S-phase cyclin-dependent kinases (CDKs) stimulate replication initiation and accelerate progression through the replication timing program, and CDK phosphorylation of the replication factor TICRR/TRESLIN is required for DNA replication. Sansam et al. show that a TICRR mutant with phosphomimetic mutations at two key CDK-phosphorylated residues (TICRRTESE) stimulates DNA synthesis and shortens S phase by increasing replication initiation. In contrast to CDK hyperactivation, the acceleration of S-phase progression by TICRRTESE does not induce DNA damage.
Keywords: DNA replication, cyclin-dependent kinase, TICRR, replication initiation, replication timing, S phase, cell cycle
Abstract
S-phase cyclin-dependent kinases (CDKs) stimulate replication initiation and accelerate progression through the replication timing program, but it is unknown which CDK substrates are responsible for these effects. CDK phosphorylation of the replication factor TICRR (TopBP1-interacting checkpoint and replication regulator)/TRESLIN is required for DNA replication. We show here that phosphorylated TICRR is limiting for S-phase progression. Overexpression of a TICRR mutant with phosphomimetic mutations at two key CDK-phosphorylated residues (TICRRTESE) stimulates DNA synthesis and shortens S phase by increasing replication initiation. This effect requires the TICRR region that is necessary for its interaction with MDM two-binding protein. Expression of TICRRTESE does not grossly alter the spatial organization of replication forks in the nucleus but does increase replication clusters and the number of replication forks within each cluster. In contrast to CDK hyperactivation, the acceleration of S-phase progression by TICRRTESE does not induce DNA damage. These results show that CDK can stimulate initiation and compress the replication timing program by phosphorylating a single protein, suggesting a simple mechanism by which S-phase length is controlled.
S-phase length in metazoans can vary by two orders of magnitude, but the mechanisms that determine the duration of S phase and the consequences of deregulating the kinetics of S-phase progression are not fully understood. Extreme variation in S-phase length is best described in egg-laying animals, but mammals also show changes in the duration of S phase during embryonic development. In Xenopus laevis and Drosophila melanogaster, life begins with a series of rapid cell cycles with S phases lasting <20 min (Blumenthal et al. 1974; Satoh 1977). In contrast, somatic cells in those species take as long as 10 h to complete S phase (Dolfini et al. 1970; Uzbekov et al. 1998). In mammals, S phase during gastrulation is as short as 2 h, whereas S phase in rapidly dividing mammalian cells later in development is ∼7–8 h long (Mac Auley et al. 1993; Alexiades and Cepko 1996). The function of prolonging S phase more than its shortest possible duration is unknown. Oncogenic mutations that abnormally accelerate DNA replication typically cause DNA damage during S phase, suggesting that the speed of S-phase progression is a hardwired property of a cell (Di Micco et al. 2006). The mechanisms that govern the speed of S-phase progression and the factors that limit that speed have implications for both embryonic development and tumorigenesis.
The genome is replicated through the concerted activity of tens of thousands of replication forks, and the rate of S-phase progression is greatly affected by the overall number of forks that initiate. Potential sites for replication initiation are marked in the G1 phase of the cell cycle, when replication origins are “licensed” through the loading of two complexes of Mcm2–7 proteins (Chong et al. 1995). The Mcm2–7 complex is an essential component of the replicative helicase, but the Mcm2–7 complexes are dormant until they are activated during S phase in a process called initiation (Li and Araki 2013). Theoretically, the maximum number of initiation events is set by the quantity of origins licensed in G1. In reality, only a small fraction of Mcm2–7 loaded onto DNA during G1 is activated, so the level of initiation plays a key role in determining fork number (Ge et al. 2007).
Initiation is a highly regulated process that requires the S-phase kinases and numerous initiation factors. In eukaryotes, two families of S-phase kinases called DDKs (Dbf4-dependent kinases) and CDKs (cyclin-dependent kinases) are required for initiation (Tanaka and Araki 2013). The DDKs phosphorylate multiple subunits of the Mcm2–7 complex to promote activation of the helicase (Tanaka and Araki 2013). MCM phosphorylation by DDKs is highly conserved in eukaryotes, but it appears that the mechanism by which CDKs stimulate initiation in higher eukaryotes has diverged significantly (Masai et al. 2000, 2006; Cho et al. 2006; Montagnoli et al. 2006). Most of what is known about the function of CDK in initiation is from studies in yeast. In Saccharomyces cerevisiae, the S-phase CDKs (S-CDKs) phosphorylate two proteins called Sld2 and Sld3 (synthetically lethal with Dpb11-1), and phosphorylation of these proteins promotes their physical interactions with a scaffolding protein called Dpb11 (Masumoto et al. 2002; Tak et al. 2006; Tanaka et al. 2007; Zegerman and Diffley 2007). Phosphorylation of the Sld2 and Sld3 proteins is both necessary and sufficient for initiation, as mutations that bypass the requirement for CDK to promote the Sld2/3–Dpb11 interactions also largely bypass the requirement for CDK in initiation (Tanaka et al. 2007; Zegerman and Diffley 2007). The biochemical function of Sld2 and Sld3 in initiation is still unclear, and although CDKs are also essential for initiation in higher eukaryotes, the function of CDK in initiation in metazoans is not well understood.
Given the importance of Sld2 and Sld3 in regulating initiation in yeast, substantial effort has been made to identify their homologs in metazoans. Sld2 is homologous to the N terminus of a vertebrate RecQ-like DNA helicase (RecQL4) (Sangrithi et al. 2005; Matsuno et al. 2006). Like Sld2, RecQL4 is required for initiation and physically interacts with TopBP1, which is the vertebrate homolog of Dpb11. The RecQL4–TopBP1 interaction is not dependent on CDK phosphorylation though, so the key functions of CDK in initiation are still not entirely known (Matsuno et al. 2006). Several metazoan proteins have been proposed to be analogs or homologs of Sld3 (Fu and Walter 2010). Recently, a protein called TICRR (TopBP1-interacting checkpoint and replication regulator)/TRESLIN was discovered in a genetic screen in zebrafish and through screening for novel TopBP1 interactors (Kumagai et al. 2010; Sansam et al. 2010). TICRR is required for replication initiation, is phosphorylated by CDK, and has weak sequence similarity to Sld3 (Sanchez-Pulido et al. 2010; Sansam et al. 2010). Like Sld3, phosphorylation of TICRR on two residues stimulates its interaction with TopBP1, and mutation of those two residues abrogates replication initiation in human cells (Boos et al. 2011; Kumagai et al. 2011). Although TICRR phosphorylation by CDK is essential for replication initiation, CDK phosphorylates many proteins, so whether TICRR is a key rate-limiting substrate for initiation is unknown (Chi et al. 2008; Pagliuca et al. 2011).
In addition to driving initiation at individual replication origins, CDK also regulates the spatiotemporal pattern of replication in the nucleus. Replication forks are spatially clustered in the nucleus into thousands of replication foci or “factories.” In somatic cells, replication factories form throughout S phase according to a stereotypical spatiotemporal pattern. Euchromatin replicates early in foci spread throughout the interior of the nucleus, whereas heterochromatin replicates later, beginning at the periphery of the nucleus and the nucleoli and then finishing in large patches in the interior of the nucleus. Although it is known that the replication timing program reflects delayed initiation of late replicating sequences, it is unknown how the replication timing program is executed. Using an in vitro system in which replication of permeabilized CHO nuclei is driven by CDK and initiation factors from X. laevis egg extracts, Thomson et al. (2010) showed that CDK activity is limiting for both the number of forks that initiate and the rate of progression through the timing program. Importantly, fork number could be uncoupled from timing program progression by titrating CDK activity to intermediate levels. This suggested that CDK might control the number of forks and the rate of progression through the timing program by phosphorylating distinct sets of substrates.
In this study, we investigated whether TICRR phosphorylation by CDK influences S-phase length and, if so, whether it does so by regulating fork number and/or progression through the replication timing program. To test the effect of deregulating TICRR phosphorylation, we expressed a phosphomimetic TICRR mutant in human cells. This mutant caused an overall increase in DNA synthesis by stimulating initiation. Expression of the TICRR phosphomimetic does not significantly alter the overall spatiotemporal patterns of replication but does increase the number of subnuclear replication foci and the rate of replication in each focus. The mutant also shortens S phase, demonstrating that increased TICRR phosphorylation by CDK both increases initiation and accelerates progression through the replication timing program. Unexpectedly, we found no sign of increased DNA damage in cells expressing the TICRR phosphomimetic. Taken together, these results reveal that S-phase length can be controlled by a rather simple signaling mechanism. Through phosphorylation of a single substrate, CDK can change the length of S phase by altering fork number and the rate of progression through the timing program.
Results
Expression of a phosphomimetic TICRR mutant stimulates DNA replication
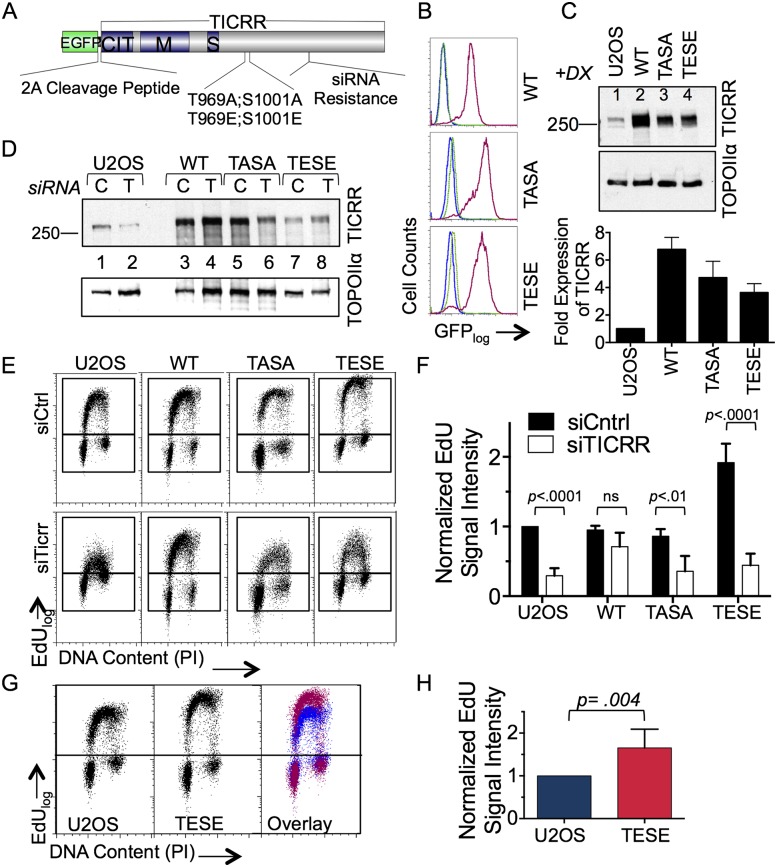
In yeast and X. laevis, homologs of TICRR are among several initiation factors that are limiting for replication initiation (Mantiero et al. 2011; Collart et al. 2013). TICRR is also phosphorylated by CDK on two residues (T969 and S1001), and CDK is limiting for S-phase progression (Boos et al. 2011; Kumagai et al. 2011). Therefore, we sought to determine the importance of TICRR expression levels and phosphorylation for S-phase length in human cells. We generated human U2OS osteosarcoma cell lines in which overexpression of wild-type TICRR, a TICRR phosphomimetic in which T969 and S1001 were mutated to glutamic acid (TICRRTESE), or a TICRR phosphomutant (TICRRTASA) could be induced with doxycycline from a single genomic locus (Fig. 1A). To provide readout of expression on a single-cell level, we fused the TICRR coding sequences to enhanced green fluorescent protein (EGFP) and the foot and mouth disease virus 2A sequence, enabling multicistronic expression of EGFP and TICRR (Fig. 1A). The 2A sequence was fused to the N terminus of TICRR, so only a single proline would be added to TICRR after cotranslational cleavage of the proteins. Clones were selected based on having a single sharp peak of EGFP expression as measured by fluorescence-activated cell sorting (FACS), and all of the clones used had similar levels of EGFP after induction (Fig. 1B). Transgene expression was further evaluated by quantitative immunoblotting (Fig. 1C). All of the transgenes used in these studies had silent mutations in an siRNA-binding site so that the ability of the exogenous proteins to rescue endogenous TICRR knockdown could be tested.
Figure 1.
Expression of a phosphomimetic TICRR mutant stimulates DNA replication. (A) Schematic of TICRR protein. (CIT) Conserved in TICRR/Treslin; (M) middle domain; (S) SLD3/TICRR/TRESLIN domain. Mutation sites at phosphorylated residues T969 and S1001 are shown. (B) Stable transgenic lines were evaluated for EGFP expression by flow cytometry 48 h after ±doxycycline (Dox) treatment. The histogram displays blue peaks (U2OS parental), green peaks (EGFP2A-TICRR −Dox), and red peaks (EGFP2A-TICRR +Dox). (C) U2OS control (lane 1) and clones expressing Dox-induced TICRR (lanes 2–4) were analyzed by immunoblotting for TICRR and TOPOIIα. (Bottom) TICRR expression was quantified and normalized to TOPOIIα. (D) TICRR protein expression levels in U2OS control and TICRR transgenic cells transfected with control siRNA (C; lanes 1,3,5,7) or TICRR siRNA (T; lanes 2,4,6,8) were analyzed by immunoblotting. (E) EdU incorporation and DNA content following siRNA knockdown of TICRR. Cells were analyzed using flow cytometry 96 h after siRNA treatment. (F) Mean EdU-positive signal intensity of replicate data from E was quantified and normalized to U2OS. Error bars represent standard deviation of the mean. P-values were calculated using two-way ANOVA. (G) TICRRTESE expression results in increased mean EdU-positive signal intensity compared with control cells. (H) Quantification of mean EdU signal intensity data from G. Error bars represent standard deviation of the mean. P-values were calculated using the paired t-test.
We first tested whether the overexpressed wild-type TICRR could rescue endogenous TICRR knockdown. Transfection with a TICRR siRNA caused a strong reduction in endogenous TICRR protein expression, whereas the expression of siRNA-resistant TICRR in the transgenic cell lines was unaffected (Fig. 1D; Supplemental Fig. S1). Consistent with published work, wild-type TICRR rescued a DNA replication defect caused by knockdown of its endogenous counterpart, showing that this protein is functional (Fig. 1E,F; Kumagai et al. 2011). To determine whether overexpression of wild-type TICRR stimulates DNA replication, we quantified the amount of incorporated ethynyl deoxyuridine (EdU) after a 15-min pulse with the thymidine analog (Fig. 1E,F). Overexpression of wild-type TICRR did not increase EdU incorporation, so expression of TICRR is not limiting for DNA replication initiation.
CDK is limiting for replication initiation, but the pivotal CDK substrates that drive DNA replication were unknown (Beck et al. 2012; Jones et al. 2013). As phosphorylation of TICRR T969 and S1001 is known to be essential for DNA replication, we asked whether increasing the level of phosphorylated TICRR would be sufficient to stimulate replication initiation and S-phase progression (Boos et al. 2011; Kumagai et al. 2011). To answer this question, we examined whether the phosphomimetic form of TICRR (TICRRTESE) could stimulate DNA replication with or without knockdown of endogenous TICRR. When endogenous TICRR was present, TICRRTESE caused a reproducible increase in EdU incorporation (1.7-fold increase) (Fig 1E–H). Unexpectedly, the siRNA-resistant TICRRTESE transgene protein was incapable of rescuing the DNA replication defect in TICRR knockdown cells (Fig. 1E–H). We were concerned that, instead of acting as an activated form of the protein, TICRRTESE could be titrating away an inhibitor of endogenous TICRR, thereby stimulating DNA replication artificially. To test this hypothesis, we generated stable cell lines expressing only the TICRR phosphomimetic region (TICRR 954–1016 and TICRRTESE 954–1016) fused to EGFP and containing a nuclear localization sequence (Supplemental Fig. S2A,B). If TICRRTESE titrates an inhibitor of DNA replication, then truncated TICRRTESE (954–1016) should act in the same manner. However, overexpression of TICRRTESE (954–1016) did not stimulate DNA replication (Supplemental Fig. S2C). To further test the hypothesis, we generated an additional set of stable cell lines overexpressing TICRR or TICRRTESE with a smaller deletion (Supplemental Fig. S2A,B). Boos et al. (2013) showed that deletion of the TICRR M domain (TICRR ΔM) abrogates TICRR function. We found that TICRRTESE ΔM also fails to stimulate DNA replication (Supplemental Fig. S2C). These results demonstrate that TICRRTESE does not stimulate replication by sequestering an inhibitor of replication and instead acts as an activated form of the protein. Nonetheless, the failure of TICRRTESE to rescue the TICRR knockdown phenotype indicates that the protein is not fully functional and that the dephosphorylated form of the protein may also be necessary for DNA replication. However, the increase in EdU incorporation caused by the TICRRTESE transgene indicates that the levels of CDK-phosphorylated TICRR are rate-limiting for DNA replication.
We then examined whether a mutation precluding phosphorylation of T969 and S1001 in TICRR would stimulate DNA replication by testing the corresponding double-alanine mutant (Fig. 1A). As previously reported, the TICRRTASA mutant failed to rescue the DNA replication defect caused by knockdown of endogenous TICRR (Fig. 1E,F; Boos et al. 2011; Kumagai et al. 2011). Unlike TICRRTESE, TICRRTASA does not stimulate DNA replication, demonstrating that this is an effect specific to the phosphomimetic mutant (Fig. 1E,F). Importantly, the failure of wild-type or TICRRTASA to cause increased DNA replication was not due to lower expression levels of those proteins, as we quantified the TICRR levels in all three clones and found that TICRRTESE was expressed at a slightly lower level than wild-type TICRR or TICRRTASA (Fig. 1C). Overall, these results demonstrate that the amount of phosphorylated TICRR in the cell is limiting for the rate of DNA replication. This result prompted us to examine the mechanism by which TICRRTESE stimulates replication initiation and the effect of the TICRRTESE transgene on overall S-phase progression.
Fork rate is unchanged in TICRRTESE cells
TICRR is essential for replication initiation, so we hypothesized that the increased DNA synthesis in TICRRTESE cells was caused by the initiation of more forks (Kumagai et al. 2010; Sansam et al. 2010). An alternate model is that TICRRTESE increases fork rate. To test this, we used DNA fiber labeling. Newly replicated DNA was labeled by sequentially treating cells with two different thymidine analogs: iododeoxyuridine (IdU) followed by chlorodeoxyuridine (CldU). Ongoing replication tracts were identified as adjacent IdU–CldU signals on individual DNA strands. Fork rate should be proportional to the length of the IdU label in tracts from ongoing replication forks (Fig. 2A). The mean IdU tract length in U2OS cells with or without TICRRTESE expression is indistinguishable, demonstrating that TICRRTESE does not increase synthesis by increasing fork rate (Fig. 2B). We next scored our labeled DNA fibers for percent initiation events out of total labeled fibers. We found that TICRRTESE cells had a significantly higher percentage of initiation events compared with control cells (3.6% U2OS vs. 7.0% TESE; P = .0003, two-tailed t-test) (Fig. 2C). Importantly, the level of increase in initiation is consistent with the increased level of EdU incorporation that we measured by FACS (1.7-fold increase) (Fig 1E–H).
Figure 2.
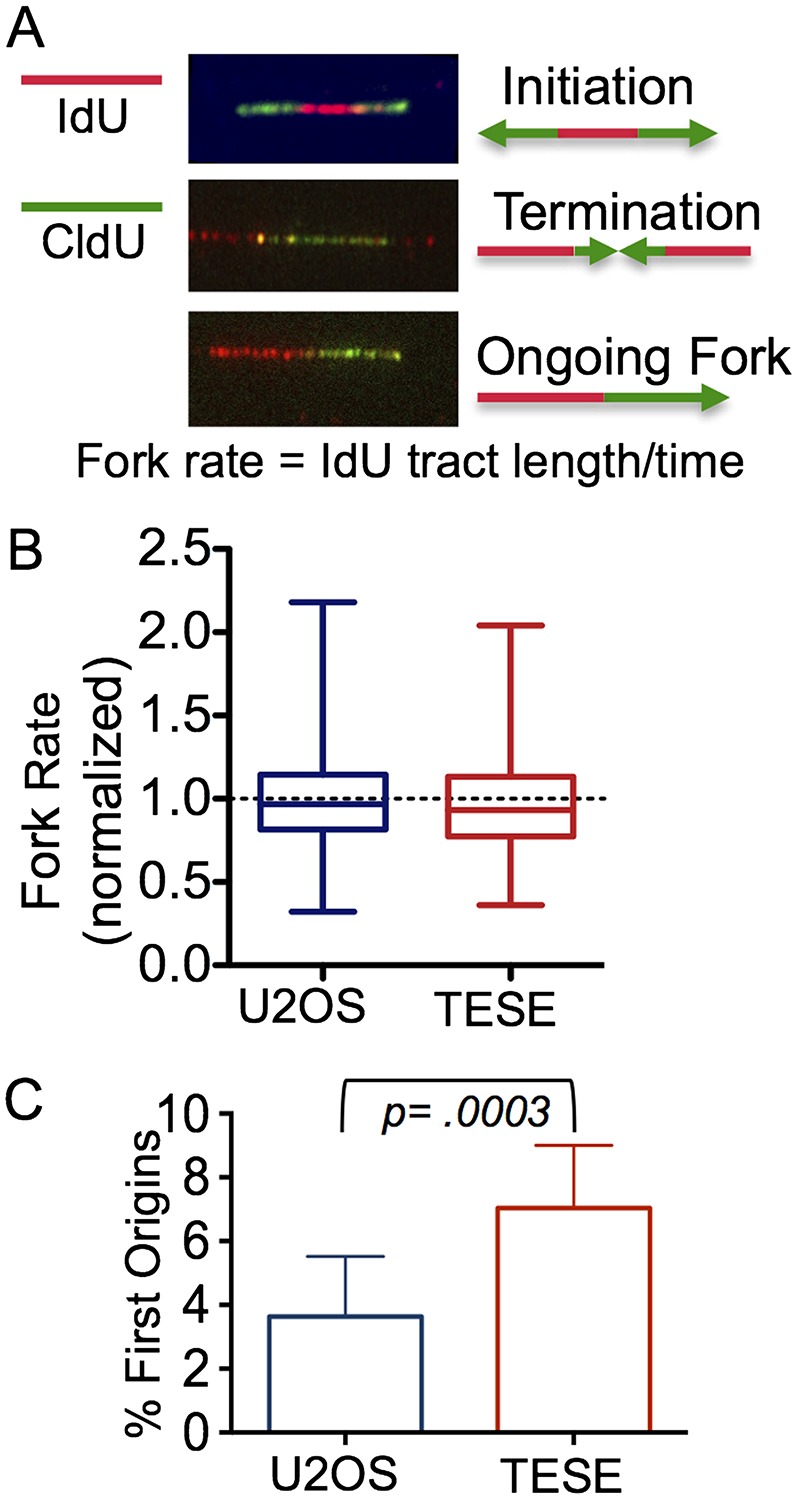
The fork rate is unchanged in TICRRTESE cells. (A) Fork rates were quantified by measuring IdU tract lengths on DNA fibers. (B) Fork speed (kilobases per minute) is normalized to U2OS. The whiskers display minimum and maximum values. (C) Initiation events were identified as adjacent CldU–IdU–CldU signals on individual DNA strands and are quantified as a percentage of all labeled structures (% First Origin). P-values were calculated using the paired t-test.
TICRRTESE increases the quantity of both replication factories and forks
Our DNA fiber labeling data are consistent with TICRRTESE stimulating replication initiation to increase the number of simultaneously active replication forks. Replication forks fire in clusters that are organized into subnuclear foci called replication factories. Most replication factories contain between five and 50 forks (Berezney et al. 2000). Little is known about what regulates the number of foci or the number of forks within each focus. CDK has been proposed to regulate foci number and fork number separately, but the CDK substrates controlling the organization of forks into foci are unknown (Thomson et al. 2010). Therefore, to understand whether CDK controls the spatiotemporal pattern of initiation through TICRR, we asked whether the number and/or intensity of EdU foci was increased in TICRRTESE-expressing cells.
First, we examined whether TICRRTESE altered the spatiotemporal pattern of replication factories. We counted asynchronously growing EdU-labeled TICRRTESE cells that have each of the five patterns characteristic of early, early/mid, mid, mid/late, or late S-phase. We found that TICRRTESE caused no difference in the proportions of cells with each of these patterns (Fig. 3A). We then asked whether TICRRTESE caused a change in the intensity or number of replication foci. EdU-labeled nuclei were imaged using confocal microscopy, and the mean fluorescence intensity of individual EdU-labeled foci was quantified. Consistent with TICRRTESE increasing the rate of DNA replication, we found that the intensity of individual foci was higher in cells expressing TICRRTESE (1.4-fold) compared with control U2OS or wild-type TICRR-expressing cells (1.0), indicating that there is a higher number of replication forks clustered within each focus (Fig. 3B). We then counted the number of foci and found that the average number of foci per nucleus was greater for TICRRTESE cells (459) than for U2OS (388) or wild-type TICRR (393) cells (Fig. 3C). These results indicate that altering the level of phosphorylated TICRR does not affect the higher-order nuclear organization of replication factories. In contrast, phosphomimetic TICRR stimulates both factory activation and fork initiation within a factory.
Figure 3.
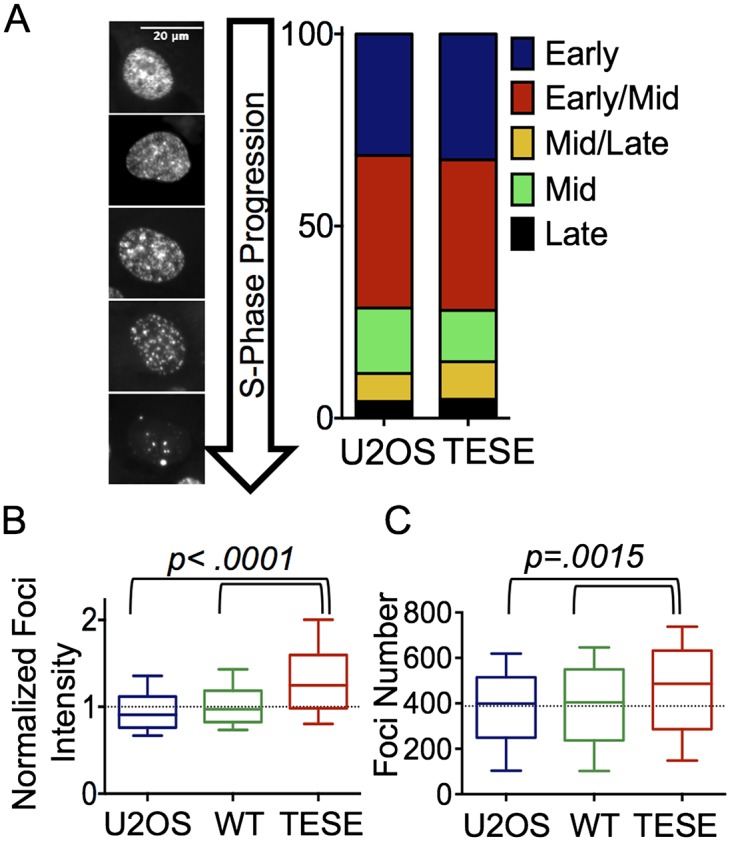
TICRRTESE cells have more sites of DNA replication and increased DNA replication within each site. Asynchronous U2OS, wild-type (WT) TICRR, or TICRRTESE-expressing cells were pulse-labeled with EdU for 15 min and stained using click chemistry on slides to identify replication foci. Confocal images of at least 500 individual cells for each type were analyzed for replication pattern (A), mean focus intensity normalized to U2OS (B), and average focus number per nucleus (C). The box-and-whisker plot displays minimum to maximum data. P-values were calculated using ANOVA.
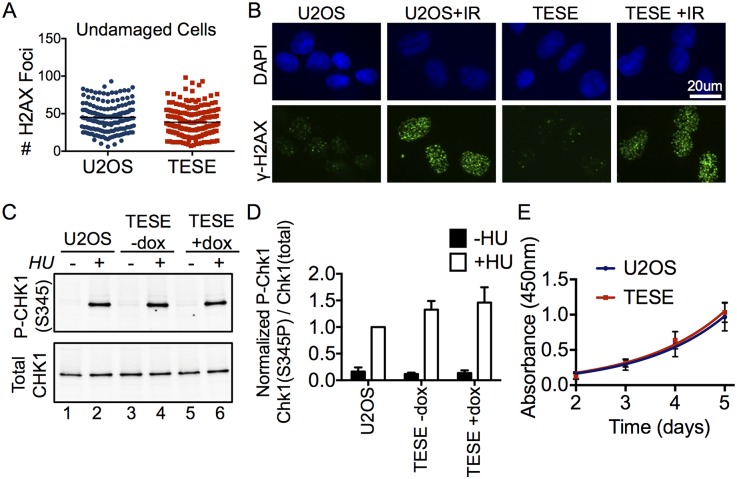
TICRRTESE does not cause DNA damage
Broad deregulation of CDK in mammalian cells generally causes DNA damage that impedes S-phase progression. One mechanism by which CDK causes DNA damage is through increased DNA replication initiation, so we predicted that TICRRTESE would also cause DNA damage. To determine whether TICRRTESE-expressing cells showed signs of DNA damage checkpoint activation, we first counted the number of foci of S139-phosphorylated Histone H2AX (γH2AX), which is a marker of dsDNA breaks. In the absence of any exogenous damage, both U2OS and TICRRTESE-expressing cells showed a very low level of γH2AX foci, indicating that TICRRTESE expression does not cause increased DNA damage or replication stress (Fig. 4A). We and others have previously shown that TICRR loss causes a DNA damage checkpoint defect, so we wanted to test whether the failure to see increased H2AX phosphorylation was due to TICRRTESE interfering with the checkpoint response (Sansam et al. 2010; Hassan et al. 2013). Exposure of U2OS cells with or without TICRRTESE to ionizing radiation (IR) to cause dsDNA breaks resulted in a similar increase in γH2AX foci, indicating that TICRRTESE does not interfere with H2AX phosphorylation in response to DNA damage (Fig. 4B). We then examined the phosphorylation of Chk1 on Ser345, a second marker for activation of checkpoint responses to DNA damage or replication stress (Fig. 4C,D). The amount of Chk1(S345) phosphorylation was barely detectable in U2OS cells with or without TICRRTESE. Replication stress triggered by hydroxyurea treatment reproducibly increased Chk1(S345) phosphorylation in TICRRTESE cells, demonstrating that Chk1 phosphorylation was not impaired by TICRRTESE expression (Fig. 4C,D). In fact, TICRRTESE caused a modest increase in the amount of Chk1 phosphorylation upon hydroxyurea exposure, possibly because the increased number of replication forks generates a stronger checkpoint response (Fig. 4D). Consistent with no observed DNA damage checkpoint activation, the viability and proliferation rate of TICRRTESE cells over 5 d did not differ from U2OS cells without the transgene (Fig. 4E). The lack of DNA damage and normal proliferation of TICRRTESE cells is in stark contrast to damage and impaired cell cycle progression caused by oncogenes and general S-CDK deregulation (Bartkova et al. 2005; Di Micco et al. 2006; Tane and Chibazakura 2009; Beck et al. 2012; Jones et al. 2013).
Figure 4.
Expression of TICRRTESE does not lead to DNA damage and does not impair checkpoint function. (A) Unperturbed U2OS and TICRRTESE cells were stained for phosphorylated H2AX (S139). The total number of individual foci in each cell was counted (U2OS mean = 45; TESE mean = 39). (B) H2AX phosphorylation in both U2OS and TICRRTESE were analyzed following exposure to 2.5 Gy of ionizing radiation. (C) TICRRTESE cells respond to replication stress by inducing phosphorylated Chk1 (S345). Immunoblot of phospho-Chk1 (S345) and total Chk1 from U2OS (lanes 1,2), uninduced TICRRTESE (lanes 3,4), and induced TICRRTESE (lanes 5,6) cells with or without treatment with hydroxyurea. (D) Graph of replicate experiments from C. Error bars represent standard deviation. (E) U2OS and TICRRTESE cells were plated at low density and monitored daily for proliferation using the WST-1 reagent.
TICRRTESE shortens S phase
The effect of increasing CDK activity on S-phase length has been difficult to study in mammalian cells because the DNA damage caused by increasing CDK activity impairs S-phase progression. We showed that TICRRTESE stimulates replication without causing DNA damage, suggesting that the level of phosphorylated TICRR might be a key determinant of S-phase length. In fact, one explanation for the higher quantity of replication factories in TICRRTESE-expressing cells could be that TICRRTESE is shortening S phase by compressing the spatiotemporal replication program. Given that CDK has been shown to increase both the number of origins that fire and the rate at which cells progress through the replication timing program in vitro, we hypothesized that both of these aspects of S-phase control occur through TICRR phosphorylation (Thomson et al. 2010). To test this hypothesis, we measured the rate of S-phase entry and exit in TICRRTESE-expressing cells. If TICRR phosphorylation is the mechanism by which CDK controls both fork initiation and progression through the timing program, then TICRRTESE should shorten S phase. Alternatively, if CDK controls progression through the timing program via other substrates, then the timing of late origin firing would not be advanced; thus, TICRRTESE would have minimal effect on S-phase length.
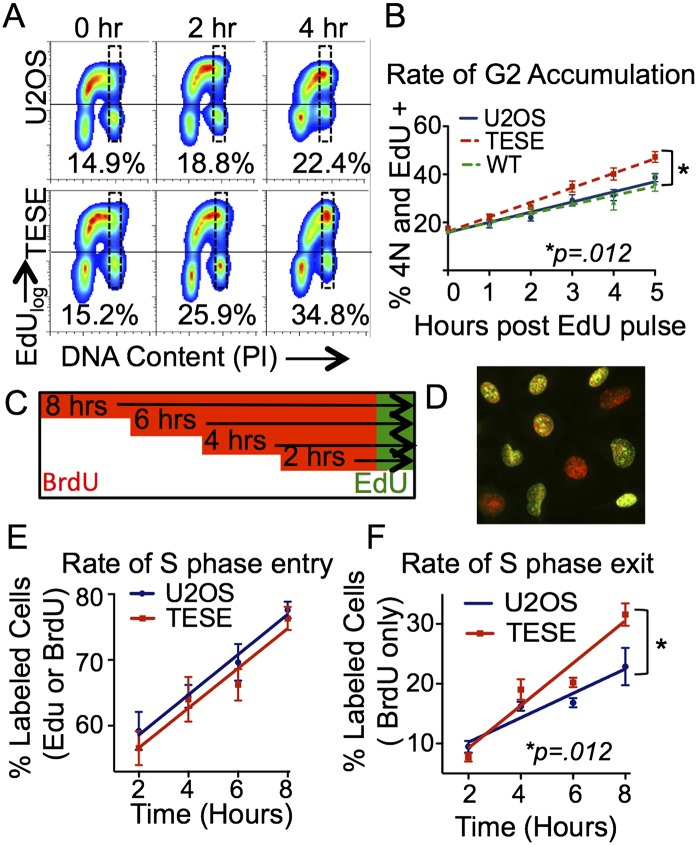
Consistent with cells progressing more rapidly through S phase, we observed an overall decrease in the percentage of cells in S phase and a corresponding increase in the percentage of cells in G1 (Supplemental Fig. S3). To more accurately measure the effect of TICRRTESE on the duration of S phase, we first monitored the progression of EdU-labeled cells through S phase and into G2 using flow cytometry. Asynchronously growing U2OS cells with or without TICRRTESE were pulse-labeled with EdU for 15 min and then incubated for up to 5 h before FACS analysis (Fig. 5A). The majority of EdU-positive cells reached 4N DNA content by 5 h following the EdU pulse, and the rate at which TICRRTESE-expressing cells accumulated 4N DNA appeared faster than for U2OS cells. To quantify this effect, we measured the percentage of EdU-positive cells that were within a narrow 4N DNA content gate at each time point. Five hours after the pulse, 47% of TICRRTESE cells were within the 4N gate, whereas only 40% of the U2OS cells were scored as 4N. Importantly, the rate at which EdU-labeled TICRRTESE cells became 4N was 1.4-fold greater than that of the U2OS cells (6.1 ± 0.46 vs. 4.3 ± 0.53; P = 0.012 ANCOVA) (Fig. 5B).
Figure 5.
TICRRTESE cells display accelerated S-phase progression. (A) U2OS, TICRRWT, and TICRRTESE cells were pulsed with EdU and collected at 1-h intervals for cell cycle analysis by FACS. EdU profiles for U2OS and TICRRWT cells at 0, 2, and 4 h following pulse. The percent of EdU-positive cells falling within a narrow 4N gate are shown. (B) Quantification of FACS data shown in A. Error bars represent standard deviation of the mean. (C) Schematic of a BrdU/EdU double-label experiment. Cells were cultured for 0–8 h in BrdU to continuously label all S-phase cells and then pulsed with EdU for 15 min prior to fixation for immunofluorescence. (D) Example of BrdU/EdU double-labeled cells. BrdU-positive cells are labeled in red; EdU-positive cells are labeled in green. Red-only cells have exited S phase, whereas green or red/green cells are in S phase. (E) The rate of S-phase entry is not affected by TICRRTESE, as measured by the accumulation of all labeled cells over time. (F) The rate of S-phase exit is accelerated in TICRRTESE cells, as measured by the percentage of BrdU-only positive cells over time. Error bars represent standard deviation. P-values were calculated using ANCOVA.
We were concerned that simply having 4N DNA content may be an inaccurate way of scoring S-phase completion, especially considering that very late replicating DNA might make up a small fraction of the genome. Thus, we took a second approach to measuring the rate of S-phase exit, in which cells were sequentially labeled with two thymidine analogs: BrdU and EdU. First, cells were continuously labeled with BrdU for up to 8 h and then were pulse-labeled with EdU for 20 min (Fig. 5C). BrdU was immunolabeled with an anti-BrdU antibody (Fig. 5D, red), whereas EdU was labeled using click chemistry with Alexa488-azide (Fig. 5D, green). The click reaction does not label BrdU, so the BrdU-only cells could be unambiguously scored as having exited S-phase between the first and second labeling periods (Fig. 5D, red-only cells). Importantly, each cell was visually scored at high magnification (40×), so cells in late S phase were not likely to be scored as G2 cells. To assess the rate of S-phase entry, we measured the change in the percentage of cells that were labeled with either BrdU or EdU over time (Fig. 5E). The rate at which cells became BrdU- or EdU-positive was the same with or without TICRRTESE expression, so TICRRTESE does not change the rate of S-phase entry. In contrast, the percentage of cells that exited S phase (BrdU-only cells) increased at a greater rate in the TICRRTESE cells than in the U2OS cells (3.549 ± 0.45 vs. 2.04 ± 0.39; P = 0.012 ANCOVA) (Fig. 5F). Based on the immunofluorescence data, we calculated that TICRRTESE shortened S phase 1.5-fold, from 18.8 h to 12.8 h. Based on both S-phase length experiments, we estimate that TICRRTESE-expressing cells exit S phase 1.4–1.7 times faster. Importantly, this is consistent with the 1.7-fold increase in EdU incorporation in TICRRTESE cells versus U2OS cells. These data indicate that fork number and the rate of timing program progression can be stimulated through increased phosphorylation and expression of a single CDK substrate.
TICRR is a key CDK substrate
To determine whether TICRR/Treslin phosphorylation is a key regulator of CDK, we challenged phosphomimetic TICRRTESE cells to acute CDK2 inhibition with 20 μM NU6102 and asked whether TICRRTESE could sustain DNA replication. Although control U2OS and overexpressing wild-type TICRR cells displayed sharp reductions in EdU incorporation within 1 h of NU6102 treatment (reduced 40% [P < 0.01, ANOVA] and 60% [P < 0.0001], respectively), TICRRTESE cells largely bypassed the requirement for CDK2 and displayed only a modest reduction in EdU incorporation (20% reduction [P = 0.028]) (Fig. 6A,B).
Figure 6.
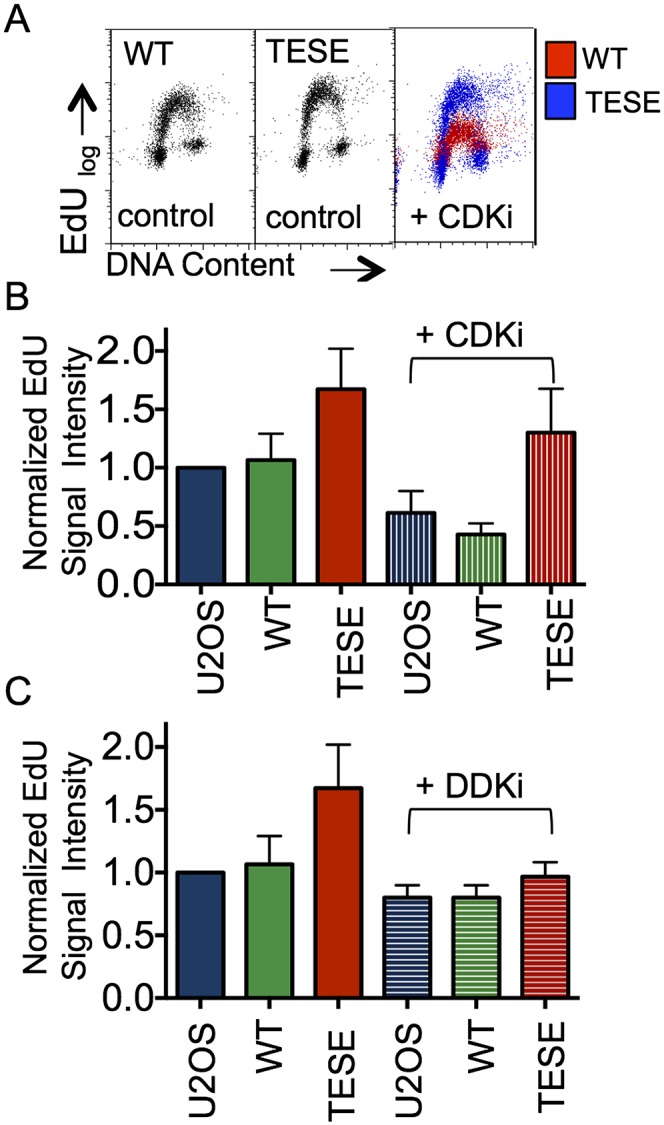
CDK2 inhibition of DNA replication is largely bypassed by TICRRTESE. (A) EdU profiles for untreated TICRRWT and TICRRTESE cells (left panels) and cells treated with the CDK2 inhibitor NU6102 (right panel, overlay). (B) Replicate data from A displayed graphically. Mean EdU-positive signal intensity was normalized to U2OS control. Error bars represent standard deviation. (C) Replicate data of EdU profiles for untreated U2OS, TICRRWT, and TICRRTESE cells and cells treated with the DDK inhibitor PHA76749. Mean EdU-positive signal intensity was normalized to U2OS control. Error bars represent standard deviation.
Replication initiation requires the activity of both CDK2 and DDK. DDK has been shown to be essential for loading Sld3 onto origins in budding yeast (Heller et al. 2011; Tanaka et al. 2011). To test whether TICRRTESE generally deregulates initiation, we treated cells with a DDK inhibitor and evaluated them for EdU incorporation. Treatment with 5 μM PHA767491 for 1 h did not significantly reduce EdU incorporation in U2OS (P = 0.92, ANOVA) or wild-type TICRR (P = 0.73) cells. In contrast, DDK inhibition reduced EdU incorporation by 40% in TICRRTESE cells (P = 0.0006, ANOVA) (Fig. 6C). These results demonstrate that TICRRTESE specifically deregulates CDK control over DNA replication initiation, supporting that TICRR is a pivotal substrate for CDK during S phase.
CDK deregulation causes hyperreplication and DNA damage through the phosphorylation of multiple substrates
Others have reported that increased S-CDK activity stimulates initiation and causes DNA damage, yet we found that TICRRTESE stimulates initiation without causing damage (Bartkova et al. 2005; Beck et al. 2012; Jones et al. 2013). We wanted to directly compare the effects of TICRRTESE with general CDK deregulation, so we used an inhibitor of the Wee1 kinase on wild-type and TICRRTESE-expressing U2OS cells. Wee1 normally restrains CDK2 activity by phosphorylating its ATP-binding domain, and inhibition of Wee1 rapidly stimulates CDK2 and causes hyperreplication (Beck et al. 2012). By using a highly specific Wee1 inhibitor (MK-1775), we acutely stimulated CDK and measured the immediate effects on DNA replication, TICRR phosphorylation, and DNA damage. Wee1 also negatively regulates mitotic CDK1, so prolonged Wee1 inhibition might deregulate mitotic entry. Importantly, we did not observe a gross increase in the number of rounded semiattached cells after 1 h of MK-1775 treatment, indicating that brief treatment with the drug does not cause high levels of premature mitotic entry in U2OS cells (data not shown). We treated U2OS cells with MK-1775 for 1 h only and measured EdU incorporation during the final 15 min. Consistent with previous reports, MK-1775 treatment rapidly increases EdU incorporation (Fig. 7A). MK-1775 also causes an increase in the electrophoretic mobility of TICRR, indicating that the protein is hyperphosphorylated (Fig. 7B). Unlike expression of phosphomimetic TICRR protein, MK-1775 treatment rapidly induces H2AX phosphorylation in S-phase cells, demonstrating that the increased replication is associated with DNA damage (Fig. 7C).
Figure 7.
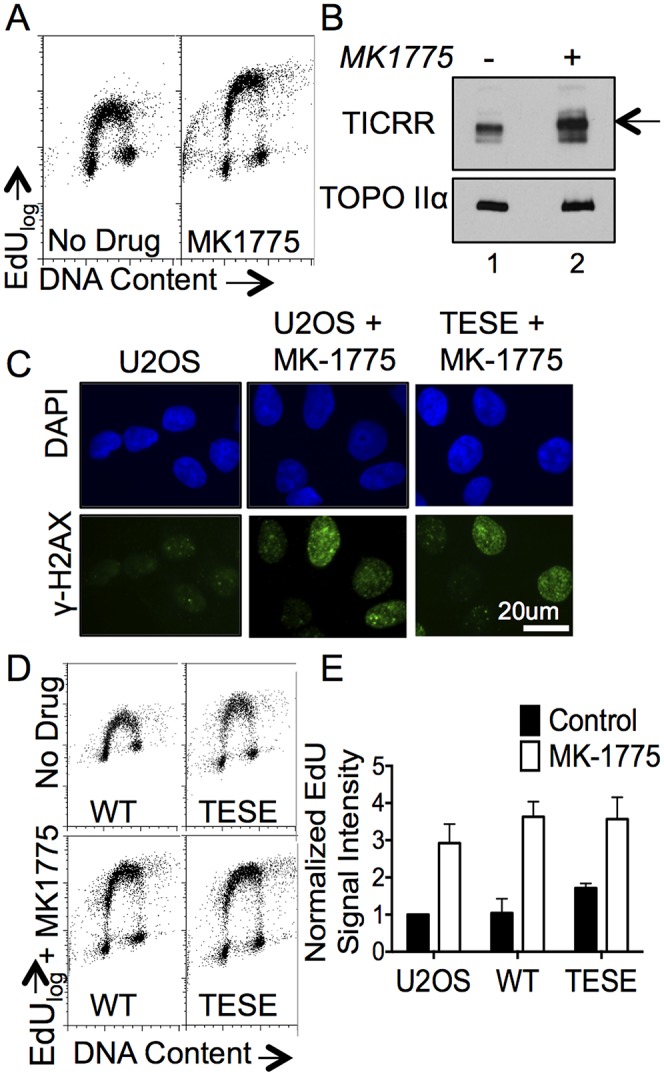
Hyperactivation of S-CDK through wee1 inhibition is associated with DNA damage. (A) EdU profiles for untreated and MK-1775-treated U2OS cells. (B) Immunoblotting of endogenous TICRR protein in untreated (lane 1) and MK-1775-treated (lane 2) U2OS cells. An electrophoretic mobility shift is indicated. (C) U2OS control, U2OS + MK-1775 (1 h), and TICRRTESE + MK-1775 (1 h) were immunostained for H2AX (S139). (D) Comparison of TICRRWT and TICRRTESE alone or following MK-1775 treatment for 1 h. EdU cell cycle profiles show that the increased EdU intensity observed in TICRRTESE can be further enhanced by the 1-h treatment with MK-1775. No difference was observed between TICRRWT and TICRRTESE cells treated with MK-1775. (E) Replicate data from D displayed graphically. Mean EdU-positive signal intensity was normalized to U2OS control. Error bars represent standard deviation.
One possibility is that the DNA damage caused by high CDK activity is due to hyperphosphorylation of multiple substrates, and deregulation of TICRR alone is insufficient to cause DNA damage. We measured the amount of EdU incorporation in cells treated with MK-1775 versus cells overexpressing wild-type TICRR or TICRRTESE. First, Wee1 inhibition caused a greater increase in EdU incorporation than that caused by TICRRTESE expression (2.9-fold and 1.7-fold, respectively) (Fig. 7D,E). This indicates that while TICRR (T969;S1001) phosphorylation is limiting for initiation, the hyperphosphorylation of additional proteins or additional TICRR residues by CDK is required for maximal DNA replication. Wild-type TICRR or TICRRTESE expression with Wee1 inhibition does not stimulate EdU incorporation more than Wee1 inhibition alone, so another factor besides TICRR may become limiting for replication upon Wee1 inhibition (Fig. 7D,E). These results show that cells are highly sensitive to S-CDK hyperactivation, but the rate of S-phase progression can be safely controlled through varying the levels of phosphorylated TICRR alone.
Discussion
Despite the central importance of CDK in DNA replication initiation, we know little about the substrates that are critical for its function. The length of S phase is affected by CDK in two ways: First, CDK activity determines the number of forks that fire, and second, CDK controls the timing of origin firing. We expressed a phosphomimetic TICRR mutant in cell culture and showed how TICRR phosphorylation is pivotal for both of these two aspects of CDK function. Remarkably, TICRRTESE expression is sufficient to stimulate DNA synthesis. DNA fiber labeling revealed that the increase in DNA synthesis is not due to an increase in fork rate, so it must be caused by an increased number of simultaneously active forks, consistent with the observed increase in initiation events. The general spatiotemporal patterns of replication foci are not grossly perturbed by TICRRTESE, but both focus intensity and number are increased. The increase in focus intensity suggests that more forks are clustered in each replication factory. The increased number of simultaneously active foci indicates that the spatiotemporal replication program is compressed. Consistent with the increase in fork number and compression of the spatiotemporal timing program, S phase is shortened in TICRRTESE-expressing cells. These effects on the replication timing program all occur without increased DNA damage. In contrast, general deregulation of CDK, through the inhibition of Wee1, causes higher levels of DNA synthesis and DNA damage. Taken together, these data suggest that TICRR is a key CDK target and that TICRR phosphorylation by itself is a mechanism by which CDK can drive the number and timing of forks without causing DNA damage and thereby vary the length of S phase.
Does TICRRTESE overexpression mimic naturally CDK-phosphorylated TICRR?
We cannot yet reconcile the ability of TICRRTESE to stimulate DNA synthesis with its inability to rescue endogenous TICRR knockdown. We speculate that TICRR has two roles in DNA replication initiation, and TICRRTESE is active in only one of those roles. Consistent with this hypothesis, yeast Sld3 functions in two separable steps. First, Sld3 binds to Cdc45 and is recruited to replication origins in a DDK-dependent manner (Kamimura et al. 2001; Heller et al. 2011; Tanaka et al. 2011). Next, CDK phosphorylates Sld3, causing it to associate with Dpb11 (Tanaka et al. 2007; Zegerman and Diffley 2007; Fukuura et al. 2011). In S. cerevisiae, if preinitiation complexes are exposed to CDK prior to DDK, DNA replication does not occur (Heller et al. 2011). It has been proposed that this requisite sequential DDK–CDK activity ensures that helicases assemble only on origins and not in solution (Heller et al. 2011). It is unknown how CDK inhibits the later activity of DDK. Possibly, phosphorylation of Sld3 by CDK prevents the DDK-dependent recruitment of Sld3–Cdc45 onto origins. If TICRR also acts in sequential DDK- and CDK-dependent steps, then the TESE mutations might mimic the effect of adding CDK prior to DDK and would be unable to load CDC45 onto origins. This would explain why TICRRTESE does not rescue TICRR knockdown. Future work is needed to determine whether TICRR is loaded in G1 by DDK and whether this loading is inhibited by prior phosphorylation of S1001 and T969 by CDK.
CDK can control factory activation and fork initiation through phosphorylation of a single substrate
Our results provide key insight into how CDK controls the rate at which cells progress through the spatiotemporal timing program. A previous study suggested that CDK might control the activation of replication foci or “factories” through a different substrate than that required to induce initiation at individual origins, yet our experiments reveal that TICRR phosphorylation controls the quantities of both replication forks and foci (Thomson et al. 2010). Thomson et al. (2010) used X. laevis extracts to drive an accelerated S phase in mammalian nuclei and showed that both the number of simultaneously active replication factories and the number of forks clustered in each factory were dependent on CDK activity levels in the extracts. Intriguingly, factory number was more sensitive to changes in CDK levels than fork number, demonstrating that these two aspects of replication control could be uncoupled. Our results show that TICRR phosphorylation controls the number of forks per focus and the overall number of foci. The fact that a single CDK substrate controls initiation at individual origins and focus number strongly suggests that fork initiation drives focus activation. Our results do not fully explain the differential sensitivity of foci and forks to CDK levels though. Thomson et al. (2010) proposed that their results could also be explained if the first forks to initiate in a focus required the highest CDK activity, possibly because a single fork changes the surrounding chromatin in a way that makes further initiation occur more easily. Our results are consistent with this hypothesis, but we cannot rule out the possibility that TICRR controls fork initiation and factory activation through distinct mechanisms.
TICRR and the control of S-phase length
Although we found that deregulation of TICRR phosphorylation by itself can significantly affect S-phase length, previous studies have suggested that S-phase length control in vertebrates is more complex. In X. laevis, a sharp extension in S-phase length occurs during early embryonic development, and, at the same time, the level of four proteins necessary for DDK and CDK signaling are reduced (Collart et al. 2013). This suggests that S-phase lengthening is caused by the reduced expression of these four factors. Consistent with this hypothesis, by increasing the expression of any three of those four initiation factors, S-phase lengthening in X. laevis embryos is prevented. In contrast, overexpression of any one of the four limiting factors has no effect on S-phase length. Three of those four factors are TICRR, RecQL4, and TopBP1. Given that TopBP1 physically interacts with TICRR and RecQL4, overexpression of these three factors is likely to drive the formation of these protein complexes. Like in X. laevis, we show that, in human cells, overexpression of wild-type TICRR by itself is insufficient to stimulate DNA replication. In contrast, the TICRRTESE phosphomimetic mutant can shorten S phase. Our results, consistent with the work of Collart et al. (2013), support a model in which S-phase length can be controlled through TICRR phosphorylation, and the important effect of TICRR phosphorylation is to promote the formation of TICRR–TopBP1 complexes (Kumagai et al. 2011). Notably, TICRRTESE overexpression can suppress the effects of CDK2 inhibition but not DDK inhibition. This suggests that DNA replication could be further stimulated by increasing DDK activity. Although we show that shortening S phase through TICRRTESE overexpression has no appreciable harmful effect on human cells in culture, this is not likely to be the case during embryonic development. Overexpression of the four limiting initiation factors in X. laevis leads to gastrulation defects and embryonic lethality (Collart et al. 2013). TICRRTESE was also overexpressed in our system, so it is still unknown whether increasing the pool of phosphorylated TICRR expressed at normal levels would have the same effect. Nonetheless, overexpression of TICRRTESE has revealed how critical phosphorylated TICRR is for controlling DNA replication initiation, and the TICRRTESE mutant will likely serve as a useful tool for studying the effects of deregulating CDK control over S-phase length during embryonic development.
Investigations of S-phase length control in early D. melanogaster embryos also show that CDK plays a central role in regulating the duration of S phase during development but possibly does so through a different mechanism. S phase is lengthened during early D. melanogaster development through a delay in the initiation of the replication of satellite sequences (Shermoen et al. 2010). Surprisingly, the onset of this delay depends on the down-regulation of CDK1, a form of CDK that is normally thought to function during mitosis (Farrell et al. 2012). The mechanism by which CDK1 can advance the replication of satellite sequences is unknown, but one possibility is that late replication origins are resistant to activation by CDK2 and are instead activated by a distinct CDK1-dependent mechanism. This hypothesis is not supported by our data, as TICRRTESE can shorten S phase. This indicates that the timing of both early and late origins is controlled through TICRR phosphorylation, which can be catalyzed by CDK2. We propose that CDK1 also works through an Sld3/TICRR homolog in flies by either priming Sld3/TICRR to be activated by CDK2 or phosphorylating that protein at the same sites as CDK1 but with higher efficiency. Indeed, CDK1 can substitute for CDK2 in replication initiation in mice, but there is no evidence that CDK1 stimulates initiation more efficiently than CDK2 (Berthet et al. 2003; Ortega et al. 2003). Unfortunately, a homolog of Sld3/TICRR has not been identified in D. melanogaster, so testing this hypothesis in flies awaits its discovery.
The existence of an intrinsic replication ‘speed limit’
Hyperactivation of CDK stimulates replication and causes DNA damage, so it was unexpected that TICRRTESE could stimulate replication without damaging DNA. There are at least two explanations for this discrepancy. First, the DNA damage caused by CDK deregulation is unlikely to be caused by increased initiation by itself. For example, hyperactivation of CDK can suppress homologous recombination repair of DNA breaks, so deregulating CDK might cause DNA damage by both stimulating replication and inhibiting repair (Krajewska et al. 2013). Of course, the level of increased replication is likely to be an important factor. Our results suggest that cells have a safe range of replication speed, and exceeding that speed limit results in DNA damage. TICRRTESE stimulates the rate of DNA replication nearly twofold and does not cause DNA damage. In contrast, Wee1 inhibition stimulates replication threefold and causes DNA damage, and the rate of replication cannot be further stimulated by additional TICRRTESE expression. We propose that TICRRTESE stimulates replication to a level that is safe for U2OS cells, whereas Wee1 inhibition exceeds that level.
The existence of an intrinsic replication “speed limit” raises several questions. First, what determines the speed limit? The limit is likely set by the abundance of certain limiting factors in the cell. One candidate for such a factor would be licensed origins. Excess licensed origins act as a safeguard against fork collapse, and even a modest (50%) reduction in licensed origins causes cells to be sensitive to replication stress (Ge et al. 2007). It is still unclear exactly how many excess licensed origins a mammalian somatic cell has, but based on estimates of Orc and MCM protein levels in mammalian cells, a threefold increase in replication initiation might deplete spare licensed origins, thereby preventing the rescue of collapsed forks (Wong et al. 2011). Another candidate for a limiting replication factor is nucleotide levels, as increasing nucleotide levels has been shown to suppress the DNA damage associated with higher than normal rates of DNA replication in mammalian cells and yeast (Mantiero et al. 2011). The U2OS cells used in this study have oncogenic mutations (such as p16 deficiency) and may have adapted to the harmful effects of hyperreplication. Thus, a second interesting question is: Do different cell types have different levels of that limiting factor and hence different replication speed limits? If so, this could have important implications for tumorigenesis, since hyperreplication-induced DNA damage is thought to be an important early step in tumor progression. Thus, the replication speed limit of a cell could be a critical factor influencing its susceptibility to oncogenic transformation.
Materials and methods
Plasmid construction
For the full-length and ΔM TICRR constructs, overlapping DNA fragments encoding EGFP, the foot and mouth disease virus 2A (TLNFDLLKLAGDVESNP) sequence, and the human TICRR cDNA (Sansam et al. 2010) were cloned into pcDNA5/FRT/TO (Invitrogen) using isothermal assembly (Gibson et al. 2009). The 954–1016 wild-type and TESE constructs were generated with isothermal assembly to combine EGFP, wild-type, or TESE synthetic 954–1016 TICRR fragments with a C-terminal SV-40 nuclear localization signal and pcDNA5/FRT/TO. Point mutations or deletions to disrupt the TICRR siRNA target sequence, alter the T969 and S1001 phosphorylation sites, or delete the M domain (amino acids 300–592) were generated using synthetic DNA constructs (G blocks) (IDT), which were inserted into pcDNA5/FRT/TO-EGFP-2A-TICRR using isothermal assembly.
Cell lines and transfections
For stable TICRR mutant lines, U2OS Flp-In TRex cells (gift of Jeffrey Parvin) were cotransfected with Flp recombinase (Invitrogen, pOG44) and TICRR constructs using TransIT-LT1 reagent (Mirus Bio). Stably integrated clones were grown in DMEM with 10% FBS and selected with 100 μg/mL hygromycin. To induce expression of TICRR constructs, 2.5 μg/mL doxycycline (Enzo) was added to the medium for 24–96 h. The TICRR siRNA (CCUGUUACGCCAAAGAAACUGUUUA) (Kumagai et al. 2010) or a control siRNA (lowGC) (Stealth siRNAs) were purchased from Life Technologies and transfected with RNAimax (Life Technologies) according to the manufacturers’ instructions.
FACS analysis
For EGFP signal evaluation, cells were harvested and resuspended in PBS. For EdU labeling experiments, cells were pulsed with 20 μM EdU for 15 min, harvested, and prepared for flow cytometry using click chemistry with Alexa647 (Life Technologies). DNA was stained with propidium iodide (PI) (Sigma). FACS acquisition was conducted on a FACSCalibur (Becton-Dickinson, BD) and analyzed by FlowJo (Tree Star, Inc.).
Immunofluorescence
For EdU label experiments, cells were pulsed with 20 μM EdU (15 min), harvested, fixed in 4% paraformaldehyde (PFA), and processed using click chemistry. Images were acquired with a Zeiss AxioObserver inverted microscope equipped with a Hamamatsu ORCA-ER camera and a Yokogawa CSU confocal scanner. EdU foci were counted in unprocessed 0.28-μm z-stacks using the FociPicker3D algorithm (Du et al. 2011). The focus intensity was calculated as the mean intensity of all pixels within a focus. Each focus intensity was normalized to the mean intensity of all foci from a match sample of parental U2OS cells. For EdU/BrdU double-label experiments, cells were pulsed with 20 μM BrdU (15 min), washed with PBS, and maintained in medium for the designated times. Cells were then pulsed with 20 μM EdU (15 min), washed, and fixed in 4% PFA. Cells were permeabilized with 0.05% Triton X-100, blocked with 1% bovine serum albumin (BSA), and treated with 2N HCl for 15 min prior to antibody labeling (BD, clone 44). EdU was detected using click chemistry (Alexa488). For H2AX, cells were incubated on ice in permeabilization buffer (0.5% Triton X-100, 20 mM HEPES at pH 7.9, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) fixed with 2% PFA for 20 min, fixed with methanol for 10 min, and blocked with 3% BSA prior to antibody labeling (Upstate Biotechnology, H2AX S139). DNA was stained with DAPI (Molecular Probes). Image acquisition was done on a Zeiss Axioplan2.
Fiber labeling
Cells were labeled for 20 min with 25 μM IdU followed by 250 μM CldU for an additional 20 min, harvested, and placed on ice. They were then washed in ice-cold PBS and resuspended at 250 cells per microliter. Labeled cells were diluted 1:8 with unlabeled cells, and 2 μL of the mixture was lysed in 5 μL of buffer (0.5% SDS, 50 mM EDTA, 200 mM Tris-HCl at pH 7.4) on the slide. DNA fibers were stretched by putting the slides at 21°–45°, ensuring that the drop reached the bottom of the slide within 5 min. Slides were air-dried, fixed in 3:1 methanol/acetic acid solution, and stored overnight. Next, slides were placed in 2.5N HCl for 80 min prior to antibody labeling. Anti-BrdU (BD) was used at 1:25 to detect IdU, and anti-BrdU (Abcam, ab6326) was used at 1:400 to detect CldU. ssDNA was counterstained using Millipore mAB3034 antibody. Slides were imaged at 63× (Zeiss).
Antibodies used for immunoblotting
Antibodies used for immunoblotting were Ticrr (Bethyl Laboratories), TopoIIα (Enzo Life Sciences), anti-phospho-Chk1(S345) (Cell Signaling), and Chk1 (Santa Cruz Biotechnology).
Selective inhibitors used in cell culture
Selective inhibitors used in cell culture were MK-1775 (Selleck Chemical) and NU6102 and PHA767491 (Santa Cruz Biotechnology).
Supplementary Material
Acknowledgments
We thank Linda Thompson, Gary Gorbsky, Susannah Rankin, Dean Dawson, and Donald Capra for helpful discussions about this work. We thank Jeffrey Parvin for the U2OS Flp-In T-Rex cells. This publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health through grants 8P20GM103447 and 5P20GM103636-02 (including imaging core support) and a Health Research Program Award from the Oklahoma Center for the Advancement of Science and Technology.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.246827.114.
References
- Alexiades MR, Cepko C. 1996. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn 205: 293–307. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870. [DOI] [PubMed] [Google Scholar]
- Beck H, Nahse-Kumpf V, Larsen MS, O’Hanlon KA, Patzke S, Holmberg C, Mejlvang J, Groth A, Nielsen O, Syljuasen RG, et al. 2012. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol 32: 4226–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484. [DOI] [PubMed] [Google Scholar]
- Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. 2003. Cdk2 knockout mice are viable. Curr Biol 13: 1775–1785. [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol 38: 205–223. [DOI] [PubMed] [Google Scholar]
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF. 2011. Regulation of DNA replication through Sld3–Dpb11 interaction is conserved from yeast to humans. Curr Biol 21: 1152–1157. [DOI] [PubMed] [Google Scholar]
- Boos D, Yekezare M, Diffley JF. 2013. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science 340: 981–984. [DOI] [PubMed] [Google Scholar]
- Chi Y, Welcker M, Hizli AA, Posakony JJ, Aebersold R, Clurman BE. 2008. Identification of CDK2 substrates in human cell lysates. Genome Biol 9: R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. 2006. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci 103: 11521–11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JP, Mahbubani HM, Khoo CY, Blow JJ. 1995. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature 375: 418–421. [DOI] [PubMed] [Google Scholar]
- Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. 2013. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 341: 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642. [DOI] [PubMed] [Google Scholar]
- Dolfini S, Courgeon AM, Tiepolo L. 1970. The cell cycle of an established line of Drosophila melanogaster cells in vitro. Experientia 26: 1020–1021. [DOI] [PubMed] [Google Scholar]
- Du G, Drexler GA, Friedland W, Greubel C, Hable V, Krucken R, Kugler A, Tonelli L, Friedl AA, Dollinger G. 2011. Spatial dynamics of DNA damage response protein foci along the ion trajectory of high-LET particles. Radiat Res 176: 706–715. [DOI] [PubMed] [Google Scholar]
- Farrell JA, Shermoen AW, Yuan K, O’Farrell PH. 2012. Embryonic onset of late replication requires Cdc25 down-regulation. Genes Dev 26: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Walter JC. 2010. DNA replication: metazoan Sld3 steps forward. Curr Biol 20: R515–R517. [DOI] [PubMed] [Google Scholar]
- Fukuura M, Nagao K, Obuse C, Takahashi TS, Nakagawa T, Masukata H. 2011. CDK promotes interactions of Sld3 and Drc1 with Cut5 for initiation of DNA replication in fission yeast. Mol Biol Cell 22: 2620–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. 2007. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Hassan BH, Lindsey-Boltz LA, Kemp MG, Sancar A. 2013. Direct role for the replication protein treslin (Ticrr) in the ATR kinase-mediated checkpoint response. J Biol Chem 288: 18903–18910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. 2011. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, Petermann E. 2013. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene 32: 3744–3753. [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H. 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J 20: 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska M, Heijink AM, Bisselink YJ, Seinstra RI, Sillje HH, de Vries EG, van Vugt MA. 2013. Forced activation of Cdk1 via wee1 inhibition impairs homologous recombination. Oncogene 32: 3001–3008. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Dunphy WG. 2010. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140: 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. 2011. Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol 193: 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Araki H. 2013. Loading and activation of DNA replicative helicases: the key step of initiation of DNA replication. Genes Cells 18: 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Auley A, Werb Z, Mirkes PE. 1993. Characterization of the unusually rapid cell cycles during rat gastrulation. Development 117: 873–883. [DOI] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P. 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K. 2000. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 bY Cdks. J Biol Chem 275: 29042–29052. [DOI] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et al. 2006. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem 281: 39249–39261. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H. 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415: 651–655. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. 2006. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase α in the initiation of DNA replication. Mol Cell Biol 26: 4843–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, Molinari A, Santocanale C. 2006. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem 281: 10281–10290. [DOI] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M. 2003. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet 35: 25–31. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Collins MO, Lichawska A, Zegerman P, Choudhary JS, Pines J. 2011. Quantitative proteomics reveals the basis for the biochemical specificity of the cell-cycle machinery. Mol Cell 43: 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Diffley JF, Ponting CP. 2010. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol 20: R509–R510. [DOI] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. 2005. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121: 887–898. [DOI] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA. 2010. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev 24: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh N. 1977. Metachronous cleavage and initiation of gastrulation in amphibian embryos. Dev Growth Differ 19: 111–117. [DOI] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O’Farrell PH. 2010. Developmental control of late replication and S phase length. Curr Biol 20: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YS, Tanaka Y, Endo S, Kamimura Y, Araki H. 2006. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2–Dpb11. EMBO J 25: 1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H. 2013. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol 5: a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. 2011. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol 21: 2055–2063. [DOI] [PubMed] [Google Scholar]
- Tane S, Chibazakura T. 2009. Cyclin A overexpression induces chromosomal double-strand breaks in mammalian cells. Cell Cycle 8: 3900–3903. [DOI] [PubMed] [Google Scholar]
- Thomson AM, Gillespie PJ, Blow JJ. 2010. Replication factory activation can be decoupled from the replication timing program by modulating Cdk levels. J Cell Biol 188: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov R, Chartrain I, Philippe M, Arlot-Bonnemains Y. 1998. Cell cycle analysis and synchronization of the Xenopus cell line XL2. Exp Cell Res 242: 60–68. [DOI] [PubMed] [Google Scholar]
- Wong PG, Winter SL, Zaika E, Cao TV, Oguz U, Koomen JM, Hamlin JL, Alexandrow MG. 2011. Cdc45 limits replicon usage from a low density of preRCs in mammalian cells. PLoS ONE 6: e17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.