Abstract
Objective: O6-methylguanine DNA methyl-transferase gene (MGMT) is a central DNA repair mechanism with a significant role in removing DNA damage caused by alkylating agents and inhibiting human oncogenesis. Two single polymorphisms in the MGMT gene, Leu84Phe and Ile143Val, have been reported to affect DNA repair capability and enzymic activity, thereby leading to formation of different cancers. In this work, we quantitatively assess the associations between MGMT polymorphisms and risk of colorectal cancer (CRC), as previous studies has implicated inconsistencies in their results. Methods: Analysis was performed on all usable data collected from the eligible studies that were searched in multiple bibliographical databases (PubMed, SCOPUS, and Embase). Results: We obtained studies on Leu84Phe and Ile143Val, providing 6,154 and 7,371 samples, respectively. In the analysis on Leu84Phe, the SNP presented no global association with CRC at both the genotypic and the allelic level, but a trend towards an increased or decreased risk was shown in the models examined. Stratification by ethnicity revealed a significant increase in risk of CRC related to the Phe/Phe genotype in Caucasian samples (homozygote genetic model: OR=1.70, 95% CI=1.06-2.72; recessive genetic model: OR=1.80, 95% CI=1.12-2.87). Conclusions: Based on the statistical data, our meta-analysis indicates that Leu84Phe polymorphism in the MGMT gene may predispose Caucasians to CRC.
Keywords: MGMT, Leu84Phe, Ile143Val, colorectal cancer, risk
Introduction
DNA repair pathway is a known defense mechanism with a fundamental role in maintenance of genomic integrity and resistance to human carcinogenesis [1]. This defensive system functions via reducing deleterious effects of DNA damage, blocking undesired mutation in cancer-related genes, and enhancing replication capability [2]. Replication of the DNA with O6-methylguanine and O6-pyridyloxobutylguanine may stimulate GC to AT conversion mutations, in a very similar fashion to p53 mutations that have been associated with many cancers [3].
The O6-methylguanine DNA methyl-transferase gene (MGMT), also named AGT, AGAT and ATase, is critical for modulating the direct damage reversal pathway, a central DNA repair mechanism by which we can see minimized DNA damage and inactivated oncogenesis. The MGMT serves as a ubiquitous repair protein that eliminates DNA alkyl adducts resulting from alkylating exposures, including dietary N-nitroso compounds and tobacco consumption [4,5]. Such unrepaired DNA breakage likely leads to germline genetic variations and thereby affects the enzymic effectiveness of DNA repair genes against DNA damage, facilitating formation of various types of cancer [6-8]. Characterized by irreversible inactivation due to the inability to dealkylate itself, MGMT is instead involved in the change of inactive guanine to cysteine in alkyl groups [9,10]. In human colorectal cell lines, researchers have detected mutagenic and cytotoxic adducts that are reportedly caused by the combination of O6-, O4- alkylguanine and aforementioned alkylating agents [11,12].
An increasing body of evidence has shown that MGMT inhibits mutagenesis and carcinogenesis, whereas it makes chemotherapy less effective in cancer patients harboring alkylating agents [13,14]. Currently, the role of MGMT gene playing in the etiology of malignant human cancers, such as colorectal cancer (CRC), remains largely unexplained. A number of studies in recent years have examined the associations of MGMT gene single nucleotide polymorphisms (SNPs, Leu84Phe, Ile143Val) and CRC, with mixed findings generated [15-17]. A possible reason to explain the noted discrepancies in results is the inadequate statistical power of the individual studies where non-homogeneous populations and ethnically different individuals were included.
In this article, we performed a meta-analysis, an analytic approach with maximum estimation power, to quantitatively assess the associations between MGMT polymorphisms and risk of CRC.
Materials and methods
Study identification
Using the combinations of (polymorphism) OR (polymorphisms) AND (AGT) OR (GDF5) OR (MGMT) OR (O6-methylguanine-DNA methyltransferase) OR (alkylguanine-DNA alkyltransferase) AND (colorectal cancer), we carried out a comprehensive search in multiple bibliographical databases (PubMed, SCOPUS, and Embase). Additional publications concerning MGMT polymorphisms and CRC were identified by checking all references of review articles, meta-analysis and the studies we finally considered eligible. No restrictions were used throughout the search completed on February, 2014.
Eligible studies and data abstraction
We selected eligible studies according to:
It must be an association study with a case-control or cohort design.
SNPs (Leu84Phe and Ile143Val) and inherited susceptibility to CRC must be investigated.
Genotype data must be sufficiently offered, such that we could estimate CRC risk [odds ratios (OR) and 95% confidence intervals (95% CI)].
It must be a unique study without subsequent update. If any, we considered the largest study.
The study that failed to comply with any of the above conditions was eventually removed. For each eligible publication, we collected major authors, publication year, geographical location of each study, ethnicity or racial descent, genotype counts between cases and controls, SNP studied, source of controls and matching characteristics, and method utilized to genotype the two MGMT polymorphisms.
Statistical analysis
To estimate risk of CRC associated with MGMT SNPs, we calculated OR and its 95% CI for homozygote, dominant, recessive, allele frequency, and heterozygote model. Z test was used to test the significance of the pooled ORs, with a P value below 0.05 indicating a statistical significance. Statistical heterogeneity was detected using the Q-test and I2 index [18]. There was significant heterogeneity in case of PQ-test < 0.05 or I2 > 50%. The Mantel-Haenszel method was used to combine the ORs in the absence of heterogeneity [19]; otherwise, the DerSimonian and Laird method was considered [20]. A funnel plot and Egger’s linear regression test were utilized to examine the potential publication bias in this study [21,22]. Sensitivity analysis was performed to identify the studies conferring disproportional influence on the combined estimates.
All tests were two-sided, and statistical data were analyzed using STATA software (version 12.0, StataCorp, College Station, TX, USA). A significance level of 0.05 was set for the analyses.
Results
Study selection
As detailed in Figure 1, we identified 451 relevant records through the bibliographical database search. An initial exclusion of 403 records was carried out after title and abstract evaluation. We then screened the whole texts of the remainder, and further excluded 43 publications not in compliance with the inclusion criteria. These exclusions resulted in five eligible publications [16,17,23-25]. Moreover, the manual search yielded an additional article (Tranah et al.), in which two subpopulations were investigated [15]. We thus had a total of seven case-control studies in this analysis.
Figure 1.
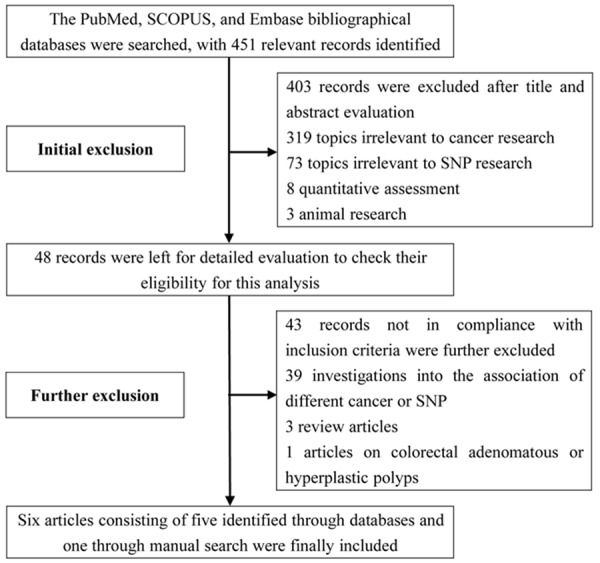
Flow diagram detailing procedures of selecting eligible studies.
Study characteristics
Among the seven studies, there were six studies on SNP Leu84Phe and five on SNP Ile143Val, as shown in Table 1. For SNP Leu84Phe, both Caucasian and Asian data were involved, while only Caucasian samples were included for SNP Ile143Val. Other characteristics, such as genotyped cases and controls, source of controls, matching properties, and genotyping methods are all presented in Table 1.
Table 1.
Basic information of the original articles included in this meta-analysis
| First author-year | Country-ethnicity | Cases (Ile143Val) | Controls (Ile143Val) | Source of control | Genotyping method | SNP studied |
|---|---|---|---|---|---|---|
| Moreno | Spain-Caucasian | 272 (359) | 299 (323) | Hospital, age and sex matched | Arrayed primer extension | Leu84Phe, Ile143Val |
| Tranah (NHS) | USA-Caucasian | 186 (190) | 2137 (2151) | Hospital, age and smoking matched | TaqMan | Leu84Phe, Ile143Val |
| Tranah (PHS) | USA-Caucasian | 257 (260) | 429 (431) | Hospital, age matched | TaqMan | Leu84Phe, Ile143Val |
| Stern | Singapore-Asian | 292 | 1166 | Population, age matched | TaqMan | Leu84Phe |
| Hazra | USA-Caucasian | 358 | 357 | Population, age matched | Multiplexed GoldenGate assay | Leu84Phe |
| Khatami | Iran-Caucasian | 200 (200) | 201 (200) | Hospital, age and sex matched | Pyrosequencing | Leu84Phe, Ile143Val |
| Loh | USA-Caucasian | 273 | 2984 | No related description | Pyrosequencing | Ile143Val |
Major findings
Effects of SNP Leu84Phe on CRC
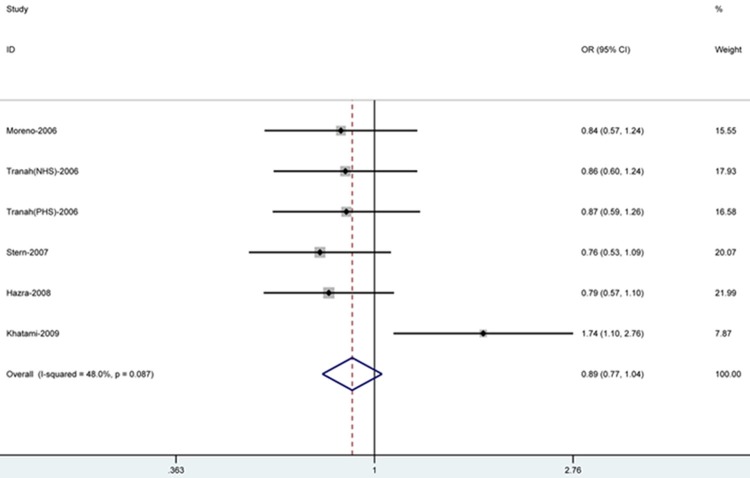
We pooled all data in the analysis of 6,154 samples (1,565 cases and 4,589 controls), as shown in Table 2. Although the SNP Leu84Phe presented no global association with CRC at both the genotypic and the allelic level, the homozygote (OR=1.46, 95% CI=0.93-2.27) and recessive model (OR=1.54, 95% CI=0.99-2.40) showed a potential increase in risk of CRC, and the dominant (OR=0.89, 95% CI=0.77-1.04) (Figure 2), allele frequency (OR=0.95, 95% CI=0.84-1.09) and heterozygote model (OR=0.87, 95% CI=0.68-1.10) revealed a possibly decreased risk.
Table 2.
Summary ORs of MGMT polymorphisms and CRC risk
| Analysis | Case/control | Homozygote model | Dominant model | Recessive model | Allele model | Heterozygote model | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| OR (95% CI) | PHet | OR (95% CI) | PHet | OR (95% CI) | PHet | OR (95% CI) | PHet | OR (95% CI) | PHet | ||
| Leu84Phe | |||||||||||
| All | 1565/4589 | 1.46 (0.93, 2.27) | 0.371 | 0.89 (0.77, 1.04) | 0.087 | 1.54 (0.99, 2.40) | 0.347 | 0.95 (0.84, 1.09) | 0.352 | 0.87 (0.68, 1.10) | 0.048 |
| Ethnicity | |||||||||||
| Caucasian | 1273/3423 | 1.70 (1.06, 2.72) | 0.691 | 0.93 (0.78, 1.10) | 0.071 | 1.80 (1.12, 2.87) | 0.660 | 1.00 (0.87, 1.15) | 0.534 | 0.89 (0.66, 1.20) | 0.027 |
| Asian | 292/1166 | 0.29 (0.04, 2.26) | / | 0.76 (0.53, 1.09) | / | 0.30 (0.04, 2.34) | / | 0.74 (0.53, 1.05) | / | 0.79 (0.55, 1.14) | / |
| Ile143Val | |||||||||||
| All | 1282/6089 | 1.07 (0.60, 1.91) | 0.171 | 1.00 (0.71, 1.41) | 0.002 | 1.10 (0.61, 1.96) | 0.188 | 1.00 (0.73, 1.38) | 0.001 | 1.00 (0.73, 1.37) | 0.008 |
Figure 2.
Fixed-effects meta-analysis on lung cancer risk and MGMT Leu84Phe polymorphism for the dominant model. Each box represents the OR point estimate, and its area is proportional to the weight of the study. The diamond (and broken line) represents the overall summary estimate, with CI representing its width.
Stratifying the data according to ethnicity revealed a 70% increased risk associated with the Phe/Phe genotype in homozygote genetic model (OR=1.70, 95% CI=1.06-2.72), and a 80% increased risk in relation to the same genotype in recessive genetic model (OR=1.80, 95% CI=1.12-2.87), only in Caucasian samples.
Effects of SNP Ile143Val on CRC
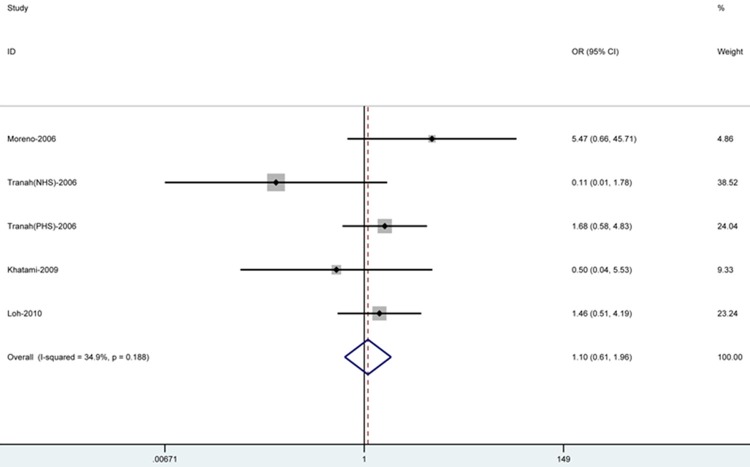
Analysis of 7,371 samples (1,282 cases and 6,089 controls) using several genetic models showed that SNP Ile143Val was not significantly associated with CRC risk. All summary ORs are listed in Table 2. Figure 3 shows the forest plots for recessive model.
Figure 3.
Fixed-effects meta-analysis on lung cancer risk and MGMT Ile143Val polymorphism for the recessive model. Each box represents the OR point estimate, and its area is proportional to the weight of the study. The diamond (and broken line) represents the overall summary estimate, with CI representing its width.
Heterogeneity test and sensitivity analysis
We found moderate between-study heterogeneity for both SNPs. Using sensitivity analysis, we identified the Iranian and NHS4 studies representing major heterogeneity sources of SNP Leu84Phe and SNP Ile143Val, respectively. Absence of heterogeneity was seen when these studies were excluded (data not shown). Interestingly, meta-analysis of the homozygous studies on SNP Leu84Phe showed a substantially altered OR (OR=0.77, 95% CI=0.65-0.91).
Publication bias
Both the funnel plots and Egger’s test revealed no evidence of significant publication bias in this analysis (SNP Leu84Phe: P=0.099; SNP Ile143Val: P=0.279; model: homozygote genetic model).
Discussion
The first investigation looking at the association between MGMT polymorphisms and CRC susceptibility was carried out in a group of Spanish population [23]. In this work, neither of the SNPs being examined was validated as predisposing risk factors of CRC. In an effort to replicate the initial finding, Tranah and co-workers employed two large independent American populations and the 143Val allele was found to be protective against CRC in the female population (NHS), while both of the Leu84Phe and Ile143Val did not show any significant association with CRC in the male population (PHS) [15]. This finding, however, appears to contradict that suggested in a subsequent Singaporean study, where the researchers found reduced CRC risk attributed to the Leu/Phe or Phe/Phe genotypes of Leu84Phe [16]. A most recent replication concerning Ile143Val only demonstrated that this SNP conferred either increased or decreased susceptibility to the patients in the USA [25]. The likely reasons for these inconsistencies in observations include ethnic differences and lack of detection power due to the limited number of subjects included in each of the independent studies.
Meta-analysis is thought of as a powerful quantitative tool that can provide a precise estimation of the association between SNPs and common diseases by combining all single studies. In the overall analysis on Leu84Phe, we observed very interesting results. A trend towards an increased risk of CRC was seen in Phe/Phe genotypes compared with Leu/Leu or combined Leu/Leu and Leu/Phe genotypes. In contrast, the combined Phe/Phe and Leu/Phe, Leu/Phe and Phe allele showed a reduced risk relative to the Leu/Leu, and Leu allele, respectively. As the associations are not statistically significant, further larger investigations are necessary to validate these findings. Stratification according to ethnicity revealed a significant association in Caucasian populations. For Ile143Val, none of the genetic models tested showed an increase or decrease in the risk of CRC, a finding that is inconsistent with a previous study [25]. A plausible explanation is that the individual study has reached a false positive conclusion as a result of the small sample size.
Previous meta-analyses examining the association of MGMT polymorphisms and susceptibility towards CRC have presented inconsistent findings. The earliest meta-analysis published in 2010 reported protective effects of Leu84Phe polymorphism in CRC [26]. This finding was replicated in two recent analyses [27,28]. The inclusion of a single polymorphism, pooling incorrect data or failure to collect all usable data may lead to underestimated associations as suggested in the aforementioned studies. We excluded the non-CRC study [29] and included two additional populations [17,24], which minimizes the possibility of biased results and maximizes the reliability of the combined estimates. The very differing effects of Leu84Phe on the development of CRC found in our analysis highlighted the importance of a large-scale study in future.
Epidemiological data have demonstrated that MGMT gene polymorphisms may affect the enzyme activity, resulting in reduced capability in response to DNA repair and increased the likelihood of developing cancer [30]. Ile143Val has been shown to modulate the biological function of the protein, as in the active site of MGMT there is an alkyl receptor at codon 145, very close to residue 143 [31-33]. Several lines of evidence have described affected MGMT function, suppressed estrogen receptor cell proliferation and reduced DNA repair capability in relation to Leu84Phe [15,17]. These data point to a conclusion that the MGMT may be a possible CRC susceptibility locus, supporting the findings in our study.
Similar to many meta-analyses, our study has some limitations. First, we noted that there was substantial heterogeneity in the analysis of Leu84Phe and Ile143Val. Most importantly, exclusion of the influence study has changed the primary pooled effects of Leu84Phe on CRC. The true association therefore requires further investigations. Second, publication bias can be minimized if small studies are excluded and unpublished data are incorporated. The failure to satisfy the two conditions makes publication bias possible in this analysis. Third, it is possible that the effects of a susceptibility gene on the development of a common disease are confounded by environmentally carcinogenic exposures and other disease-related genes via gene-to-environment or gene-to-gene interactions. Investigation into the role of the confounding factors in CRC seems practical if the sample is sufficiently large.
In conclusion, we found evidence that MGMT gene Leu84Phe, but not the Ile143Val, was a susceptibility risk factor of CRC in Caucasian populations. Considering the limited sample, further larger studies are necessary to clarity whether the two polymorphisms play a major role in the development of CRC.
Disclosure of conflict of interest
None.
References
- 1.Hung RJ, Baragatti M, Thomas D, McKay J, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, Bencko V, Chabrier A, Moullan N, Canzian F, Hall J, Boffetta P, Brennan P. Inherited predisposition of lung cancer: a hierarchical modeling approach to DNA repair and cell cycle control pathways. Cancer Epidemiol Biomarkers Prev. 2007;16:2736–2744. doi: 10.1158/1055-9965.EPI-07-0494. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Crosbie PA, McGown G, Thorncroft MR, O’Donnell PN, Barber PV, Lewis SJ, Harrison KL, Agius RM, Santibanez-Koref MF, Margison GP, Povey AC. Association between lung cancer risk and single nucleotide polymorphisms in the first intron and codon 178 of the DNA repair gene, O6-alkylguanine-DNA alkyltransferase. Int J Cancer. 2008;122:791–795. doi: 10.1002/ijc.23059. [DOI] [PubMed] [Google Scholar]
- 4.Margison GP, Santibanez-Koref MF. O6-alkylguanine-DNA alkyltransferase: role in carcinogenesis and chemotherapy. Bioessays. 2002;24:255–266. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- 5.Loh YH, Mitrou PN, Wood A, Luben RN, McTaggart A, Khaw KT, Rodwell SA. SMAD7 and MGMT genotype variants and cancer incidence in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Cancer Epidemiol. 2011;35:369–374. doi: 10.1016/j.canep.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Hu JJ, Mohrenweiser HW, Bell DA, Leadon SA, Miller MS. Symposium overview: genetic polymorphisms in DNA repair and cancer risk. Toxicol Appl Pharmacol. 2002;185:64–73. doi: 10.1006/taap.2002.9518. [DOI] [PubMed] [Google Scholar]
- 7.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 8.Mirvish SS. Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 1995;93:17–48. doi: 10.1016/0304-3835(95)03786-V. [DOI] [PubMed] [Google Scholar]
- 9.Samson L. The suicidal DNA repair methyltransferases of microbes. Mol Microbiol. 1992;6:825–831. doi: 10.1111/j.1365-2958.1992.tb01533.x. [DOI] [PubMed] [Google Scholar]
- 10.Pegg AE, Byers TL. Repair of DNA containing O6-alkylguanine. FASEB J. 1992;6:2302–2310. doi: 10.1096/fasebj.6.6.1544541. [DOI] [PubMed] [Google Scholar]
- 11.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Povey AC, Badawi AF, Cooper DP, Hall CN, Harrison KL, Jackson PE, Lees NP, O'Connor PJ, Margison GP. DNA alkylation and repair in the large bowel: animal and human studies. J Nutr. 2002;132:3518S–3521S. doi: 10.1093/jn/132.11.3518S. [DOI] [PubMed] [Google Scholar]
- 13.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1–8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- 15.Tranah GJ, Bugni J, Giovannucci E, Ma J, Fuchs C, Hines L, Samson L, Hunter DJ. O6-methylguanine-DNA methyltransferase Leu84Phe and Ile143Val polymorphisms and risk of colorectal cancer in the Nurses’ Health Study and Physicians' Health Study (United States) Cancer Causes Control. 2006;17:721–731. doi: 10.1007/s10552-006-0005-y. [DOI] [PubMed] [Google Scholar]
- 16.Stern MC, Conti DV, Siegmund KD, Corral R, Yuan JM, Koh WP, Yu MC. DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese Health Study. Cancer Epidemiol Biomarkers Prev. 2007;16:2363–2372. doi: 10.1158/1055-9965.EPI-07-0268. [DOI] [PubMed] [Google Scholar]
- 17.Khatami F, Noorinayer B, Mohebi SR, Ghiasi S, Mohebi R, Hashemi M, Zali MR. Effects of amino acid substitution polymorphisms of two DNA methyltransferases on susceptibility to sporadic colorectal cancer. Asian Pac J Cancer Prev. 2009;10:1183–1188. [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 20.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 21.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 22.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, Gonzalez S, Guino E, Capella G, Canzian F. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- 24.Hazra A, Chanock S, Giovannucci E, Cox DG, Niu T, Fuchs C, Willett WC, Hunter DJ. Large-scale evaluation of genetic variants in candidate genes for colorectal cancer risk in the Nurses’ Health Study and the Health Professionals’ Follow-up Study. Cancer Epidemiol Biomarkers Prev. 2008;17:311–319. doi: 10.1158/1055-9965.EPI-07-0195. [DOI] [PubMed] [Google Scholar]
- 25.Loh YH, Mitrou PN, Bowman R, Wood A, Jeffery H, Luben RN, Lentjes MA, Khaw KT, Rodwell SA. MGMT Ile143Val polymorphism, dietary factors and the risk of breast, colorectal and prostate cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk study. DNA Repair (Amst) 2010;9:421–428. doi: 10.1016/j.dnarep.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Zhong Y, Huang Y, Zhang T, Ma C, Zhang S, Fan W, Chen H, Qian J, Lu D. Effects of O6-methylguanine-DNA methyltransferase (MGMT) polymorphisms on cancer: a meta-analysis. Mutagenesis. 2010;25:83–95. doi: 10.1093/mutage/gep050. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Zhang R, Chen F, Yu C, Sun Y, Jia C, Zhang L, Salahuddin T, Li X, Lang J, Song X. MGMT Leu84Phe polymorphism contributes to cancer susceptibility: evidence from 44 case-control studies. PLoS One. 2013;8:e75367. doi: 10.1371/journal.pone.0075367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Zhou M, Li K, Zhang K, Kong X, Zheng Y, Li J, Liu L. Two DNA repair gene polymorphisms on the risk of gastrointestinal cancers: a meta-analysis. Tumour Biol. 2014;35:1715–1725. doi: 10.1007/s13277-013-1320-z. [DOI] [PubMed] [Google Scholar]
- 29.Bigler J, Ulrich CM, Kawashima T, Whitton J, Potter JD. DNA repair polymorphisms and risk of colorectal adenomatous or hyperplastic polyps. Cancer Epidemiol Biomarkers Prev. 2005;14:2501–2508. doi: 10.1158/1055-9965.EPI-05-0270. [DOI] [PubMed] [Google Scholar]
- 30.Kaur TB, Travaline JM, Gaughan JP, Richie JP, Stellman SD, Lazarus P. Role of polymorphisms in codons 143 and 160 of the O6-alkylguanine DNA alkyltransferase gene in lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2000;9:339–342. [PubMed] [Google Scholar]
- 31.Chueh LL, Nakamura T, Nakatsu Y, Sakumi K, Hayakawa H, Sekiguchi M. Specific amino acid sequences required for O6-methylguanine-DNA methyltransferase activity: analyses of three residues at or near the methyl acceptor site. Carcinogenesis. 1992;13:837–843. doi: 10.1093/carcin/13.5.837. [DOI] [PubMed] [Google Scholar]
- 32.Margison GP, Heighway J, Pearson S, McGown G, Thorncroft MR, Watson AJ, Harrison KL, Lewis SJ, Rohde K, Barber PV, O’Donnell P, Povey AC, Santibanez-Koref MF. Quantitative trait locus analysis reveals two intragenic sites that influence O6-alkylguanine-DNA alkyltransferase activity in peripheral blood mononuclear cells. Carcinogenesis. 2005;26:1473–1480. doi: 10.1093/carcin/bgi087. [DOI] [PubMed] [Google Scholar]
- 33.Mijal RS, Kanugula S, Vu CC, Fang Q, Pegg AE, Peterson LA. DNA sequence context affects repair of the tobacco-specific adduct O(6)-[4-Oxo-4-(3-pyridyl)butyl] guanine by human O(6)-alkylguanine-DNA alkyltransferases. Cancer Res. 2006;66:4968–4974. doi: 10.1158/0008-5472.CAN-05-3803. [DOI] [PubMed] [Google Scholar]