaps in single or multiple sequences were not considered for creating the phylogenetic tree.
Data access
All sequencing data have been submitted to the SRA database (http://www.ncbi.nlm.nih.gov/sra) under the accession number SRP040019.
Supplementary Material
Acknowledgments
We thank members of the Macfarlan and Pedersen laboratories for useful discussions, and Lone Højgaard, Alex Grinberg, Shery Ralls, Lisa Williams-Simons, and Victoria Carter for technical assistance. We thank the Molecular Genomics Laboratory of the Eunice Kennedy Shriver National Institute of Child Health and Human Development for SOLiD sequencing and computational support (Steve Coon, Alan Pang, and James Iben). We thank Daniel Wolf and Stephen Goff for ZFP809 antibodies, and Yoichi Shinkai and Matthew Lorincz for Setdb1 conditional knockout MEFs. This work was supported by The Danish Council for Independent Research/Natural Sciences, The Lundbeck Foundation, The Danish Genetically Modified Animal Resource (DAGMAR), and the Danish Research School for Gene Medicine. National Institutes of Health grants to Velocigene at Regeneron, Inc. (U01HG004085) and the CSD (CHORI [Children's Hospital Oakland Research Institute], the Wellcome Trust Sanger Institute, and the University of California at Davis) Consortium (U01HG004080) funded the generation of gene-targeted ESCs in the KOMP Program, which are archived and distributed by the KOMP Repository at the University of California at Davis and CHORI (U42RR024244). T.S.M. is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development DIR grant HD008933.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.252767.114.
References
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. [DOI] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. 2010. Bcl-6 and NF-κB cistromes mediate opposing regulation of the innate immune response. Genes Dev 24: 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barklis E, Mulligan RC, Jaenisch R. 1986. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell 47: 391–399. [DOI] [PubMed] [Google Scholar]
- Baylac-Kalabokias H, Astier-Gin T, Moynet D, Hernould M, Mamoun R, Legrand E, Guillemain B. 1991. A new leukemogenic retrovirus isolated from tumor cells derived from a radio-induced lymphoma of C57BL/6 mice: analysis of the env and LTR sequences. Virus Res 18: 117–134. [DOI] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. 2014. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28: 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. 2013. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet 45: 325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsinotti A, Kapopoulou A, Gubelmann C, Imbeault M, Santoni de Sio FR, Rowe HM, Mouscaz Y, Deplancke B, Trono D. 2013. Global and stage specific patterns of Kruppel-associated-box zinc finger protein gene expression in murine early embryonic cells. PLoS ONE 8: e56721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalsgaard T, Moldt B, Sharma N, Wolf G, Schmitz A, Pedersen FS, Mikkelsen JG. 2009. Shielding of sleeping beauty DNA transposon-delivered transgene cassettes by heterologous insulators in early embryonal cells. Mol Ther 17: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching L, Kapopoulou A, Sachdeva R, Petri R, Jonsson ME, Manne C, Turelli P, Jern P, Cammas F, Trono D, et al. 2015. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell Reports 10: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford WD, Pfaff SL, Macfarlan TS. 2013. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol 23: 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46. [DOI] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. 2010. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet 6: e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas DL, Lutzko C, Logan AC, Cho GJ, Skelton D, Jin Yu X, Pepper KA, Kohn DB. 2003. The Moloney murine leukemia virus repressor binding site represses expression in murine and human hematopoietic stem cells. J Virol 77: 9439–9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herquel B, Ouararhni K, Martianov I, Le Gras S, Ye T, Keime C, Lerouge T, Jost B, Cammas F, Losson R, et al. 2013. Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nat Struct Mol Biol 20: 339–346. [DOI] [PubMed] [Google Scholar]
- Iacovino M, Bosnakovski D, Fey H, Rux D, Bajwa G, Mahen E, Mitanoska A, Xu Z, Kyba M. 2011. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29: 1580–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansa SA, Weksler M. 2004. Phylogeny of muroid rodents: relationships within and among major lineages as determined by IRBP gene sequences. Mol Phylogenet Evol 31: 256–276. [DOI] [PubMed] [Google Scholar]
- Jern P, Coffin JM. 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42: 709–732. [DOI] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiotis G. 2014. Endogenous retroviruses and the development of cancer. J Immunol 192: 1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Kochert K, Bouhlel MA, Richter J, Soler E et al. 2010. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med 16: 571–579. [DOI] [PubMed] [Google Scholar]
- Le Tissier P, Stoye JP, Takeuchi Y, Patience C, Weiss RA. 1997. Two sets of human-tropic pig retrovirus. Nature 389: 681–682. [DOI] [PubMed] [Google Scholar]
- Leung DC, Lorincz MC. 2012. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci 37: 127–133. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. 2011. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 25: 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. 2006. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modin C, Lund AH, Schmitz A, Duch M, Pedersen FS. 2000. Alleviation of murine leukemia virus repression in embryonic carcinoma cells by genetically engineered primer binding sites and artificial tRNA primers. Virology 278: 368–379. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Musser GG, Carleton MD. 2005. Superfamily Muroidea. In Mammal species of the world: a taxonomic and geographic reference (ed. Wilson DE, Reeder DM), pp 894–1531. Johns Hopkins University Press, Baltimore. [Google Scholar]
- Nexo BA, Christensen T, Frederiksen J, Moller-Larsen A, Oturai AB, Villesen P, Hansen B, Nissen KK, Laska MJ, Petersen TS, et al. 2011. The etiology of multiple sclerosis: genetic evidence for the involvement of the human endogenous retrovirus HERV-Fc1. PLoS ONE 6: e16652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J 18: 6385–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H, Frietze S, Farnham PJ. 2010. Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods Mol Biol 649: 437–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnam S, Engler P, Bozek G, Mao L, Podlutsky A, Austad S, Martin T, Storb U. 2014. Identification of Ssm1b, a novel modifier of DNA methylation, and its expression during mouse embryogenesis. Development 141: 2024–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D. 2011. Dynamic control of endogenous retroviruses during development. Virology 411: 273–287. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. 2013a. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140: 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. 2013b. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 23: 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S, Goff SP. 2013. Silencing of proviruses in embryonic cells: efficiency, stability and chromatin modifications. EMBO Rep 14: 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S, Meshorer E, Goff SP. 2014. Asynchronous transcriptional silencing of individual retroviral genomes in embryonic cells. Retrovirology 11: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC. 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol 26: 8623–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J. 2004. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol 53: 533–553. [DOI] [PubMed] [Google Scholar]
- Stocking C, Kozak CA. 2008. Murine endogenous retroviruses. Cell Mol Life Sci 65: 3383–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. 2011. Coevolution of retroelements and tandem zinc finger genes. Genome Res 21: 1800–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. 2014. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res 24: 1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R. 2003. KRAB-containing zinc-finger repressor proteins. Genome Biol 4: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RA. 2006. The discovery of endogenous retroviruses. Retrovirology 3: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2008. Host restriction factors blocking retroviral replication. Annu Rev Genet 42: 143–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Cammas F, Losson R, Goff SP. 2008. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J Virol 82: 4675–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Nielsen AL, Mikkelsen JG, Pedersen FS. 2013. Epigenetic marking and repression of porcine endogenous retroviruses. J Gen Virol 94: 960–970. [DOI] [PubMed] [Google Scholar]
- Xue X, Huang X, Nodland SE, Mates L, Ma L, Izsvak Z, Ivics Z, LeBien TW, McIvor RS, Wagner JE, et al. 2009. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood 114: 1319–1330. [DOI] [PubMed] [Google Scholar]
- Yamauchi M, Freitag B, Khan C, Berwin B, Barklis E. 1995. Stem cell factor binding to retrovirus primer binding site silencers. J Virol 69: 1142–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340. [DOI] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. 2009. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 23: 2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.