Abstract
To investigate association between the three single nucleotide polymorphisms (SNPs, rs146456111, rs143154304 and rs147260142) in cathepsin S (Cat S) and the risks of allergic asthma attack induced by the acaroid mites in the Chinese population. A case-control study was performed in 412 cases and 454 volunteers/controls to evaluate the effects of three SNPs in Cat S on the risks of asthma attack. The genotypes were determined using polymerase chain reaction (PCR) and cleaved amplification polymorphism sequence-tagged sites (PCR-RFLP). The frequencies of genotypes and alleles in these SNPs in the asthmatic group were also analyzed between the two groups. The locus of rs146456111 in Cat S gene, the allele frequency of A and C in asthmatic group were significantly different from the control group (χ2 = 184.425, P = 0.000), and the difference was significant regarding the distribution of the genotypes (AA, AC, and CC) between asthmatic subjects and normal controls (χ2 = 177.915, P = 0.000). Logistic regression analysis revealed that the AC, CC, and AC + CC genotypes were significantly increased with the risk of asthma (AC vs. AA, OR = 4.013, 95% CI = 2.989-4.751, P = 0.000; CC vs. AA, OR = 3.167, 95% CI = 2.483-3.785, P = 0.000; AC + CC vs. AA, OR = 3.418, 95% CI = 2.381-4.214, P = 0.000, respectively), compared with AA genotype. Moreover, by comparison with allele A, allele C (OR = 2.187, 95% CI = 1.743-2.281, P < 0.001) tended to increase the risk of asthma; For the locus of rs143154304, compared with the allele frequency G with A in control group, there was no difference (χ2 = 1.434, P = 0.231) in that of asthmatic group, as well as the distributions of the genotypes (AA, AG, and GG) between asthmatic subjects and normal controls (χ2 = 1.997, P = 0.369); Logistic regression analysis showed that the AG, GG, and AG + GG genotypes were no risk to asthma (AG vs. AA, OR = 0.991, 95% CI = 0.625-1.507, P = 0.968; GG vs. AA, OR = 0.812, 95% CI = 0.525-1.258, P = 0.352; AG + GG vs. AA, OR = 0.914, 95% CI = 0.612-1.366, P = 0.660, respectively) as compared with AA genotype. By comparison with allele A, G allele (OR = 0.888, 95% CI: 0.732-1.078, P = 0.231) failed to increase the risk of asthma; The allele frequencies A and G of rs147260142 in asthma group and control group showed no significant difference (χ2 = 0.162, P = 0.688), yet the distribution of the genotypes (AA, AG, and GG) in control group was different from that in asthmatic subjects (χ2 = 7.520, P = 0.023); Logistic regression analysis suggested that, in compassion with AA genotype, the AG, and AG + GG genotypes appeared to increases the risk of asthma (AG vs. AA, OR = 0.630, 95% CI = 0.439-0.903, P = 0.012; AG + GG vs. AA, OR = 0.710, 95% CI = 0.507-0.996, P = 0.047); However, the GG genotype (OR = 0.843, 95% CI = 0.576-1.234, P = 0.286) and the allele G (OR = 1.040, 95% CI = 0.860-1.258, P = 0.688) were not related to the risk of asthma. The SNP of rs146456111A/C may be susceptible to the risks of asthma in the Chinese population; however, The SNPs of rs143154304A/G and rs147260142A/G may be less likely associated with the asthma attack.
Keywords: Cathepsin S, acaroid mites, allergic asthma, single nucleotide polymorphism
Introduction
Acaroid mites belong to the kingdom of arthropod, class of Arachnida (Acari), Acarina (acaridida), including a total of 7 families of Acaridae, demodicidae, arrenuridae, chortoglyphidae, carpoglyphidae, pyrogyphidae and Histiostomatidae. Acaroid mites hold a number of species and are widely distributed in the nature, and their secretions, excretions and lysates are strong allergens that potentially cause a variety of human allergic diseases, such as allergic asthma, allergic rhinitis and urticaria. Of these disorders, the mite’s allergic asthma can be greater risks to human health.
The cathepsin in its antigen processing may play a key role in inducing an allergic disease. The cathepsin, a member of papain family, is a lysosomal protease that can be activated at a certain pH values in its surroundings. This protease known to date include Cat A, Cat B, Cat D, Cat E, Cat F, Cat G, Cat H, Cat K, Cat L, Cat S and Cat V [1], and some reports [2-5] described that the Cat S is critical in the allergic asthma attack induced by the acaroid mites.
Single nucleotide polymorphism (SNP), one of the important interests involved in the study of the association between genetic phenotypic and acarid mite allergic asthma, is defined as the DNA sequence polymorphism in genomic level resulted from a single base change and occurrence. At least one SNP may occur averagely in 500-1000 base pairs in the human genome, and the SNP, a genetic marker for the third generation, may approximately amount to 3,000,000 in number. Typically, a SNP presents with biallelic variation and susceptibility to allelic conversion, such as C→T. Therefore, SNP can be used to mark the difference of human population and individual dissimilarity, and investigate the phenotypic differences among individuals, especially the susceptibility to complex genetic diseases and the difference of individual with diverse phenotypes susceptible to dissimilar drug response as well as response to the environmental factor reaction.
In view of allergic effects of acaroid mites on humans and the pathogenesis role of Cat S in mites allergic asthma, the current study was undertaken to investigate the association of allele loci rs146456111, rs143154304 and rs147260142 in Cat S and SNPs, based on case-control profile (allergic patients and healthy subjects in Chinese population) and detection and comparison of the differences with such genotype and allele frequencies, to supply evidence for early screening of the allergic asthma in Chinese population as well as early intervention and novel gene therapy for allergic asthmatics.
Subjects and methods
Sample collection
412 patients with allergic asthma were recruited from the outpatients or in-hospital from the Department of Respiratory Diseases of the Affiliated Yijishan Hospital of Wannan Medical College and the No. 1 Xuancheng Municipal People’s Hospital. The patients were aged 47.03 ± 14.26 years old, and their disease confirmation was complying with the Criteria for Asthma Diagnosis by the Chinese Medical Association (2008 version) [6]. Skin prick was performed in all patients for further confirmation. Another 454 subjects, aged 48.20 ± 14.20 years and free of asthma symptoms or allergic disease history as well as history of hepatic and renal diseases, were included from the Medical Examination Center of the Affiliated Yijishan Hospital of Wannan Medical College as healthy controls. The participants in the two groups were totally non-sibship and from southern areas of Anhui province, China (Table 1). All subjects signed the informed consents before study.
Table 1.
Demographic information for the total subjects
| Items | Asthma group (n = 412) | Control group (n = 454) | P-value |
|---|---|---|---|
| Gender, n (male/female) | 232/180 | 257/197 | 0.930a |
| Age, years (mean ± SD) | 47.03 ± 14.26 | 48.20 ± 14.20 | 0.973b |
| FVC (mean ± SD) | 2.81 ± 1.25 | 4.02 ± 0.97 | 0.043a |
| FEV1/FVC (%) (mean ± SD) | 63.52 ± 12.13 | 79.75 ± 13.81 | 0.029a |
| FEF25%-75% (mean ± SD) | 1.40 ± 0.48 | 3.18 ± 1.12 | 0.010a |
| PEF (mean ± SD) | 4.15 ± 1.90 | 7.13 ± 2.45 | 0.001a |
Note: FVC, Forced vital capacity; FEV1, Forced expiratory volume in 1 s; FEF, Forced expiratory flow; PEF, Peak expiratory flow.
P-value was calculated by t-test for the categorical data;
P-value was calculated by Chi-square test for the categorical data.
Main instruments and reagents
Micro pipette (BIOHIT 720 series of 10 μL, 100 μL, 200 μL and 1000 μL) was manufactured by BIOHIT Co. Ltd (Holland); The desktop high speed refrigerated centrifuge (Backman Microfuge type 22R) was a product of Beckman company (USA); ABI 2720 PCR instrument (Applied Biosystems, USA); BS 224S electric balance (Sartorius, Germany); Horizontal multi-electrophoresis apparatus (BG-subMIDI type) (BAYGENE, USA); Ultra low temperature freezer (DW-HL388 type) (Meiling Cryogenic Technology, China); G:BOX gel imaging system (SYNGENE, UK); Super clean bench (SW-CJ-2A type) (Wujiang Xinchangcheng Air Purification Co. Ltd., China); and HH-1 digital display thermostat water bath pot (Jiangsu Ronghua Instrument Manufacturing Co., Ltd., China).
Dream Taq DNA polymerase (5 U/μL) was a product of MBI Inc. (UK) company; 10 × PCR buffer, 25 mM MgCl2, sucrose, SanPrep Column PCR Purification Kit, and Blood Genome Extraction Kit were purchased from Sangon Biotech (Shanghai) Co., Ltd. (China); dNTP Mixture was from TrakaBa (Dalian, China); 200 bp DNA Marker; EDTA, Agarose, Tris I, and bromophenol blue were purchased from BIO BASIC INC. (Canada); and Nde I, BspE I and BseR I were from Fermentas Inc. (Canada). The acetic acid (analytical grade) was a procurement from Suzhou Chemical Reagent Factory (China).
Methods
Sample collection
Venous blood was obtained by 5 mL for each aliquot from both asthmatic patients and healthy control subjects, maintained in the tube containing EDTA-Na2 and stored at -80°C for following use.
Genomic DNA extraction
The genomic DNA extraction was performed as follows: ① 500 μL anticoagulated blood was taken and 800 μL TBP buffer solution was added. The mixture was fully homogenized and centrifuged at 4000 × g for 5, and the supernatant was removed; ② Precipitation resuspension was performed by adding 800 μL TBP buffer (Caution: the solution shall be carefully mixed with the head of a pipettor to prevent rupture of DNA) that was centrifuged 4000 × g for 5 min, and the supernatant was removed. When the mixture was precipitated to colorless or slight redness, 200 μL TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8) was added for resuspension; ③ 500 μL TBM buffer was added into the above 200 μL samples, and mixed gently. Then 4 μL protease K was added, mixed gently, and incubated at 55°C for 10-20 min to remove residual protein; ④ 260 μL absolute ethanol was added, with gentle mixing. The total samples were transferred to the UNIQ-10 column that was sheathed in a 2 mL collection tube. Centrifugation was performed again at 8000 × g, for 1 min, and the remaining liquid was removed; ⑤ The UNIQ-10 column was moved to the same collection tube, and 500 μL washing buffer was added, centrifuged at 10000 × g at room temperature for 1 min before removal of the residual solution; ⑥ 500 μL washing buffer was added, and centrifugation was performed at 10000 × g at room temperature for 1 min, and the liquid was removed. The UNIQ-10 column was moved back to the same collection tube that underwent centrifugation at 10000 × g at room temperature for 15 s to remove the residual buffer solution; ⑦ The UNIQ-10 column was transferred into a 1.5 mL sterile centrifuge tube, and 40 μL eluting buffer was injected at the central bottom of the column, a site near the DNA membrane binding, and stored at 37°C to 55°C for 2 min. Then centrifugation was performed at 10000 × g at room temperature for 1 min. Finally, genomic DNA was obtained and stored at -80°C for the following use.
Primers were designed and synthesized
The primers used in this study were complying with the human DNA sequences released by NCBI (http://www.ncbi.nlm.nih.gov/) and undergone re-design. DNA synthesis was performed by Sangon Biotech (Shanghai) Co., Ltd. (China). The primers were as follows:
(1) rs146456111: AATGATTTCCTTTATTTCTTGCCAT[A/C]CGAATATATCCTTCTTCACCAAAGT. Amplified fragment: TTTCCTTTATTTCTTGCCAT[A/C]CGAATATATCCTTCTTCACCAAAGTTGTGGCCCCAGCTTTAGAAAAAGAAATAATACAAAATTACAAATGCGTACAATC. Primer sequences (The lower-case font indicates the joint sequence to increase the enzyme fragment length): rs146456111-F: 5’-gcggtcccaaaagggtcagtTTTCCTTTATTTCTTGCCATATGAA-3’; rs146456111-R: 5’-gcggtcccaaaagggtcagtGATTGTACGCATTTGTAATTTTGTAT-3’. The base C at the fourth from the right forward primer was mutated into T, where Nde I restriction enzyme sites was introduced. Nde I enzyme recognition sequence included: CA^TATG; GTAT^AC. Genotype: C/C: 140bp; A/A:39bp, 101bp; A/C: 140bp, 39bp, 101bp.
(2) rs143154304: TGATTTCCTTTATTTCTTGCCATCC[A/G]AATATATCCTTCTTCACCAAAGTTG. Amplified fragments: AATGATTTCCTTTATTTCTTGCCATCC[A/G]AATATATCCTTCTTCACCAAAGTTGTGGCCCCAGCTTTAGAAAAAGAAATAATACAAAATTAC- AAATGCGTACAATCAAACCATGTTATC. Primer sequences (The lower-case font indicates the joint sequence to extend the enzyme fragment length): rs143154304-F: 5’-gcggtcccaaaagggtcagtCAATGATTTCCTTTATTTCTTGCCATCCGGA-3’; rs143154304-R: 5’-gcggtcccaaaagggtcagtGATAACATGGTTTGATTGTACGCATT-3’. The base A at the second from the right forward primer was mutated into G, where BspE I restriction enzyme sites was introduced. BspE I enzyme recognition sequence included: T^CCGGA; AGGCC^T. The genotyp: A/A: 139 bp; G/G: 46 bp, 93 bp; A/G: 139 bp, 46 bp, 93 bp.
(3) rs147260142: CAGGACATCTTCTCTGCCATAAGGA[A/G]GTTCAGTGTACTTTGAACATGTGGC. Amplified fragments: CATCTTCTCTGCCATAAGGA[A/G]GTTCAGTGTACTTTGAACATGTGGCAGCACGATATTTTGAGTCATATTGACATTTCTGATCCTGCAAAAAGAAGTATAAATATGATG. Primer sequences (The lower-case font indicates the joint sequence to increase the enzyme fragment length): rs147260142-F: 5’-gcggtcccaaaagggtcagtCACTTCTCTGCCATGAGGA-3’; rs147260142-R: 5’-gcggtcccaaaagggtcagtCATCATATTTATACTTCTTTTTGCAGG-3’. The base A at the fifth from the right forward primer was mutated into G, where BseR I restriction enzyme sites was introduced. BseR I enzyme recognition sequence included: GAGGAG(N)10^…; CTCCTC(N)8^…. The genotype: A/A: 148 bp; G/G: 51 bp, 97 bp; A/G: 148 bp, 51 bp, 97 bp.
PCR amplification
PCR reaction system (total volume of 10 μL) consisted of 10 × PCR buffer (1.0 μL) and dNTP (2.5 mM each) 0.5 μL. The forward primer (50 pmol/μL) 0.1 μL; reverse primers (50 pmol/μL) 0.1 μL; Dream Taq DNA polymerase (5 U/L) 0.06 μL; and 8 μL ddH2O.
The reaction conditions were set for degeneration at 95°C for 5 min and for 30 s, respectively, annealing at 68°C-0.5°C for 45 s (Caution: the annealing at each cycle shall be decreased by 0.5°C). Extension reaction was performed at 72°C for 60 s, and repeated for 20 cycles. Then degenerated at 95°C for 30 s, and annealed at 58°C for 30 s, with extension at 72°C for 40 s by repetition for 20 cycles. Finally, extension reaction was performed at 72°C for 6 min.
Restriction enzyme analysis of PCR products
Enzyme systems (final volume: 15 μL) consisted of 1.5 L 10 × enzyme buffer, 0.1 μL restriction endonuclease (10 U/μL, 3.5 μL ddH2O), and the products were incubated in the enzyme at 55°C overnight.
Product treatment by agarose gel electrophoresis
A total of 15 μL enzyme products were evenly mixed in 3 μL 6 × DNA buffer sample, then added to sample well containing 4% agarose gel, and run in the 1 × TAE DNA electrophoresis buffer solution at 150 V for 0.5 h. The photo was taken for observation.
Data analysis
The genotype and allele count were performed using gene counting method for the case group and the healthy controls on the electrophoresis finding basis. χ2 test was used to compare the genotypes and allele frequencies between the groups, and univariate Logistic regression analysis for estimation of the relative risks of locus polymorphism to asthma attack. All statistics was performed with SPSS version 16.0 for windows, and P < 0.05 was accepted as statistical significance.
Results
Findings for the subjects
By the previous experimental protocol and three repeated tests, rs146456111 was still found partially deleted in the DNA samples from the 412 allergic asthmatic patients and healthy controls. The deletion occurred in 7 for the former and 12 for the latter. The remaining two loci were totally detected in the two groups of participants. The statistical results showed that the gender and ages were not associated with asthma attack (P = 0.930, P = 0.973) (Table 1). However, the case group were statistically different from the controls regarding the forced vital capacity (FVC), FEV1/FVC (%), FEF25-75% and PEF (P = 0.043, P = 0.029; P = 0.010, P = 0.001, respectively).
PCR-RFLP and electrophoresis findings for rs146456111A/C, rs143154304A/G and rs147260142A/G in Cat S
By PCR-RFLP detection and treatment in 4% agarose gel electrophoresis, the gene fragments were amplified for the three loci rs146456111A/C, S rs143154304A/G and rs147260142A/G in Cat S in either patients with asthma or healthy controls, and the fragment length was consistent with the expected size. Parts of the PCR amplification and enzyme digestion results are shown in Figure 1. Taking the rs147260142A/G site as an example, the first lane indicates the DNA marker; the second, genotype AA; the third, AG genotype; and the fourth, genotype GG. The principle to estimate the genotype was: if an AA genotype in rs147260142 occurred in the Cat S, the BseR I restriction enzyme sites would not be exposed in the double stranded PCR products (the restriction enzyme digesting site is located at GAGGAG (N) 10^), therefore, the double stranded PCR products cannot be digested with the enzyme, which resulted in only one band after electrophoresis treatment (the size is 148 bp, and the enzyme digestion was designed according to the necessary primers needed in current work) as shown in the second lane. Contrarily, if genotype AG occurred in rs147260142, BseR I restriction enzyme site was possibly appeared in one of double stranded PCR products and absent in another, thus three bands will be developed on the electropherogram after endonuclease digestion. The size was 148 bp, 51 bp and 97 bp as shown in the third lane. Similarly, if GG genotype in rs147260142 occurred, the BseR I restriction enzyme sites would be exposed in the the double stranded PCR products, which led to two bands on the electropherogram, with a size of 51 bp and 97 bp, respectively (Figure 1).
Figure 1.
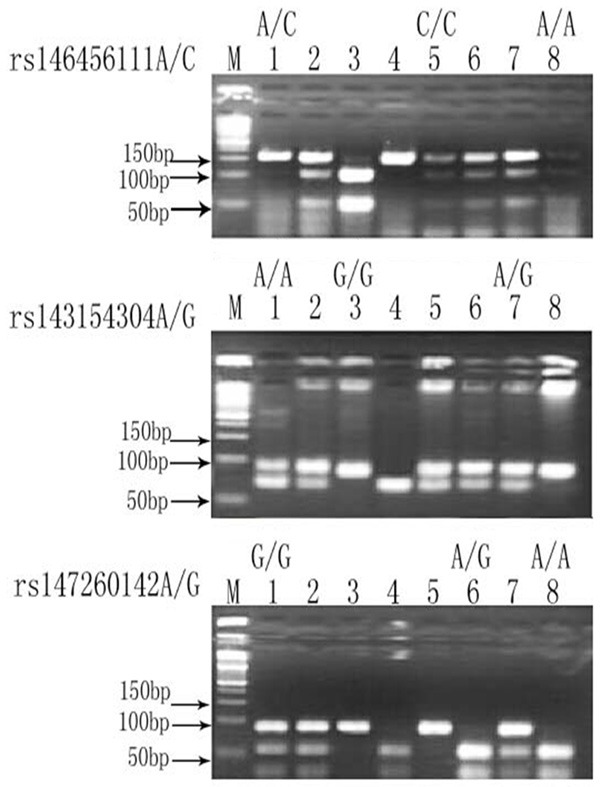
The genotype of rs146456111A/C, rs143154304A/G and rs147260142A/G
Comparison of the genotypes and allele frequencies of rs146456111A/C, rs143154304A/G and rs147260142A/G in the Cat S
Comparison of the genotype and allele frequency in rs146456111A/C
As is shown in Table 2, rs146456111A/C locus in 405 asthmatic patients induced by the acaroid mites presented with genotype AA in 43 cases, AC in 189 and CC in 173. Allele A frequency was 34% and allele C 66%. In the 442 healthy subjects, genotype AA occurred in 188, AC in 216, and CC in 36. Allele A frequency accounted for 67%, and C allele for 43%. Compared with the control group, the distribution of genotype AA, AC and CC in the rs146456111A/C locus as well as allele A and C was statistically different (χ2 = 177.915, P = 0.000; χ2 = 184.425, P = 0.000, respectively).
Table 2.
The genotype an allele distribution frequencies of rs146456111 in Cat S in both groups
| Groups | Genotype | χ 2 | P | Alleles | χ 2 | P | |||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| AA | AC | CC | A | C | |||||
| Asthma (n, %) | 43 (10.6) | 189 (46.7) | 173 (42.7) | 177.915 | 0.000 | 275 (34.0) | 535 (66.0) | 184.425 | 0.000 |
| Control (n, %) | 188 (42.5) | 216 (48.9) | 38 (8.6) | 592 (67.0) | 292 (43.0) | ||||
Comparison of the genotype and allele frequency in rs143154304A/G
As is shown in the Table 3, the rs143154304A/G locus in 412 allergic asthmatic patients from acaroid mites was associated with genotype AA (n = 55), AG (n = 220) and GG (n = 138). Allele A frequency was 39.8% and allele G, 60.2%. In the 454 controls, genotype AA took place in 55, AG in 226 and GG in 173. The allele A was 77% and allele G, 63%. However, no statistical difference was found in the case group concerning the distribution of genotype in rs143154304A/G locus and alleles as compared with the healthy controls (χ2 = 1.997, P = 0.369; χ2 = 1.434, P = 0.231, respectively).
Table 3.
The genotype an allele distribution frequencies of rs143154304 in Cat S in both groups
| Groups | Genotype | χ 2 | P | Alleles | χ 2 | P | |||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| AA | AG | GG | A | G | |||||
| Asthma | 54 (13.1%) | 220 (53.4%) | 138 (33.5%) | 1.997 | 0.369 | 328 (39.8%) | 496 (60.2%) | 1.434 | 0.231 |
| Control | 55 (12.1%) | 226 (50.0%) | 173 (37.9%) | 336 (37.0%) | 572 (63.0%) | ||||
Comparison of the genotype and allele frequency in rs147260142A/G
The Table 4 showed that the rs147260142A/G locus detected in 412 allergic asthma patients from acaroid mites and in 454 control subjects was involved in genotype AA (n = 92 vs. n = 77), AG (n = 234 vs. n = 234) and GG (n = 144 vs. n=143). The difference was significant (χ2 = 7.520, P = 0.023). The frequency of allele A occurred in both groups by 43.7% vs. 42.7%, and allele G by 56.3% vs. 57.3%, which suggested no significance (χ2 = 0.162, P = 0.688).
Table 4.
The genotype an allele distribution frequencies of rs147260142 in Cat S in both groups
| Groups | Genotype | χ 2 | P | Alleles | χ 2 | P | |||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| AA | AG | GG | A | G | |||||
| Asthma | 92 (22.3%) | 176 (42.7%) | 144 (35.0%) | 7.520 | 0.023 | 360 (43.7%) | 464 (56.3%) | 0.162 | 0.688 |
| Control | 77 (17.0%) | 234 (51.5%) | 143 (31.5%) | 388 (42.7%) | 520 (57.3%) | ||||
The association of asthma attack risks and rs146456111A/C, rs143154304A/G and rs147260142A/G in Cat S
The results by univariate logistic regression analysis on the genotypes and alleles in the rs146456111A/C, rs143154304A/G and rs147260142A/G loci are respectively shown in Tables 5 and 6. ① rs146456111A/C gene locus: As compared with genotype AA, AC, CC and AC + CC were likely to increase the risks of asthma development (OR = 4.013, 95% CI: 2.989-4.751; OR = 3.167, 95% CI: 2.483-3.785; OR = 3.418, 95% CI: 2.381-4.214, respectively, and all P < 0.001); Genotype CC had a higher risk to develop an asthma than AA + AC genotype did (OR = 4.320, 95% CI: 2.989-4.762; P < 0.001). The two alleles C and A were higher risks for asthma (OR = 2.187, 95% CI: 1.743-2.281; P < 0.001); ② The rs143154304A/G locus: Genotype AG, GG and AG + GG was less associated with the risks of asthma attack than genotype AA [OR = 0.991 (0.625-1.507), OR = 0.812, 95% CI: 0.525-1.258; OR = 0.914, 95% CI: 0.612-1.366; P < 0.968, P < 0.352, P < 0.660, respectively], and composite genotype AG + GG had a higher risk than the single GG genotype in asthma development (OR = 0.818, 95% CI: 0.619-1.081; P = 0.158), and its corresponding allele G and A were also negatively related to the asthma risks (OR = 0.888; 95% CI: 0.732-1.078; P = 0.231); ③ The rs147260142A/G locus: By comparison with genotype AA, AG and AG + GG genotypes were more predisposed to asthma attack (OR = 0.630, 95% CI: 439-0.903; OR = 0.710, 95% CI: 0.507-0.996; P = 0.012, P = 0.047, respectively), yet GG genotype had no relation with the risks of asthma (OR = 0.843, 95% CI: 0.576-1.234, P = 0.286). In addition, genotype GG had no risks to asthma compared with composite AA + AG genotype (OR = 1.169, 95% CI: 0.880-1.551, P = 0.281), and the alleles A and G were also less effects (OR = 1.040, 95% CI: 0.860-1.258, P = 0.688).
Table 5.
Associations between SNPs in Cat S and risks of asthma attack by univariate logistic regression analysis
| SNP | Genotype | Cases | Controls | OR (95% CI) | P value | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| n | % | n | % | ||||
| rs146456111 | AA | 43 | 10.6 | 188 | 42.5 | 1.00 | - |
| Asthma, n = 405 | AC | 189 | 46.7 | 216 | 48.9 | 4.013 (2.989-4.751) | 0.000 |
| Controls, n = 442 | CC | 173 | 42.7 | 38 | 8.6 | 3.167 (2.483-3.785) | 0.000 |
| AC + CC | 362 | 89.4 | 254 | 57.5 | 3.418 (2.381-4.214) | 0.000 | |
| AA + AC | 232 | 57.3 | 404 | 91.4 | 1.00 | 0.000 | |
| CC | 173 | 42.7 | 38 | 8.6 | 4.320 (2.989-4.762) | 0.000 | |
| rs143154304 | AA | 54 | 13.1 | 55 | 12.1 | 1.00 | - |
| Asthma, n = 412 | AG | 220 | 53.4 | 226 | 49.8 | 0.991 (0.625-1.507) | 0.968 |
| Controls, n = 454 | GG | 138 | 33.5 | 173 | 38.1 | 0.812 (0.525-1.258) | 0.352 |
| AG + GG | 358 | 86.9 | 399 | 87.9 | 0.914 (0.612-1.366) | 0.660 | |
| AG + GG | 274 | 66.5 | 281 | 61.9 | 1.00 | - | |
| GG | 138 | 33.5 | 173 | 38.1 | 0.818 (0.619-1.081) | 0.158 | |
| rs147260142 | AA | 92 | 22.3 | 77 | 17.0 | 1.00 | - |
| Asthma, n = 412 | AG | 176 | 42.7 | 234 | 51.5 | 0.630 (0.439-0.903) | 0.012 |
| Controls, n = 454 | GG | 144 | 35.0 | 143 | 31.5 | 0.843 (0.576-1.234) | 0.286 |
| AG + GG | 320 | 77.7 | 377 | 83.0 | 0.710 (0.507-0.996) | 0.047 | |
| AA + AG | 268 | 65.0 | 311 | 68.5 | 1.00 | - | |
| GG | 144 | 35.0 | 143 | 31.5 | 1.169 (0.880-1.551) | 0.281 | |
Table 6.
Allele frequencies of SNPs in Cat S between cases and controls
| SNP | Allele | Asthma number (%) | Controls number (%) | OR (95% CI) | P-value |
|---|---|---|---|---|---|
| rs146456111 | n = 405 | n = 442 | |||
| A | 275 (34.0) | 592 (67.0) | 1.00 | - | |
| C | 535 (66.0) | 292 (43.0) | 2.187 (1.743-2.281) | < 0.001 | |
| rs143154304 | n = 412 | n = 454 | |||
| A | 328 (39.8) | 336 (37.0) | 1.00 | - | |
| G | 496 (60.2) | 572 (63.0) | 0.888 (0.732-1.078) | 0.231 | |
| rs147260142 | n = 412 | n = 454 | |||
| G | 464 (56.3) | 520 (57.3) | 1.00 | - | |
| A | 360 (43.7) | 388 (42.7) | 1.040 (0.860-1.258) | 0.688 |
Discussion
Allergic asthma induced by the acaroid mites allergen, a chronic airway inflammatory disorder characterized in clinic by the pulmonary eosinophilic granulocyte infiltration and airway hyper-responsiveness, is a type I hypersensitivity. This entity is commonly associated with the genetic susceptibility facors, routes of exposure, allergen dose intake and allergen configuration properties in some cases [7]. The pathogenesis of allergic asthma of acarid mites is primarily involved in the inflammatory response and airway hyper-responsiveness, mainly caused by imbalanced Th1/Th2, CD4+T lymphocyte subsets, and other factors. Once the T cells are activated by the antigens delivered by the antigen-presenting cells (APC), the activated T helper cells (mainly Th2) are likely to boost IL-4, IL-5, IL-10 and IL-13 production, and further generate activation of B cells, which synthesize a specific IgE and bind with the IgE receptors at the surfaces of mast cells and basophils. If the same allergen goes again into the body, the allergen can be in conjunction and synthesis with the specific IgE bound to the cells to release various active media, eventually leading to contraction of smooth muscles, increased mucous secretion and vascular permeability as well as inflammatory cell infiltration. Moreover, inflammatory cells regulated by the medium may further secrete various media that are capable of worsening the airways and aggravating the inflammatory infiltration, manifested as immediate allergy and allergic inflammation of the target organs [8] .
Cathepsin is an endopeptidase or exonuclease, synthesized on the rough endoplasmic reticulum. Most of the members become activated at the low pH found in lysosomes. Specific activity of the cathepsin commonly occurs in lysosomes and endosomes to break down proteins in certain abnormal conditions and after death. Yet Cat K, Cat L and Cat S work extracellularly after secretion by the mononuclear macrophages. Under pathological conditions, the cathepsin level secreted intracellularly or extracellularly tends to increase, suggesting its involvement in various physiological and pathological processes [5,9-11]. Cat S activity is important for the antigen presenting through MHC II, and the expression of Cat S is mediated by some cytokines such as IFN-γ and IL-1β, which indicates that Cat S functions a lot in immune response [12].
The acaroid mite antigen, once entering the organism, can be degraded through the endocytic pathway in APCs to form a short peptide, which will be presented to CD4+T cell for recognition and activation by APCs via MHC II molecular antigen peptide complex, thus the immune responses occurs on specific antigen basis. Conversion of Ig class in B cells and production of the antibody require cooperative signal stimulation from Th cells and cytokines. The APCs expressed in the Cat S from bone marrow is involved in the degradation of Ii and maturation of MHC-II as well as its transportation. Therefore, decreased activity of Cat S tends to reduce the number of MHC-II antigen peptide complexes, leading to failed presenting of some epitopes in T cells to the antigen [3]. This shows that Cat S plays an important role in the immune system based on the CD4+T cell activation via MHC-II mediation [13-15].
Genome screening and linkage analysis confirmed that asthma is a complex polygenic disorder, yet its genetic mechanism remains unclear. Besides, diverse genes and dissimilar environmental factors may play a role in the asthma attack in different patients due to cooperative action of the previous factors [16-20]. This necessitates accurate identification of susceptible genes to asthma development as one of the important research targets for diagnosis and treatment of such entity allergic asthma. Collaborative Studies on the Genetics of Asthma (CSGA) has finished the entire genome screening in more than 140 families from three racial groups and identified 4 important candidate gene and novel sequence variants associated with asthma by using positional candidate gene approaches. These genetic influences of chromosomes on the asthma include 5q23-31, 6p21.3, 11q12, and 12q14.3-24.1. So far, 165 genes have been identified [21]. With the completion of project The International HapMap Project (HapMap), the research on the susceptibility genes to asthma will be advanced.
In recent years, the research on pathogenesis of asthma on SNPs basis is mainly targeted at identification of the related genes, and several candidate genes with this entity have been identified. SNP analysis is helpful to determination of the individual phenotypic differences in disease susceptibility, especially investigation on the response of SNP to complex genetic disorders, sensitivity of various drugs and different environmental factors [22]. Minematsu et al described the association of the polymorphism at the promoter region in Cat S and COPD phenotype. The results showed that the genotype-768 A-(CA)11-(TCCC) ins, -768G-(CA)11-(TCCC) del and -768G-(CA)12-(TCCC) ins respectively accounted for 98% and 92% in loci in population from smokers and the controls, which further confirms the relationship of upregulation of Cat S level with COPD development [3]. Because of the role Cat S in COPD and pneumonia development, Cat S may also be related to the lung diseases such as asthma. Cimerman et al described that the Cat S levels varied a lot in the sera from patients with asthma and healthy subjects, and the levels tended to significantly decrease in serum concentration in asthmatic patients [23], for which we hypothesized that this difference may be caused by the polymorphism of Cat S gene. Accordingly, a certain haplotype may serve as a bio-marker in estimation of the asthma progress or therapy outcomes. However, little research has ever been done on the association of the asthma attack with the genotype in the gene loci rs146456111A/C, rs143154304A/G and rs147260142A/G in Cat S.
Our detection on the locus rs146456111A/C showed that AC, CC and AC + CC as well as AA + AC genotype may be higher risks than the genotype AA and CC for allergic asthma from acaroid mites. Similarly, allele C has higher risks than allele A. The results by detecting rs143154304A/G loci revealed that AG, GG, AG + GG genotype had no association with the allergic asthma of mites as compared with genotype AA, and genotype GG had less risk to the asthma attack than the AG + GG genotype did. The allele G had no relation to the allergic asthma compared with allele A, either. In detection of rs147260142A/G locus, AG and AG + GG genotype tended to increase the risk of allergic asthma of acaroid mites by comparison with the genotype AA, yet simple GG genotype, or compared to composite AA + AG genotype, may be freely associated with the allergic asthma risks. Still, allele A seemed less likely to increase the asthma risk than allele G. In summary, the polymorphism in the rs146456111A/C gene has potential effects on the mite allergic asthma, whereas rs143154304A/G and rs147260142A/G genes may be free.
In the current study, we introduced the conventional PCR-RFLP approach to determination of the single nucleotide polymorphisms. This technique has its advantages in lower experimental cost and easier genotyping on agarose gel electrophoresis basis in direct sequencing of the target genes, and its defects are in a lot of manpower involvement and relatively longer time in testing, which is not workable in large sample size needed high-throughput screening. This investigation included only 412 cases and 454 healthy subjects, because of the limited human resources and material supply, more reliable conclusion on the Cat S and asthma attack shall be confirmed by sample expansion in the following study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81270091; No. 81172790; No. 30872367) and Project of Anhui Provincial Educational Administration (No. 2006KJ101A).
Disclosure of conflict of interest
None.
References
- 1.Conus S, Simon HU. Cathepsins and their involvement in immune responses. Swiss Med Wkly. 2010;140:w13042. doi: 10.4414/smw.2010.13042. [DOI] [PubMed] [Google Scholar]
- 2.Turkenburg JP, Lamers MB, Brzozowski AM, Wright LM, Hubbard RE, Sturt SL, Williams DH. Structure of a Cys25-->Ser mutant of human cathepsin S. Acta Crystallogr D Biol Crystallogr. 2002;58:451–455. doi: 10.1107/s0907444901021825. [DOI] [PubMed] [Google Scholar]
- 3.Minematsu N, Nakamura H, Furuuchi M, Nakajima T, Takahashi S, Tsujimura S, Tateno H, Ishizaka A. Common functional polymorphisms in the cathepsin S promoter in Japanese subjects: possible contribution to pulmonary emphysema. Respirology. 2008;13:498–504. doi: 10.1111/j.1440-1843.2008.01280.x. [DOI] [PubMed] [Google Scholar]
- 4.Storm van’s Gravesande K, Layne MD, Ye Q, Le L, Baron RM, Perrella MA, Santambrogio L, Silverman ES, Riese RJ. IFN regulatory factor-1 regulates IFN-gamma-dependent cathepsin S expression. J Immunol. 2002;168:4488–4494. doi: 10.4049/jimmunol.168.9.4488. [DOI] [PubMed] [Google Scholar]
- 5.Kim N, Bae KB, Kim MO, Yu DH, Kim HJ, Yuh HS, Ji YR, Park SJ, Kim S, Son KH, Yoon D, Lee DS, Lee S, Lee HS, Kim TY, Ryoo ZY. Overexpression of cathepsin S induces chronic atopic dermatitis in mice. J Invest Dermatol. 2012;132:1169–1176. doi: 10.1038/jid.2011.404. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 7.Akdis M, Akdis CA. Mechanisms of allergen-specific immunotherapy. J Allergy Clin Immunol. 2007;119:780–791. doi: 10.1016/j.jaci.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Lu ZY, Zhong NS. Internal Medicine. Beijing: People’s Medical Publishing House; 2008. [Google Scholar]
- 9.Ai-Ming Z, Hai-Ling L, Ping H. Study progress on the interleukin-4 receptor gene polymorphism of allergy and asthma. Section Genet Foreign Med Sci. 2005;2005:187–190. [Google Scholar]
- 10.Qin Y, Shi GP. Cysteinyl cathepsins and mast cell proteases in the pathogenesis and therapeutics of cardiovascular diseases. Pharmacol Ther. 2011;131:338–350. doi: 10.1016/j.pharmthera.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sina C, Lipinski S, Gavrilova O, Aden K, Rehman A, Till A, Rittger A, Podschun R, Meyer-Hoffert U, Haesler R, Midtling E, Putsep K, McGuckin MA, Schreiber S, Saftig P, Rosenstiel P. Extracellular cathepsin K exerts antimicrobial activity and is protective against chronic intestinal inflammation in mice. Gut. 2013;62:520–530. doi: 10.1136/gutjnl-2011-300076. [DOI] [PubMed] [Google Scholar]
- 12.Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544–551. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 13.Riese RJ, Mitchell RN, Villadangos JA, Shi GP, Palmer JT, Karp ER, De Sanctis GT, Ploegh HL, Chapman HA. Cathepsin S activity regulates antigen presentation and immunity. J Clin Invest. 1998;101:2351–2363. doi: 10.1172/JCI1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi GP, Villadangos JA, Dranoff G, Small C, Gu L, Haley KJ, Riese R, Ploegh HL, Chapman HA. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 1999;10:197–206. doi: 10.1016/s1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 15.Riese RJ, Wolf PR, Bromme D, Natkin LR, Villadangos JA, Ploegh HL, Chapman HA. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity. 1996;4:357–366. doi: 10.1016/s1074-7613(00)80249-6. [DOI] [PubMed] [Google Scholar]
- 16.Jimenez-Morales S, Gamboa-Becerra R, Baca V, Del Rio-Navarro BE, Lopez-Ley DY, Velazquez-Cruz R, Saldana-Alvarez Y, Salas-Martinez G, Orozco L. MiR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens. 2012;80:317–321. doi: 10.1111/j.1399-0039.2012.01929.x. [DOI] [PubMed] [Google Scholar]
- 17.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, Thannickal VJ, Abraham E, Liu G. Participation of miR-200 in pulmonary fibrosis. Am J Pathol. 2012;180:484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milosevic J, Pandit K, Magister M, Rabinovich E, Ellwanger DC, Yu G, Vuga LJ, Weksler B, Benos PV, Gibson KF, McMillan M, Kahn M, Kaminski N. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;47:879–887. doi: 10.1165/rcmb.2011-0377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Zhao M, Lv Z, Zhang X, Qin X, Wang H, Wang S, Su J, Lv X, Liu H, Du W, Zhou W, Chen X, Fei K. MiR-138 inhibits tumor growth through repression of EZH2 in non-small cell lung cancer. Cell Physiol Biochem. 2013;31:56–65. doi: 10.1159/000343349. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q, Si Q, Xiao S, Xie Q, Lin J, Wang C, Chen L, Wang L. Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol. 2013;30:353. doi: 10.1007/s12032-012-0353-2. [DOI] [PubMed] [Google Scholar]
- 23.Cimerman N, Brguljan PM, Krasovec M, Suskovic S, Kos J. Circadian and concentration profile of cathepsin S in sera from healthy subjects and asthmatic patients. Pflugers Arch. 2001;442:R204–206. doi: 10.1007/s004240100026. [DOI] [PubMed] [Google Scholar]


