Abstract
Lung cancer is one of the most common cancers in the world, especially in China. It is believed that genetic polymorphisms played a role in cancer susceptibility. Here we investigated the association of interleukin-6 (IL-6) and interleukin-10 (IL-10) gene polymorphisms with the susceptibility of lung cancer in never-smoking Chinese Han population. In this study, we performed a case-control study including 330 cases of never-smoking lung cancer patients and 336 cancer-free never-smoking controls in Chinese Han population. We used polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method to identify gene polymorphisms, and then verified by sequencing method. The results indicated that the four single nucleotide polymorphisms (IL-6 -1363T/G and -572G/C, IL-10 -819T/C and -592A/C) were genotyped by PCR-RFLP and confirmed by sequencing, and we found that the allelic frequencies of G in IL-6 -1363T/G, C in IL-10 -819T/C and C in IL-10 -592A/C were significantly increased in lung cancer patients, by comparing with the control group. However, there was no significant difference in the distribution of the IL-6 572G/C polymorphisms between patients and controls. In conclusion, the IL-6 -1363T/G, IL-10 -819T/C and IL-10 -592A/C polymorphisms are closely related to genetic susceptibility to lung cancer in never-smoking Chinese Han population, and these genetic variants might be used as molecular markers for detecting lung cancer susceptibility.
Keywords: Lung cancer, IL-6 gene, IL-10 gene, single-nucleotide polymorphisms, cancer susceptibility
Introduction
Lung cancer is a very common and growing health problem, which causes high morbidity and mortality in the world, especially in China. According to the recent studies, more than one million deaths are caused by lung cancer worldwide [1,2], and the incidence and mortality rate has increased rapidly. Take Chinese as an example, the overall five-year survival rate is lower than 15%, and the mortality number is estimated to exceed one million by 2025 [3,4]. Many environmental and occupational factors, such as exposure to tobacco smoke as well as other exogenous carcinogens, are clearly related to the development of lung cancer [5]. However, only 11% of tobacco smokers ultimately develop lung cancer, and many genetic factors have been suggested to modulate the pathogenesis of lung cancer [6].
Although smoking is the principal risk factor for lung cancer, many common gene variants have been recently identified to involve in the lung cancer tumorigenesis and development [7-10]. Identification of genes involved in the occurrence and development of lung cancer could contribute to further understanding of the underlying mechanisms, and even a very important additional appropriate prevention strategies and targeted treatments for reducing lung cancer burden [5]. Chronic inflammation is likely to be closely related to the development of lung cancer. As reported based on the genetic and molecular studies, many inflammatory markers such as cytokines showed differential expressions and gene polymorphisms in lung tumor tissues and in peripheral blood [11,12]. Cytokines are important mediators involved in the inflammatory response. More and more studies demonstrated that human lung cancer risk could be modified by polymorphisms of interleukins, for example, the genetic polymorphism of IL-1 beta had influence on the risk of non-small cell lung cancer [13], IL-4 -590T/C influenced the susceptibility to non-small cell lung cancer [14], IL-8 gene polymorphisms was associated with non-small lung cancer in Tunisia [15], IL-18 promoter polymorphisms was associated with lung cancer [16]. Interleukin-6 (IL-6) was a prevalent pro-inflammatory cytokine which plays a central role in regenerative or anti-inflammatory processes [17]. IL-6 has been reported to be the predominant cytokine elevated expressed in lung tumor tissue [18], several single–nucleotide polymorphisms in the IL-6 gene have been described to involve in its transcriptional regulation, such as the -174G/C polymorphism [19], the -634C/G polymorphism [20], the -572G/C polymorphism and the -1363G/T polymorphism [21]. Interleukin-10 (IL-10) is another important prevalent cytokine, which influences many aspects of the immune response through having pleiotropic effects in immunoregulation and inflammation [22]. Many previous studies have indicated that many IL-10 polymorphisms, especially in the promoter region, were associated with the susceptibility of cancer, especially the lung cancer, and affected the expression level of IL-10 [11,23,24]. Based the results of many recent studies, of the IL-10 SNPs, the -1082A/G, -819C/T, -592G/A in the promoter have been confirmed to play important function in immunoregulation and inflammation by functional assays in cell models [11,25,26].
Although many polymorphism sites of IL-6 and IL-10 were suggested to associate with the risk of lung cancer [11,18,20,25,27,28], the potential association analysis for IL-6 -1363T/G and -572G/C, IL-10 -819T/C and -592A/C genetic polymorphism with lung cancer in never-smoking Chinese Han population has not been reported. Thus, in this study, we focused on investigating the distribution of these four SNPs and determining the influence on lung cancer susceptibility in never-smoking Chinese Han population.
Materials and methods
Clinical samples
All the 330 patient specimens were obtained from never-smoking adults diagnosed and/or treated with lung cancer in the First Affiliated Hospital of Henan University of Science and Technology, during June 1, 2010 to May 30, 2013, and all the patients were the Chinese Han people. Accordingly, 336 healthy age-matched never-smoking Chinese Han people who had no history of any lung diseases were enrolled as controls in this study. There was no significant difference regarding age and gender between case and control groups (P > 0.05). Patients’ data about risk factors were obtained from questionnaires, including living environment conditions, occupation, cancer stage, tumor size and family history of cancer. This study was approved by the local ethics committee, and the informed consent form was obtained by each subject.
PCR amplification and genotyping
Genomic DNA was extracted from peripheral venous blood by using the Axygen DNA isolation kit (Axygen, CA), according to the recommended protocol, and then stored at -80°C for analyzed. All polymerase chain reaction (PCR) primers were synthesized by TaKaRa Biotechnology Co., Ltd (Dalian, China) as references listed in Table 1, and the theoretical annealing temperature, fragment region, and size were also listed in Table 1. All PCRs were carried out in 25 μL of reaction mixture containing 50 ng template DNA, 1×buffer (Tris–HCl 100 mmol/L, pH 8.3; KCl 500 mmol/L), 0.25 μmol/L primers, 2.0 mmol/L MgCl2, 0.25 mmol/L dNTPs and 0.5U rTaq DNA polymerase (Promega, Madison, WI, USA). The PCRs were performed on 94°C for 5 min, followed by 35 cycles of 94°C for 30s, annealing at the corresponding temperature (shown in Table 1) for 30s and 72°C for 30s, and a final extension at 72°C for 8 min. All amplified PCR products were preliminary checked by electrophoresis on 2.0% agarose gel and then analyzed under UV light. All SNPs of IL-6 and IL-10 gene were genotyped by the created restriction site-polymerase chain (CRS-PCR) method. Aliquots of 5 μL amplified PCR products were digested with 2U selected restriction enzymes (MBI Fermentas, St. Leon-Rot, Germany, Table 1) at the corresponding temperatures for 10h following the recommended supplier’s manual. Digested products were separated by 2.5% agarose gel electrophoresis and analyzed under UV light. 10% of random samples were re-analyzed by DNA sequencing method (ABI3730xl DNA Analyzer, Applied Biosystems, Foster City, CA, USA) to make sure concordance with the genotyping results from CRS-PCR.
Table 1.
Primer pairs, PCR and CRS-PCR analysis for IL-6 and IL-10 gene
| SNP | Primer sequence | Tm (°C) | PCR Products (bp) | Restriction enzyme | Genotype (bp) | References |
|---|---|---|---|---|---|---|
| IL-6 -1363T/G | 5’-CGGGTCCTGAAATGTTAT-3’ | 59 | 222 | Tag I | G: 156, 66 | [21,30] |
| 5’-GTTGTCCCTCCAGTCTCC-3’ | C: 222 | |||||
| IL-6 -572G/C | 5’-CTCCTCTAAGTGGGCTGAAG-3’ | 56 | 212 | BsrB I | G: 139, 73 | [21,30] |
| 5’-GTTGTCCCTCCAGTCTCC-3’ | C: 212 | |||||
| IL-10 -819T/C | 5’-TCATTCTATGTGCTGGAGATGG-3’ | 59 | 209 | Msl I | C: 116, 93 | [31,32] |
| 5’-TGGGGGAAGTGGGTAAGAGT-5 | T: 209 | |||||
| IL-10 -592A/C | 5’-GGTGAGCACTACCTGACTAGC-3’ | 58 | 412 | Rsa I | A: 236, 176 | [31,32] |
| 5’-CCTAGGTCACAGTGACGTGG-3’ | C: 412 |
Statistical analysis
All statistical analyses were performed by using the Statistical Package for Social Sciences software (SPSS, Windows version release 15.0; SPSS Inc.; Chicago, IL, USA). The chi-squared (χ2) test was utilized to evaluate the Hardy-Weinberg equilibrium in genotypic distributions and clinical characteristics. A level of P < 0.05 was considered statistically significant.
Results
Population characteristics
The demographic and clinical characteristics of the study population were summarized in Table 2. There were no significant statistical different of age and gender between the lung cancer group and the control group. The age range was 30 to 77 years for the lung cancer group, and 31 to 74 years for the control group. The mean of in cancer year was 3.72±1.06 years for the lung cancer group, while 0 year for the control group. The percentage of those who have historical lung cancer was 13.9% in the lung cancer group, and 0% in the control group.
Table 2.
Characteristics of the lung cancer group and the control group in this study
| Characteristics | Lung cancer group (n=330) | The control group (n=336) | P-value |
|---|---|---|---|
| Age mean (Range) (years) | 50.11±11.72 (30-77) | 50.32±10.98 (31-74) | NS |
| Gender (male/female) | 110/220 | 116/220 | NS |
| In cancer years (Range) | 3.72±1.06 (1.2-7.1) | 0 | P<0.001* |
| Historical lung cancer (Y/N)b | 46/324 | 0/336 | P<0.001* |
Abbreviations: NS, Not significant; Y/N, Has historical lung cancer/Not has historical lung cancer;
Represents statistically significant.
IL-6 and IL-10 SNPs identification and genotyping
Through CRS-PCR and DNA sequencing, we investigated the -1363T>G, and -572G>C SNPs of IL-6 and -819T>C, and -592A>C SNPs of IL-10 gene. The PCR-amplified products were digested with Tag I or BsrB I or Msl I or Rsa I restriction enzyme, and divided into two or three genotypes as shown in Table 1.
Genotypic and allelic frequencies
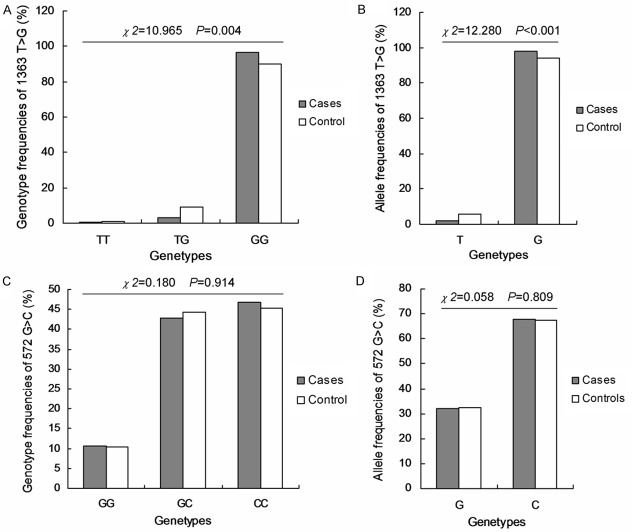
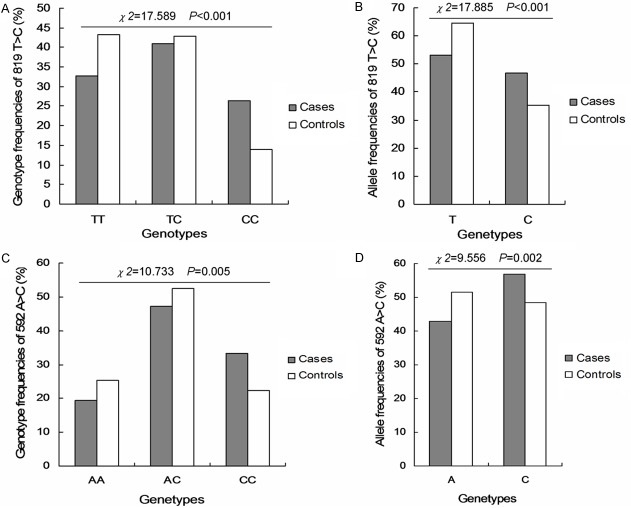
All allelic and genotypic frequencies in the studied populations were investigated. For the -1363 T>G of IL-6, the genotypic frequencies of TT, TG and GG were 10.6%, 42.7% and 46.7% in the patients group, and 10.4%, 44.3% and 45.2% in the controls group (Figure 1A). And the corresponding allelic frequencies in the patients and controls group were 32.0% and 32.6% for G allele, and 68.0% and 67.4% for C allele, respectively (Figure 1B). For the -572G>C of IL-6, the genotypic frequencies of GG, GC and CC were 0.3%, 3.3% and 96.4% in the patients group, and 1.2%, 8.9% and 89.9% in the controls group (Figure 1C). And the corresponding allelic frequencies in the patients and controls group were 2.0% and 5.7% for T allele, and 98.0% and 94.3% for A allele, respectively (Figure 1D). For the -819 T>C of IL-10, the genotypic frequencies of TT, TC and CC were 32.7%, 40.9% and 26.4% in the patients group, and 43.2%, 42.9% and 14.0% in the controls group (Figure 2A). And the corresponding allelic frequencies in the patients and controls group were 53.2% and 64.6% for T allele, and 46.8% and 35.4% for C allele, respectively (Figure 2B). For the -592A>C of IL-10, the genotypic frequencies of AA, AC and CC were 19.4%, 47.3% and 33.3% in the patients group, and 25.3%, 52.4% and 22.3% in the controls group (Figure 2C). And the corresponding allelic frequencies in the patients and controls group were 43.0% and 51.5% for A allele, and 57.0% and 48.5% for A allele, respectively (Figure 2D).
Figure 1.
The genotypic frequencies of IL-6-1363 T>G and IL6-572 G>C in patients and controls group. A. Genotype frequency of 1363 T>G in patients and controls group. B. Allele frequency of 1363 T>G in patients and controls group. C. Genotype frequency of 572 G>C in patients and controls group. D. Allele frequency of 572 G>C in patients and controls group. The different value of χ2 and P were illustrated in the figures.
Figure 2.
The genotypic frequencies of IL-10-819 T>C and IL-10-592 A>C in patients and controls group. A. Genotype frequency of 819 T>C in patients and controls group. B. Allele frequency of 819 T>C in patients and controls group. C. Genotype frequency of 592 A>C in patients and controls group. D. Allele frequency of 592 A>C in patients and controls group. The different value of χ2 and P were illustrated in the figures.
Association between the IL-6, IL-10 SNPs and lung cancer
Table 3 shows the patients’ risk factor characteristics of -1363T>G, and -572G>C SNPs of IL-6 and -819T>C, and -592A>C SNPs of IL-10 gene. As for SNPs of IL-6, the T-allele of IL-6 -1363T>G variant genotype is associated with a much lower lung cancer risk than the G-allele variant genotype (χ2 = 10.965, P = 0.004). There is no statistical different of the allelic gene distributions of IL-6 -572G>C (χ2 = 0.180, P = 0.914). As for SNPs of IL-10, the T-allele of IL-10 -819T>C variant genotype is associated with a much lower lung cancer risk than the C-allele variant genotype (χ2 = 17.589, P < 0.001), and the A-allele of IL-10 -592A>C variant genotype is associated with a much lower lung cancer risk than the C-allele variant genotype (χ2=10.733, P = 0.005).
Table 3.
Allele frequencies of IL-6 and IL-10 genetic polymorphisms in cases and controls
| Genotype frequencies (%) | Allele frequencies (%) | ||||
|---|---|---|---|---|---|
| IL-6 | |||||
| 1363 T>G | TT | TG | GG | T | G |
| Cases VS. Controls | OR=0.237, 95% CI (0.026-2.136) | OR=0.335, 95% CI (0.177-0.635) | |||
| 572 G>C | GG | GC | CC | G | C |
| Cases VS. Controls | OR=0.987, 95% CI (0.587-1.659) | OR=0.972, 95% CI (0.773-1.223) | |||
| IL-10 | |||||
| 819 T>C | TT | TC | CC | T | C |
| Cases VS. Controls | OR=0.402, 95% CI (0.261-0.621) | OR=0.623, 95% CI (0.500-0.776) | |||
| -592 A>C | AA | AC | CC | A | C |
| Cases VS. Controls | OR=0.513, 95% CI (0.331-0.795) | OR=0.712, 95% CI (0.573-0.883) | |||
Discussion
As a result of complex interactions between environmental and genetic factors, lung cancer evolves with highly variable patterns of genetic diversity, and cause increasing rate of morbidity and mortality in both developed and developing countries. It is generally accepted that the genetic variants of candidate genes which influence the development of lung cancer play key roles in the pathogenesis of lung cancer. In this case–control study, we analyzed the effects of genetic polymorphisms of the prevalent pro-inflammatory cytokine genes IL-6 and IL-10 on lung cancer susceptibility in never-smoking Chinese Han population. We investigated the possible association of -1363T>G, and -572G>C SNPs of IL-6 and -819T>C, and -592A>C SNPs of IL-10 gene on the risk factors of lung cancer. Our data indicated that the distribution of allele and genotype frequencies of IL-6 -1363T>G, IL-10 -819T>C, and IL-10 -592A>C in lung cancer patients were statistical different from lung cancer-free controls (P < 0.05, Table 3). For more specifically, the T-allele polymorphism of IL-6 -1363T>G, the T-allele polymorphism of IL-10 -819T>C and the A-allele polymorphism of IL-10 -592A>C were associated with significantly decreased lung cancer risk (χ2 = 12.280, P < 0.001; χ2 = 17.885, P < 0.001 and χ2 = 9.556, P = 0.002). While for the IL-6 -572G>C, results from our study suggested that no statistically significant differences were found in the distribution of allele and genotype frequencies between the lung cancer patients group and the lung cancer-free group (χ2 = 0.058, P = 0.809). As some research groups showed that the genetic variants of IL-6 and IL-10 were associated with the susceptibility of breast cancer and gastric cancer [17,29-33], our results also suggested that many sites of the genetic variants of IL-6 and IL-10 were associated with the susceptibility of lung cancer in never-smoking Chinese Han population. However, as reported by Jiao et al [20], not all the reported SNPs sites of IL-6 have a relevance on the susceptibility of lung cancer. The genetic variants of IL-6 -572G>C may not have a correlation on the occurrence and development of lung cancer in never-smoking Chinese Han Population.
Results from this study may suggest a role of cytokine genes polymorphisms in lung cancer development. However, what and how environmental factors induced the cancer-related cytokine genes polymorphisms are still unknown. Further study should confirm the association between IL-6 -1363T>G, IL-10 -819T>C, and IL-10 -592A>C or other SNPs of IL-6 and IL-10 and lung cancer risk in larger different populations and elucidate the underlying molecular mechanisms in the lung cancer development.
Disclosure of conflict of interest
None.
References
- 1.Kandemir O, Karakus K, Katrancioglu O, Sarikaya A. Semi-quantitative investigation of primary tumor and bone metastasis in lung cancer patients using the PET-CT approach. Int J Clin Exp Med. 2014;7:2624–2631. [PMC free article] [PubMed] [Google Scholar]
- 2.Guilbert J. The world health report 2002-reducing risks, promoting healthy life. Educ Health (Abingdon) 2003;16:230–230. doi: 10.1080/1357628031000116808. [DOI] [PubMed] [Google Scholar]
- 3.Ma X, Lin C, Zhen W. Cancer care in China: A general review. Biomed Imaging Interv J. 2008;4:e39. doi: 10.2349/biij.4.3.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan Q, Huang J, Ding Z, Lin H, Lu S, Luo Q. Meta-analysis for curative effect of lobectomy and segmentectomy on non-small cell lung cancer. Int J Clin Exp Med. 2014;7:2599–2604. [PMC free article] [PubMed] [Google Scholar]
- 5.Tardon A. Genetic polymorphisms and lung cancer risk. Med Clin (Barc) 2014;143:113–114. doi: 10.1016/j.medcli.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Amos C, Xu W, Spitz MR. Is there a genetic basis for lung cancer susceptibility? Recent Results Cancer Res. 1999;151:3–12. doi: 10.1007/978-3-642-59945-3_1. [DOI] [PubMed] [Google Scholar]
- 7.Jiang YQ, Zhou ZX, Ji YL. Suppression of EGFR-STAT3 signaling inhibits tumorigenesis in a lung cancer cell line. Int J Clin Exp Med. 2014;7:2096–2099. [PMC free article] [PubMed] [Google Scholar]
- 8.Hussein AG, Pasha HF, El-Shahat HM, Gad DM, Toam MM. CYP1A1 gene polymorphisms and smoking status as modifier factors for lung cancer risk. Gene. 2014;541:26–30. doi: 10.1016/j.gene.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Jiao F, Xu D, Li Q, Liu G, Liu H, Ren T. Lack of association between -174G>C and -634C>G polymorphisms in interleukin-6 promoter region and lung cancer risk: a meta-analysis. Tumour Biol. 2014;35:5021–7. doi: 10.1007/s13277-014-1662-1. [DOI] [PubMed] [Google Scholar]
- 10.Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes-Gibby CC, Wang J, Spitz M, Wu X, Yennurajalingam S, Shete S. Genetic variations in interleukin-8 and interleukin-10 are associated with pain, depressed mood, and fatigue in lung cancer patients. J Pain Symptom Manage. 2013;46:161–172. doi: 10.1016/j.jpainsymman.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai S, Laskar S, Pandey B. Autocrine IL-8 and VEGF mediate epithelial–mesenchymal transition and invasiveness via p38/JNK-ATF-2 signalling in A549 lung cancer cells. Cell Signal. 2013;25:1780–1791. doi: 10.1016/j.cellsig.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 13.Wu KS, Zhou X, Zheng F, Xu XQ, Lin YH, Yang J. Influence of interleukin-1 beta genetic polymorphism, smoking and alcohol drinking on the risk of non-small cell lung cancer. Clin Chim Acta. 2010;411:1441–1446. doi: 10.1016/j.cca.2010.05.035. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Shi W, Yu G, Lin L, Yang B, Li J, Guo W, Tang C, Wang H, Gao H, Qin H, Liu Y, Liu X. Interleukin-4 -590T/C polymorphism influences the susceptibility to nonsmall cell lung cancer. DNA Cell Biol. 2012;31:797–800. doi: 10.1089/dna.2011.1425. [DOI] [PubMed] [Google Scholar]
- 15.Rafrafi A, Chahed B, Kaabachi S, Kaabachi W, Maalmi H, Hamzaoui K, Sassi FH. Association of IL-8 gene polymorphisms with non small cell lung cancer in Tunisia: A case control study. Hum Immunol. 2013;74:1368–1374. doi: 10.1016/j.humimm.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 16.Farjadfar A, Mojtahedi Z, Ghayumi MA, Erfani N, Haghshenas MR, Ghaderi A. Interleukin-18 promoter polymorphism is associated with lung cancer: a case-control study. Acta Oncol. 2009;48:971–976. doi: 10.1080/02841860902878145. [DOI] [PubMed] [Google Scholar]
- 17.Ruzzo A, Catalano V, Canestrari E, Giacomini E, Santini D, Tonini G, Vincenzi B, Fiorentini G, Magnani M, Graziano F. Genetic modulation of the interleukin 6 (IL-6) system in patients with advanced gastric cancer: a background for an alternative target therapy. BMC Cancer. 2014;14:357. doi: 10.1186/1471-2407-14-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seifart C, Plagens A, Dempfle A, Clostermann U, Vogelmeier C, von Wichert P, Seifart U. TNF-α, TNF-β, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers. 2005;21:157–165. doi: 10.1155/2005/707131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matos MF, Lourenco DM, Orikaza CM, Bajerl JA, Noquti MA, Morelli VM. The role of IL-6, IL-8 and MCP-1 and their promoter polymorphisms IL-6-174GC, IL-8-251AT and MCP-1-2518AG in the risk of venous thromboembolism: A case-control study. Thromb Res. 2011;128:216–220. doi: 10.1016/j.thromres.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Jiao F, Xu D, Li Q, Liu G, Liu H, Ren T. Lack of association between- 174G> C and- 634C> G polymorphisms in interleukin-6 promoter region and lung cancer risk: a meta-analysis. Tumour Biol. 2014;35:5021–7. doi: 10.1007/s13277-014-1662-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhang HY, Feng L, Wu H, Xie XD. The association of IL-6 and IL-6R gene polymorphisms with chronic periodontitis in a Chinese population. Oral Dis. 2014;20:69–75. doi: 10.1111/odi.12075. [DOI] [PubMed] [Google Scholar]
- 22.Shao N, Xu B, Mi YY, Hua LX. IL-10 polymorphisms and prostate cancer risk: a meta-analysis. Prostate Cancer Prostatic Dis. 2011;14:129–35. doi: 10.1038/pcan.2011.6. [DOI] [PubMed] [Google Scholar]
- 23.Zhang G, Manaca MN, McNamara-Smith M, Mayor A, Nhabomba A, Berthoud TK, Khoo SK, Wiertsema S, Aguilar R, Barbosa A, Quinto L, Candelaria P, Schultz EN, Hayden CM, Goldblatt J, Guinovart C, Alonso PL, Lesouef PN, Dobano C. Interleukin-10 (IL-10) polymorphisms are associated with IL-10 production and clinical malaria in young children. Infect Immun. 2012;80:2316–2322. doi: 10.1128/IAI.00261-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres-Poveda K, Burguete-Garcia AI, Cruz M, Martinez-Nava GA, Bahena-Roman M, Ortiz-Flores E, Ramirez-Gonzalez A, Lopez-Estrada G, Delgado-Romero K, Madrid-Marina V. The SNP at -592 of human IL-10 gene is associated with serum IL-10 levels and increased risk for human papillomavirus cervical lesion development. Infect Agents Cancer. 2012;7:32. doi: 10.1186/1750-9378-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khaghanzadeh N, Samiei A, Ramezani M, Mojtahedi Z, Hosseinzadeh M, Ghaderi A. Umbelliprenin induced production of IFN-γ and TNF-α, and reduced IL-10, IL-4, Foxp3 and TGF-β in a mouse model of lung cancer. Immunopharmacol Immunotoxicol. 2014;36:25–32. doi: 10.3109/08923973.2013.863912. [DOI] [PubMed] [Google Scholar]
- 26.Ramkumar HL, Shen de F, Tuo J, Braziel RM, Coupland SE, Smith JR, Chan CC. IL-10-1082. SNP and IL-10 in primary CNS and vitreoretinal lymphomas. Graefes Arch Clin Exp Ophthalmol. 2012;250:1541–8. doi: 10.1007/s00417-012-2037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Fu P, Pang Y, Liu C, Shao Z, Zhu J, Li J, Wang T, Zhang X, Liu J. TERT rs2736100T/G polymorphism upregulates interleukin 6 expression in non-small cell lung cancer especially in adenocarcinoma. Tumour Biol. 2014;35:4667–72. doi: 10.1007/s13277-014-1611-z. [DOI] [PubMed] [Google Scholar]
- 28.Benveniste H, Zhang S, Reinsel RA, Li H, Lee H, Rebecchi M, Moore W, Johansen C, Rothman DL, Bilfinger TV. Brain metabolomic profiles of lung cancer patients prior to treatment characterized by proton magnetic resonance spectroscopy. Int J Clin Exp Med. 2012;5:154–164. [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan LJ, Jin TB, Yin JK, Du XL, Wang Q, Dong R, Wagn SZ, Cui Y, Chen C, Lu JG. Polymorphisms of tumor-related genes IL-10, PSCA, MTRR and NOC3L are associated with the risk of gastric cancer in the Chinese Han population. Cancer Epidemiol. 2012;36:e366–e372. doi: 10.1016/j.canep.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Galicia JC, Tai H, Komatsu Y, Ikezawa I, Yoshie H. Interleukin-6 receptor gene polymorphisms and periodontitis in a non-smoking Japanese population. J Clin Periodontol. 2006;33:704–709. doi: 10.1111/j.1600-051X.2006.00978.x. [DOI] [PubMed] [Google Scholar]
- 31.Yao CJ, Du W, Chen HB, Xiao S, Wang CH, Fan ZL. Associations of IL-10 gene polymorphisms with acute myeloid leukemia in Hunan, China. Asian Pac J Cancer Prev. 2013;14:2439–2442. doi: 10.7314/apjcp.2013.14.4.2439. [DOI] [PubMed] [Google Scholar]
- 32.Lin WP, Lin JH, Chen XW, Wu CY, Zhang LQ, Huang ZD, Lai JM. Interleukin-10 promoter polymorphisms associated with susceptibility to lumbar disc degeneration in a Chinese cohort. Genet Mol Res. 2011;10:1719–1727. doi: 10.4238/vol10-3gmr1283. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Yang J, Wang C, Yang Q, Zhou X. SB-273005, an antagonist of alphaV-beta 3 integrin, reduces the production of Th2 cells and cytoine IL-10 in pregnant mice. Exp Ther Med. 2014;7:1677–1682. doi: 10.3892/etm.2014.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]