Abstract
Background: Glioma is the most devastating type of malignant brain tumors in adults. Genetic factors play important roles in the pathogenesis of glioma. In recent years, some studies found that there were significant association between regulator of telomere elongation helicase 1 rs6010620 polymorphism and glioma susceptibility, however, the results were controversial. The aim of this study was to obtain a more exact estimation of the association between regulator of telomere elongation helicase 1 rs6010620 polymorphism and glioma through a meta-analysis. Methods: The meta-analysis included 19 published case-control studies involving 8541 cases and 14226 controls. The included papers were searched from PubMed and Embase database. Odds ratio (OR) with 95% confidence interval (95% CI) were used to evaluate the association of regulator of telomere elongation helicase 1 rs6010620 polymorphism with glioma. Results: A significant association between regulator of telomere elongation helicase 1 rs6010620 polymorphism and glioma susceptibility was observed for GG vs. AA+AG (OR=1.28, 95% CI=1.14-1.43) and G vs. A (OR=1.07, 95% CI=1.03-1.10). Further subgroup analysis based on ethnicity showed similar results in Asians and Caucasians. In the subgroup analysis of source of control, a significant association between the G allele and glioma susceptibility were found in population-based group and hospital-based group. Conclusions: The meta-analysis suggested that regulator of telomere elongation helicase 1 rs6010620 polymorphism was a risk factor for glioma. And this study also suggested that rs6010620 GG genotype and G allele may be indicators for the risk of glioma.
Keywords: Regulator of telomere elongation helicase 1, glioma, polymorphism, susceptibility
Introduction
Glioma, the most common type of brain tumors in adults, arises from glial cells and starts in the brain or spine, which accounts for about 30% of all tumors in central nervous system and 80% of malignant tumors in brain [1]. The pathogenesis of glioma involves various factors including pollution, living environment, electromagnetic radiation (cells phones), infection and genetic factor. Among the factors, genetic factors play an important role in the pathogenesis of glioma [2,3].
Regulator of telomere elongation helicase 1 gene, located in 20q13.3, plays an important role in DNA repair, ATP-dependent DNA helicase activity, acid binding, and apotosis [4,5]. Previous researches have suggested that RTELI contributes to genomic stability, DNA replication and telomere maintenance [4,6]. Moreover, the inactivation of regulator of telomere elongation helicase 1 could cause chromosome breaks, fusions and telomere loss [7]. In addition, the increasing studies have showed that there exists significant association between regulator of telomere elongation helicase 1 polymorphisms and glioma susceptibility.
Among the polymorphisms, regulator of telomere elongation helicase 1 rs6010620 is the most studied single nucleotide polymorphism (SNP). However, the relationship of regulator of telomere elongation helicase 1 rs6010620 and glioma was still inconclusive. One meta-analysis conducted by Zhao et al., have suggested that regulator of telomere elongation helicase 1 rs6010620 is associated with the increased risk for glioma under four genetic models [8]. While, Li et al. have reported that the GG genotype of rs6010620 acts as the protective genotype for glioma [9]. Therefore, we conducted a meta-analysis with 8541 cases and 14226 controls to derive a more precise estimation of the correlation between regulator of telomere elongation helicase 1 rs6010620 and glioma risk.
Materials and methods
Search strategy and inclusion criteria
We searched in PubMed and Embase databases with the following key words “regulator of telomere elongation helicase 1”, “RTEL1”, “polymorphism” and “glioma”.
Inclusion criteria were defined as follows: (1) case-control studies estimating the relationship of regulator of telomere elongation helicase 1 rs6010620 with glioma risk; (2) sufficient data for evaluating the odds ratio (OR) with 95% CI; (3) data collection and analysis must be statistically acceptable. If the studies with overlapping data published by the same investigators, we included the most recent or complete study.
Data extraction
The data were extracted by two investigators according to the inclusion criteria. For controversial evaluation, the investigators should discuss with other members of the team until a consensus was reached.
The data extracted from the articles included the name of first author, publication date, ethnicity, country of origin, number of cases and controls, genotyping method, genotype frequencies in cases and controls and source of the control and Hardy-Weinberg equilibrium (HWE).
Statistical analysis
Pooled ORs with 95% CIs were conducted to assess the strength of the association between regulator of telomere elongation helicase 1 rs6010620 and glioma risk. The pooled ORs were performed for GG vs. AA, GG+AG vs. AA, GG vs. AA+AG, G vs. A, AG vs. AA. In the subgroup analysis, statistical analysis was conducted in Asians and Caucasians. Z test was used to evaluate whether the pooled ORs were significant. P<0.05 was considered statistically significant. Heterogeneity assumption was testified by Q test. The pooled ORs were calculated by the fixed-effects model when P (heterogenity) >0.05. Otherwise, the random-effects model was used. We adopted Begg’s funnel plots and Egger’s test to assess the publication bias. HWE was checked by χ2 test. The sensitivity analysis was conducted repeatedly by precluding a single study every time in multiple genetic models. Statistical analysis was performed with STATA version 12.0 (Stata Corporation, College Station, TX, USA).
Results
Articles search and the characteristics of the studies
As listed in Figure 1, a total of 143 relevant articles were identified. According to the inclusion criteria, 124 studies were excluded: 6 studies for overlapping data, 79 studies for unrelated research, 17 studies for no control group and 22 studies for no locus. Finally, 19 studies were considered acceptable and included into our meta-analysis. The characteristics of nineteen studies were shown in Table 1.
Figure 1.
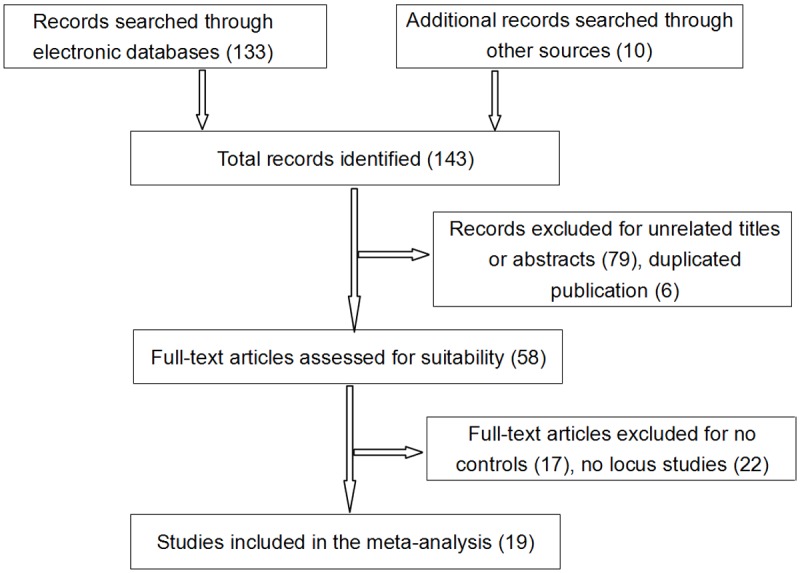
Flow diagram of the study selection process for the meta-analysis.
Table 1.
Principle characteristics of the studies included in the meta-analysis
| First author | Year | Country | Ethnicity | Control source | Genotyping method | Case | Control | HWE | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||
| Sample size | AA | AG | GG | A | G | Sample size | AA | AG | GG | A | G | |||||||
| Shete (England) | 2009 | America | Caucasian | PB | PCR/MALDI-TOF-MS | 631 | 26 | 179 | 426 | 231 | 1031 | 1433 | 82 | 533 | 818 | 697 | 2169 | 0.69 |
| Shete (America) | 2009 | America | Caucasian | HB | PCR/MALDI-TOF-MS | 1247 | 46 | 405 | 796 | 497 | 1997 | 2235 | 123 | 785 | 1327 | 1031 | 3439 | 0.62 |
| Shete (France) | 2009 | America | Caucasian | PB | PCR/MALDI-TOF-MS | 1332 | 34 | 386 | 912 | 454 | 2210 | 1545 | 59 | 508 | 978 | 626 | 2464 | 0.49 |
| Shete (German) | 2009 | America | Caucasian | PB | PCR/MALDI-TOF-MS | 499 | 16 | 147 | 336 | 179 | 819 | 557 | 28 | 177 | 352 | 233 | 881 | 0.35 |
| Shete (Sweden) | 2009 | America | Caucasian | PB | PCR/MALDI-TOF-MS | 645 | 20 | 195 | 430 | 235 | 1055 | 774 | 54 | 264 | 456 | 372 | 1176 | 0.07 |
| Schoemaker (Denmark) | 2010 | England | Caucasian | PB | PCR | 122 | 1 | 38 | 83 | 40 | 204 | 147 | 8 | 56 | 83 | 72 | 222 | 0.72 |
| Schoemaker (Finland) | 2010 | England | Caucasian | PB | PCR | 95 | 4 | 22 | 69 | 30 | 160 | 96 | 3 | 30 | 63 | 36 | 156 | 0.80 |
| Schoemaker (Sweden) | 2010 | England | Caucasian | PB | PCR | 200 | 4 | 52 | 144 | 60 | 340 | 371 | 25 | 116 | 230 | 166 | 576 | 0.05 |
| Schoemaker (UK-North) | 2010 | England | Caucasian | PB | PCR | 376 | 18 | 106 | 252 | 142 | 610 | 632 | 44 | 212 | 376 | 300 | 964 | 0.07 |
| Schoemaker (UK-South) | 2010 | England | Caucasian | PB | PCR | 232 | 8 | 65 | 159 | 81 | 383 | 390 | 28 | 129 | 233 | 185 | 595 | 0.09 |
| Chen | 2011 | China | Asian | HBl | MassARRAY | 958 | 411 | 454 | 93 | 1276 | 640 | 1040 | 547 | 438 | 55 | 1532 | 548 | 0.01 |
| Wang | 2011 | America | Caucasian | Mixed | HumanHap | 332 | 15 | 99 | 218 | 129 | 535 | 817 | 49 | 296 | 472 | 394 | 1240 | 0.77 |
| Li | 2013 | China | Asian | PB | MassARRAY | 629 | 293 | 261 | 75 | 847 | 411 | 644 | 337 | 267 | 40 | 941 | 347 | 0.18 |
| Safaeian (NCI) | 2013 | America | Caucasian | PB | HumanHap | 322 | 11 | 93 | 218 | 115 | 529 | 385 | 20 | 134 | 231 | 174 | 596 | 0.92 |
| Safaeian (NIOSH) | 2013 | America | Caucasian | PB | HumanHap | 300 | 12 | 106 | 182 | 130 | 470 | 539 | 25 | 175 | 339 | 225 | 853 | 0.69 |
| Safaeian (PLCO) | 2013 | America | Caucasian | PB | HumanHap | 133 | 0 | 29 | 104 | 29 | 237 | 855 | 51 | 323 | 481 | 425 | 1285 | 0.74 |
| Safaeian (ATBC) | 2013 | Finland | Caucasian | PB | HumanHap | 37 | 1 | 11 | 25 | 13 | 61 | 1269 | 51 | 374 | 844 | 476 | 2062 | 0.24 |
| Safaeian (AHS) | 2013 | America | Caucasian | PB | HumanHap | 18 | 2 | 6 | 10 | 10 | 26 | 35 | 1 | 8 | 26 | 10 | 60 | 0.69 |
| Jin | 2013 | China | Asian | PB | MassARRAY | 433 | 48 | 181 | 204 | 277 | 589 | 462 | 24 | 202 | 236 | 250 | 674 | 0.02 |
PCR: polymerase chain reaction; MALDI-TOF: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; TaqMan: TaqManSNP; NCI: the National Cancer Institute; NIOSH: the National Institute for Occupational Safety and Health; PLCO: the Prostate, Lung, Colorectal and Ovarian; ATBC: the Alpha-Tocopherol, Beta-Carotene; AHS: the Agricultural Health Study; HWE: Hardy-Weinberg equilibrium.
Meta analysis results
As shown in Table 2, the overall ORs with 95% CIs demonstrated that the presence of regulator of telomere elongation helicase 1 rs6010620 polymorphism with GG genotype or G allelle was an increased factor for glioma risk (GG vs. AA+AG: OR=1.28, 95% CI=1.14-1.43; G vs. A: OR=1.07, 95% CI=1.03-1.10). In the subgroup analysis by ethnicity, findings were similar in Asians (GG vs. AA: OR=1.31, 95% CI=1.09-1.57; G vs. A: OR=1.13, 95% CI=1.04-1.23) and Caucasians (GG vs. AA+AG: OR=1.25, 95% CI=1.11-1.41; G vs. A: OR=1.06, 95% CI=1.02-1.09). In the subgroup analysis by source of control, elevated risk was observed with G allele based on population (G vs. A: OR=1.06, 95% CI=1.02-1.10) and hospital (G vs. A: OR=1.09, 95% CI=1.02-1.16) (Figures 2, 3).
Table 2.
Regulator of telomere elongation helicase 1 rs6010620 polymorphism and glioma risk
| GG versus AA | GG+AG versus AA | GG versus AA+AG | G versus A | AG versus AA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| OR (95% CI) | Ph | OR (95% CI) | Ph | OR (95% CI) | Ph | OR (95% CI) | Ph | OR (95% CI) | Ph | ||
| Ethnicity | Caucasian | 1.04 (0.98, 1.10) | 1.000 | 1.02 (0.98, 1.07) | 1.000 | 1.25 (1.11, 1.41) | 0.000 | 1.06 (1.02, 1.09) | 0.998 | 1.04 (0.96, 1.12) | 1.000 |
| Asian | 1.31 (1.09, 1.57) | 0.000 | 1.10 (1.00, 1.22) | 0.125 | 1.46 (0.86, 2.45) | 0.000 | 1.13 (1.04, 1.23) | 0.005 | 1.09 (0.97, 1.22) | 0.195 | |
| Source of control | Population | 1.05 (0.98, 1.11) | 0.775 | 1.02 (0.98, 1.07) | 1.000 | 1.28 (1.12, 1.47) | 0.000 | 1.06 (1.02, 1.10) | 0.864 | 1.03 (0.95, 1.12) | 1.000 |
| Hospital | 1.11 (0.99, 1.25) | 0.000 | 1.07 (0.99, 1.16) | 0.069 | 1.37 (0.81, 2.31) | 0.004 | 1.09 (1.02, 1.16) | 0.010 | 1.11 (0.99, 1.24) | 0.275 | |
| Mixed | 1.03 (0.83, 1.29) | 0.000 | 1.02 (0.85, 1.22) | 0.000 | 1.14 (0.93, 1.40) | 0.000 | 1.06 (0.93, 1.22) | 0.000 | 1.01 (0.74, 1.38) | 0.000 | |
| Total | 1.06 (1.00, 1.11) | 0.160 | 1.03 (0.99, 1.08) | 0.995 | 1.28 (1.14, 1.43) | 0.000 | 1.07 (1.03, 1.10) | 0.547 | 1.05 (0.99, 1.12) | 0.999 | |
Ph: P-value of heterogeneity test.
Figure 2.
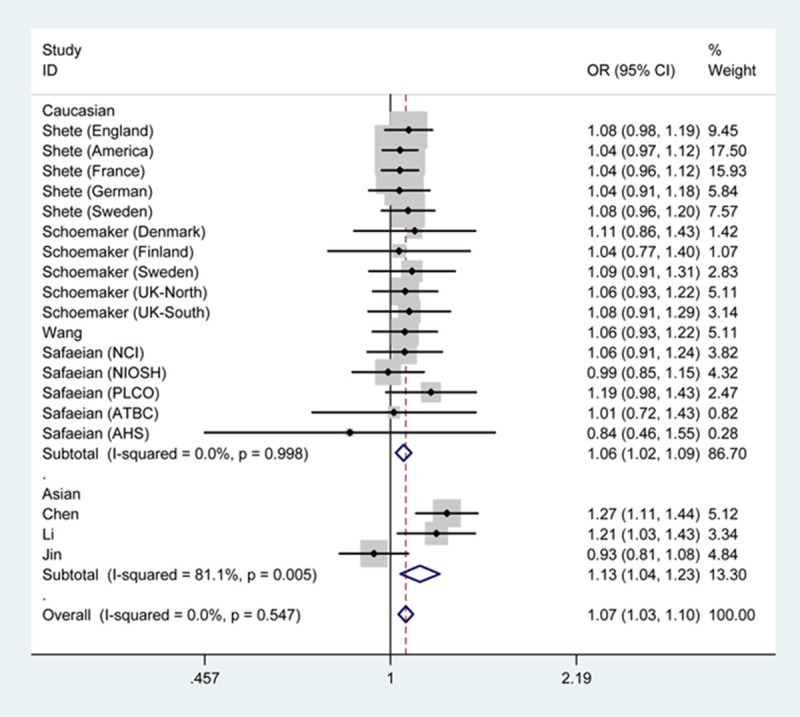
Forest plot of glioma risk associated with regulator of telomere elongation helicase 1 rs6010620 under G vs. A genetic model by ethnicity.
Figure 3.
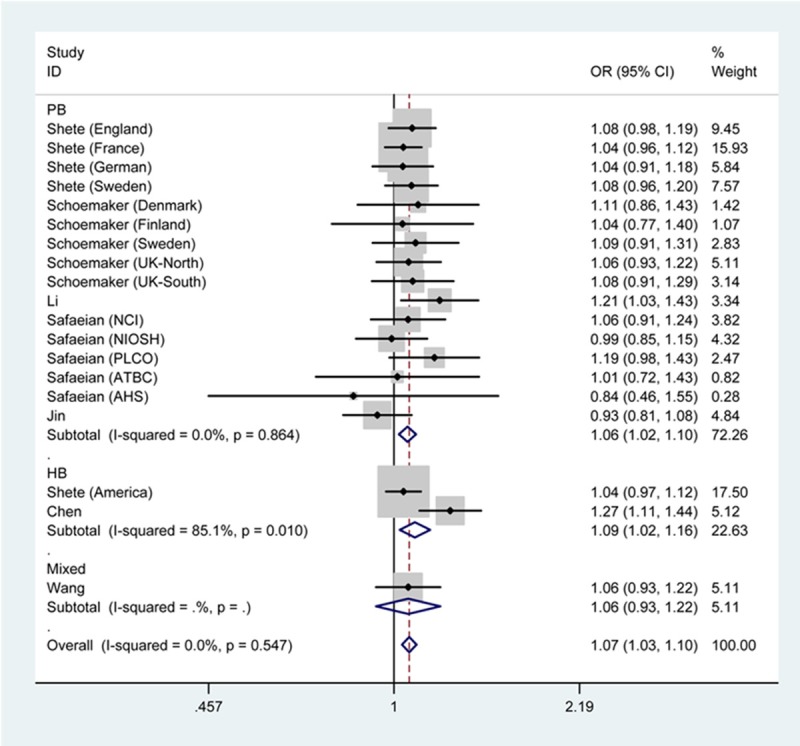
Forest plot of glioma risk associated with regulator of telomere elongation helicase 1 rs6010620 under G vs. A genetic model by source of control.
Sensitivity analysis
The sensitivity analysis was conducted repeatedly by excluding a single study every time in multiple genetic models. The results showed that the corresponding pooled ORs were not altered, suggesting that our meta-analysis results were reliable (data not shown).
Publication bias
We used funnel plot and Egger’s test to estimate the publication bias of literature. As shown in Figure 4, the shape of the funnel plot seemed symmetrical, and Egger’s test had no statistical evidence for publication bias (P=0.791). Thus, there existed no apparent publication bias in the present meta-analysis.
Figure 4.
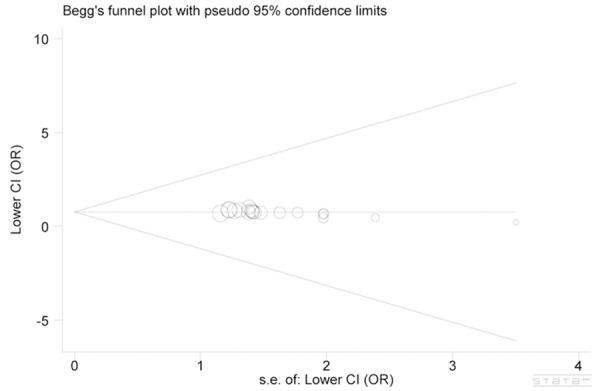
Begg’s funnel plot of publication bias test.
Discussion
Glioma orginating from glial cells is the most common primary tumors of the central nervous system, and it accounts for the vast majority of the malignant brain tumors [10-14]. The incidence rate of glioma is increasing in a number of Asian countries, especially in China. According to the Health Statistics Yearbook 2009 of China, the annual mortality rate of glioma in China was approximately 3.13 per 100,000 population during 2008 [15]. For the etiology, genetic factors had strong effects on the development of glioma [16-21]. Regulator of telomere elongation helicase 1 is an essential DNA helicase that disassembles a variety of DNA secondary structures to maintain telomere integrity [22-25]. As we all know, regulator of telomere elongation helicase 1 had many polymorphic sites and rs6010620 was the most widely studied one. Regulator of telomere elongation helicase 1 polymorphisms play important roles in cancer pathogenesis, since the mutation in regulator of telomere elongation helicase 1 could cause telomere dysfunction [26] that was associated with risk of various cancers [27]. For regulator of telomere elongation helicase 1 polymorphisms, Jin et al. have reported that three locus (rs2297440, rs2853676, and rs6010620) of regulator of telomere elongation helicase 1 are associated with the increased risk of glioma [28]. Walsh et al. also have found that regulator of telomere elongation helicase 1 rs6010620 serves as risk factor for glioma based on Caucasian population [29].
The present meta-analysis, with 8541 cases and 14226 controls, was conducted to derive a more precise assessment between regulator of telomere elongation helicase 1 rs6010620 and glioma susceptibility. The results suggested that the GG genotype of regulator of telomere elongation helicase 1 rs6010620 was significantly associated with the risk for glioma under recessive model. Further subgroup analysis was based on ethnicity and source of control, and the G allele also played an important role in the pathogenesis of glioma.
Overall, our meta-analysis presented regulator of telomere elongation helicase 1 rs6010620 as the genetic-susceptibility factor for glioma. However, there were several limitations that should be addressed. Firstly, we failed to discuss how the risk G allele of rs6010620 polymorphism affect the development of glioma. Further studies are required to research this issue. Secondly, the results were based on unadjusted estimates, which might affect the validity of the association. Finally, lack of considering the effects of other genetic or environmental factors on glioma risk might make our results biased. Therefore, further well-designed investigations based on larger scales are needed to clarify this point of view.
Disclosure of conflict of interest
None.
References
- 1.Mamelak AN, Jacoby DB. Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601) Expert Opin Drug Deliv. 2007;4:175–186. doi: 10.1517/17425247.4.2.175. [DOI] [PubMed] [Google Scholar]
- 2.Foster KA, Jane EP, Premkumar DR, Morales A, Pollack IF. Co-administration of ABT-737 and SAHA induces apoptosis, mediated by Noxa upregulation, Bax activation and mitochondrial dysfunction in PTEN-intact malignant human glioma cell lines. J Neurooncol. 2014;120:459–72. doi: 10.1007/s11060-014-1575-2. [DOI] [PubMed] [Google Scholar]
- 3.Pandey JP, Kaur N, Costa S, Amorim J, Nabico R, Linhares P, Vaz R, Viana-Pereira M, Reis RM. Immunoglobulin genes implicated in glioma risk. Oncoimmunology. 2014;3:e28609. doi: 10.4161/onci.28609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uringa EJ, Lisaingo K, Pickett HA, Brind’Amour J, Rohde JH, Zelensky A, Essers J, Lansdorp PM. RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol Biol Cell. 2012;23:2782–2792. doi: 10.1091/mbc.E12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 6.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, Boulton SJ. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao W, Bian Y, Zhu W, Zou P, Tang G. Regulator of telomere elongation helicase 1 (RTEL1) rs6010620 polymorphism contribute to increased risk of glioma. Tumour Biol. 2014;35:5259–5266. doi: 10.1007/s13277-014-1684-8. [DOI] [PubMed] [Google Scholar]
- 9.Li G, Jin T, Liang H, Zhang Z, He S, Tu Y, Yang H, Geng T, Cui G, Chen C, Gao G. RTEL1 tagging SNPs and haplotypes were associated with glioma development. Diagn Pathol. 2013;8:83. doi: 10.1186/1746-1596-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, Kollmeyer T, Kosel ML, Molinaro AM, McCoy LS, Bracci PM, Cabriga BS, Pekmezci M, Zheng S, Wiemels JL, Pico AR, Tihan T, Berger MS, Chang SM, Prados MD, Lachance DH, O’Neill BP, Sicotte H, Eckel-Passow JE, van der Harst P, Wiencke JK, Samani NJ, Jenkins RB, Wrensch MR. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgasov PN. [Anti-alcohol education of the population] . Sov Zdravookhr. 1986:17–22. [PubMed] [Google Scholar]
- 12.Orlova NV. [Bradycardia arrhythmia in children] . Pediatriia. 1987:79–83. [PubMed] [Google Scholar]
- 13.Noll KM, Rinehart KL Jr, Tanner RS, Wolfe RS. Structure of component B (7-mercaptoheptanoylthreonine phosphate) of the methylcoenzyme M methylreductase system of Methanobacterium thermoautotrophicum. Proc Natl Acad Sci U S A. 1986;83:4238–4242. doi: 10.1073/pnas.83.12.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renard P, Pasqual JC, Minault S, Blaison D, Demange L, Zeitoun P. [Cancer of the esophagus after mediastinal radiotherapy for Hodgkin’s disease. Apropos of a case] . Ann Gastroenterol Hepatol (Paris) 1989;25:299–300. [PubMed] [Google Scholar]
- 15.Bondy ML, Scheurer ME, Malmer B, Barnholtz-Sloan JS, Davis FG, Il’yasova D, Kruchko C, McCarthy BJ, Rajaraman P, Schwartzbaum JA, Sadetzki S, Schlehofer B, Tihan T, Wiemels JL, Wrensch M, Buffler PA. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113:1953–1968. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radner H, el-Shabrawi Y, Eibl RH, Brustle O, Kenner L, Kleihues P, Wiestler OD. Tumor induction by ras and myc oncogenes in fetal and neonatal brain: modulating effects of developmental stage and retroviral dose. Acta Neuropathol. 1993;86:456–465. doi: 10.1007/BF00228580. [DOI] [PubMed] [Google Scholar]
- 17.Fan W, Chen X, Li C, Chen L, Liu P, Chen Z. The analysis of deregulated expression and methylation of the PER2 genes in gliomas. J Cancer Res Ther. 2014;10:636–640. doi: 10.4103/0973-1482.138202. [DOI] [PubMed] [Google Scholar]
- 18.Hu B, Emdad L, Bacolod MD, Kegelman TP, Shen XN, Alzubi MA, Das SK, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) interacts with Akt isoform 2 to control glioma growth, survival and pathogenesis. Cancer Res. 2014;74:7321–32. doi: 10.1158/0008-5472.CAN-13-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Tong X, Gao H, Yan X, Xu X, Sun S, Wang Q, Wang J. Silencing HIWI suppresses the growth, invasion and migration of glioma cells. Int J Oncol. 2014;45:2385–2392. doi: 10.3892/ijo.2014.2673. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Li X, Zhu Y, Yang P. MicroRNA-16 suppresses epithelial-mesenchymal transitionrelated gene expression in human glioma. Mol Med Rep. 2014;10:3310–3314. doi: 10.3892/mmr.2014.2583. [DOI] [PubMed] [Google Scholar]
- 21.Melin B. Genetic causes of glioma: new leads in the labyrinth. Curr Opin Oncol. 2011;23:643–647. doi: 10.1097/CCO.0b013e32834a6f61. [DOI] [PubMed] [Google Scholar]
- 22.Vannier JB, Sarek G, Boulton SJ. RTEL1: functions of a disease-associated helicase. Trends Cell Biol. 2014;24:416–425. doi: 10.1016/j.tcb.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Frizzell A, Nguyen JH, Petalcorin MI, Turner KD, Boulton SJ, Freudenreich CH, Lahue RS. RTEL1 inhibits trinucleotide repeat expansions and fragility. Cell Rep. 2014;6:827–835. doi: 10.1016/j.celrep.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Guen T, Jullien L, Schertzer M, Lefebvre A, Kermasson L, de Villartay JP, Londono-Vallejo A, Revy P. [RTEL1 (regulator of telomere elongation helicase 1), a DNA helicase essential for genome stability] . Med Sci (Paris) 2013;29:1138–1144. doi: 10.1051/medsci/20132912018. [DOI] [PubMed] [Google Scholar]
- 25.Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 26.Gramatges MM, Bertuch AA. Short telomeres: from dyskeratosis congenita to sporadic aplastic anemia and malignancy. Transl Res. 2013;162:353–363. doi: 10.1016/j.trsl.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Z, Glousker G, Molczan A, Fox AJ, Lamm N, Dheekollu J, Weizman OE, Schertzer M, Wang Z, Vladimirova O, Schug J, Aker M, Londono-Vallejo A, Kaestner KH, Lieberman PM, Tzfati Y. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proc Natl Acad Sci U S A. 2013;110:E3408–3416. doi: 10.1073/pnas.1300600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh KM, Rice T, Decker PA, Kosel ML, Kollmeyer T, Hansen HM, Zheng S, McCoy LS, Bracci PM, Anderson E, Hsuang G, Wiemels JL, Pico AR, Smirnov I, Molinaro AM, Tihan T, Berger MS, Chang SM, Prados MD, Lachance DH, Sicotte H, Eckel-Passow JE, Wiencke JK, Jenkins RB, Wrensch MR. Genetic variants in telomerase-related genes are associated with an older age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. Neuro Oncol. 2013;15:1041–1047. doi: 10.1093/neuonc/not051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y, Tong X, Tang LL, Zhou K, Zhong CH, Jiang S. Associations between the rs6010620 polymorphism in RTEL1 and risk of glioma: a meta-analysis of 20,711 Participants. Asian Pac J Cancer Prev. 2014;15:7163–7167. doi: 10.7314/apjcp.2014.15.17.7163. [DOI] [PubMed] [Google Scholar]


