Abstract
Osteoarthritis (OA) is characterized by degeneration of articular cartilage, limited intraarticular inflammation with synovitis, and changes in peri-articular and subchondral bone. In recent years, more and more evidence demonstrated that microRNAs (miRNAs) play important roles in the molecular mechanisms in OA by suppressing gene expression at the post-transcriptional level. In current study, histological staining of toluidine blue and cartilage-specific gene express revealed that the bone matrix gelatin (BMG) rat model could demonstrate the different development of cartilage. In current study, we tested whether some miRNAs associated with OA differently expressed in BMG rat model. We verified that miR-140 and miR-455 were associated with cartilage development, and further revealed that miR-140-5p and miR-455-3p might play more important function than miR-140-3p and miR-455-5p in the BMG rat model. Moreover, we found that miR-9 and miR-98 were involved in the endochondral ossification, suggesting they may be also the key regulators in the process of endochondral ossification. In fact, many miRNAs worked as a miRNA-mediated regulatory network in the process of cartilage development and OA. Further functional discovery will clarify the roles of individual miRNAs and their targets, and serve as a strong foundation for translating these findings to the clinic therapy for OA.
Keywords: Osteoarthritis, microRNA, BMG rat model
Introduction
Osteoarthritis (OA) is characterized by degeneration of articular cartilage, limited intraarticular inflammation with synovitis, and changes in peri-articular and subchondral bone [1]. It is widely accepted that the pathogenesis of OA is the loss of the homeostatic balance between anabolic factors and catabolic factors in articular cartilage, leading to the degradation of the extracellular matrix (ECM) and the damage of articular cartilage [2-4]. Some studies documented that OA is related to cartilage development [5]. In the development of growth plate, the chondrocytes undergo a process of proliferation, hypertrophy, mineralization, and programmed cell death [6,7]. Chondrocytes in articular cartilage are constrained from completing this program, allowing maintenance of a functional cartilage layer. A variety of biological macromolecules such as type II collagen (Col2) and proteoglycans are dysregulated in the initiation and progress of OA. In recent years, more and more evidence demonstrated that microRNAs play important roles in the molecular mechanisms in OA by suppressing gene expression at the post-transcriptional level.
MicroRNAs (miRNAs) are a class of endogenous and non-coding single-strand RNAs with a length of about 22 nucleotides, and many of them are evolutionarily conserved. Hundreds of miRNAs have been found in various organisms, and more than half of all human protein-coding genes are potentially regulated by miRNAs [8]. The functions of regulated genes involve almost all aspects of cells, such as proliferation, differentiation, motivation, communication, senescence and apoptosis. miRNAs have been implicated in the process of development and pathogenesis of diseases, such as cancer and cardiovascular diseases [9,10]. Some miRNAs are differentially expressed in cartilage of OA patients compared to normal tissue, and associated with the initiation and/or progression with the disease, such as miR-9, miR-98, miR-146, miR-22 [11]. Similarly, miRNAs are regulated across cartilage development, and their function is beginning to be delineated [12]. While the cartilage-specific miR-140 was associated with both cartilage homeostasis and development [13-15].
In our previous study, we collected femoral head cartilage of rats at postnatal day 0, day 21 and day 42 and sequenced them by the method of Solexa sequencing. We found that about 30 miRNAs are differently expressed during articular cartilage development [16]. When the bone matrix gelatin (BMG), which produced by allogeneic demineralized bone matrix, was implanted in muscle in adult rats, it induced chondrogenesis and bone information [17]. Using BMG rat model, our further study demonstrated that miR-337 was directly implicated in chondrogenesis and Aggrecan was differentially expressed in both the gain and loss of function of miR-337 experiments, providing evidence that miR-337 could influence cartilage-specific gene expression in chondrocytes [18]. In current study, we tested whether some miRNAs associated with OA differently expressed in BMG rat models, which further disclose the potential mechanisms of some miRNAs and provide potential combined therapies of some molecules for OA treatment.
Material and method
Animals
This study was approved by the Institutional Animal Ethics Committee of the University. Dark Agouti (DA) rats were bred in a climate controlled environment, housed in polystyrene cages containing wood shavings, and fed standard rodent chow and water ad libitum in the SPF animal house of Department of Biochemistry and Molecular Biology, Xi’an Jiaotong University Health Science Center. The rats originated from the Section of Medical Inflammation Research, Lund University, Sweden.
Bone matrix gelatin induced endochondral ossification (BMG-ECO)
The bone matrix gelatin (BMG) was prepared from bone of DA rats as previously reported [17]. DA rats at age of 12 weeks were killed by lethal dose of aether. Diaphyseal shafts of femur and tibia were collected and dissected free of muscles and connective tissues. After removed the bone marrow, bones were cut into chips about the size of about 1 cm. Liquid nitrogen was used to freeze the bone shafts in this procedure to avoid possible denaturation of proteins. Next, the mixture of chloroform and methanol (1:1) was used to remove the lipid of bones within 1 hour and the bones was demineralized in 0.6M HCL within 24 hours. Then, BMG was extracted successively with the solution of 2.0M CaCl2, 0.5 M ethylenediamine tetra acetic acid (EDTA), 0.8 M LiCl and 55°C water to remove soluble proteins. And then, the bone chips were dried by vacuum freeze-drying and stored in -20°C for preparing the implantation. Twenty-four adult DA rats including 12 females and 12 males received the syngeneic BMG implantation in supraspinatus and rhomboid muscles. Each rat could be planted in six pieces of BMG with the size of 1 cm. The rats were randomly divided into four groups and each group was made up of 3 female rats and 3 male rats. After 1, 2, 3, and 4 weeks of implantation, the generated cartilaginous tissues were harvested from six rats of each time point, respectively. The partial tissue was subjected to histological examination to observe the cartilage development and the other was used for RNA extraction and quantitation.
Tissue collection and histological staining of toluidine blue
The tissue pieces were obtained from back muscles of DA rats after minimal invasion surgery and fixed in 4% paraformaldehyde solution for 24 hours. Then the pieces were dehydrated in a graded series of ethanol (80%, 95%, and 100%), embedded in paraffin agent and cut into 8 mm sections. Slides carrying sections were coated with poly-L-lysine, deparaffinized in xylene, and rehydrated in graded ethanol dilutions (100%, 95%, 80%). The rehydrated sections were stained with 0.1% toluidine blue (1 g sodium borate and 10 g toluidine blue were dissolved in 100 ml ultrapure water) for 10 to 12 minutes. After rinsing in water, the slides were routinely dehydrated in a graded series of ethanol (80%, 95%, and 100%) and in xylene. Finally, all slides were mounted in rhamsan gum and analyzed in bright-field under a microscope. The resulting toluidine blue-stained histological slides of BMG pieces were used to detect the distribution of sulfated glycosaminoglycan (pink staining) on a blue background staining.
RNA isolation
Total RNA was extracted from the cartilage with TRIzol® (Invitrogen, USA) according to the manufacturer’s instructions and modified method [19]. Briefly, 30 μg regeneration tissue was cut as quickly as possible, placed in the DEPC treated cryogenic vials, and then stored in liquid nitrogen. The tissue was homogenized by grinding in liquid nitrogen, and the tissue powder was transferred into DEPC-treated 1.5 mL EP tubes filled with 1 mL of TRIzol® reagent. Afterward, the procedure followed the introduction described by the TRIzol® reagent manufacturer as usual and the RNA was dissolved in RNase-free water for reverse transcription.
cDNA synthesis, mRNA and microRNA expression detection by quantitative real-time PCR
The expression of aggrecan, Col2a1 has been documented to be the hallmark of skeletal development and chondrocyte differentiation [20]. The mRNA-cDNAs were synthesized from total RNA by utilizing RevertaidTM First Strand cDNA Synthesis kit according to the manufacturer’s instructions (Fermentas, Canada). Briefly, 1 μg total RNA and OligoT was added into a 12 μL reaction volume system and was incubated at 65°C for 5 minutes, then immediately place in the ice. And then 5×reaction buffer, RiboLockTM inhibitor and Moloney Murine Leukemia Virus (MMLV) reverse transcriptase were added and the mixture was incubated at 42°C for 60 minutes. The reaction was stopped by heating at 70°C for 5 minutes and the cDNA was stored at -20°C. According to the previous research, miR-1, miR-337 were associated with cartilage development [18,21]; miR-9, miR-22, miR-27b, miR-98, miR-145, miR-146 and miR-455 were involved in OA [11,22-25]; miR-140 and miR-455 played both roles in the cartilage development and homeostasis [13,14,26]. These miRNAs were all demonstrated change in different development stages of rat articular cartilage identified by Solexa sequencing in our previous study [16]. Thus, we selected them to do the further research. The miRNA-cDNAs were synthesized by utilizing one step PrimeScript® miRNA cDNA Synthesis Kit (TaKaRa, Japan) Briefly, 20 μL reaction volume system including 2×miRNA Reaction Buffer Mix (for real time), 1 g/L BSA, miRNA PrimerScriopt® RT Enzyme Mix, total RAN 1 μg and some DEPC water. Then the mixture was incubated at 37°C for 60 minutes, 85°C for 5 seconds and the cDNA was stored at -20°C.
The information of mRNA primer sequences was shown in Table 1. The sequences of miRNA forward primers were shown in Table 2 and the reverse primer sequence was provided by the one step PrimeScript® miRNA cDNA Synthesis Kits. Primers were synthesized by GenScript company. All qPCR was performed with SYBR Premix Ex TaqTM II (TaKaRa, Japan) as the dsDNA-specific binding dye by a standardized protocol in iCycler iQ5 real-time PCR Detection system (Bio-Rad, CA, USA). Briefly, 1 μL cDNA (1:20 dilution) was used for amplification, and the mixture of real-time PCR contained 4 μL cDNA, 5 μL 2×PCR SYBR Premix Ex TaqTM II (TaKaRa, Japan) mix, 0.5 μL each primer. Triplicates were used for each sample. The reactions were incubated in a 96-well optical plate and mRNA and miRNA expression was normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and rno-let-7a, respectively. Purity of PCR products was confirmed using a melting curve, and all data were analyzed using the 2-ΔΔCt (relative quantification) method.
Table 1.
Information of rat Aggrecan and Col2a1 primers for Real-time PCR
| Gene | Accession No. | Sequence (5’-3’) | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| Aggrecan | NM_022190.1 | For: TCCACATCAGAAGAGCCATAC | 196 | 62 |
| Rev: AGTCAAGGTCGCCAGAGG | ||||
| Col2a1 | NM_053304 | For: GCTGTGGAAGTGGATGAAGA | 174 | 62 |
| Rev: TGAGGAACTGTGGAGAGACG | ||||
| Gapdh | NM_017008.4 | For: TGAGGACCAGGTTGTCTCCT | 162 | 60 |
| Rev: ATGTAGGCCATGAGGTCCAC |
Table 2.
Information of rat miRNA primers for Real-time PCR
| miRNAs | Accession No. | Forward primer |
|---|---|---|
| rno-miR-1-3p | MIMAT0003125 | TGGAATGTAAAGAAGTGTGTAT |
| rno-miR-22-3p | MIMAT0000791 | AAGCTGCCAGTTGAAGAACTGT |
| rno-miR-25-3p | MIMAT0000795 | CATTGCACTTGTCTCGGTCTGA |
| rno-miR-27b-3p | MIMAT0000798 | TTCACAGTGGCTAAGTTCTGC |
| rno-miR-29a-3p | MIMAT0000802 | TAGCACCATCTGAAATCGGTTA |
| rno-miR-98-5p | MIMAT0000819 | TGAGGTAGTAAGTTGTATTGTT |
| rno-miR-103-3p | MIMAT0000824 | AGCAGCATTGTACAGGGCTATGA |
| rno-miR-140-5p | MIMAT0000573 | CAGTGGTTTTACCCTATGGTAG |
| rno-miR-140-3p | MIMAT0000574 | TACCACAGGGTAGAACCACGG |
| rno-miR-145-5p | MIMAT0000851 | GTCCAGTTTTCCCAGGAATCCCT |
| rno-miR-146a-5p | MIMAT0000852 | TGAGAACTGAATTCCATGGGTT |
| rno-miR-150-5p | MIMAT0000853 | TCTCCCAACCCTTGTACCAGTG |
| rno-miR-195-5p | MIMAT0000870 | TAGCAGCACAGAAATATTGGC |
| rno-miR-223-3p | MIMAT0000892 | TGTCAGTTTGTCAAATACCCC |
| rno-miR-337-3p | MIMAT0000577 | TTCAGCTCCTATATGATGCCTTT |
| rno-miR-377-3p | MIMAT0003123 | TGAATCACACAAAGGCAACTTTT |
| rno-miR-455-5p | MIMAT0005316 | TATGTGCCTTTGGACTACATCG |
| rno-miR-455-3p | MIMAT0017308 | GCAGTCCACGGGCATATACACT |
Statistics
Quantitative data were expressed as mean ± standard error of the mean (SEM), and statistical analysis of differences between experimental groups was performed by the Mann-Whitney U-test. Differences with P-values less than 0.05 were considered as statistically significant.
Result
Histological staining of toluidine blue and the expression of cartilage-specific genes
The matrix around chondrocytes could be stained purple with toluidine blue which is a kind of phenothiazine cationic dye, and the chondrocytes were almost not stained. In 1 week after implantation in BMG rat, the chondrocytes was produced and continued to proliferate. The purple color around the chondrocytes were concentrated and deep (Figure 1A). In 2 weeks after surgery, the abundant chondrocyte was appeared and a small amount of chondrocytes began to become hypertrophic. The stained purple was not so deep but the coloring areas were a little more and scattered (Figure 1B). At 3rd and 4th week, hypertrophic chondrocytes were gradually reduced and replaced by blood vessels and bone tissues (Figure 1C, 1D). Accordingly, the stained purple were lighter and almost disappeared. The expression of aggrecan and Col2a1 were shown as Figure 1E and 1F. Aggrecan and Col2a1 were up-regulated at 1st and 2nd week, and down-regulated at 3rd and 4th week. The expression level of these two genes were very low at the last two weeks and they both showed differently expressed in the different time point. The major components of cartilage extracellular matrix are the fibrillar cartilage-specific Col2a1, and large aggregates of hyaluronan with the cartilage-specific proteoglycan aggrecan. In the process of endochondral ossification, Col2 and aggrecan were abundant through the chondrocyte formation, proliferation and matrix production. With the chondrocytes undergoing hypertrophy and apoptotic cell death, Aggrecan, especially Col2 were sharply decreased [21]. The results showed that the expression of the two genes was both significantly different between the first two weeks and the late weeks, which was consistent with the histological staining results of toluidine blue. Therefore, the BMG rat model in current study was suitable for further research.
Figure 1.
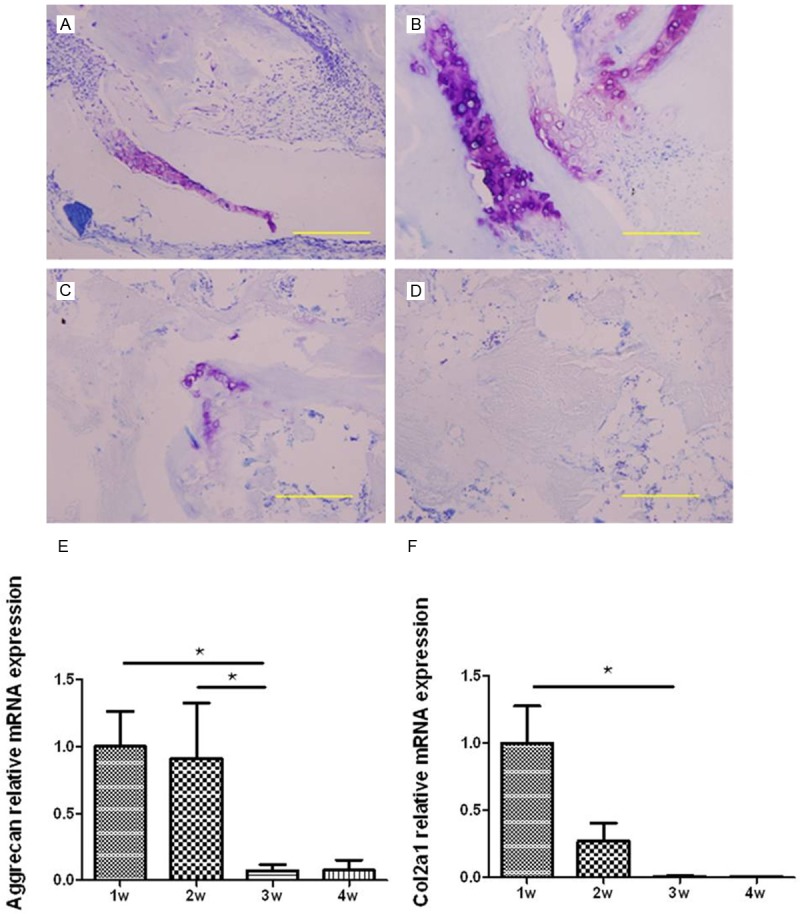
The histology of BMG stained with toluidine blued at four time points and the expression of cartilage-specific genes. Representative images at different point of time: (A) 1 week, (B) 2 weeks, (C) 3 weeks and (D) 4 weeks (bar = 200 μm). Purple areas represent the typical cartilage extracellular matrix components. The relative expression level of Aggrecan and Col2a1 was evaluated by RT-qPCR (bar represents standard error of the mean). N = 6 for each time point, *represent P < 0.05.
The expression of miRNAs
According to the previous researches, we selected some miRNAs to test the expression in the four time point in BMG rat model. The results were shown on Figure 2. miR-1, miR-140, and miR-337 were related with cartilage development, and miR-1-3p, miR-140-5p and miR-337-3p demonstrated differently expressed in the rat model. To the OA associated miRNAs, the expression of miR-9a-5p, miR-98-5p, and miR-445-5p was significantly different, while the expression of miR-22-3p, miR-27-3p, miR-145-5p, miR-146a-5p and miR-455-3p in different time point was not. All the significant difference existed between the first two weeks and the late two weeks. On the basis of the histological staining of toluidine blue, 1w and 2w in BMG rat model represented the chondrogenesis and proliferation stage of cartilage, and 3w and 4w represented the hypertrophy and mineralization stage in late cartilage development. Thus, the different expression of miRNAs in four time points represented the different expression in the process from chondrogenesis to endochondral ossification.
Figure 2.
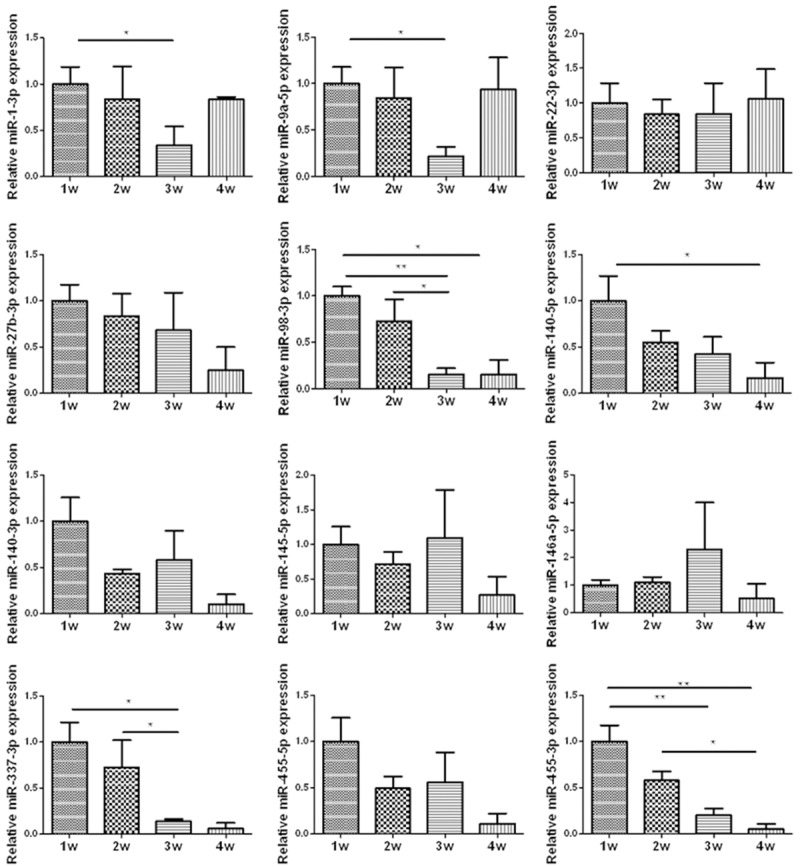
The relative expression level of some miRNAs was evaluated by RT-qPCR (bar represents standard error of the mean). N = 6 for each time point, * and **represent P < 0.05 and P < 0.01 respectively.
Discussion
Rats are generally accepted as a useful model for studying human diseases. the BMGs that implanted in the rats present all the repertoire from chondrongenesis to the endochondral ossification. In the current research, we performed the study of histological staining of toluidine blue and cartilage-specific gene express in the BMG rat model, and the significant difference between the early two weeks and late two weeks after implant surgery revealed that the BMG rat model could demonstrate the different development of cartilage.
Sumiyoshi et al. reported that miR-1 was downregulated upon hypertrophic differentiation of chondrocyte. miR-1 was found to be involved in the regulation of the chondrocytic phenotype and play an important role in chondrocytes during the late stage of the differentiation process, maintaining the integrity of the cartilage tissue [21]. Using rat articular cartilage at different development stages and three rat models, Zhong et al. demonstrated that miR-337 was directly bound up with chondrogenesis [18]. In the BMG model, the expression of miR-1 showed significant difference between 1w and 3w, and miR-337 was differently expressed between 1w and 3w, 2w and 3w, which were coincided with the previous study. Therefore, these two miRNA further showed that the rat model could be used to test some miRNAs associated with cartilage development.
Recently, the cartilage-specific miR-140 has been reported in many studies to play significant roles in OA pathogenesis. Although the expression change of miR-140 was not coincided in articular cartilage in OA patients compared with health controls, the dual function of miR-140 in cartilage homeostasis and development have been established in previous studies [13,14,26]. In current research, the expression level of miR-140-5p in 1w and 2w was higher than 3w and 4w. The significant difference expression between 1w and 3w suggested that the OA associated miR-140-5p played a role in the cartilage development, which was accordance with the previous studies [13,14]. Using microarray and RT-qPCR in the ATDC5 cell model of chondrogenesis, Swingler et al. found that the expression of both miR-455-3p and miR-140-5p increased over the course of chondrogenesis. When the human OA was compared cartilage with cartilage obtained from patients with femoral neck fractures, they observed that the expression of both miR-140-5p and miR-455-3p was increased in OA cartilage [26]. In our study, miR-455-3p was highly expressed in 1w and 2w but it was lowly expressed in 3w and 4w. The statistical analysis showed the significantly different expression between multiple groups which were 1w and 3w groups, 1w and 4w groups, and 2w and 3w groups. Thus, it was verified that the differently expressed miR-455-3p was correlated with cartilage development. At the same time, it was interesting that the expression of miR-140-3p and miR-455-5p in different time point was not significantly different, but the expression trade was the same as miR-140-5p and miR-455-3p respectively. They might play a subsidiary function, but the real role was unknown.
Iliopoulos et al. measured the expression of 365 miRNAs and identified 16 miRNAs expressed differentially in OA cartilage compared with normal controls. Functional experiments implicated miR-9 in the regulation of MMP-13 expression, as well as miR-9, miR-98, and miR-146 in the control of tumor necrosis factor α (TNFα) expression, suggesting that these miRNAs may play a protective role in OA. Moreover, enforced miR-22 overexpression resulted in increases in IL-1β and MMP-13 protein levels, suggesting that this miRNA deregulation can affect OA [20]. When comparing normal versus late-stage OA cartilage, Jones et al. identified 17 miRNAs which expressed differently not less than 4-fold. The altered expression of miR-9, miR-98, and miR-146 in OA cartilage were highlighted. The overexpression of these miRNAs also reduced IL-1β-induced TNFαproduction, while the changed expression of miR-9 modulated MMP-13 secretion [27,28]. Song et al. reported a significant decrease of miR-9 expression in OA chondrocytes which was contradicted with the previous studies. This probably because both studies used human OA samples and human samples were heterogeneous. In our study, miR-9 expression varied significantly between the chondrogenesis stage and the hypertrophic stage, and the expression of miR-98 was significantly different between chondrogenesis and the late cartilage development including the hypertrophy and mineralization. These miRANs may be two of the key regulators in the process of endochondral ossification. The expression of miR-146 and miR-22 did not show significant difference at different point in the BMG rat model, suggesting their role may be not as important as the aforementioned two miRNAs in the cartilage development. The same as miR-146 and miR-22, miR-27b and miR-145 have been documented that they were both response to IL-1β stimulation and associated with OA [22,24,25], while their expression showed no significant in the BMG rat model.
In summary, the BMG rat model was used to test the expression of some miRNAs associated with OA. We verified that miR-140 and miR-455 was associated with cartilage development, and further revealed that miR-140-5p and miR-455-3p might play more important function than miR-140-3p and miR-455-5p in the BMG rat model. Moreover, we found that miR-9 and miR-98 were involved in the endochondrol ossification, suggesting they may be also the key regulators in the process of endochondral ossification. In fact, many miRANs worked as a miRNA-mediated regulatory network in the process of cartilage development and OA. The damage of cartilage was accompanied with the pathogenesis of OA. The current study revealed the potential roles of some miRNAs. Further functional discovery will clarify the roles of individual miRNAs and their targets, and serve as a strong foundation for translating these findings to the clinic therapy for OA.
Acknowledgements
This work was supported by funding from the National Natural Science Foundation of China (No. 81272023, No. 31371298, No. 81301151, No. 81301598, No. 81201426); the Key Project of International Scientific Cooperation of Shaanxi Province (No. 2013KW25-02) and the China Postdoctoral Science Foundation (2013M542337).
Disclosure of conflict of interest
None.
References
- 1.Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213:626–634. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 2.Lotz MK, Kraus VB. New developments in osteoarthritis. Posttraumatic osteoarthritis: pathogenesis and pharmacological treatment options. Arthritis Res Ther. 2010;12:211. doi: 10.1186/ar3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashimoto M, Nakasa T, Hikata T, Asahara H. Molecular network of cartilage homeostasis and osteoarthritis. Med Res Rev. 2008;28:464–481. doi: 10.1002/med.20113. [DOI] [PubMed] [Google Scholar]
- 4.Goldring MB, Marcu KB. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res Ther. 2009;11:224. doi: 10.1186/ar2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drissi H, Zuscik M, Rosier R, O’Keefe R. Transcriptional regulation of chondrocyte maturation: potential involvement of transcription factors in OA pathogenesis. Mol Aspects Med. 2005;26:169–179. doi: 10.1016/j.mam.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Mackie EJ, Tatarczuch L, Mirams M. The skeleton: a multi-functional complex organ: the growth plate chondrocyte and endochondral ossification. J Endocrinol. 2011;211:109–121. doi: 10.1530/JOE-11-0048. [DOI] [PubMed] [Google Scholar]
- 7.Goldring MB, Goldring SR. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann N Y Acad Sci. 2010;1192:230–237. doi: 10.1111/j.1749-6632.2009.05240.x. [DOI] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 10.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3:e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol. 2012;8:543–552. doi: 10.1038/nrrheum.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK, Asahara H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, Takada S, Lotz MK, Ueno-Kudo H, Asahara H. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Ma J, Yao J. Molecular mechanisms of the cartilage-specific microRNA-140 in osteoarthritis. Inflamm Res. 2013;62:871–877. doi: 10.1007/s00011-013-0654-8. [DOI] [PubMed] [Google Scholar]
- 16.Sun J, Zhong N, Li Q, Min Z, Zhao W, Sun Q, Tian L, Yu H, Shi Q, Zhang F, Lu S. MicroRNAs of rat articular cartilage at different developmental stages identified by Solexa sequencing. Osteoarthritis Cartilage. 2011;19:1237–1245. doi: 10.1016/j.joca.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Urist MR, Iwata H, Ceccotti PL, Dorfman RL, Boyd SD, McDowell RM, Chien C. Bone morphogenesis in implants of insoluble bone gelatin. Proc Natl Acad Sci U S A. 1973;70:3511–3515. doi: 10.1073/pnas.70.12.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong N, Sun J, Min Z, Zhao W, Zhang R, Wang W, Tian J, Tian L, Ma J, Li D, Han Y, Lu S. MicroRNA-337 is associated with chondrogenesis through regulating TGFBR2 expression. Osteoarthritis Cartilage. 2012;20:593–602. doi: 10.1016/j.joca.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Ren W, Wang X, Wang F, Gao Y, Ning Q, Han Y, Song T, Lu S. A modified method using TRIzol reagent and liquid nitrogen produces high-quality RNA from rat pancreas. Appl Biochem Biotechnol. 2009;158:253–261. doi: 10.1007/s12010-008-8391-0. [DOI] [PubMed] [Google Scholar]
- 20.Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40:46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Sumiyoshi K, Kubota S, Ohgawara T, Kawata K, Nishida T, Shimo T, Yamashiro T, Takigawa M. Identification of miR-1 as a micro RNA that supports late-stage differentiation of growth cartilage cells. Biochem Biophys Res Commun. 2010;402:286–290. doi: 10.1016/j.bbrc.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Akhtar N, Rasheed Z, Ramamurthy S, Anbazhagan AN, Voss FR, Haqqi TM. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010;62:1361–1371. doi: 10.1002/art.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamasaki K, Nakasa T, Miyaki S, Ishikawa M, Deie M, Adachi N, Yasunaga Y, Asahara H, Ochi M. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60:1035–1041. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287:916–924. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang B, Kang X, Xing Y, Dou C, Kang F, Li J, Quan Y, Dong S. Effect of microRNA-145 on IL-1beta-induced cartilage degradation in human chondrocytes. FEBS Lett. 2014;588:2344–2352. doi: 10.1016/j.febslet.2014.05.033. [DOI] [PubMed] [Google Scholar]
- 26.Swingler TE, Wheeler G, Carmont V, Elliott HR, Barter MJ, Abu-Elmagd M, Donell ST, Boot-Handford RP, Hajihosseini MK, Munsterberg A, Dalmay T, Young DA, Clark IM. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64:1909–1919. doi: 10.1002/art.34314. [DOI] [PubMed] [Google Scholar]
- 27.Song J, Kim D, Chun CH, Jin EJ. MicroRNA-9 regulates survival of chondroblasts and cartilage integrity by targeting protogenin. Cell Commun Signal. 2013;11:66. doi: 10.1186/1478-811X-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones SW, Watkins G, Le Good N, Roberts S, Murphy CL, Brockbank SM, Needham MR, Read SJ, Newham P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage. 2009;17:464–472. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]


