Abstract
Objective: This study was to explore the expression of human cervical cancer oncogene-1 (HCCR-1) in colon cancer and its clinical significance. Methods: RT-PCR, immunohistochemistry and Western blot assay were employed to detect HCCR-1 expression in 152 colon cancers, 43 adjacent non-cancerous tissues and 37 normal tissues. In addition, immunohistochemistry was done to detect CEA in colon cancers. Results: The mRNA expression of HCCR-1 in colon cancers was higher than that in the adjacent non-cancerous tissues (P < 0.05), and the mRNA expression of HCCR-1 in adjacent non-cancerous tissues was higher than that in normal tissues (P < 0.05). The positive rate of HCCR-1 in colon cancers was 80.9%, which was higher than that in adjacent non-cancerous tissues (P < 0.05). Almost no HCCR-1 expression was observed in normal tissues, weak expression in adjacent non-cancerous tissues and strong expression in colon cancers. The positive rate of HCCR-1 in colon cancer at Duke stage B-C was 87.3%, which was higher than that in stage A colon cancer (63.6%, P < 0.05). Conclusion: HCCR-1 is over-expressed in colon cancers, indicating that HCCR-1 may participate in occurrence and development of colon cancer and has a correlation with the pathological progress of colon cancer progression.
Keywords: Colonic cancer, gene expression, gene, human cervical cancer oncogene-1
Introduction
Colon cancer is one of the most common and highly aggressive tumors. In 2005, about 760 million people died of cancer world wide, of which colon cancer accounted for 655000 deaths, and was the fourth leading cause of cancer related death following lung cancer, stomach cancer and liver cancer. In recent years, with the improvement of living standard and the change in diet habit, the incidence of colon cancer shows an increasing trend. In the United States, the incidence of colon cancer has become the third common malignancy [1]. With the continuous improvement of Chinese economic development and living standard, as well as the changes in lifestyle and diet habit, the incidence of colorectal cancer increases over year in China [2]. Although the diagnosis and surgical treatment of colon cancer have achieved considerable progress, the 5-year survival rate is still around 50% in China and remains stable in past 10 years in Western countries [3]. Therefore, improving the early diagnosis is crucial for the improvement of prognosis. Electronic colonoscopy is the gold standard in the diagnosis of colon cancer. However, it is an invasive examination, needs plentiful experience and has risks for bleeding and perforation, which limits its wide application in extensive clinical screening. In recent years, clinicians attempt to develop sensitive, reliable and non-invasive methods to improve the diagnosis of early-stage colon cancer.
Human cervical cancer oncogenes (HCCRs), which are over-expressed in primary cervical cancer and cervical cancer cell lines [4], were first identified in primary cervical cancers and cervical cancer cell lines by differential display RT-PCR [4-6]. HCCR locates in chromosome 12 q and encodes HCCR-1 and HCCR-2 proteins. Studies have shown that HCCR inhibits p53 activity in several ways and plays an important role in the tumorigenesis [5]. Cho et al [7] reported that HCCR expression was regulated by phosphatidylinositol 3-kinase (PI3K/Akt) signal pathway, and its over-expression led to mitochondrial dysfunction, thereby preventing ultraviolet-induced apoptosis [8]. Most studies mainly focus on the relationship between HCCR-1 and cancers. Besides its over-expression in cervical cancer, over-expression of HCCR-1 is also found in other cancers, such as liver cancer, breast cancer, gastric cancer, colon cancer, pancreatic cancer, kidney cancer and leukemia [4,9,10]. Moreover, HCCR-1 can be used as bioremark for the diagnosis of liver cancer and breast cancer [6,11]. This study aimed to explore the relationship between HCCR-1 expression and clinicopathological features of colon cancer by detecting HCCR-1 expression in human colon cancer.
Materials and methods
Patients and collection of general information
Colon cancer tissues were collected from 152 patients with colon cancer during surgery in Central Hospital of Tai’an City from March 2012 to May 2013. These patients were pathologically diagnosed with colon cancer. There were 87 males and 65 females with the median age of 58 years (range: 36 to 83 years). According to the Duke’s staging system on colorectal cancer in 2003, colon cancer was classified as stage A in 22 patients, stage B-C in 118 and stage D in 12. In addition, adjacent non-cancerous tissues were collected within 3 cm away from the colon cancer of 4 patients. There were 27 males and 16 females with the median age of 61 years (range: 41 to 72 years). Moreover, normal colon tissues were collected from 37 patients with benign lesions of the colon who received surgery in the same period. There were 14 males and 23 females with medium age of 55 years (range: 32 to 76 years). All the specimens were fixed in 4% formaldehyde and embedded in paraffin.
Reagents
Rabbit anti-human HCCR-1 antibody and immunohistochemical kits were purchased from Beijing Bios Biological Technology Co., Ltd; Trizol reagent was from Invitrogen Biological Company. RNA reverse transcription kit, dNTPs, Taq DNA polymerase, and PVDF membrane were purchased from Nanjing Keygen Biotech P.R. China. Primers were synthesized in Invintrogen Company. TRIzol Reagent was purchased from Invintrogen Company. First-Strand cDNA Synthesis Kit was purchased from Biotech Co., Shanghai, PR China. PCR amplification kit (2720) was from ABI company (USA). Automatic Gel Imaging analyzer (GDS 8000) was from UVP company (USA).
RT-PCR
Premier 5.0 was employed to design primers according to the sequence in Gene Bank. The primers were as follows: forward: 5’-ACCCCAAAACAACAAACTG-3’; reverse: 5’-ATGGTTCATTCAGCGCCTT-3’. The anticipated length of PCR product was 579 bp. β-actin was used as an internal reference. Primers were synthesized in Invintrogen company. Trizol was used to extract total RNA from different tissue (colon cancers, adjacent non-cancerous tissues and normal colon tissues) according to manufacturer’s instructions. Reverse transcription of total RNA (10 μg) was performed for each sample using a First-Strand cDNA Synthesis Kit, and cDNA was subjected to PCR for 30 cycles of amplification described as follows: 30 sec at 94°C, 30 sec at 56°C and 30 sec at 72°C. The absorbance was detected to determine the concentration of RNA. 5 μl of PCR products were resolved on a 1% agarose gel. DNA bands were visualized by UV light. Semi quantitative analysis of Band intensity was performed with Gene Tools software (UVP, Inc., Upland, CA).
Immunohistochemistry (IHC)
Immunohistochemical (IHC) analysis was performed as previously reported [13-15]. In brief, 4-μm paraffin-embedded colon cancer tissues were placed in an oven overnight at 37°C. The slides were dewaxed in xylene, rehydrated in graded alcohol, and incubated in 0.01 M citrate buffer at 95°C for 20 min as antigen retrieval. After blocking for nonspecific antibody binding, the HCCR-1 monoclonal antibody (1:200) was applied. Signal visualization was developed by the avidin- biotin- peroxidase method using DAB. Following immunohistochemical staining assay, the slides were placed in hematoxylin for 1 min. The primary antibody was replaced with PBS in negative control.
All of the fields in the selected block were taken into consideration for assessment of immunostaining. Representative areas of each section were selected, and cells were counted in at least four fields at 400-fold magnification. Evaluations of immunoreactivity of IHC were performed according to previous description [13-15]. Cells were scored according to the staining intensity of the cytoplasm and the percentage of positive cells. Staining intensity: grade 0: basically non-staining; grade 1: pale yellow; grade 2: yellow; grade 3: brown. Percentage of positive cells: grade 0: < 5%; grade 1: 5%-25%; grade 2: > 25%-50%; grade3: > 50%. The final score was a product of both scores. The final score of ≥ 4 refers to positive expression, while 0-3 as negative expression.
Western blot assay
Western blotting was performed according to standard methods as described previously [16,17]. In brief: 1) Proteins were extracted from different tissues; 2) Proteins were separated by 10% SDS-PAGE, with 50 μg proteins being loaded onto each lane; 3) Proteins were electro-transferred onto PVDF membrane, at 100 V for 1 h; 4) PVDF membrane was washed in TBST for 5 min, and then blocked in 5% non-fat milk for 1 h; 5) PVDF membrane was washed in TBST thrice (5 min for each), and then incubated with primary antibody (1:400) overnight at 4°C; 6) PVDF membrane was washed in TBST thrice (5 min for each) and then incubated with HRP-conjugated secondary antibody (1:10 000) for 1 h; 7) PVDF membrane was washed in TBST thrice (5 min for each). Visualization was done with electrochemical luminescence.
Statistical analysis
SPSS version 10.0 was used for statistical analysis. Chi-square test was employed for comparison between groups, and a value of P < 0.05 was considered statistically significant.
Results
Expression of HCCR-1 gene
The mRNA expression of HCCR-1 was 0.766 ± 0.101 in colon cancers, 0.586 ± 0.121 in adjacent non-cancerous tissues, and 0.430 ± 0.154 in normal colon tissues (P < 0. 01) (Figure 1).
Figure 1.
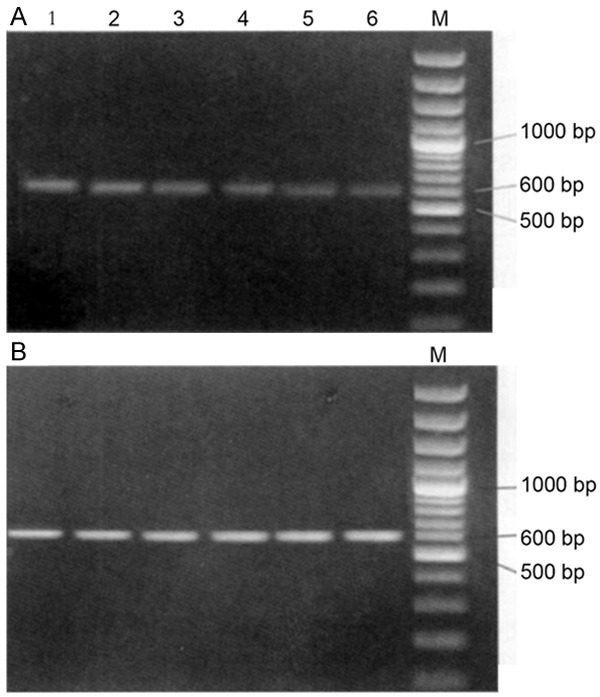
mRNA expression of HCCR-1 in colon cancers, adjacent non-cancerous tissues, and normal colon tissues. M: Marker; A. 1 and 2: colon cancers; 3 and 4: adjacent non-cancerous tissues; 5 and 6: normal colon tissues. B. β-actin.
Immunohistochemistry
Immunohistochemistry showed HCCR-1 expression was mainly found in the cytoplasm. Positive rate of HCCR-1 was 80.9% (123/152) in colon cancers and 48.8% (21/43) in adjacent non-cancerous tissues, as shown in Figure 2. The positive rate of HCCR-1 in colon cancers was significantly higher than that in adjacent non-cancerous tissues (X2 = 17.86, P < 0.01).
Figure 2.
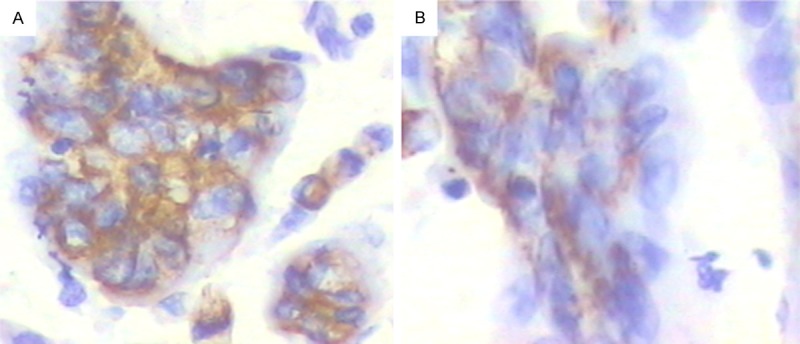
Positive expression of HCCR-1 in colon cancer tissues and adjacent non-cancerous tissues (DAB × 400). A. Colon cancer tissues. B. Adjacent non-cancerous tissues.
Relationship between HCCR-1 expression and clinicopathological features of colon cancer
Among 152 colon cancer tissues, 97 were positive for CEA and 123 positive for HCCR-1; among 55 colon cancer tissues negative for CEA, 38 were positive for HCCR-1; among 22 Duke’ s stage A colon cancers, 10 were positive for CEA and 14 were positive for HCCR-1 (X2 = 11.12, P < 0.05). Positive rate of HCCR-1 in Duke’ stage B-C colon cancers [87.3%, (103/118)] was higher than that in stage A colon cancers [63.6%, (14/22)] (X2 = 7.55, P < 0.05). However, the positive rate of HCCR-1 had no relationship with gender, age, tumor size and lymph node metastasis (LNM) (Table 1).
Table 1.
Correlation between HCCR expression and clinicopathological features of colon cancer
| Clinicopathological features | Number | HCCR expression | χ2 | P | |
|---|---|---|---|---|---|
|
| |||||
| Positive | Negative | ||||
| Age | |||||
| ≤ 60 | 69 | 56 | 13 | 0.372 | > 0.05 |
| > 60 | 83 | 64 | 19 | ||
| Gender | |||||
| Male | 95 | 73 | 22 | 0.312 | > 0.05 |
| Female | 57 | 46 | 11 | ||
| Pathological stage | |||||
| A | 22 | 14 | 8 | 7.55 | < 0.05 |
| B-C | 118 | 103 | 15 | ||
| Tumor size | |||||
| ≤ 3 | 89 | 71 | 18 | 0.03 | > 0.05 |
| > 3 | 56 | 44 | 12 | ||
| Lymph node metastasis | |||||
| Yes | 37 | 29 | 8 | 0.02 | > 0.05 |
| No | 112 | 89 | 23 | ||
Western blot assay
HCCR-1 was not expressed in normal colon tissues, weakly expressed in adjacent non-cancerous tissues, and strongly expressed in colon cancer tissues (Figure 3).
Figure 3.
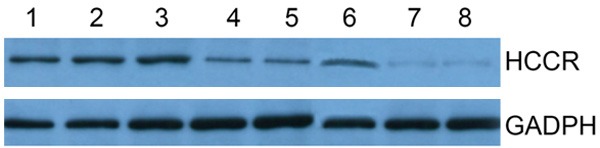
HCCR-1 was not expressed in normal colon tissues, weakly expressed in adjacent non-cancerous tissues, and strongly expressed in colon cancer tissues.
Discussions
HCCR-1 gene was first identified in cervical cancer, and studies have shown that HCCR-1 expression increases in the serum of colon cancer patients, suggesting that HCCR-1 is closely related to colon cancer [12]. Our study showed that the positive rate of HCCR-1 increased from normal colon tissues, adjacent non-cancerous tissues to colon cancer tissues, showing that HCCR-1 over-expression is closely related to the early development of colon cancer. Experiments have revealed that the positive rate of HCCR-1 in Duke’s stage B-C colon cancer is significantly higher than in stage A colon cancer, suggesting there is a relationship between HCCR-1 over-expression and invasion and metastasis of colon cancer. However, according to the analysis with exact probabilities, HCCR-1 expression in colon cancers was not associated with the gender, age, tumor size and LNM of colon cancer patients. This may be ascribed to the small sample size. More trials with a larger sample size are needed to confirm our findings.
The survival rate of patients with early stage colon cancer following surgical treatment may reach 95%. However, at present, the early diagnosed colorectal cancer accounts for only 12.5% of all colorectal cancers [13]. CEA is one of the most widely used tumor marks with significantly elevated expression in patients with colorectal cancer. It has been reported that the positive rate of CEA in patients with colon cancer is as high as 40%-70% [14]. However, it has a poor specificity, and thus CEA detection is mainly used in the evaluation of prognosis and therapeutic efficacy and the monitoring of relapse and metastasis in patients with colon cancer [15]. Our results showed that, among 152 human colon cancer tissues, 97 were positive for CEA, and 123 positive for HCCR-1; among 55 patients negative for CEA, 38 were positive for HCCR-1; in 22 Duke’s stage A colon cancers, 10 were positive for CEA and 14 positive for HCCR-1, suggesting that the diagnostic accuracy according to HCCR-1 is higher than that on the basis of CEA in early stage colon cancer. Thus, HCCR-1 detection may become a promising strategy for the early diagnosis of colon cancer.
Disclosure of conflict of interest
None.
References
- 1.Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, Dash C, Giardiello FM, Glick S, Johnson D, Johnson CD, Levin TR, Pickhardt PJ, Rex DK, Smith RA, Thorson A, Winawer SJ. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134:1570–1595. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Ju JH, Chang SC, Wang HS, Yang SH, Jiang JK, Chen WC, Lin TC, Hung H, Wang FM, Lin JK. Changes in disease pattern and treatment outcome of colorectal cancer: a review of 5,474 cases in 20 years. Int J Colorectal Dis. 2007;22:855–862. doi: 10.1007/s00384-007-0293-z. [DOI] [PubMed] [Google Scholar]
- 3.Rayter Z, Leicester RJ, Mansi JL. Adjuvant chemotherapy for colorectal cancer. Ann R Coll Surg Engl. 1995;77:81–84. [PMC free article] [PubMed] [Google Scholar]
- 4.Ko J, Lee YH, Hwang SY, Lee YS, Shin SM, Hwang JH, Kim J, Kim YW, Jang SW, Ryoo ZY, Kim IK, Namkoong SE, Kim JW. Identification and differential expression of novel human cervical cancer oncogene HCCR-2 in human cancers and its involvement in p53 stabilization. Oncogene. 2003;22:4679–4689. doi: 10.1038/sj.onc.1206624. [DOI] [PubMed] [Google Scholar]
- 5.Ko J, Shin SM, Oh YM, Lee YS, Ryoo ZY, Lee YH, Na DS, Kim JW. Transgenic mouse model for breast cancer: induction of breast cancer in novel oncogene HCCR-2 transgenic mice. Oncogene. 2004;23:1950–1953. doi: 10.1038/sj.onc.1207356. [DOI] [PubMed] [Google Scholar]
- 6.Chung YJ, Kim JW. Novel oncogene HCCR: its diagnostic and therapeutic implications for cancer. Histol Histopathol. 2005;20:999–1003. doi: 10.14670/HH-20.999. [DOI] [PubMed] [Google Scholar]
- 7.Cho GW, Shin SM, Namkoong H, Kim HK, Ha SA, Hur SY, Kim TE, Chai YG, Kim JW. The phosphatidylinositol 3-kinase/Akt pathway regulates the HCCR-1 oncogene expression. Gene. 2006;384:18–26. doi: 10.1016/j.gene.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Cho GW, Shin SM, Kim HK, Ha SA, Kim S, Yoon JH, Hur SY, Kim TE, Kim JW. HCCR-1, a novel oncogene, encodes a mitochondrial outer membrane protein and suppresses the UVC-induced apoptosis. BMC Cell Biol. 2007;8:50. doi: 10.1186/1471-2121-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon SK, Lim NK, Ha SA, Park YG, Choi JY, Chung KW, Sun HS, Choi MJ, Chung J, Wands JR, Kim JW. The human cervical cancer oncogene protein is a biomarker for human hepatocellular carcinoma. Cancer Res. 2004;64:5434–5441. doi: 10.1158/0008-5472.CAN-03-3665. [DOI] [PubMed] [Google Scholar]
- 10.Xu Z, Zhang Y, Jiang J, Yang Y, Shi R, Hao B, Zhang Z, Huang Z, Kim JW, Zhang G. Epidermal growth factor induces HCCR expression via PI3K/Akt/mTOR signaling in PANC-1 pancreatic cancer cells. BMC Cancer. 2010;10:161. doi: 10.1186/1471-2407-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung SS, Park HS, Lee IJ, Namkoong H, Shin SM, Cho GW, Ha SA, Park YG, Lee YS, Ko J, Kim JW. The HCCR oncoprotein as a biomarker for human breast cancer. Clin Cancer Res. 2005;11:7700–7708. doi: 10.1158/1078-0432.CCR-04-2609. [DOI] [PubMed] [Google Scholar]
- 12.Shin SM, Chung YJ, Oh ST, Jeon HM, Hwang LJ, Namkoong H, Kim HK, Cho GW, Hur SY, Kim TE, Lee YS, Park YG, Ko J, Kim JW. HCCR-1-interacting molecule “deleted in polyposis 1” plays a tumor-suppressor role in colon carcinogenesis. Gastroenterology. 2006;130:2074–2086. doi: 10.1053/j.gastro.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YL. The suggestions for early colorectal cancer endoscopic biopsy diagnostic criteria. Chin J Digest Endo. 2001;18:135–138. [Google Scholar]
- 14.Gordon PH. Mallignant neoplasm of the colon. In: Gordon PH, Nivatvong S, editors. Principles of surgery for the colon, retune and anus. St Louis: Quality Medicelpublis; 1992. p. 501. [Google Scholar]
- 15.Gebauer G, Muller-Ruchholtz W. Tumor marker concentrations in normal and malignant tissues of colorectal cancer patients and their prognostic relevance. Anticancer Res. 1997;17:2939–2942. [PubMed] [Google Scholar]
- 16.Liu Y, Li K, Ren Z, Li S, Zhang H, Fan Q. Clinical implication of elevated human cervical cancer oncogene-1 expression in esophageal squamous cell carcinoma. J Histochem Cytochem. 2012;60:512–20. doi: 10.1369/0022155412444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguilar C, Aguilar C, Lopez-Marure R, Jiménez-Sánchez A, Rocha-Zavaleta L. Co-stimulation with stem cell factor and erythropoietin enhances migration of c-Kit expressing cervical cancer cells through the sustained activation of ERK1/2. Mol Med Rep. 2014;9:1895–902. doi: 10.3892/mmr.2014.2044. [DOI] [PubMed] [Google Scholar]


