Abstract
Background: Glutathione S-transferase (GST) family genes are of vital importance in maintaining cellular defence systems, protecting cells against the toxic effects of reactive oxygen produced during the synthesis of melanin, and detoxifying environmental mutagens and chemical or synthetic drugs. As no previous meta-analyses have examined the association of polymorphisms at GSTT1, GSTP1 Ile105Val with skin cancer risk and independently published studies have produced inconsistent conclusions, we were promoted to estimate the associations in the largest study to date. Methods: Computer-assisted searches were carried out to systematically identify the studies of GST polymorphisms and skin cancer. The eligibility of studies was evaluated following the requirements of inclusion criteria. Risk of skin cancers (OR and 95% CI) was assessed with the fixed or random effects meta-analysis. Major findings: The fixed effects meta-analysis of 15 studies suggested no overall association between GSTT1 null and skin cancer. Nor was there a significant association in any subgroup. However, in the stratified analysis by histologic type for GSTP1 Ile105Val, we found 1.56 times higher risk of malignant melanoma (MM) among people with the 105-Val/Val genotype (Val/Val vs. Ile/Ile: OR = 1.56, 95% CI = 1.05-2.32, pheterogeneity = 0.584). Conclusions: These statistical data demonstrate that Ile105Val polymorphism of the GSTP1 gene may have genetic contribution to the development of skin cancer, MM in particular.
Keywords: GSTT1, GSTP1, skin cancer, polymorphism, genetic contribution
Introduction
Melanoma and non-melanoma are two major types of skin cancer, a most common form of cancer with an increasingly higher incidence across the global in recent years [1,2]. Non-melanoma histologically subdivided into squamous cell carcinoma (SCC), basal cell carcinoma (BCC) and many other types is almost twenty times more prevalent than melanoma [3]. There is a realization that light skin pigmentation and frequent ultraviolet exposure which has caused 80% of melanoma should account a large part for the occurrence of various skin cancers [4-6]. However, as more and more susceptibility loci for skin cancer are detected by means of genome-wide association studies (GWAS), including the low-penetrance MC1R gene, and the high-penetrance CDK4 and CDKN2, a number of candidate gene studies, another primary approach widely used for genetic research, are successively carried out and identified a long list of genes involved in proliferation, DNA-repair, inflammatory processes, oxidative stress, pigmentation, telomere maintenance, and tumorigenesis [7-13]. Despite the previous efforts, knowledge of genetic contribution of many susceptibility genes to the invasive disease is still limited.
A fundamental mechanism in defence against DNA damage from carcinogenic compounds relates to glutathione S-transferase (GST) enzymes, a supergene family important for the detoxification of exogenous substances, such as chemotherapeutic agents, and reactive oxygen species [14,15]. Lack of GST arises because of inherited or somatic mutations facilitates the malignant progression of human cancer, including malignant melanoma (MM) [16,17]. Existing data have shown the genetic variants in GSTT1 eliminate molecular functions of gene products and that the GSTP1 105-val is related to decreased enzyme activity [18-20].
People who harbor GSTT1, -P1 genotypes or alleles might vary considerably in the ability to metabolize mutagenic compounds and hence have different susceptibility to skin cancer. A case-control study in samples of Caucasian ethnicity suggested no association between GSTT1 null and BCC risk [21]. The original report was followed by many publications which have produced mixed findings: elevated or reduced risk of MM [17,22]. Likewise, for GSTP1 Ile105Val, a 10 times lower risk for SCC was associated with 105-Val/Val, whereas the same genotype was found to increase the risk for melanoma [23,24]. These studies characterized by different sample size, various study designs, and non-homogeneous populations may be underpowered to detect the true associations. Herein, we performed a meta-analysis to supply strong evidence for the association between GSTT1, -P1 polymorphisms and genetic risk of skin cancer.
Methods
Search strategy and study selection
Multiple known databases (the Cochrane Library, EBSCO, BIOSIS, PubMed, Embase, CNKI, and WANFANG) were searched up to April 15, 2014. The identification of relevant published papers was conducted by using MeSHs and keywords: ‘skin cancer’, ‘melanoma’, ‘basal cell carcinoma’, ‘squamous cell carcinoma’, ‘glutathione S-transferase’, ‘glutathione S-transferase T1’, ‘glutathione S-transferase P1’, ‘genotypes’, ‘variants’ and ‘polymorphism’. The additional papers that may have been left out during computer-based searches were identified via manual searches of human case-control studies or meta-analyses examining the association of GST polymorphism with any type of skin cancer. We also consulted experts in this domain to obtain new information or unpublished data. This study was performed complying with the guidelines of Preferred Reporting Items for Systematic Reviews and Meta-Analyses [25].
Eligible papers were selected based on: a) a case-controlled skin cancer study for either GSTT1 or GSTP1 Ile105Val polymorphism; and b) information on genotype distribution was presented in detail. We also included the studies with a cross-section or cohort design. When the same patients were included in two or more studies concerning the association of interest, we selected the largest study.
The research articles that failed to meet the pre-described requirements were not considered in further meta-analysis.
Data extraction
Having singled out all eligible studies, two independent investigators set out to record information on first author, year of publication, genotyped cases and controls, phenotype of cases (SCC, BCC, or MM), ethnicity of each population being analyzed (Caucasians or East Asians), matching criteria, polymorphism investigated, genotyping assay, genotype count of cases and controls, source of controls and study country. Any discrepancy was resolved though consultation with a third investigator.
Statistical analysis
Using the genotypic data extracted from each study, we assessed the risk of skin cancer in relation to GST polymorphisms by calculating an odds ratio (OR) and its 95% confidence interval (95% CI). For GSTT1, the combined effects were estimated assuming the Null vs. Active model; for GSTP1 Ile105Val, Val/Val vs. Ile/Ile, Ile/Val vs. Ile/Ile, dominant model and recessive model were tested. Stratified analyses by ethnicity and histologic type were performed for GSTT1, while data for GSTP1 Ile105Val were only stratified by histologic type.
The Chi square based Q test was used to detect the heterogeneity across studies considered in the meta-analysis, with P values less than 0.05 being deemed significant. The I2 statistic described by Higgins and Thompson was used to quantify the variance between studies and I2 values excessing 50% corresponded to large heterogeneity [26]. We selected the Mantel-Haenszel method (M-H, the fixed effects model) to evaluate the risk of skin cancer if P > 0.05 or I2 < 50%; alternatively, the DerSimonian and Laird method (D-L, the random effect model) was employed.
To test the Hardy-Weinberg equilibrium (HWE) in control populations, we chose the X2 test. The leave-one-out sensitivity analyses were performed to reflect the influence from the independent studies on pooled effect estimations. Publication bias was inspected by the funnel plot and the Egger’s test. Statistical data were analyzed using STATA software (Version 12.0, STATA Corp, College Station, TX). All P values < 0.05 was judged as statistically significant.
Results
Meta-analysis database
The computer-assisted searches using the aforementioned strategy resulted in 105 papers. We initially eliminated 32 duplicates and then 49 papers due to obvious irrelevance. 24 papers remained and we scanned the full-texts to evaluate their eligibility. Of these, 9 were excluded because of review articles [27-30], articles of GSTM1 polymorphism or unrelated disease [31-33], and meta-analyses [34,35]. The final pooling dataset consisted of 15 studies: 15 for GSTT1 and 6 for GSTP1 Ile105Val [17,21-24,36-45]. A flow chart showing the literature selection is presented in Figure 1.
Figure 1.
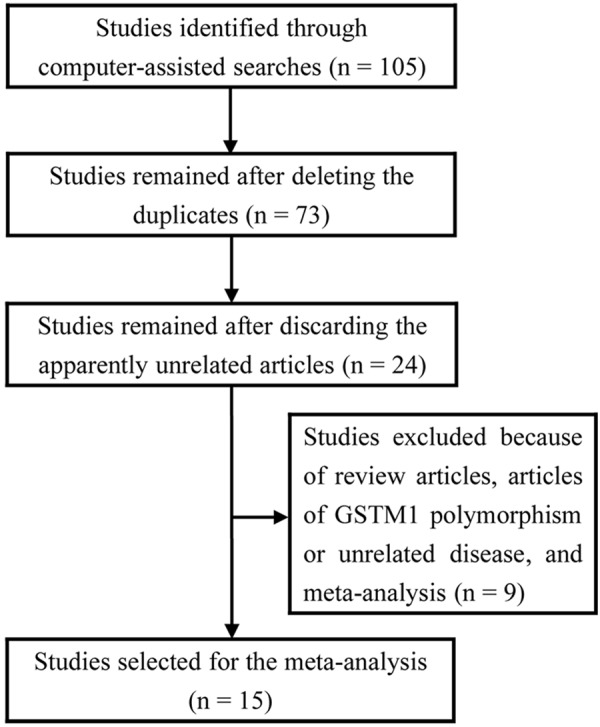
Flow diagram of study exclusion and inclusion.
Summary description of studies
As described in Table 1, the studies identified were published between 1996 and 2013. Studies of Caucasian samples accounted for about 86.6% and only 13.3% employed East Asian samples. Original data were sufficient for three types of skin cancer: BCC, SCC, MM. Most studies did not state information on matching criteria, even did, they were not uniformly defined. In addition, the included studies differed considerably in control source, ranging from healthy subjects without any disease to cancer-free renal transplant recipients. The genotype distribution of GSTP1 Ile105Val studies were in HWE with the exception of Lira et al. from Italy (P = 0.018). Fryer et al. provided data for BCC and SCC, but not for total skin cancer, this study therefore was merged into each subgroup.
Table 1.
Main characteristics of the meta-analysis studies
| First Author | Year | Study Country | Ethnicity | Case/Control | Phenotype of Cases | Matching Status | SNP |
|---|---|---|---|---|---|---|---|
| Heagerty | 1996 | UK | Caucasian | 584/484 | BCC | Matched for age, gender, ethnicity | GSTT1 |
| Yengi | 1996 | UK | Caucasian | 259/284 | BCC | Not stated | GSTT1 |
| Marshall | 2000 | UK | Caucasian | 48/174 | SCC, BCC | Not stated | GSTT1 |
| Kanetsky | 2001 | USA | Caucasian | 362/271 | MM | Not stated | GSTT1 |
| Ramsay | 2001 | UK | Caucasian | 29/151 | SCC, BCC | Not stated | GSTT1, GSTP1 Ile105Val |
| Fryer | 2005 | UK | Caucasian | 135/198 | SCC, BCC | Not stated | GSTT1, GSTP1 Ile105Val |
| Dolzan | 2006 | Slovenia | Caucasian | 137/116 | MM | Unmatched fo age, gender | GSTT1 |
| Lira | 2006 | Italy | Caucasian | 106/131 | NMSC | Matched for type of transplanted organ, duration of transplantation, gender and age | GSTT1, GSTP1 Ile105Val |
| Bu | 2007 | Sweden | Caucasian | 154/203 | MM | Matched for gender and age | GSTT1, GSTP1 Ile105Val |
| Leite | 2007 | Brazil | Caucasian | 105/124 | SCC, BCC, MM | Not stated | GSTT1, GSTP1 Ile105Val |
| Mossner | 2007 | Germany | Caucasian | 319/346 | MM | Not stated | GSTT1 |
| Xie | 2010 | China | East Asian | 77/107 | NNM | Not stated | GSTT1 |
| Chiyomaru | 2011 | Japan | East Asian | 115/92 | SCC, BCC, AK, BD | Matched for gender and age | GSTT1, GSTP1 Ile105Val |
| Ibarrola-Villava | 2012 | Spain | Caucasian | 560/337 | MM | Not stated | GSTT1, GSTP1 Ile105Val |
| Fortes | 2013 | Italy | Caucasian | 188/152 | MM | Matched for gender and age | GSTT1 |
BCC, basal cell carcinoma; SCC, squamous cell carcinoma; MM, malignant melanoma; NMSC, non-melanoma skin cancer; NNM, naevi of the nail matrix; AK, actinic keratosis; BC, Bowen’s disease.
GSTT1 polymorphism and skin cancer
Fourteen studies, with 3,043 cases and 2,972 controls, were combined to evaluate the association of GSTT1 polymorphism with skin cancer. No statistical evidence of a significant relation was shown in the overall analysis (OR = 1.01, 95% CI = 0.89-1.14, pheterogeneity = 0.996) (Figure 2; Table 2). In further stratified analyses by ethnicity, we did not find an elevated risk of skin cancer among people carrying the null genotype relative to those carrying the active genotype. We saw the same trend when stratifying the data by histologic type (Table 2).
Figure 2.
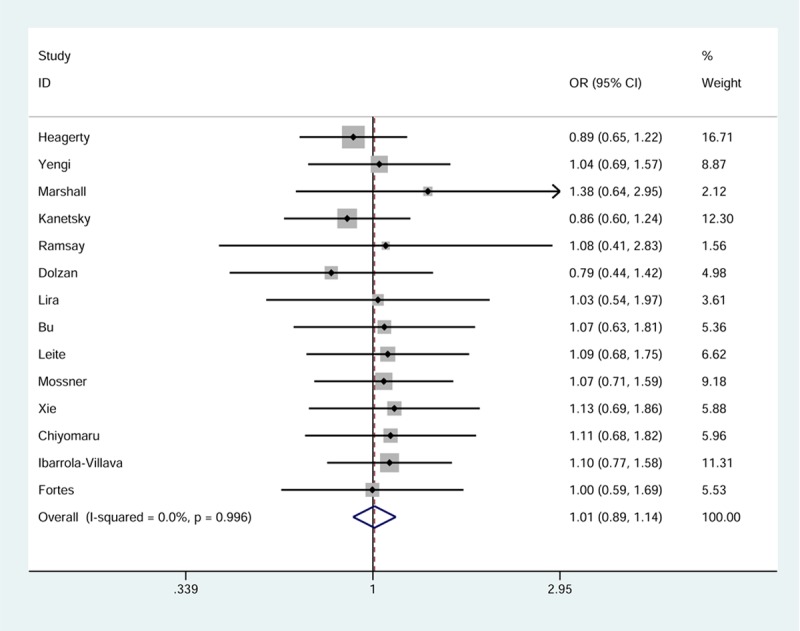
Forest plot of skin cancer risk related to GSTT1 null genotype. Each box corresponds to the OR point estimate, and its area is proportional to the weight of the study. The diamond (and broken line) represents the overall summary estimate, with CI represented by its width. The unbroken vertical line is set at the null value (OR =1.0).
Table 2.
Meta-analysis GSTT1 polymorphism
| Variables | Geentic Model | N | Cases/Controls | Test of Association | Test of Heterogeneity | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| OR | 95% CI | PHet | I2 (%) | ||||
| ALL | NULL vs. ACTIVE | 14 | 3,043/2,972 | 1.01 | (0.89, 1.14) | 0.996 | 0.0 |
| CAUCASIAN | NULL vs. ACTIVE | 12 | 2,851/2,773 | 0.99 | (0.87, 1.13) | 0.990 | 0.0 |
| EAST ASIAN | NULL vs. ACTIVE | 2 | 192/199 | 1.12 | (0.79, 1.59) | 0.951 | 0.0 |
| BCC | NULL vs. ACTIVE | 8 | 1,168/1,673 | 1.01 | (0.85, 1.19) | 0.806 | 0.0 |
| SCC | NULL vs. ACTIVE | 6 | 295/900 | 1.03 | (0.81, 1.32) | 0.579 | 0.0 |
| MM | NULL vs. ACTIVE | 7 | 1,725/1,549 | 0.98 | (0.82, 1.17) | 0.909 | 0.0 |
GSTP1 Ile105Val and skin cancer
Six studies of GSTP1 Ile105Val provided a total of 2,076 subjects. The calculation of ORs and 95% CIs indicated no elevated or reduced risk for developing skin cancer in individuals harboring the Val/Val or Val/Val + Ile/Val genotypes (Table 3). However, according to Figure 3, which shows the pooled results in Val/Val vs. Ile/Ile model, the individuals with the 105-Val/Val had 1.56 times higher risk of MM compared to the individuals with the 105-Ile/Ile (OR = 1.56, 95% CI = 1.05-2.32, pheterogeneity = 0.584). No noteworthy associations were indicated in subgroups of BCC and SCC.
Table 3.
Meta-analysis GSTP1 polymorphism
| Variables | N | Cases/Controls | Val/Val vs. Ile/Ile | Ile/Val vs. Ile/Ile | Dominant | Recessive | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Val/Val + Ile/Val vs. Ile/Ile | Val/Val vs. Ile/Val + Ile/Ile | |||||||||
|
| ||||||||||
| OR (95% CI) | PHet/I2 (%) | OR (95% CI) | PHet/I2 (%) | OR (95% CI) | PHet/I2 (%) | OR (95% CI) | PHet/I2 (%) | |||
| All | 6 | 1,057/1,019 | 0.83 (0.43, 1.60) | 0.020/65.6 | 1.08 (0.90, 1.29) | 0.799/0.0 | 1.04 (0.89, 1.22) | 0.566/0.0 | 0.79 (0.43, 1.48) | 0.026/63.7 |
| Histologic Type | ||||||||||
| BCC | 5 | 303/691 | 0.66 (0.40, 1.08) | 0.388/0.8 | 0.90 (0.68, 1.19) | 0.981/0.0 | 0.88 (0.69, 1.13) | 0.967/0.0 | 0.69 (0.39, 1.21) | 0.075/56.6 |
| SCC | 5 | 249/697 | 0.66 (0.38, 1.13) | 0.128/47.3 | 0.97 (0.72, 1.30) | 0.978/0.0 | 0.93 (0.72, 1.20) | 0.973/0.0 | 0.47 (0.14, 1.53) | 0.075/56.6 |
| MM | 3 | 718/664 | 1.56 (1.05, 2.32) | 0.584/0.0 | 1.16 (0.94, 1.44) | 0.994/0.0 | 1.17 (0.97, 1.43) | 0.920/0.0 | 1.48 (0.98, 2.25) | 0.335/8.7 |
Figure 3.
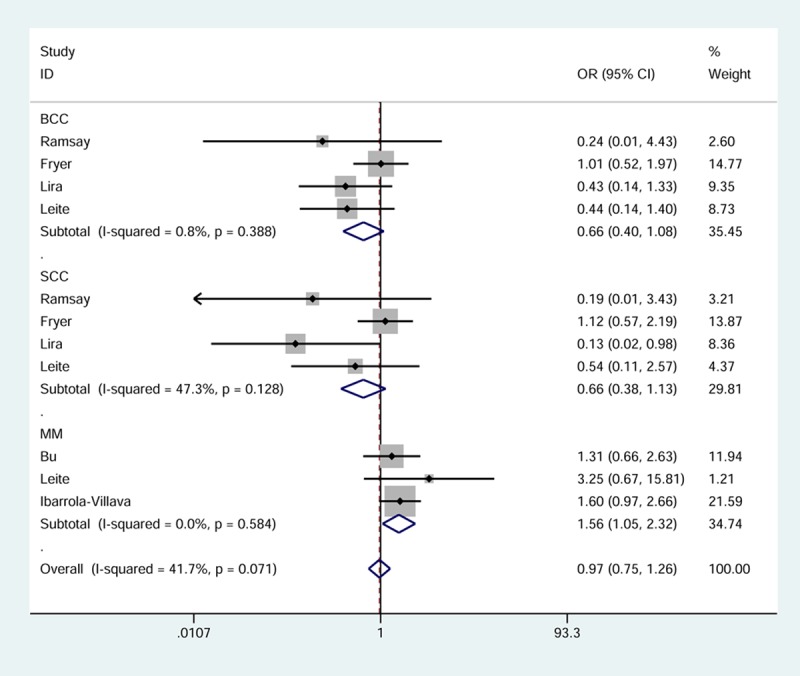
Forest plot of skin cancer risk related to GSTP1 Ile105Val stratified by histologic type (Val/Val vs. Ile/Ile). Each box corresponds to the OR point estimate, and its area is proportional to the weight of the study. The diamond (and broken line) represents the overall summary estimate, with CI represented by its width. The unbroken vertical line is set at the null value (OR = 1.0).
Heterogeneity and sensitivity analyses
We noted significant heterogeneity for GSTP1 Ile105Val in the Val/Val vs. Ile/Ile model (P = 0.020, I2 = 65.6%) and the recessive model (P = 0.026, I2 = 63.7%). Subsequent sensitivity analysis identified Ibarrola-Villava et al. and Lira et al. were outliers for the two models respectively, as the homogeneity increased remarkably when they were deleted (P = 0.775, I2 = 0.0%; P = 0.174, I2 = 39.7%). The heterogeneous studies appeared to not affect the overall results (data not shown).
Publication bias
The inspection of publication bias among the selected studies for the meta-analysis was done with the funnel plot and Egger’s test. The shape of funnel plots seemed symmetrical in all genetic models and the Egger’s test showed evidence supporting lack of publication bias in this analysis. The funnel plots in Null vs. Active model and the dominant model were displayed in Figure 4 (t = -0.31, P = 0.765) and Figure 5 (t = 0.46, P = 0.672), respectively.
Figure 4.
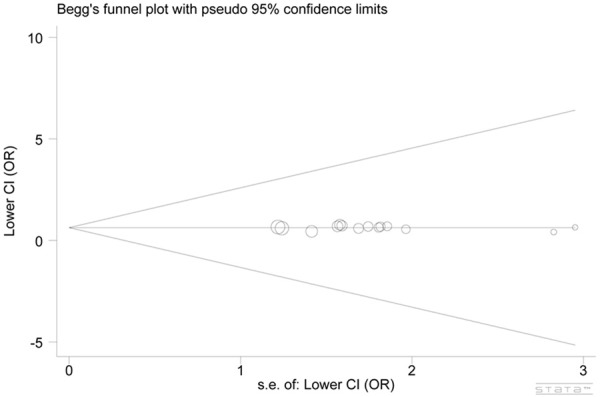
Begg’s funnel plot of GSTT1 polymorphism and skin cancer risk (Null vs. Active).
Figure 5.
Begg’s funnel plot of GSTP1 Ile105Val polymorphism and skin cancer risk (the dominant model).
Discussion
As yet, there have been no previous meta-analyses examining the association of GSTT1 and GSTP1 polymorphisms with the development of skin cancer, we hence decided to combine all published studies which have produced inconsistent conclusions. The current meta-analysis highlighted one point that the incidence of BCC, SCC, and MM was not associated with GSTT1 null and Caucasians were not seemed to be more susceptible to skin cancer as compared to East Asians, because no statistical evidence of a significantly increased risk was indicated in either the overall analysis or the subgroup analyses by ethnicity and histologic type. Meta-analysis of GSTP1 Ile105Val demonstrated that although elevation or reduction in the risk of overall skin cancer was not related to the carriage of Ile105Val genotypes, the people with the Val/Val or Val/Val + Ile/Val genotypes were more prone to MM compared to the Ile/Ile genotype. It is unclear, however, whether this significantly elevated risk detected in 1,382 subjects is a false-positive finding.
Prior findings from an earlier meta-analysis of genetic data from GSTM1 and GSTT1 studies and melanoma suggested that the null genotypes at GSTM1 or GSTT1 were not effect modifiers [34]. This lack of an association between GSTT1 null and melanoma was confirmed in this study in which three additional studies provided 1,902 new subjects. Further analyses to detect the effects on SCC and BCC showed no increased risk associated with GSTT1 null genotype. Different from the previous analysis, we identified that the null genotype did not represent an independent risk factor for skin cancer in Caucasians and East Asians, and that the GSTP1 105-Val/Val was associated with 1.56 times greater risk for developing MM. A recent meta-analysis of GSTM1 polymorphism indicated that the null genotype appeared to have no major effects on risk of BCC and SCC [35]. GSTM1, -T1, -P1 are important members of GST multigene superfamily. The potent GSTs enzymes are crucial mediators in cellular defence systems and play key roles in protecting cells against the toxic effects of reactive oxygen produced during the synthesis of melanin, and detoxifying environmental mutagens and chemical or synthetic drugs [46-48]. From this perspective, GST genes are likely to have protective effects against the progression of skin cancer, as reported by Fortes et al. [22], which was not detected in our study possibly due to the small number.
The fact of growingly higher incidence of skin cancer, melanoma in particular, in the USA and European countries in recent decades suggests that people of Caucasian ethnicity are more likely than those of other ethnicities to develop skin cancer, and that melanoma is more prevalent than BCC and SCC. Therefore, is it not surprised to find increased risk of MM in relation to GSTP1 105-Val/Val, because the high incidence may likely be caused by the polymorphism alone or the combination with a variety of modifying genes and carcinogenic exposures. Available evidence shown by Menon et al. suggested that people with darker skin complexion have more eumelanin, a better protector in defence against cellular oxidative stress as compared to pheomelanin, a greater amount of which appear in people with fair skin complexion. However, we failed to confirm the association between GSTT1 null and skin cancers in white populations (Caucasians) initially observed in a hospital-based case-control study by Kanetsk et al. and later confirmed in a Slovenian study caried out by Dolzan et al. [17,40]. Due to the unavailability of raw data, we were also unable to confirm whether the GSTP1 polymorphism has major impact in Caucasian populations, although as high as 4.5 times increased risk of non-melanoma skin cancer has been reported by Lira et al. [23]. Our results are therefore indefinitive and require further investigations.
This is the largest meta-analysis evaluating the risk of skin cancer associated with GSTT1, -P1 polymorphisms for the first time. We identified that GSTT1 null was not associated with BCC, and SCC, and that 105-Val/Val was a risk factor for MM independent of confounding variables, which have not been found in any of the previous meta-analyses concerning skin cancer.
The current study has several shortcomings. We detected large inter-study heterogeneity for GSTP1 Ile105Val, and the findings should be explained with caution even though there was no difference in the effect estimations before and after excluding the heterogeneous studies. The second shortcoming refers to the small number of subjects. The sample inadequacy may lead to decreased precision in estimations and thus we cannot exclude slightly or moderately elevated or reduced risk not suggested in our study. A final point to take into consideration in interpreting our findings is that we did not evaluate the effects of gene-gene and gene-environment interactions. GST genes tend to work in combination and published studies have shown people with both GSTM1 null and GSTT1 null had about 10-fold risk of MM and a more pronounced protective effect among people with the null genotypes [17,22]. Further studies are necessary to identify the role of potential carcinogens in skin cancers.
In conclusion, the GSTP1 Ile105Val, but not GSTT1 null appeared to be associated with elevated risk for developing MM in this largest meta-analysis of both Caucasians and Asians. In future, researchers are expected to carry out a larger study to determine the association between the GST polymorphisms and skin cancer such that we can identify the at-risk populations.
Disclosure of conflict of interest
None.
References
- 1.Leiter U, Garbe C. Epidemiology of melanoma and nonmelanoma skin cancer--the role of sunlight. Adv Exp Med Biol. 2008;624:89–103. doi: 10.1007/978-0-387-77574-6_8. [DOI] [PubMed] [Google Scholar]
- 2.Godar DE. Worldwide increasing incidences of cutaneous malignant melanoma. J Skin Cancer. 2011;2011:858425. doi: 10.1155/2011/858425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shao Y, Zhang J, Zhang R, Wan J, Zhang W, Yu B. Examination of Smad2 and Smad4 copy-number variations in skin cancers. Clin Transl Oncol. 2012;14:138–142. doi: 10.1007/s12094-012-0773-7. [DOI] [PubMed] [Google Scholar]
- 4.IARC monographs on the evaluation of carcinogenic risks to humans. Solar and ultraviolet radiation. IARC Monogr Eval Carcinog Risks Hum. 1992;55:1–316. [PMC free article] [PubMed] [Google Scholar]
- 5.Oliveria SA, Saraiya M, Geller AC, Heneghan MK, Jorgensen C. Sun exposure and risk of melanoma. Arch Dis Child. 2006;91:131–138. doi: 10.1136/adc.2005.086918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young C. Solar ultraviolet radiation and skin cancer. Occup Med (Lond) 2009;59:82–88. doi: 10.1093/occmed/kqn170. [DOI] [PubMed] [Google Scholar]
- 7.Gerstenblith MR, Shi J, Landi MT. Genome-wide association studies of pigmentation and skin cancer: a review and meta-analysis. Pigment Cell Melanoma Res. 2010;23:587–606. doi: 10.1111/j.1755-148X.2010.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han J, Colditz GA, Samson LD, Hunter DJ. Polymorphisms in DNA double-strand break repair genes and skin cancer risk. Cancer Res. 2004;64:3009–3013. doi: 10.1158/0008-5472.can-04-0246. [DOI] [PubMed] [Google Scholar]
- 9.Stacey SN, Gudbjartsson DF, Sulem P, Bergthorsson JT, Kumar R, Thorleifsson G, Sigurdsson A, Jakobsdottir M, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Scherer D, Rudnai P, Gurzau E, Koppova K, Hoiom V, Botella-Estrada R, Soriano V, Juberias P, Grasa M, Carapeto FJ, Tabuenca P, Gilaberte Y, Gudmundsson J, Thorlacius S, Helgason A, Thorlacius T, Jonasdottir A, Blondal T, Gudjonsson SA, Jonsson GF, Saemundsdottir J, Kristjansson K, Bjornsdottir G, Sveinsdottir SG, Mouy M, Geller F, Nagore E, Mayordomo JI, Hansson J, Rafnar T, Kong A, Olafsson JH, Thorsteinsdottir U, Stefansson K. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet. 2008;40:1313–1318. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- 10.Gudbjartsson DF, Sulem P, Stacey SN, Goldstein AM, Rafnar T, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Sveinsdottir SG, Magnusson V, Lindblom A, Kostulas K, Botella-Estrada R, Soriano V, Juberias P, Grasa M, Saez B, Andres R, Scherer D, Rudnai P, Gurzau E, Koppova K, Kiemeney LA, Jakobsdottir M, Steinberg S, Helgason A, Gretarsdottir S, Tucker MA, Mayordomo JI, Nagore E, Kumar R, Hansson J, Olafsson JH, Gulcher J, Kong A, Thorsteinsdottir U, Stefansson K. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 11.Raimondi S, Sera F, Gandini S, Iodice S, Caini S, Maisonneuve P, Fargnoli MC. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008;122:2753–2760. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- 12.Gu F, Qureshi AA, Kraft P, Guo Q, Hunter DJ, Han J. Polymorphisms in genes involved in DNA repair, cell growth, oxidative stress and inflammatory response, and melanoma risk. Br J Dermatol. 2009;161:209–212. doi: 10.1111/j.1365-2133.2009.09219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nan H, Qureshi AA, Hunter DJ, Han J. Genetic variants in FGFR2 and FGFR4 genes and skin cancer risk in the Nurses’ Health Study. BMC Cancer. 2009;9:172. doi: 10.1186/1471-2407-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartzbaum JA, Ahlbom A, Lonn S, Warholm M, Rannug A, Auvinen A, Christensen HC, Henriksson R, Johansen C, Lindholm C, Malmer B, Salminen T, Schoemaker MJ, Swerdlow AJ, Feychting M. An international case-control study of glutathione transferase and functionally related polymorphisms and risk of primary adult brain tumors. Cancer Epidemiol Biomarkers Prev. 2007;16:559–565. doi: 10.1158/1055-9965.EPI-06-0918. [DOI] [PubMed] [Google Scholar]
- 15.Yuan JM, Chan KK, Coetzee GA, Castelao JE, Watson MA, Bell DA, Wang R, Yu MC. Genetic determinants in the metabolism of bladder carcinogens in relation to risk of bladder cancer. Carcinogenesis. 2008;29:1386–1393. doi: 10.1093/carcin/bgn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997;6:733–743. [PubMed] [Google Scholar]
- 17.Kanetsky PA, Holmes R, Walker A, Najarian D, Swoyer J, Guerry D, Halpern A, Rebbeck TR. Interaction of glutathione S-transferase M1 and T1 genotypes and malignant melanoma. Cancer Epidemiol Biomarkers Prev. 2001;10:509–513. [PubMed] [Google Scholar]
- 18.Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300:271–276. doi: 10.1042/bj3000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275–280. doi: 10.1093/carcin/19.2.275. [DOI] [PubMed] [Google Scholar]
- 20.Sun KH, Chang KH, Clawson S, Ghosh S, Mirzaei H, Regnier F, Shah K. Glutathione-S-transferase P1 is a critical regulator of Cdk5 kinase activity. J Neurochem. 2011;118:902–914. doi: 10.1111/j.1471-4159.2011.07343.x. [DOI] [PubMed] [Google Scholar]
- 21.Heagerty A, Smith A, English J, Lear J, Perkins W, Bowers B, Jones P, Gilford J, Alldersea J, Fryer A, Strange RC. Susceptibility to multiple cutaneous basal cell carcinomas: significant interactions between glutathione S-transferase GSTM1 genotypes, skin type and male gender. Br J Cancer. 1996;73:44–48. doi: 10.1038/bjc.1996.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortes C, Mastroeni S, Boffetta P, Antonelli G, Pilla MA, Botta G, Anzidei P, Venanzetti F. The protective effect of coffee consumption on cutaneous melanoma risk and the role of GSTM1 and GSTT1 polymorphisms. Cancer Causes Control. 2013;24:1779–1787. doi: 10.1007/s10552-013-0255-4. [DOI] [PubMed] [Google Scholar]
- 23.Lira MG, Provezza L, Malerba G, Naldi L, Remuzzi G, Boschiero L, Forni A, Rugiu C, Piaserico S, Alaibac M, Turco A, Girolomoni G, Tessari G. Glutathione S-transferase and CYP1A1 gene polymorphisms and non-melanoma skin cancer risk in Italian transplanted patients. Exp Dermatol. 2006;15:958–965. doi: 10.1111/j.1600-0625.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- 24.Bu H, Rosdahl I, Holmdahl-Kallen K, Sun XF, Zhang H. Significance of glutathione S-transferases M1, T1 and P1 polymorphisms in Swedish melanoma patients. Oncol Rep. 2007;17:859–864. doi: 10.3892/or.17.4.859. [DOI] [PubMed] [Google Scholar]
- 25.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 27.Strange RC, Lear JT, Fryer AA. Polymorphism in glutathione S-transferase loci as a risk factor for common cancers. Arch Toxicol Suppl. 1998;20:419–428. doi: 10.1007/978-3-642-46856-8_37. [DOI] [PubMed] [Google Scholar]
- 28.Strange RC, Lear JT, Fryer AA. Glutathione S-transferase polymorphisms: influence on susceptibility to cancer. Chem Biol Interact. 1998;111-112:351–364. doi: 10.1016/s0009-2797(97)00172-5. [DOI] [PubMed] [Google Scholar]
- 29.Hengstler JG, Arand M, Herrero ME, Oesch F. Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Recent Results Cancer Res. 1998;154:47–85. doi: 10.1007/978-3-642-46870-4_4. [DOI] [PubMed] [Google Scholar]
- 30.Strange RC, Fryer AA. The glutathione S-transferases: influence of polymorphism on cancer susceptibility. IARC Sci Publ. 1999:231–249. [PubMed] [Google Scholar]
- 31.Shanley SM, Chenevix-Trench G, Palmer J, Hayward N. Glutathione S-transferase GSTM1 null genotype is not overrepresented in Australian patients with nevoid basal cell carcinoma syndrome or sporadic melanoma. Carcinogenesis. 1995;16:2003–2004. doi: 10.1093/carcin/16.8.2003. [DOI] [PubMed] [Google Scholar]
- 32.Lafuente A, Molina R, Palou J, Castel T, Moral A, Trias M. Phenotype of glutathione S-transferase Mu (GSTM1) and susceptibility to malignant melanoma. MMM group. Multidisciplinary Malignant Melanoma Group. Br J Cancer. 1995;72:324–326. doi: 10.1038/bjc.1995.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mertens AC, Mitby PA, Radloff G, Jones IM, Perentesis J, Kiffmeyer WR, Neglia JP, Meadows A, Potter JD, Friedman D, Yasui Y, Robison LL, Davies SM. XRCC1 and glutathione-S-transferase gene polymorphisms and susceptibility to radiotherapy-related malignancies in survivors of Hodgkin disease. Cancer. 2004;101:1463–1472. doi: 10.1002/cncr.20520. [DOI] [PubMed] [Google Scholar]
- 34.Nie F, Chen Z, Cao C, Cen Y. Absence of association between GSTM1 and GSTT1 polymorphisms and melanoma susceptibility: a meta-analysis. DNA Cell Biol. 2011;30:783–788. doi: 10.1089/dna.2010.1198. [DOI] [PubMed] [Google Scholar]
- 35.Peng H, He Q, Zhu J, Peng C. Effect of GSTM1 polymorphism on risks of basal cell carcinoma and squamous cell carcinoma: a meta-analysis. Tumour Biol. 2013;34:675–681. doi: 10.1007/s13277-012-0595-9. [DOI] [PubMed] [Google Scholar]
- 36.Yengi L, Inskip A, Gilford J, Alldersea J, Bailey L, Smith A, Lear JT, Heagerty AH, Bowers B, Hand P, Hayes JD, Jones PW, Strange RC, Fryer AA. Polymorphism at the glutathione S-transferase locus GSTM3: interactions with cytochrome P450 and glutathione S-transferase genotypes as risk factors for multiple cutaneous basal cell carcinoma. Cancer Res. 1996;56:1974–1977. [PubMed] [Google Scholar]
- 37.Marshall SE, Bordea C, Haldar NA, Mullighan CG, Wojnarowska F, Morris PJ, Welsh KI. Glutathione S-transferase polymorphisms and skin cancer after renal transplantation. Kidney Int. 2000;58:2186–2193. doi: 10.1111/j.1523-1755.2000.00392.x. [DOI] [PubMed] [Google Scholar]
- 38.Ramsay HM, Harden PN, Reece S, Smith AG, Jones PW, Strange RC, Fryer AA. Polymorphisms in glutathione S-transferases are associated with altered risk of nonmelanoma skin cancer in renal transplant recipients: a preliminary analysis. J Invest Dermatol. 2001;117:251–255. doi: 10.1046/j.0022-202x.2001.01357.x. [DOI] [PubMed] [Google Scholar]
- 39.Fryer AA, Ramsay HM, Lovatt TJ, Jones PW, Hawley CM, Nicol DL, Strange RC, Harden PN. Polymorphisms in glutathione S-transferases and non-melanoma skin cancer risk in Australian renal transplant recipients. Carcinogenesis. 2005;26:185–191. doi: 10.1093/carcin/bgh291. [DOI] [PubMed] [Google Scholar]
- 40.Dolzan V, Rudolf Z, Breskvar K. Genetic susceptibility to environmental carcinogenesis in Slovenian melanoma patients. Acta Dermatovenerol Alp Pannonica Adriat. 2006;15:69–78. [PubMed] [Google Scholar]
- 41.Leite JL, Morari EC, Granja F, Campos GM, Guilhen AC, Ward LS. Influence of the glutathione s-transferase gene polymorphisms on the susceptibility to basal cell skin carcinoma. Rev Med Chil. 2007;135:301–306. doi: 10.4067/s0034-98872007000300004. [DOI] [PubMed] [Google Scholar]
- 42.Mossner R, Anders N, Konig IR, Kruger U, Schmidt D, Berking C, Ziegler A, Brockmoller J, Kaiser R, Volkenandt M, Westphal GA, Reich K. Variations of the melanocortin-1 receptor and the glutathione-S transferase T1 and M1 genes in cutaneous malignant melanoma. Arch Dermatol Res. 2007;298:371–379. doi: 10.1007/s00403-006-0708-7. [DOI] [PubMed] [Google Scholar]
- 43.Xie F, Z Z, Li Z, Yu H, Li L, Li H. Relationship between the deletion of GSTM1, GSTT1 genes and susceptibility to malignant of nail matrix nnevus. Zhong Hua Shi Yan Wai Ke Za Zhi. 2011;27:1279–1231. [Google Scholar]
- 44.Chiyomaru K, Nagano T, Nishigori C. Polymorphisms of glutathione S-transferase in skin cancers in a Japanese population. Kobe J Med Sci. 2011;57:E11–16. [PubMed] [Google Scholar]
- 45.Ibarrola-Villava M, Martin-Gonzalez M, Lazaro P, Pizarro A, Lluch A, Ribas G. Role of glutathione S-transferases in melanoma susceptibility: association with GSTP1 rs1695 polymorphism. Br J Dermatol. 2012;166:1176–1183. doi: 10.1111/j.1365-2133.2012.10831.x. [DOI] [PubMed] [Google Scholar]
- 46.Lytvynets L. [Contribution of genes of xenobiotic detoxification in children with bronchial asthma] . Lik Sprava. 2013:65–69. [PubMed] [Google Scholar]
- 47.Xu ZB, Zou XP, Zhang N, Feng QL, Zheng SC. Detoxification of insecticides, allechemicals and heavy metals by glutathione S-transferase SlGSTE1 in the gut of Spodoptera litura. Insect Sci. 2014 doi: 10.1111/1744-7917.12142. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 48.Lusini L, Tripodi SA, Rossi R, Giannerini F, Giustarini D, del Vecchio MT, Barbanti G, Cintorino M, Tosi P, Di Simplicio P. Altered glutathione anti-oxidant metabolism during tumor progression in human renal-cell carcinoma. Int J Cancer. 2001;91:55–59. doi: 10.1002/1097-0215(20010101)91:1<55::aid-ijc1006>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]


