Abstract
Objectives: MicroRNAs (miRNAs) are small, non-coding RNAs that have been increasingly shown important roles in various classes of cancers. However, miR-1269 has not been comprehensively studied in hepatocellular carcinoma (HCC). Thus, the purpose of the study was to evaluate the relationship between the expression of miR-1269 and clinicopathological parameters in HCC patients, and to predict its potential target genes. Methods: Total RNA was extracted from 95 pairs of HCC and matching adjacent non-cancerous tissues. The level of miR-1269 expression was detected by using quantitative real-time RT-PCR and calculated with the 2-ΔCq method. Eighteen online biological databases were used for targets prediction. Results: MiR-1269 expression was up-regulated in HCC tissues (1.9264±0.7160) compared to their non-tumor livers (1.5518±0.7273, P < 0.001). Level of miR-1269 was positively correlated to tumor nodes (r = 0.206, P = 0.046), metastasis (r = 0.203, P = 0.049), portal vein tumor embolus (r = 0.247, P = 0.016), vaso-invasion (r = 0.273, P = 0.008), tumor capsular infiltration (r = 0.407, P < 0.001) and expression of MTDH (r = 0.211, P = 0.005). Finally, 7 databases could be applied for the target prediction successfully. There were 9 targeted genes which had been shown concurrently by at least 4 databases: AGAP1, AGK, BPTF, C16orf74, DACT1, LIX1L, RBMS3, ZNF706 and BMPER. Conclusions: MiR-1269 may be possibly involved in the tumorigenesis and progress of HCC. MiR-1269 could also act as a potential biomarker for the prognosis prediction for HCC.
Keywords: miR-1269, hepatocellular carcinoma, paraffin-embedded tissues, RT-qPCR, target prediction
Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs that negatively regulate target genes by binding to their 3’-untranslated region (UTR) [1]. The regulation is acted by triggering degradation or repression of translation [2]. The combination between miRNAs and their mRNA targets could be imperfect, allowing a single miRNA to potentially modulate multiple genes [3]. Although the function of most miRNAs is presently unaware, these molecules have been involved in various biological processes including cell proliferation, differentiation and progression [4].
An increasing number of articles suggested that miRNAs are implicated in the proliferation, apoptosis and metastasis of hepatocellular carcinoma (HCC) cells [4,5]. Thus, it is of great significance to discover the abnormal expressed miRNAs and their targets, which will provide corresponding theoretical basis to reveal the underlying mechanisms and to seek effective therapeutic targets for HCC.
In the present study, we examined miR-1269 expression in HCC tissues and found that miR-1269 was frequently up-regulated in HCC. Notably, the higher expression of miR-1269 in HCC group was significantly associated with its clinicopathological parameters, especially with tumor capsular infiltration, vascular invasion and portal vein tumor thrombus, which was measured by quantitative real-time polymerase chain reaction (qRT-PCR).
Materials and methods
Tissue samples
HCC and matched adjacent non-tumor tissues (n = 95) were obtained from patients who accepted routine surgery at the First Affiliated Hospital of the Guangxi Medical University (Nanning, China) between March 2010 and December 2011. Among them, 75 were males and 20 were females, with a mean age of 52 year-old, ranged from 29 to 82 year-old. None of the patients received chemotherapy or radiation therapy prior to resection. The informed consent was obtained. Their clinicopathological data, which had been collected from medical records, were summarized in Table 1.
Table 1.
Relationship between the expression of miR-1269 and clinicopathological parameters in HCC
| Clinicopathological Feature | n | miR-1269 relevant expression (2-ΔCq) | |||
|---|---|---|---|---|---|
|
| |||||
| Mean ± SD | t | P | |||
| Tissue | Adjacent non-cancerous liver | 95 | 1.5518±0.7273 | 3.578 | < 0.001 |
| HCC | 95 | 1.9264±0.7160 | |||
| Age | ≥ 50 | 46 | 1.9070±0.7156 | 0.255 | 0.799 |
| < 50 | 49 | 1.9447±0.7232 | |||
| Gender | male | 75 | 1.8645±0.7315 | 1.646 | 0.103 |
| female | 20 | 2.1585±0.6165 | |||
| Differentiation | high | 6 | 2.1183±0.8424 | *F = 0.794 | 0.455 |
| moderate | 60 | 1.9697±0.6880 | |||
| low | 29 | 1.7972±0.7535 | |||
| Size | < 5 cm | 18 | 2.1050±0.9294 | 1.178 | 0.242 |
| ≥ 5 cm | 77 | 1.8847±0.6569 | |||
| Tumor nodes | single | 52 | 1.7937±0.6762 | 2.020 | 0.046 |
| multiple | 43 | 2.0870±0.7375 | |||
| Metastasis | Without metastasis | 46 | 1.7667±0.6838 | 2.146 | 0.034 |
| With metastasis | 49 | 2.0763±0.7199 | |||
| Clinical TNM stage | I~II | 22 | 1.6609±0.7819 | 2.016 | 0.047 |
| III~IV | 73 | 2.0064±0.6804 | |||
| Portal vein tumor embolus | - | 63 | 1.8017±0.6475 | 2.444 | 0.016 |
| + | 32 | 2.1719±0.7887 | |||
| Vaso-invasion | - | 59 | 1.7817±0.6803 | 2.598 | 0.011 |
| + | 36 | 2.1636±0.7188 | |||
| Tumor capsular infiltration | With complete capsule | 45 | 1.6327±0.6334 | 4.100 | < 0.001 |
| No capsule or infiltration | 50 | 2.1908±0.6875 | |||
| HCV | - | 63 | 1.8694±0.6890 | 1.091 | 0.278 |
| + | 32 | 2.0388±0.7650 | |||
| HBV | - | 17 | 2.0994±0.9718 | 0.853 | 0.404 |
| + | 78 | 1.8887±0.6492 | |||
| AFP | - | 41 | 1.9029±0.7423 | 0.586 | 0.560 |
| + | 38 | 1.9979±0.6847 | |||
| Cirrhosis | - | 50 | 1.8636±0.7643 | 0.901 | 0.370 |
| + | 45 | 1.9962±0.6597 | |||
| nm23 | - | 20 | 1.8110±0.8699 | 0.810 | 0.420 |
| + | 75 | 1.9572±0.6725 | |||
| MTDH | - | 131 | 1.5931±0.6876 | 3.127 | 0.002 |
| + | 47 | 1.9694±0.7357 | |||
| P53 | - | 40 | 1.8470±0.7444 | 0.921 | 0.359 |
| + | 55 | 1.9842±0.6958 | |||
| P21 | - | 62 | 1.8647±0.7382 | 1.154 | 0.251 |
| + | 33 | 2.0424±0.6677 | |||
| VEGF | - | 25 | 1.8248±0.8502 | 0.735 | 0.467 |
| + | 70 | 1.9627±0.6647 | |||
| Ki-67 LI | Low | 47 | 1.8670±0.6835 | 0.799 | 0.427 |
| High | 48 | 1.9846±0.7490 | |||
| MVD | Low | 47 | 1.9555±0.7505 | 0.390 | 0.697 |
| High | 48 | 1.8979±0.6872 | |||
ANOVA was performed to analyze the difference of miR-1269 among differentiation grading.
qRT-PCR
Isolation and normalization of RNA were performed as reported [6-9]. MiR-1269 expression levels were measured by using a mirVana qRT-PCR miRNA Detection kit (Ambion Inc., Austin, TX, USA). The combination of RNU6B and RNU48 was served as internal reference. qRT-PCR was conducted using Applied Biosystems PCR7900. cDNA was synthesized using TaqMan® MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems, Life Technologies Grand Island, NY 14072 USA) in a total volume of 10 µl per reaction. The sequence of targeted miRNA and reference miRNAs in the study was as follow: miR-1269 (Applied Biosystems Cat. No. 4427975-002789): 5’-CUGGACUGAGCCGUGCUACUGG-3’; RNU6B (Applied Biosystems Cat. No. 4427975-001093): CGCAAGGAUGACACGCAAAUUCGUGAAGCGUUCCAUAUUUUU; RNU48 (Applied Biosystems Cat. No. 4427975-001006): GAUGACCCCAGGUAACUCUGAGUGUGUCGCUGAUGCCAUCACCGCAGCGCUCUGACC. The expression of miR-1269 in the FFPE experiments was calculated with the formula 2-Δcq [6,9,10].
Statistical analysis
The data are recorded as the mean ± SD. All statistical analyses were performed using SPSS 20.0 (Munich, Germany). Student’s t-test was utilized to analyze the significance of difference between two groups. One-way analysis of variance (ANOVA) test was appropriated for the data which was divided into three groups such as differentiation. Spearman correlation was applied to study the relationship between miR-1269 expression and clinicopathological parameters. Receiver operating characteristic (ROC) curve was drawn to test the effectiveness of miR-1269 when distinguishing HCC from their non-tumor livers. P < 0.05 was considered to indicate a statistically significant difference.
MiRNA target prediction
To determine the potential target genes, 18 online biological databases were attempted. However, only 7 could be processed: DIANA-MICROT (http://diana.imis.athena-innovation.gr/), MICRORNA.ORG (http://www.microrna.org/microrna/home.do), MIRDB (http://mirdb.org/miRDB/), RNA22-HSA (https://cm.jefferson.edu/), TARGETSCAN (http://www.targetscan.org/), miRWlak (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/), and PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html). We recorded the top 100 target genes in each database and made a comparison between them. Only genes emerging more than four times would be noted in the current study.
Results
miR-1269 was significantly up-regulated in HCC tissues
MiR-1269 was significantly up-regulated in HCC tissues (1.9264±0.7160) compared with their matching adjacent non-cancerous tissues (1.5518±0.7273, P < 0.001; Table 1; Figure 1A). Furthermore, ROC curve was performed to prove the diagnostic role of miR-1269. The area under curve (AUC) of miR-1269 was 0.640 (95% CI: 0.562-0.719, P = 0.001, Figure 1B).
Figure 1.
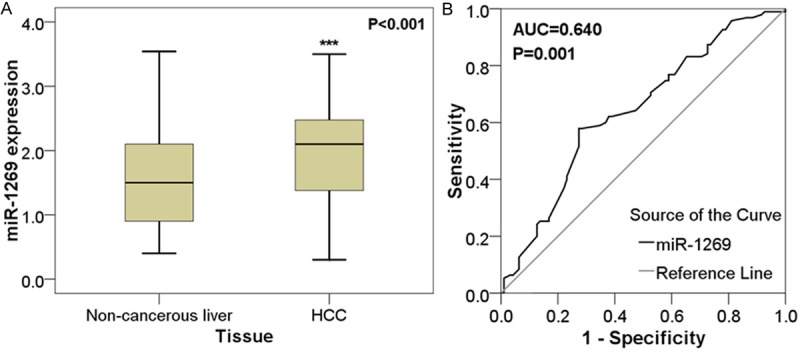
Expression of miR-1269 in adjacent non-tumor liver tissues and HCC tissues. Quantitative real-time RT-PCR was performed to detect the expression of miR-1269. A. The difference of relevant miR-1269 expression between adjacent non-tumor liver tissues and HCC tissues. ***P < 0.001; B. ROC curve of miR-1269 expression to distinguish HCC from adjacent non-tumor liver tissues. The area under curve (AUC) of miR-1269 was 0.640 (95% CI: 0.562-0.719, P = 0.001).
Relevance between miR-1269 expression levels and clinicopathological characteristics
To examine the clinicopathological significance of miR-1269 in HCC tissues, Student’s t-test analysis method was applied. The relative level of miR-1269 in HCC patients with multiple tumor nodes (2.0870±0.7375) was prominently higher than those with single tumor nodes (1.7937±0.6762, P = 0.046). miR-1269 level in patients with metastasis (2.0763±0.7199) was up-regulated by comparison with the patients without metastasis (1.7667±0.6838, P = 0.034). Compared to early stages (I & II, 1.6609 ±0.7819), the relevant level of miR-1269 in advanced stages (III & IV, 2.0064±0.6804, P = 0.047) markedly increased. Subsequently, In comparison with those with portal vein tumor embolus (2.1719±0.7887), the expression of miR-1269 was reduced in HCC patients without portal vein tumor embolus (1.8017±0.6475, P = 0.016). The relative level of miR-1269 in HCC with vaso-invasion (2.1636±0.7188) obviously increased than those without (1.7817±0.6803, P = 0.011). Additionally, miR-1269 expression levels strikingly increased in the tissues with no capsule or with infiltration (2.1908±0.6875) compared with those with complete capsule (1.6327±0.6334, P < 0.001). And level of miR-1269 was found much higher in HCC patients with MTDH positive expression (1.9694±0.7357) than those with MTDH negative expression (1.5931±0.6876, P = 0.002) (Figure 2).
Figure 2.
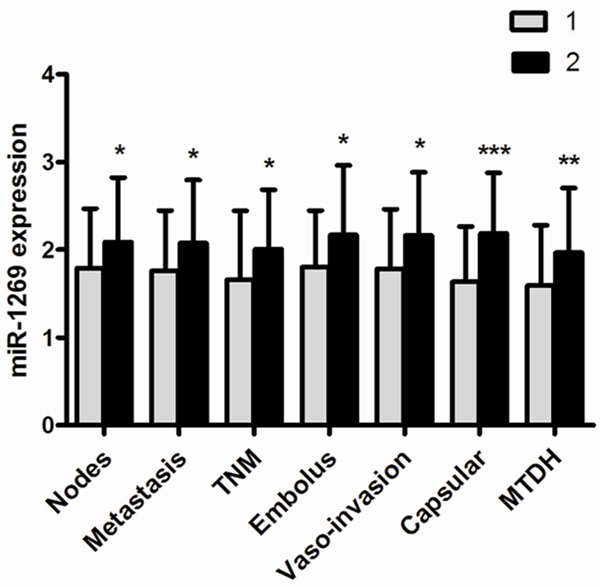
The relationship between miR-1269 and clinical parameters. A. Tumor nodes: 1. single tumor nodes; 2. multiple. B. Metastasis: 1. No; 2. Yes. C. Clinical TNM stage: 1. I-II; 2. III-IV. D. Portal vein tumor embolus: 1. No; 2. Yes. E. Vaso-invasion: 1. No; 2. Yes. F. Tumor capsular infiltration: 1. complete capsule; 2. no capsule or infiltration. G. Expression of MTDH: 1. low level; 2. high level. *P < 0.05; **P < 0.01; ***P < 0.001.
Simultaneously, the Spearman correlation test between the relative expression levels of miR-1269 and its clinicopathological features demonstrated that there were significant positive correlations between the high expression of miR-1269 and a certain of parameters, such as tumor capsular infiltration (r = 0.407, P < 0.001), vaso-invasion (r = 0.273, P = 0.008), portal vein tumor embolus (r = 0.247, P = 0.016), tumor nodes (r = 0.206, P = 0.045) and metastasis (r = 0.203, P = 0.049). However, the miR-203 expression has no association with other features.
ROC analyses of clinicopathological data
ROC curve was implemented to confirm the predictive value of miR-1269 level in HCC patients for clinicopathological characteristics. The AUC with tumor capsular infiltration was 0.735 (95% CI: 0.629~0.841, P < 0.001). The AUC in patients with portal vein tumor embolus was 0.651 (95% CI: 0.526~0.775, P = 0.017). ROC curve displayed an AUC of 0.619 (95% CI: 0.504-0.735, P = 0.046) to predict tumor nodes. However, there are no inferior diagnostic values for other features (Figure 3).
Figure 3.
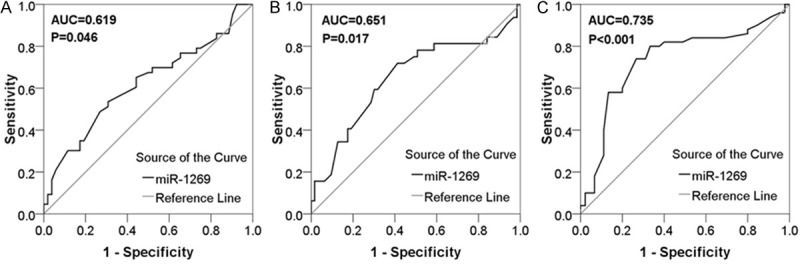
ROC curve of miR-1269 expression of clinicopathological parameters. A. ROC curve of tumor nodes. The AUC was 0.619 (95% CI: 0.504-0.735, P = 0.046). B. ROC curve of portal vein tumor embolus. The AUC was 0.651 (95% CI: 0.526~0.775, P = 0.017). C. ROC curve of tumor capsular. The area under curve (AUC) was 0.735 (95% CI: 0.629~0.841, P < 0.001).
Role of miR-1269 expression in recurrence of HCC
Among the total cases, we carried out an ideal follow-up for 70 cases whose overall recurrence time was 57.095±2.876 months. The median time of follow-up was 32.78 months (range 2.68-68.00 months). Concerning the 70 HCC patients, 31 had high miR-1269 expression while 39 had low expression. In aspect of recurrence, increased expression group of miR-1269 was 32.290±13.749 months, slightly shorter than that in the decreased expression group (33.250±14.832 months). However, no significant difference was found between recurrent time and miR-1269 expression level ( chi-square = 0.008, P = 0.929; Figure 4).
Figure 4.
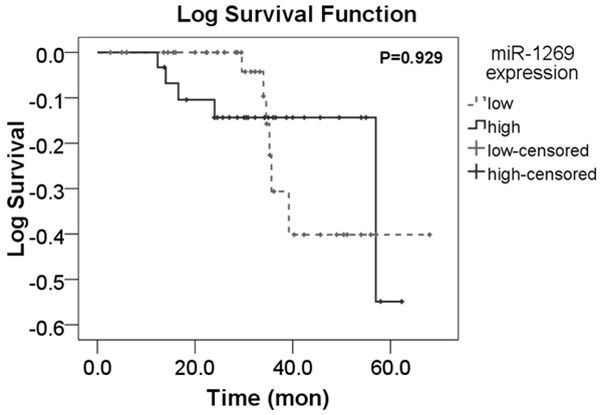
The K-M curve of recurrence between low expression and high expression group of miR-1269.
Targets prediction of miR-1269
Having been searched in 7 database (DIANA MICROT, MICRORNA.ORG, MIRBD, RNA22-HSA, TARGETSCAN, miRWlak and PITA), 9 qualified target genes were found at least in 4 databases. They were as follows: AGAP1, AGK, BMPER, BPTF, C16orf74, DACT1, LIX1L, RBMS3 and ZNF706 (Table 2).
Table 2.
Targets prediction of miR-1269
| Target gene | Biological databases | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| DINAN-MICROT | MICRORNA.ORG | MIRDB | RNA22-HSA | TARGETSCAN | miRWlak | PITA | |
| AGAP1 | √ | √ | √ | √ | |||
| AGK | √ | √ | √ | √ | |||
| BPTF | √ | √ | √ | √ | |||
| C16orf74 | √ | √ | √ | √ | |||
| DACT1 | √ | √ | √ | √ | |||
| LIX1LR | √ | √ | √ | √ | |||
| RBMS3 | √ | √ | √ | √ | |||
| ZNF706 | √ | √ | √ | √ | |||
| BMPER | √ | √ | √ | √ | |||
√: the gene appears in corresponding database.
Discussion
Since the discovery in C. elegans, miRNAs alterations have been certified to play an important role in different steps of tumor formation and progression. Although a large number of researches have analyzed the global expression pattern of miRNAs, there were only 3 reports correlated with miR-1269. Julian [11] detected the high expression of miR-1269 in colonic adenocarcinomas tissues by using the technology of high-throughput sequencing in 8 colorectal cancer patients. Nevertheless, the association between the level of this miR and clinical parameters was not available. Guillaume [12] conducted an experiment using two type cell lines, the enterocyte-like Caco2-BBE and the colonocyte-like HT29-Cl.19A, which were intestinal epithelial cell models. HT29-Cl.19A cells exhibited down-regulation of miR-1269 compared with Caco2-BBE cells. Transfection of Caco2-BBE cells with antisense of mature miR-1269 and other several miRs their shift toward HT29-Cl.19A cell phenotype. The last report was performed by Ryan [13] with Illumina sequencing technology, and miR-1269 was regarded as one of putative novel miRNAs exhibiting significant differential expression in embryonic stem cells. Noteworthily, the latter two studies were in normal cells or tissues. And there have been few, if any, references on the function and role in cancers of miR-1269.
In the present study, we analyzed the expression of miR-1269 in 95 HCC patients and demonstrated that miR-1269 was significantly higher in HCC tissues compared with adjacent non-tumor liver tissues. The phenomenon was in accordance with the result of colorectal carcinoma research. Notably, we investigated the expression of miR-1269 on clinical characteristics. Our data showed that the miR-1269 was repressed in HCC tissues with complete capsule compared to those without capsule, even infiltration (P < 0.001). As HCC developed and there appeared vaso-invasion, miR-1269 was found to be upregulated (P = 0.016). Similarly, miR-1269 was more intense in HCC with portal vein tumor embolus in comparison with those without (P = 0.011). In addition, the high levels of miR-1269 expression also presented in patients with multiple tumor nodes (P = 0.046), in advanced clinical TNM stages (P = 0.047) and with metastasis (P = 0.034). There were also significant positive correlations between miR-1269 and aforementioned clinical parameters as proved by spearman correlation. The evidence also has shown that there were significance differences in miR-1269 expression between high-MTDH-expresssors and low-expressors (P < 0.001). It is proved that MTDH was overexpressed in HCC and indicated the importance of the MDTH pathway in tumorigenesis through EMT [14]. Besides, owing to the role of MTDH in HCC in promotion cell growth and the resistance to anoikis, MTDH provided a novel mechanism supporting HCC metastasis [15]. On the whole, high expression of miR-1269 was correlated with the carcinogenesis, metastasis and invasion of HCC.
Since the mechanism of miR-1269 remained largely unknown, we then attempted to predict the potential target genes of miR-1269. We detected 9 qualified genes after searching in 7 different bioinformatics databases, including DIANA MICROT, MICRORNA.ORG, MIRBD, RNA22-HSA, TARGETSCAN, miRWlak and PITA. Some of the possible targeted genes have been studied in HCC. First, DACT1 (also called HDPR1), was downregulated in HCC through involvement of methylation-mediated gene silencing [16]. Till date, DACT1 is regarded as a tumor suppressor via inhibiting NF-κB signaling and WNT/beta-catenin signaling pathways which were associated with infiltration and metastasis of tumor. Then, RBMS3 was overexpressed in activated hematopoietic stem cells (HSCs) and fibrotic livers and increased expression of transcription factor Prx1, which is one of the factors boost fibrogenic transformation of HSCs [17]. And in nasopharyngeal carcinoma, RBMS3 inhibited its progression through inhibiting cell proliferation, angiogenesis and inducing apoptosis [18]. However, no study was aimed at the expression level of RBMS3 in HCC and other cancers. AGK, a mitochondrial membrane protein, has already been discovered overexpression in esophageal squamous cell carcinoma, breast cancer, lung cancer and prostate cancer [19,20]. BMPER (BMP binding endothelial regulator) is striking highly expressed upon malignant deterioration in lung, colon and cervix carcinomas, owning to its pro-angiogenic features in endothelial cells and their progenitors [21]. The bromodomain PHD finger transcription BPTF was found to be a negative predictor for brain metastasis in primary NSCLC [22]. The translocation breakpoint of BPTF deregulated its expression and conferred the cells with pre-malignant phenotype [23]. The decreased expression of C16orf74 was significantly associated with progression in bladder cancer [24]; Expression of ZNF706 was higher in oral squamous cell carcinoma tissues than in normal reference [25]. There is no data linking to AGAP1, LIX1L. Among these possible target genes, only ACT1 has been reported to be downregulated in HCC. The result of target prediction is consistent with clinical significance. RBMS3 has been found to be related to the liver fibrosis, and no studies have been performed to investigate the role of other genes in HCC. Thus, the genes mentioned above were just conjecture based on the theory. Experiments need to be designed and carried out to explore the contribution of miR-1269 in HCC via targeting several genes.
To our knowledge, our study is the first supported case suggesting the expression of miR-1269 in HCC tissues. To a certain degree, our present findings are in agreement with the prediction target gene, showing that miR-1269 may act as an onco-miR in tumorigenesis progression. In conclusion, our study raises miR-1269 probably as a potential regulator of HCC.
Acknowledgements
The study was supported partly by the Fund of Guangxi Provincial Health Bureau Scientific Research Project (Z2014054), Youth Science Foundation of Guangxi Medical University (GXMUYSF201311), Guangxi University Science and Technology Research Projects (LX2014075), and the Fund of National Natural Science Foundation of China (NSFC 81360327). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Phuah NH, Nagoor NH. Regulation of MicroRNAs by Natural Agents: New Strategies in Cancer Therapies. Biomed Res Int. 2014;2014:804510. doi: 10.1155/2014/804510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Leva G, Briskin D, Croce CM. MicroRNA in cancer: new hopes for antineoplastic chemotherapy. Ups J Med Sci. 2012;117:202–216. doi: 10.3109/03009734.2012.660551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iuliano R, Vismara MF, Dattilo V, Trapasso F, Baudi F, Perrotti N. The role of microRNAs in cancer susceptibility. Biomed Res Int. 2013;2013:591931. doi: 10.1155/2013/591931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J, Lu H, Wang X, Jin H. MicroRNAs in hepatocellular carcinoma: regulation, function, and clinical implications. Scientific World Journal. 2013;2013:924206. doi: 10.1155/2013/924206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibit tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 6.Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS One. 2013;8:e61054. doi: 10.1371/journal.pone.0061054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albertini MC, Olivieri F, Lazzarini R, Pilolli F, Galli F, Spada G, Accorsi A, Rippo MR, Procopio AD. Predicting microRNA modulation in human prostate cancer using a simple String IDentifier (SID1.0) J Biomed Inform. 2011;44:615–620. doi: 10.1016/j.jbi.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci. 2014;119:19–24. doi: 10.3109/03009734.2013.856970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y, Qin W, Cao H, Xu R, Tan Y, Lu T, Wang H, Tong W, Rong X, Li G, Yuan M, Li C, Abe K, Lu L, Chen G. HCV 6a prevalence in Guangdong province had the origin from Vietnam and recent dissemination to other regions of China: phylogeographic analyses. PLoS One. 2012;7:e28006. doi: 10.1371/journal.pone.0028006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One. 2012;7:e34150. doi: 10.1371/journal.pone.0034150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalmasso G, Nguyen HT, Yan Y, Laroui H, Srinivasan S, Sitaraman SV, Merlin D. MicroRNAs determine human intestinal epithelial cell fate. Differentiation. 2010;80:147–154. doi: 10.1016/j.diff.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J, Li C, Wu X, Yang Y, Hao M, Sheng S, Sun Y, Zhang H, Long J, Hu C. Astrocyte elevated gene-1 is a novel biomarker of epithelial-mesenchymal transition and progression of hepatocellular carcinoma in two China regions. Tumour Biol. 2014;35:2265–2269. doi: 10.1007/s13277-013-1300-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Z, Deng H, Yan W, Luo M, Tu W, Xia Y, He J, Han P, Fu Y, Tian D. AEG-1 promotes anoikis resistance and orientation chemotaxis in hepatocellular carcinoma cells. PLoS One. 2014;9:e100372. doi: 10.1371/journal.pone.0100372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yau TO, Chan CY, Chan KL, Lee MF, Wong CM, Fan ST, Ng IO. HDPR1, a novel inhibitor of the WNT/beta-catenin signaling, is frequently downregulated in hepatocellular carcinoma: involvement of methylation-mediated gene silencing. Oncogene. 2005;24:1607–1614. doi: 10.1038/sj.onc.1208340. [DOI] [PubMed] [Google Scholar]
- 17.Fritz D, Stefanovic B. RNA-binding protein RBMS3 is expressed in activated hepatic stellate cells and liver fibrosis and increases expression of transcription factor Prx1. J Mol Biol. 2007;371:585–595. doi: 10.1016/j.jmb.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Kwong DL, Zhu CL, Chen LL, Dong SS, Zhang LY, Tian J, Qi CB, Cao TT, Wong AM, Kong KL, Li Y, Liu M, Fu L, Guan XY. RBMS3 at 3p24 inhibits nasopharyngeal carcinoma development via inhibiting cell proliferation, angiogenesis, and inducing apoptosis. PLoS One. 2012;7:e44636. doi: 10.1371/journal.pone.0044636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Ying Z, Lin X, Lin H, Wu J, Li M, Song L. Acylglycerol kinase augments JAK2/STAT3 signaling in esophageal squamous cells. J Clin Invest. 2013;123:2576–2589. doi: 10.1172/JCI68143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Lin C, Zhao X, Liu A, Zhu J, Li X, Song L. Acylglycerol kinase promotes cell proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Mol Cancer. 2014;13:106. doi: 10.1186/1476-4598-13-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinke J, Kerber M, Rahner S, Mnich L, Lass-mann S, Helbing T, Werner M, Patterson C, Bode C, Moser M. Bone morphogenetic protein modulator BMPER is highly expressed in malignant tumors and controls invasive cell behavior. Oncogene. 2012;31:2919–2930. doi: 10.1038/onc.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajduch M, Jacob-Hirsch J, Amariglio N, Krupsky M, Simansky DA, Ram Z, Pfeffer R, Galernter I, Steinberg DM, Ben-Dov I, Rechavi G, Izraeli S. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res. 2009;15:1755–1761. doi: 10.1158/1078-0432.CCR-08-2124. [DOI] [PubMed] [Google Scholar]
- 23.Buganim Y, Goldstein I, Lipson D, Milyavsky M, Polak-Charcon S, Mardoukh C, Solomon H, Kalo E, Madar S, Brosh R, Perelman M, Navon R, Goldfinger N, Barshack I, Yakhini Z, Rotter V. A novel translocation breakpoint within the BPTF gene is associated with a pre-malignant phenotype. PLoS One. 2010;5:e9657. doi: 10.1371/journal.pone.0009657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim WT, Yun SJ, Park C, Kim IY, Moon SK, Kwon TG, Choi YH, Kim WJ. Identification of C16orf74 as a marker of progression in primary non-muscle invasive bladder cancer. PLoS One. 2010;5:e15260. doi: 10.1371/journal.pone.0015260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colombo J, Fachel AA, De Freitas Calmon M, Cury PM, Fukuyama EE, Tajara EH, Cordeiro JA, Verjovski-Almeida S, Reis EM, Rahal P. Gene expression profiling reveals molecular marker candidates of laryngeal squamous cell carcinoma. Oncol Rep. 2009;21:649–663. [PubMed] [Google Scholar]


