Abstract
Objective: To evaluate the correlation between genetic polymorphisms in x-ray repair cross complementing group 1 (XRCC1) and sensitivity to platinum-based chemotherapy drugs in patients with non-small cell lung cancer. Methods: Reports published before June 2014 were retrieved from the following databases: China Biology Medicine (CBM), China Academic Journal Full-Text Database (CNKI), China Science and Technology Journal Full-Text Database (VIP), Wanfang Data, PubMed and Excerpta Medica dataBASE (EMBASE). After extracting the data and evaluating the quality, meta-analysis was performed using RevMan5.2 software. Results: A total of 29 studies with 4807 patients were included. Two polymorphisms (Arg399Gln and Arg194Trp) were analyzed. Meta-analysis showed that the efficacy of chemotherapy for patients with the TT genotype [TT vs. CC, OR=1.66, 95% OR=1.66, 95 CI (1.30-2.14)] and the CT genotype [CT vs. CC, OR=1.62, 95% CI (1.35-1.93)] at codon 194 of the XRCC1 gene was significantly higher than that for patients with the CC genotype. The efficacy of chemotherapy for patients with mutant (CT+TT) genotypes was significantly higher than for patients with the wild-type (CC) genotype [TT+CT vs. CC, OR=1.63; 95% CI (1.38-1.92)]. The sensitivity to chemotherapy in patients with the AG genotype at codon 399 of the XRCC1 gene was lower than in patients with the GG genotype [AG vs. GG, OR=0.72, 95% CI (0.55-0.92)] in Chinese population. However, we did not found this association in Caucasus population. Conclusion: Genetic polymorphisms in the XRCC1 gene are correlated with sensitivity to platinum-based chemotherapy in patients with non-small cell lung cancer.
Keywords: Non-small cell lung cancer, platinum-based drug, XRCC1, single-nucleotide polymorphism, meta-analysis
Introduction
Lung cancer is one of the most common malignant tumors, showing the highest mortality among cancers in males and the second-highest mortality in females [1]. Lung cancer is classified either as non-small cell lung cancer (NSCLC) or as small cell lung cancer (SCLC). NSCLC is the most common type of lung cancer, accounting for 80%-85% of lung cancer. It includes squamous cell carcinoma, adenocarcinoma, adenosquamous carcinoma, and large cell carcinoma [2]. Chemotherapy is the main treatment for advanced NSCLC, and platinum-based chemotherapy regimens are commonly used today. The platinum-based compounds can damage DNA by forming intramolecular or intermolecular DNA cross-links, thereby causing the eventual death of tumor cells [3]. However, the efficacy of platinum-based chemotherapy varies largely among different individuals, and genetic polymorphisms at the x-ray repair cross complementing group 1 (XRCC1) locus are an important basis for this variation [4]. Some studies have found that polymorphisms in codons 194 and 399 of XRCC1 can play a role in predicting the efficacy of platinum-based chemotherapy, but the results in these reports are inconsistent [5-10]. Recently published meta-analyses literatures also did not reach the same conclusion [9,10]. By considering the influence of different populations and ethnic groups on different genetic polymorphisms, this study applies the basic principles and methods of evidence-based medicine to assess previous studies related to XRCC1 genetic polymorphisms and the efficacy of platinum-based chemotherapy in advanced NSCLC cases in different ethnicity population. This work is expected to provide a basis for further research on the relationship between XRCC1 gene polymorphisms and platinum drug efficacy.
Materials and methods
The inclusion criteria were as follows: the subjects of the study were patients with primary NSCLC that was diagnosed by pathology, bronchoscopy or biopsy; the study investigated the relationship between XRCC1 gene polymorphisms and the efficacy of platinum-based chemotherapy for NSCLC; the study was a retrospective study; the platinum-based chemotherapy regimen was administered for at least two courses; the original data were already published in the literature; and the study used non-conditional logistic regression to perform corrections for confounding factors of the research subjects, such as age, gender, and smoking status. The exclusion criteria included the following: the study was on animals or lung cancer cell lines; the study examined non-primary lung cancer, such as metastatic or recurrent cancer; the study addressed small cell lung cancer; the study examined non-platinum-based chemotherapy programs or chemotherapy programs that were administered for less than two courses; the study did not focus on the relationship between genetic polymorphisms of XRCC1 and the efficacy of platinum chemotherapy for NSCLC; the study was a duplicate; and the study was an abstract or review.
Search strategy
The following databases were used: China Biology Medicine (CBM, 1978-2014), China Academic Journal Full-Text Database (CNKI, 1979-2014), China Science and Technology Journal Full-Text Database (VIP, 1989-2014), Wanfang Data, PubMed and EMBASE. Publications were jointly retrieved with “lung cancer or lung carcinoma or lung neoplasms” and “XRCC1 or X-ray cross-complementing group 1 or base excision repair or BER” and “polymorphisms or SNPs” as the searching keywords.
Quality assessment
Because of limitations imposed by the nature of the studies, quality assessment could not be achieved using the Jadad scale. Instead, quality was evaluated by two reviewers according to the following quality standards [11]: whether the experimental design was scientific; whether the basic characteristics of the subjects were clear; whether the inclusion and exclusion criteria for the subjects in the study were specified; whether the criteria of efficacy were specified; whether some cases were deleted and withdrawn, and if so, whether the number and reason were reported; whether the statistical methods were appropriate; and whether bias in the study was discussed. Meeting the above 7 standards earned 1 point each, so that a perfect score was 7 points. Publications with a score of 4 or more points were considered reliable. When disagreement occurred, the discrepancy was discussed to resolve it, or the study was submitted to a third consulting evaluator to achieve a resolution.
Statistical analysis
Statistical testing was conducted using RevMan5.2 software that was provided by the Cochrane Collaboration. According to the results of a heterogeneity test, an appropriate type of statistical model was selected. If no statistical heterogeneity was found among the studies within a group (I2<50%), a fixed-effect model was selected for analysis. Conversely, if significant heterogeneity was found (I2>50%), the sources of the heterogeneity were first analyzed and ruled out; a random-effect model was selected for analysis if the heterogeneity could not be ruled out. The difference between two categorical variables was presented using an OR (odds ratio), with a confidence interval (CI) estimate of 95%. The difference between continuous variables was presented using the weighted mean difference (WMD) with a 95% CI estimate; the results were presented in a forest plot, and a two-sided test was performed for all data. P<0.05 was considered statistically significant.
Results
General characteristics and quality assessment of the included studies
A total of 158 published studies on the relationship between XRCC1 gene polymorphisms and the efficacy of platinum-based chemotherapy were retrieved from the literature databases. Among them, 106 duplicate studies and reviews were excluded on the basis of their abstracts. After further reading the full text of each study, 23 additional studies were excluded for not meeting the inclusion criteria. As a result, 29 studies were eventually included [5-8,12-37], among which 14 were in Chinese and 15 were in English; they cumulatively included 4708 cases. The general characteristics of the included studies are shown in Table 1. The quality assessment demonstrated that the 29 included studies had scientific experimental designs, clear inclusion and exclusion criteria, rational genetic testing methods, and reasonable efficacy evaluation.
Table 1.
The characteristics of included studies
| Authors | Publication year | Ethnicity | N | Stages | Quality Scores | Arg39-9G1n | Arg19-4Trp | Genotyping methods |
|---|---|---|---|---|---|---|---|---|
| Dong et al. | 2012 | Chinese | 564 | III-IV | 5 | Yes | No | PCR-RFLP |
| Joerger et al., | 2012 | Caucasian | 131 | IIIB-IV | 7 | Yes | Yes | PCR-RFLP |
| Sullivan et al. | 2014 | Caucasian | 161 | III-IV | 7 | Yes | Yes | TaqMan |
| Zhao et al. | 2013 | Chinese | 147 | IIIB-IV | 7 | Yes | Yes | TaqMan |
| Jin ZY et al. | 2014 | Chinese | 378 | IIIB-IV | 6 | Yes | Yes | PCR-RFLP |
| Liu D et al. | 2014 | Chinese | 378 | IIIB-IV | 6 | Yes | Yes | SpectroCHIP microarray |
| Zhang et al. | 2014 | Chinese | 375 | IIIB-IV | 6 | Yes | Yes | PCR-RFLP |
| Liao et al. | 2012 | Chinese | 62 | IIIB-IV | 5 | Yes | No | PCR-RFLP |
| Xu CA et al. | 2011 | Chinese | 130 | IIIB-IV | 4 | Yes | Yes | PCR-RFLP |
| Zhao W et al. | 2011 | Chinese | 151 | IIIB-IV | 6 | Yes | Yes | PCR-RFLP |
| Cheng HY et al. | 2011 | Chinese | 120 | IIIB-IV | 5 | Yes | No | PCR |
| Zhou et al. | 2011 | Chinese | 111 | IIIB-IV | 7 | Yes | No | PCR |
| Han Y et al. | 2011 | Chinese | 91 | IIIB-IV | 4 | Yes | No | PCR-RFLP |
| Li DR et al. | 2011 | Chinese | 89 | IIIB-IV | 6 | Yes | No | PCR |
| Ding CL et al. | 2010 | Chinese | 54 | IIIB-IV | 4 | Yes | Yes | PCR |
| Qian XP et al. | 2010 | Chinese | 107 | IIIB-IV | 5 | Yes | No | PCR-RFLP |
| Ying RB et al. | 2010 | Chinese | 80 | IIIB-IV | 4 | No | Yes | PCR-RFLP |
| Sun XC et al. | 2009 | Chinese | 82 | IIIB-IV | 7 | Yes | Yes | PCR |
| Hong CY et al. | 2009 | Chinese | 164 | III-IV | 6 | Yes | Yes | PCR-RFLP |
| Qiu LX et al. | 2009 | Chinese | 107 | IIIB-IV | 5 | No | Yes | PCR-RFLP |
| Song DG et al. | 2007 | Chinese | 97 | IIIB-IV | 6 | Yes | Yes | PCR-RFLP |
| Gao CM et al. | 2006 | Chinese | 57 | IIIB-IV | 5 | Yes | Yes | PCR-RFLP |
| Jin YW et al. | 2006 | Chinese | 162 | IIIB-IV | 6 | No | Yes | PCR-RFLP |
| Yuan P et al. | 2006 | Chinese | 200 | IIIB-IV | 6 | No | Yes | PCR-RFLP |
| Wang ZH et al. | 2004 | Chinese | 105 | IIIB-IV | 6 | Yes | No | |
| Kalikaki A et al. | 2009 | Caucasian | 119 | III- IV | 6 | Yes | No | PCR-RFLP |
| Giachino et al. | 2007 | Caucasian | 248 | IIIA- IV | 7 | Yes | Yes | PCR-RFLP |
| de las Penas et al. | 2006 | Caucasian | 135 | IIIB-IV | 6 | Yes | Yes | PCR-RFLP |
| Gurubhagavatula et al. | 2004 | Caucasian | 103 | IIIB-IV | 7 | Yes | Yes | PCR-RFLP |
Polymorphism of Arg194Trp in XRCC1
A total of 15 studies reported on the relationships between the TT, CT and CC genotypes at codon 194 of XRCC1, which can cause Arg194Trp substitution, and sensitivity to platinum-based chemotherapy drugs. Heterogeneity analysis revealed no statistically significant heterogeneity among the results of these studies (I2=0%, P=0.78), Therefore, a fixed-effect model was used for the meta-analysis. The combined OR was 1.66 [95% CI (1.30, 2.14)], as shown in Figure 1. The results showed that the chemotherapy sensitivity of the patients with the TT genotype at codon 194 of XRCC1 was greater than that of patients with the CC genotype. The chemotherapy sensitivity of patients with the CT genotype was also greater than that of the CC genotype carriers [OR=1.62, 95% CI (1.35-1.93)], as shown in Figure 2. Overall, the chemotherapy sensitivity of patients carrying the TT or CT genotypes was significantly increased compared with patients carrying the wild-type CC genotype [OR=1.63, 95% CI (1.38-1.92)], as shown in Figure 3.
Figure 1.
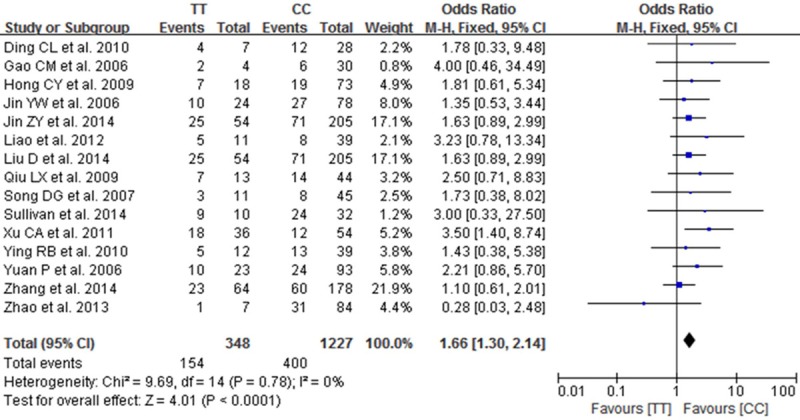
Forest plot of Arg194Trp polymorphism of the XRCC1 gene and sensitivity to platinum-based drugs in non-small cell lung cancer, the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95% CI. (TT vs. CC).
Figure 2.
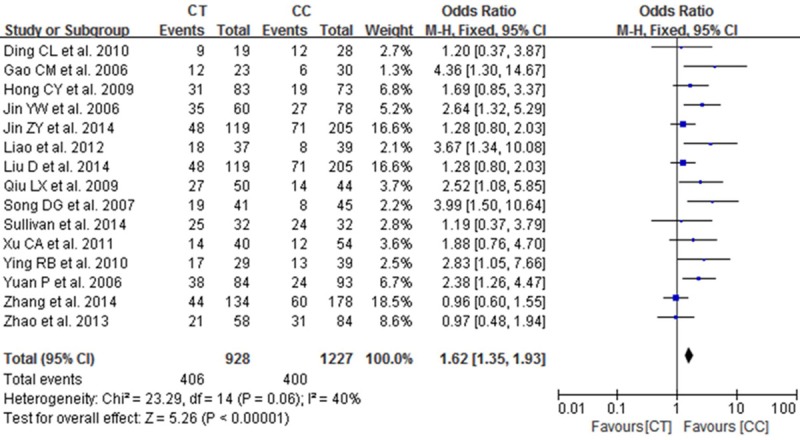
Forest plot of Arg194Trp polymorphism of the XRCC1 gene and sensitivity to platinum-based drugs in non-small cell lung cancer, the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95% CI (CT vs. CC).
Figure 3.
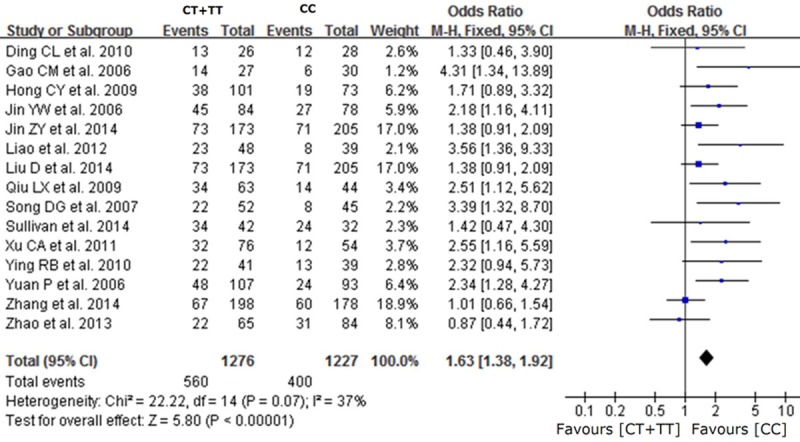
Forest plot of Arg194Trp polymorphism of the XRCC1 gene and sensitivity to platinum-based drugs in non-small cell lung cancer, the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95% CI (CT+TT vs. CC).
Polymorphism encoding Arg399Gln in XRCC1
A total of 23 studies reported on the relationships between the AA, AG and GG genotypes at codon 399 of XRCC1, which can cause an Arg399Gln substitution, and sensitivity to platinum-based chemotherapy drugs. Heterogeneity analysis revealed no statistically significant heterogeneity among the results of these studies in Caucasus population (I2=42%, P=0.12), but a significant heterogeneity was found in Chinese population. Therefore, a fixed-effect model was used in Caucasus population and a random-effect mode was used in Chinese population. The results showed that the chemotherapy sensitivity of patients with the AA genotype at codon 399 of XRCC1 was not significant difference compared to that of patients with the GG genotype [Chinese population: OR=0.90, 95% CI (0.55-1.48); Caucasus population: OR=0.96, 95% CI (0.59-1.56)], as shown in Figure 4. The chemotherapy sensitivities of the AG genotype was lower than that of patients with GG genotypes in Chinese population [OR=0.9, 95% CI (0.55-0.92)] but not in Caucasus population [OR=0.70, 95% CI (0.45-1.09)], as shown in Figure 5. We did not found the chemotherapy sensitivities of the mutant (AA+AG) genotypes at codon 399 of XRCC1 were different from that of the wild-type (GG) genotype [Chinese population: OR=0.82, 95% CI (0.56-1.21); Caucasus population: OR=0.82, 95% CI (0.54-1.44)] Figure 6.
Figure 4.
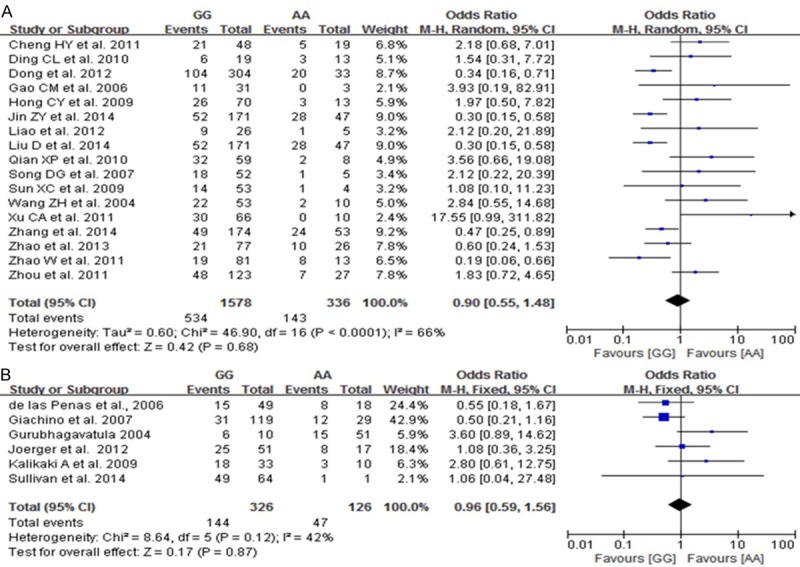
Forest plot of Arg399Gln polymorphism of the XRCC1 gene and sensitivity to platinum-based drugs in non-small cell lung cancer, the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95% CI (GG vs. AA). A: Chinese population; B: Caucasus population.
Figure 5.
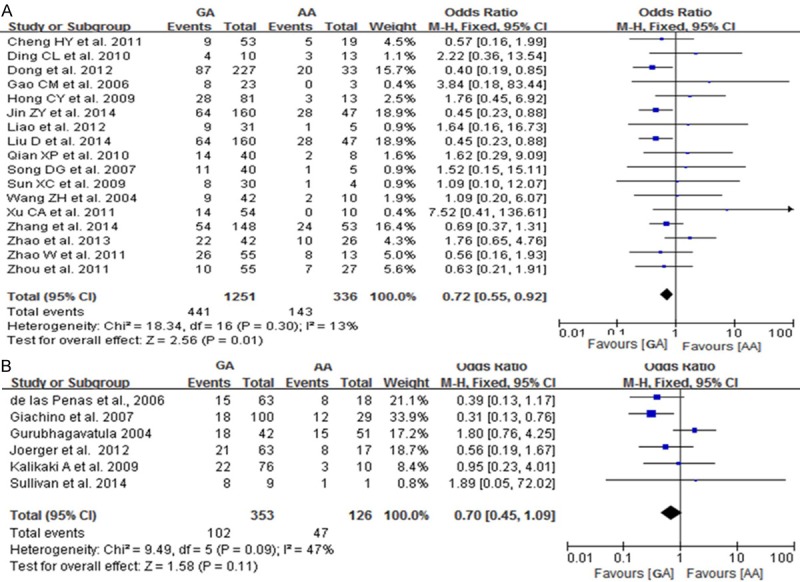
Forest plot of Arg399Gln polymorphism of the XRCC1 gene and sensitivity to platinum-based drugs in non-small cell lung cancer, the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95% CI (AG vs. AA). A: Chinese population; B: Caucasus population.
Figure 6.
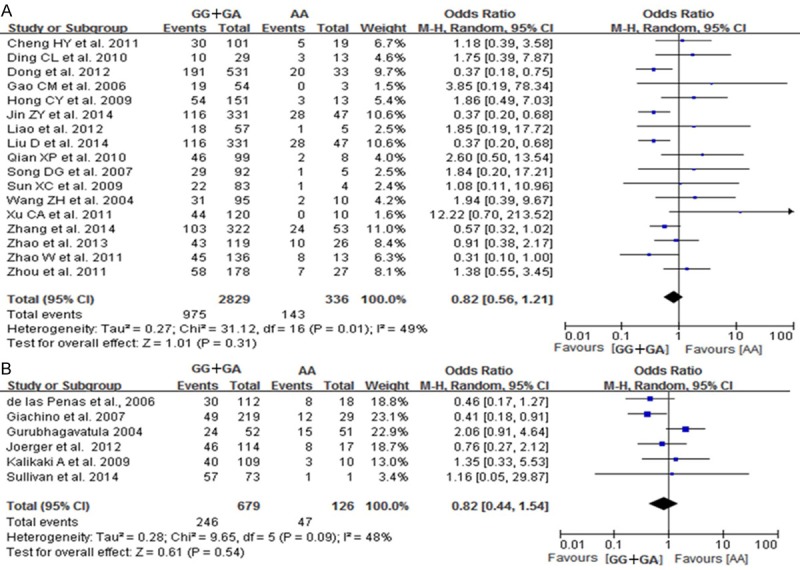
Forest plot of Arg399Gln polymorphism of the XRCC1 gene and sensitivity to platinum-based drugs in non-small cell lung cancer, the horizontal lines correspond to the study-specific OR and 95% CI, respectively. The area of the squares reflects the study-specific weight. The diamond represents the pooled results of OR and 95% CI (AG+GG vs. AA). A: Chinese population; B: Caucasus population.
Assessment of publication bias
Assessment of publication bias was conducted with a funnel plot prepared by statistical software using XRCC1 Arg194Trp (TT vs. CC) and XRCC1 Arg399Gln (AA vs. GG) as the examples. As shown in Figure 7, the resulting funnel plots are basically horizontally symmetric, preliminarily indicating that the existing data of the current studies had no significant publication bias and that the overall results of the included studies are reliable.
Figure 7.
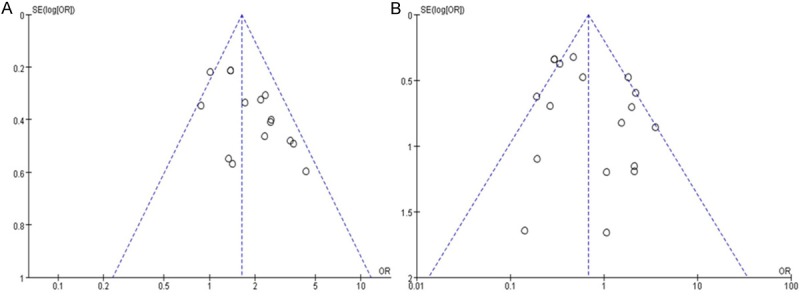
Funnel plot for publication bias tests. Each point represents a separate study for the indicated association. Log or represents natural logarithm of OR. Vertical line represents the mean effects size. A: Arg194Trp; B: Arg399Gln.
Sensitivity analysis
In the meta-analysis of the gene polymorphisms encoding XRCC1 Arg194Trp and XRCC1 Arg399Gln and their relationship with sensitivity to platinum-based chemotherapy, one study was retrospectively excluded from the included literature, and the integrated effect of the exclusion was assessed. The results showed that excluding one study did not significantly change the effect, indicating that the assessment results of this system are reliable.
Discussion
DNA repair gene plays important roles in avoiding genetic mutations and in maintaining the stability and integrity of the human genome. Studies have shown that genetic polymorphisms in DNA repair genes represent one important factor creating differences in individual sensitivity to platinum-based drugs. The pathways for DNA repair mainly include base excision repair (BER), DNA double-strand-break repair (DDSBR), mismatch repair (MMR), and nucleotide excision repair (NER). Among these, NER participates in the repair process of platinum drug-induced DNA damage, and hence, it is closely related to sensitivity to platinum-based chemotherapy.
The XRCC1 gene is a key gene in the NER pathway, and the relationship of genetic polymorphisms in this gene with sensitivity to platinum-based chemotherapy has attracted much interest. The C→T point mutation in codon 194 of XRCC1 results in a substitution of the encoded amino acid (Arg→Trp), which affects the normal function of XRCC1 protein and leads to a reduction in DNA repair capability. However, the findings regarding the correlation between genetic polymorphisms at codon 194 of XRCC1 and sensitivity to platinum-based chemotherapy have not been consistent. Marsh et al. [38] found no correlation between genetic polymorphisms at codon 194 of XRCC1 and sensitivity to platinum-based chemotherapy. A retrospective study by Kim et al. [39] showed that sensitivity to platinum-based chemotherapy was higher for the mutant genotype at codon 194 of XRCC1 than for the wild-type genotype. A study by Zhao et al [37]. Also found that sensitivity to chemotherapy was higher for the mutant genotype at codon 194 of XRCC1 than for the wild-type genotype. The results of the meta-analysis in this study showed that the efficacy of chemotherapy for patients carrying the mutant TT or CT genotypes at codon 194 of the XRCC1 gene was significantly higher than that for patients with the wild-type CC genotype. A G→A transition at codon 399 of the XRCC1 gene results in a substitution of the encoded amino acid (Arg→Gln), which affects the normal function of XRCC1 protein. The results of the meta-analysis in this study also showed that the sensitivity to chemotherapy in patients with the AG genotype at codon 399 of the XRCC1 gene was lower than that in the patients with the GG genotype. However, this association was only found in Chinese population but not in Caucasus population. Among the 29 included studies, the sample size of some studies was too small; the experiments were also designed with different factors, such as different chemotherapy regimens, test methods, histological type, tumor stage, gender, age, and smoking history; therefore, the experimental results should be carefully evaluated. Meanwhile, more high-quality clinical studies are expected to provide high-quality clinical evidence that will lead to more effective treatment options for NSCLC patients.
In conclusion, this meta-analysis clarified that the Arg194Trp polymorphism is associated with the sensitivity to platinum-based drugs in non-small cell lung cancer. However, the heterozygotes of Arg399Gln have reduced sensitivity to platinum-based drugs in non-small cell lung cancer in Chinese population. We did not found any association of Arg399Gln in Caucasus population.
Disclosure of conflict of interest
None.
References
- 1.Hancock J, Rosen J, Moreno A, Kim AW, Detterbeck FC, Boffa DJ. Management of clinical stage IIIA primary lung cancers in the National Cancer Database. Ann Thorac Surg. 2014;98:424–32. doi: 10.1016/j.athoracsur.2014.04.067. [DOI] [PubMed] [Google Scholar]
- 2.Szasz A. Current status of oncothermia therapy for lung cancer. Korean J Thorac Cardiovasc Surg. 2014;47:77–93. doi: 10.5090/kjtcs.2014.47.2.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaguchi M, Sugio K. Current status of induction treatment for N2-Stage III non-small cell lung cancer. Gen Thorac Cardiovasc Surg. 2014;62:651–9. doi: 10.1007/s11748-014-0447-1. [DOI] [PubMed] [Google Scholar]
- 4.Lee SY, Kang HG, Yoo SS, Kang YR, Choi YY, Lee WK, Choi JE, Jeon HS, Shin KM, Oh IJ, Kim KS, Lee J, Cha SI, Kim CH, Kim YC, Park JY. Polymorphisms in DNA repair and apoptosis-related genes and clinical outcomes of patients with non-small cell lung cancer treated with first-line paclitaxel-cisplatin chemotherapy. Lung Cancer. 2013;82:330–9. doi: 10.1016/j.lungcan.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Jin ZY, Zhao XT, Zhang LN, Wang Y, Yue WT, Xu SF. Effects of polymorphisms in the XRCC1, XRCC3, and XPG genes on clinical outcomes of platinum-based chemotherapy for treatment of non-small cell lung cancer. Genet Mol Res. 2014;13:7617–25. doi: 10.4238/2014.March.31.13. [DOI] [PubMed] [Google Scholar]
- 6.Peng Y, Li Z, Zhang S, Xiong Y, Cun Y, Qian C, Li M, Ren T, Xia L, Cheng Y, Wang D. Association of DNA base excision repair genes (OGG1, APE1 and XRCC1) polymorphisms with outcome to platinum-based chemotherapy in advanced nonsmall-cell lung cancer patients. Int J Cancer. 2014;135:2687–96. doi: 10.1002/ijc.28892. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Ma W, Li Y, Wu J, Shi GY. Pharmacogenetics of DNA repair gene polymorphisms in non-small-cell lung carcinoma patients on platinum-based chemotherapy. Genet Mol Res. 2014;13:228–36. doi: 10.4238/2014.January.14.2. [DOI] [PubMed] [Google Scholar]
- 8.Zhao W, Hu L, Xu J, Shen H, Hu Z, Ma H, Shu Y, Shao Y, Yin Y. Polymorphisms in the base excision repair pathway modulate prognosis of platinum-based chemotherapy in advanced non-small cell lung cancer. Cancer Chemother Pharmacol. 2013;71:1287–95. doi: 10.1007/s00280-013-2127-8. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Zhao QW, Shi GM, Wang LR. XRCC1 Arg399Gln and clinical outcome of platinum-based treatment for advanced non-small cell lung cancer: a meta-analysis in 17 studies. J Zhejiang Univ Sci B. 2012;13:875–83. doi: 10.1631/jzus.B1200083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Liu J, Zhou Y, Ying J, Zou H, Guo S, Wang L, Zhao N, Hu J, Lu D, Jin L, Li Q, Wang JC. Predictive value of XRCC1 gene polymorphisms on platinum-based chemotherapy in advanced non-small cell lung cancer patients: a systematic review and meta-analysis. Clin Cancer Res. 2012;18:3972–81. doi: 10.1158/1078-0432.CCR-11-1531. [DOI] [PubMed] [Google Scholar]
- 11.Little J, Bradley L, Bray MS, Clyne M, Dorman J, Ellsworth DL, Hanson J, Khoury M, Lau J, O’Brien TR, Rothman N, Stroup D, Taioli E, Thomas D, Vainio H, Wacholder S, Weinberg C. Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations. Am J Epidemiol. 2002;156:300–10. doi: 10.1093/oxfordjournals.aje.a000179. [DOI] [PubMed] [Google Scholar]
- 12.Xu C, Wang X, Zhang Y, Li L. Effect of the XRCC1 and XRCC3 genetic polymorphisms on the efficacy of platinum-based chemotherapy in patients with advanced non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2011;14:912–7. doi: 10.3779/j.issn.1009-3419.2011.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou F, Yu Z, Jiang T, Lv H, Yao R, Liang J. Genetic polymorphisms of GSTP1 and XRCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer (NSCLC) patients. Swiss Med Wkly. 2011;141:w13275. doi: 10.4414/smw.2011.13275. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Zhou Q, Liu Y, Yang Y, Li Q. DNA repair gene polymorphism associated with sensitivity of lung cancer to therapy. Med Oncol. 2012;29:1622–8. doi: 10.1007/s12032-011-0033-7. [DOI] [PubMed] [Google Scholar]
- 15.Dong J, Hu Z, Shu Y, Pan S, Chen W, Wang Y, Hu L, Jiang Y, Dai J, Ma H, Jin G, Shen H. Potentially functional polymorphisms in DNA repair genes and non-small-cell lung cancer survival: a pathway-based analysis. Mol Carcinog. 2012;51:546–52. doi: 10.1002/mc.20819. [DOI] [PubMed] [Google Scholar]
- 16.Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, Georgoulias V, Mavroudis D. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10:118–23. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 17.Sun X, Li F, Sun N, Shukui Q, Baoan C, Jifeng F, Lu C, Zuhong L, Hongyan C, YuanDong C, Jiazhong J, Yingfeng Z. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer. 2009;65:230–6. doi: 10.1016/j.lungcan.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Giachino DF, Ghio P, Regazzoni S, Mandrile G, Novello S, Selvaggi G, Gregori D, DeMarchi M, Scagliotti GV. Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin Cancer Res. 2007;13:2876–81. doi: 10.1158/1078-0432.CCR-06-2543. [DOI] [PubMed] [Google Scholar]
- 19.Yuan P, Miao XP, Zhang XM, Wang ZH, Tan W, Sun Y, Zhang XR, Xu BH, Lin DX. XRCC1 and XPD genetic polymorphisms predict clinical responses to platinum-based chemotherapy in advanced non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi. 2006;28:196–9. [PubMed] [Google Scholar]
- 20.Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC. XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J. Clin. Oncol. 2004;22:2594–601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan I, Salazar J, Majem M, Pallarés C, Del Río E, Páez D, Baiget M, Barnadas A. Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Lett. 2014;353:160–6. doi: 10.1016/j.canlet.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Song D, Liu J, Wang Z, Song B, Li C. Single nucleotide polymorphisms in XRCC1 and XPD and clinical response to platin-based chemotherapy in advanced non-small cell lung cancer. Chin J Gerontol. 2007;27:1684–6. [PubMed] [Google Scholar]
- 23.de las Peñas R, Sanchez-Ronco M, Alberola V, Taron M, Camps C, Garcia-Carbonero R, Massuti B, Queralt C, Botia M, Garcia-Gomez R, Isla D, Cobo M, Santarpia M, Cecere F, Mendez P, Sanchez JJ, Rosell R Spanish Lung Cancer Group. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol. 2006;17:668–75. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 24.Sun X, Li F, Sun N, Shukui Q, Baoan C, Jifeng F, Lu C, Zuhong L, Hongyan C, YuanDong C, Jiazhong J, Yingfeng Z. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer. 2009;65:230–6. doi: 10.1016/j.lungcan.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Gao C, Shi M, Wu J, Cao H, Feng J, Xu L. Polymorphisms in XRCC1 gene and sensitivity to gemcitabine/cisplatin chemotherapy in non-small cell lung cancer. Pract J Cancer. 2006;21:351–3. [Google Scholar]
- 26.Qian X, Qiu L, Yang Y, Jiang M, Zhang Y, Yu L, et al. Predictive value of base-excision repair gene polymorphisms in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. J Modern Oncol. 2010;18:1303–5. [Google Scholar]
- 27.Wang Z, Miao X, Tan W, Zhang X, Xu B, Lin D. Single nucleotide polymorphisms in XRCC1 and clinical response to platin-based chemotherapy in advanced non-small cell lung cancer. Chin J Cancer. 2004;23:865–8. [PubMed] [Google Scholar]
- 28.Cheng HY, Chen BA, Sun XC, Sun N, Cheng HY, Li F, Zhang HM, Feng JF, Qin SK, Cheng L, Lu ZH. Relationship of single nucleotide polymorphisms and clinical response to platinum-based chemotherapy in advanced non-small cell lung cancer. Jiang Su Med J. 2011;37:272–274. [Google Scholar]
- 29.Ding CL, Liu LH, Song H. Polymorphism in XRCC1 and sensitivity to platin-based chemotherapy in advanced non-small cell lung cancer. China Pharm. 2010;13:1399–1401. [Google Scholar]
- 30.Hong CY, Xu Q, Yue Z, Zhang Y, Yuan Y. Correlation of the sensitivity of NP chemotherapy in non-small lung cancer with DNA repair gene XRCC1 polymorphism. Ai Zheng. 2009;28:1291–1297. doi: 10.5732/cjc.009.10139. [DOI] [PubMed] [Google Scholar]
- 31.Jin YW, Liu J, Wang ZH. Prediction of XPD and XRCC1 gene polymorphisms in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. Shandong Yi Yao. 2006;46:42–43. [Google Scholar]
- 32.Wu J, Liu J, Zhou Y, Ying J, Zou H, Guo S, Wang L, Zhao N, Hu J, Lu D, Jin L, Li Q, Wang JC. Predictive value of XRCC1 polymorphisms in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. Mod Oncol. 2009;17:263–265. [Google Scholar]
- 33.Liao WY, Shih JY, Chang GC, Cheng YK, Yang JC, Chen YM, Yu CJ. Genetic polymorphism of XRCC1 Arg399Gln is associated with survival in non-small-cell lung cancer patients treated with gemcitabine/platinum. J Thorac Oncol. 2012;7:973–81. doi: 10.1097/JTO.0b013e31824fe98c. [DOI] [PubMed] [Google Scholar]
- 34.Jin ZY, Zhao XT, Zhang LN, Wang Y, Yue WT, Xu SF. Effects of polymorphisms in the XRCC1, XRCC3, and XPG genes on clinical outcomes of platinum-based chemotherapy for treatment of non-small cell lung cancer. Genet Mol Res. 2014;13:7617–25. doi: 10.4238/2014.March.31.13. [DOI] [PubMed] [Google Scholar]
- 35.Liu D, Wu J, Shi GY, Zhou HF, Yu Y. Role of XRCC1 and ERCC5 polymorphisms on clinical outcomes in advanced non-small cell lung cancer. Genet Mol Res. 2014;13:3100–7. doi: 10.4238/2014.April.17.6. [DOI] [PubMed] [Google Scholar]
- 36.Han Y, Lv HY, Yao RY. Effects of polymorphisms in the XRCC1, ERCC1 genes on clinical outcomes of platinum-based chemotherapy for treatment of non-small cell lung cancer. Zhongguo Shi Yong Nei Ke Za Zhi. 2011;31:638–639. [Google Scholar]
- 37.Zhao W, Hu LM, Wang Q. Effects of polymorphisms in the XRCC1, PARP1, and APE1 genes on clinical outcomes of platinum-based chemotherapy for treatment of non-small cell lung cancer. Journal of Nanjing Medical University. 2011;31:638–639. [Google Scholar]
- 38.Marsh S, Paul J, King CR, Gifford G, McLeod HL, Brown R. Pharmacogenetic assessment of toxicity and outcome after platinum plus taxane chemotherapy in ovarian cancer: the Scottish Randomised Trial in Ovarian Cancer. J. Clin. Oncol. 2007;25:4528–35. doi: 10.1200/JCO.2006.10.4752. [DOI] [PubMed] [Google Scholar]
- 39.Kim K, Kang SB, Chung HH, Kim JW, Park NH, Song YS. XRCC1 Arginine194Tryptophan and GGH-401Cytosine/Thymine polymorphisms are associated with response to platinum-based neoadjuvant chemotherapy in cervical cancer. Gynecol Oncol. 2008;111:509–15. doi: 10.1016/j.ygyno.2008.08.034. [DOI] [PubMed] [Google Scholar]


