Abstract
Aflatoxin B1 (AFB1), resulting in the formation of AFB1-DNA adducts, is a known human carcinogen. AFB1-exposure individuals with inherited susceptible carcinogen-repairing genotypes may experience an increased risk of genotoxicity. This study was aimed to investigate whether DNA repair gene xerodermapigmentosum complementation group C codon 939 polymorphism (rs2228001) affected the levels of AFB1-DNA adducts in Guangxi Population (n = 2558), from an AFB1-exposure area. AFB1-DNA adducts were measured by ELISA, and XPC codon 939 genotypes were identified by TaqMan-PCR. We found that longer AFB1-exposure years significantly increased XPC genotypes with codon 939 Gln alleles (namely, XPC-LG and -GG, odds ratios [95% confidence intervals] were 1.37 (1.15-1.63) and 1.99 (1.55-2.55), respectively) was significantly associated with higher levels of AFB1-DNA adducts. Furthermore, there was a positive joint effect between XPC genotypes and long-year AFB1 exposure in the formation of AFB1-DNA adducts. These results suggest that individuals with susceptible genotypes XPC-LG and -GG may experience an increased risk of DNA damage elicited by AFB1 exposure.
Keywords: AFB1, AFB1-DNA adducts, XPC, polymorphism
Introduction
Aflatoxin B1 (AFB1) is an important toxin produced by Aspergillus fungi. This toxin is mainly metabolized by cytochrome P450 into the genotoxic metabolic 8,9-epoxide-AFB1 (AFBO). AFBO can bind to DNA, and cause the formation of AFB1-DNA adducts that may be removed or repaired by DNA repair enzymes [1,2]. While xerodermapigmentosum complementation group C (XPC) is required for the efficient repair of this DNA adducts [3-5]. The XPC codon 939 polymorphic locus (rs2228001) has been of particular interest in molecular epidemiology studies [6-11]. Increasing evidence has shown this polymorphism may be associated with decreased DNA repair capacity and increased tumor risk [5-8,12-21]. This suggests that reduced DNA repair capacity may result in the high risk of AFB1-DNA adducts [9,10]. Therefore, we specifically conducted a study to examine whether XPC codon 939 polymorphism influences the levels of AFB1-DNA adducts among Guangxi population from an AFB1 exposure area.
Materials and methods
Study subjects
A total of 2558 healthy adults (27-78 yrs of age)population who were residence of Guangxi Zhuang Autonomous Regionwere enrolled from affiliated hospitals of Youjiang Medical College for Nationalities and Guangxi Medical University between January 2002 and December 2009. All study subjects, including 1600 individuals previously studied [11], were without any clinical evidence of liver diseases. The characteristic information of all study subjects, including sex, age, ethnicity, hepatitis B virus (HBV) infection, and HCV infection were ascertained as described previously [22]. These having hepatitis B surface antigen (HBsAg)-positive or anti-HCV-positive in their peripheral serum were defined as groups infected HBV and HCV, respectively. Additionally, after informed consent was obtained, each subject donated 4 mL of peripheral blood for AFB1-DNA adducts and XPC codon 939 genotypes analysis. One hundred percent of people asked to participate in this study who did enroll agreed to participate in the investigative study, and no one dropped out. The protocol of this study was approved by the Ethic Committees of the hospitals involved in the study.
DNA isolation
Leukocytes from peripheral venous blood samples were isolated by standard procedures. DNA was then extracted from leukocyte samples by standard phenol-chloroform extraction and ethanol precipitation. DNA was stored at -20°C until additional analysis.
AFB1-exposure years assay
AFB1-exposure years was ascertained by our previously published methods [22]. To analyze, AFB1-exposure years were divided into two groups: short-AFB1 exposure (< 40 yrs) and long-AFB1 exposure (≥ 40 yrs), according to the value of AFB1-exposure years, with one cutoff points of 40 yrs, the average AFB1-exposure time.
AFB1-DNA adducts assay
AFB1-DNA adducts levels of DNA samples from leucocytes were measured by competitive enzyme-linked immunosorbent assay (ELISA) using monoclonal antibody 6A10 and 50 μg of DNA as described by Hsieh LL, et al [23]. The quality control for adduct assays was administered by blank and positive controls. To analyze, AFB1-DNA adduct levels were divided into two groups: low level (≤ 1.00 μmol/mol DNA) and high level (≥ 1.01 μmol/mol DNA), according to the value of AFB1-DNA adduct levels, with one cutoff points of 1.00 μmol/mol DNA, the average adducts levels among study subjects.
XPC genotype analysis
Gene polymorphism analysis of XPC codon 939 was detected by using a previously published TaqMan-PCR [11]. Briefly, PCR reactions were run in a 25 μL final volume containing 1 × Premix Ex TaqTM (TaKaRa, catalog # DRR039A), 0.2 μM of each probe, 0.2 μM of each primer, and 50-100 ng of genomic DNA. Cycling conditions were 95°C for 2 min, and 45 cycles of 95°C for 10 s, 60°C for 1 min and 72°C 10 s. Controls were included in each run and repeated genotyping of a random 10% subset yielded 100% identical genotypes. Data analysis for allele discrimination was performed with the iCycleriQ software. The quality control for genotypic assays was administered by blank and positive controls.
Statistical analysis
The association between XPC codon 939 genotypes and the levels of AFB1-DNA adducts was analyzed using t test. Logistic regression with an adjustment for age, sex, ethnicity, HBsAg, and anti-HCV was used to estimate the odds ratio (OR) and the 95% confidence interval (CI) for the XPC codon 939 genotypes. In this analysis, genotype variable was treated as an ordinal variable (XPC-LL coded as 1, XPC-LG as 2, and as 3), and the corresponding risk value was calculated using the additive model. Additionally, the combinative analysis of risk genotypes (XPC-LG + XPC-GG, also called XPC-LG/GG) was accomplished compared with XPC-LL in dominant model. The interactions between XPC codon 939 genotypes and modified factors (including race, sex, HBV and HCV infection, and AFB1-exposure years) on the levels of AFB1-DNA adducts were also estimated and tested on a multiplicative scale by combining genotypes and adding a multiplicative term in the logistic regression model. The statistical significance of the term interaction of genotype-modified factors was evaluated through Likelihood ratio test. All tests were two-tailed. A P-value of < 0.05 was considered statistically significant in this study. All the analysis was performed by the statistical package for social science (SPSS) version 18.0 (SPSS Institute, Chicago, IL).
Results
Demographic characteristics for subjects
Table 1 showed the demographic data of all study subjects. The mean age study subjects were 49.29 years. While HBV and HCV infective rates were 72.2% and 18.2%, respectively. These results were in accord with our previously published data [11,22,24].
Table 1.
Characteristics of study population
| Characteristics | |
|---|---|
| Age, yr | |
| Mean ± SD | 49.29 ± 11.34 |
| Range | 27-78 |
| Sex | |
| Male n (%) | 1930 (75.4) |
| Female n (%) | 628 (24.6) |
| Ethnicity | |
| Han n (%) | 1175 (45.9) |
| Minority n (%) | 1383 (54.1) |
| Years of AFB1 exposure, yr | |
| Mean ± SD | 40.23 ± 11.51 |
| Range | 5-76 |
| HBV infection | |
| HBsAg (-) n (%) | 712 (27.8) |
| HBsAg (+) n (%) | 1846 (72.2) |
| HCV infection | |
| Anti-HCV (-) n (%) | 2092 (81.8) |
| Anti-HCV (+) n (%) | 466 (18.2) |
AFB1-exposure years increased AFB1-DNA adducts levels
The average AFB1-exposure years were 40.23 (Table 1). We also found those individuals featuring long-AFB1 exposure time were likely to have higher levels of AFB1-DNA adducts in their peripheral blood white blood cells (adjusted OR = 1.45, P < 0.01, Table 2 and Figure 1A).
Table 2.
Years of AFB1 exposure and the levels of AFB1-DNA adducts
| Exposure years | Low-level adducts (n = 1537) | High-level adducts (n = 1021) | P | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| n | % | n | % | OR (95% CI)a | ||
| Short | 871 | 56.7 | 488 | 47.8 | Reference | |
| Long | 666 | 43.3 | 533 | 52.2 | 1.45 (1.23-1.70) | 7.00 × 10-7 |
Adjusted for age, sex, ethnicity, HBsAg, and anti-HCV.
Figure 1.
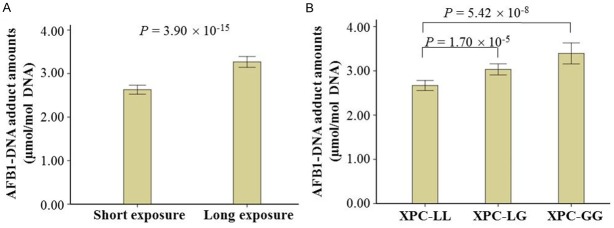
The effects of AFB1-exposure years and XPC codon 939 polymorphism on AFB1-DNA adduct levels. In this study, AFB1-DNA adducts levels in the peripheral blood leukocytes were evaluated using ELISA. Longer AFB1-exposure years (A) and risk genotypes of XPC codon 939 (B) increased AFB1-DNA adduct amounts. Data were analyzed using t test and shown as means ± S.E.
XPC codon 939 polymorphism increased AFB1-DNA adducts levels
To investigate whether the XPC codon 939 polymorphisms were associated with difference in detoxification and DNA repair, which might be reflected in levels of genotoxic damage, we compared this polymorphism with levels of AFB1-DNA adducts (Figure 1B and Table 3). The data exhibited that the adjusted OR for those individuals carrying the heterozygotes for Lys and Gln allele of XPC codon 939 (XPC-LG) compared with those exhibiting the homozygote for Lys alleles (XPC-LL) was 1.37 (95% CI, 1.15-1.63), and the corresponding OR for those featuring the homozygote for Gln alleles (XPC-GG) was 1.99 (95% CI, 1.55-2.55), which showed the risk of high AFB1-DNA adduct levels was related with the number of codon 939 Gln alleles. The genotype distributions of XPC codon 939 polymorphisms in the subjects were consistent with Hardy-Weinberg equilibrium.
Table 3.
XPC genotypes and the levels of AFB1-DNA adducts
| XPC genotype | Low-level adducts (n = 1537) | High-level adducts (n = 1021) | OR (95% CI)a | P | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| n | % | n | % | |||
| LL | 700 | 45.5 | 369 | 36.1 | 1 | |
| LG | 699 | 43.5 | 478 | 46.8 | 1.37 (1.15-1.63) | 3.74 × 10-4 |
| GG | 168 | 10.9 | 174 | 17.0 | 1.99 (1.55-2.55) | 5.62 × 10-8 |
| LG/GG | 837 | 54.5 | 654 | 63.9 | 1.49 (1.27-1.76) | 1.00 × 10-6 |
Adjusted for age, sex, ethnicity, HBsAg, and anti-HCV.
Abbreviations: LL, XPC genotype with codon 939 Lys alleles; LG, XPC genotype with codon 939 Lys and Gln alleles; GG, XPC genotype with codon 939 Gln alleles; LG/GG, the combination of LG and GG genotypes.
The XPC codon 939 genotype distribution stratified by age, race, gender, HBV infection, and HCV infection was shown in Table 4. The results demonstrated similar risk estimates of around 1.5-fold increased high-level AFB1-DNA adducts risk with XPC genotypes with codon 939 Gln alleles (XPC-LG/GG, P interaction > 0.05). Interestedly, we found those individuals featuring HBV- or HCV-infection history and carrying XPC-GG had higher risk of increasing AFB1-DNA adducts levels compared to those having XPC-LL (OR = 2.45 for positive-HBsAg status and 2.39 for positive-anti-HCV status). The hepatitis virus infection-gene interactive analysis, however, did not show statistically significant effects on the levels of AFB1-DNA adducts (P interaction > 0.05).
Table 4.
XPC polymorphism and associated OR in relation to ethnicity, sex, HBsAg and anti-HCV
| Low-level adducts | High-level adducts | OR (95% CI) | P | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| n | % | n | % | ||||
| Ethnicitya | XPC | ||||||
| Han | LL | 319 | 45.6 | 174 | 36.6 | Reference | |
| LG | 296 | 42.3 | 218 | 45.8 | 1.35 (1.05-1.74)b | 0.02 | |
| GG | 84 | 12.0 | 84 | 17.6 | 1.84 (1.29-2.63)b | 7.70 × 10-4 | |
| LG/GG | 380 | 54.4 | 302 | 63.4 | 1.46 (1.15-1.85)b | 2.04 × 10-3 | |
| Minority | LL | 381 | 45.5 | 195 | 35.8 | Reference | |
| LG | 373 | 44.5 | 260 | 47.7 | 1.41 (1.11-1.79)b | 4.86 × 10-3 | |
| GG | 84 | 10.0 | 90 | 16.5 | 2.10 (1.48-2.97)b | 3.40 × 10-5 | |
| LG/GG | 457 | 54.5 | 350 | 64.2 | 1.54 (1.23-1.93)b | 1.94 × 10-4 | |
| Age (yrs)c | XPC | ||||||
| ≤ 49 | LL | 366 | 44.5 | 194 | 35.7 | Reference | |
| LG | 369 | 44.9 | 254 | 46.8 | 1.34 (1.05-1.70)d | 0.02 | |
| GG | 87 | 10.6 | 95 | 17.5 | 2.12 (1.51-2.99)d | 1.70 × 10-5 | |
| LG/GG | 456 | 55.5 | 349 | 64.3 | 1.49 (1.16-1.86)d | 5.93 × 10-4 | |
| ≥ 50 | LL | 334 | 46.7 | 175 | 36.6 | Reference | |
| LG | 300 | 42.0 | 224 | 46.9 | 1.41 (1.10-1.83)d | 0.01 | |
| GG | 81 | 11.3 | 79 | 16.5 | 1.90 (1.32-2.74)d | 6.30 × 10-4 | |
| LG/GG | 381 | 53.3 | 303 | 63.4 | 1.51 (1.19-1.93)d | 7.40 × 10-4 | |
| Sexe | XPC | ||||||
| Male | LL | 167 | 43.4 | 94 | 25.5 | Reference | |
| LG | 178 | 46.2 | 111 | 30.1 | 1.07 (0.75-1.53)f | 0.70 | |
| GG | 40 | 10.4 | 38 | 10.3 | 1.69 (1.01-2.85)f | 0.04 | |
| LG/GG | 218 | 56.6 | 275 | 74.5 | 1.52 (1.20-1.93)f | 7.40 × 10-4 | |
| Female | LL | 533 | 46.3 | 275 | 35.3 | Reference | |
| LG | 491 | 42.6 | 367 | 47.2 | 1.46 (1.20-1.78)f | 2.10 × 10-4 | |
| GG | 128 | 11.1 | 136 | 17.5 | 2.10 (1.58-2.79)f | 3.03 × 10-7 | |
| LG/GG | 619 | 53.7 | 503 | 64.7 | 1.59 (1.32-1.92)f | 1.00 × 10-6 | |
| HBsAgg | XPC | ||||||
| Positive | LL | 196 | 42.9 | 96 | 37.6 | Reference | |
| LG | 195 | 42.7 | 120 | 47.1 | 1.41 (1.15-1.72)h | 9.72 × 10-4 | |
| GG | 66 | 14.4 | 39 | 15.3 | 2.45 (1.82-3.30)h | 3.56 × 10-9 | |
| LG/GG | 261 | 57.1 | 159 | 62.4 | 1.59 (1.32-1.93)h | 2.00 × 10-6 | |
| Negative | LL | 504 | 46.7 | 273 | 35.6 | Reference | |
| LG | 474 | 43.9 | 358 | 46.7 | 1.32 (0.94-1.86)h | 0.11 | |
| GG | 102 | 9.4 | 135 | 17.6 | 1.13 (0.70-1.83)h | 0.62 | |
| LG/GG | 576 | 53.3 | 493 | 64.4 | 1.27 (0.92-1.76)h | 0.15 | |
| Anti-HCVi | XPC | ||||||
| Positive | LL | 562 | 44.2 | 296 | 36.1 | Reference | |
| LG | 567 | 44.6 | 382 | 46.5 | 1.82 (1.20-2.74)j | 4.42 × 10-3 | |
| GG | 143 | 11.2 | 143 | 17.4 | 2.39 (1.29-4.43)j | 5.67 × 10-3 | |
| LG/GG | 710 | 55.8 | 525 | 63.9 | 1.93 (1.31-2.85)j | 8.87 × 10-4 | |
| Negative | LL | 138 | 52.1 | 73 | 36.3 | Reference | |
| LG | 102 | 38.5 | 96 | 47.8 | 1.28 (1.05-1.55) j | 0.01 | |
| GG | 25 | 9.4 | 32 | 15.9 | 1.91 (1.45-2.51)j | 4.00 × 10-4 | |
| LG/GG | 127 | 47.9 | 128 | 63.7 | 1.40 (1.17-1.68)j | 2.53 × 10-4 | |
Likelihood ration test for interaction of the stratified variable (Han and Minority) and XPC genotype was calculated as test for the heterogeneity of ORs across strata (P interaction = 0.876).
Adjusted for age, sex, HBsAg, anti-HCV, and years of AFB1 exposure.
Likelihood ration test for interaction of the stratified variable (Age) and XPC genotype was calculated as test for the heterogeneity of ORs across strata (P interaction = 0.695).
Adjusted for race, sex, HBsAg, anti-HCV, and years of AFB1 exposure.
Likelihood ration test for interaction of the stratified variable (Male and Female) and XPC genotype was calculated as test for the heterogeneity of ORs across strata (P interaction = 0.168).
Adjusted for age, ethnicity, HBsAg, anti-HCV, and years of AFB1 exposure.
Likelihood ration test for interaction of the stratified variable (HBsAg-positive and negative) and XPC genotype was calculated as test for the heterogeneity of ORs across strata (P interaction = 0.192).
Adjusted for age, sex, ethnicity, anti-HCV, and years of AFB1 exposure.
Likelihood ration test for interaction of the stratified variable (Anti-HCV-positive and negative) and XPC genotype was calculated as test for the heterogeneity of ORs across strata (P interaction = 0.241).
Adjusted for age, sex, ethnicity, HBsAg, and years of AFB1 exposure.
Abbreviations: LL, XPC genotype with codon 939 Lys alleles; LG, XPC genotype with codon 939 Lys and Gln alleles; GG, XPC genotype with codon 939 Gln alleles; LG/GG, the combination of LG and GG genotypes.
Joint effects of AFB1-exposure years and XPC codon 939 polymorphism on AFB1-DNA adducts levels
We next analyzed the combination effects of AFB1-exposure years and XPC codon 939 polymorphism on the levels of AFB1-DNA adducts (Table 5). In this analysis, we used reference the lowest risk group: those who had short AFB1-exposure years and XPC-LL. The results showed those with long AFB1-exposure years and XPC-GG were more likely to have AFB1-DNA adducts in their peripheral blood white blood cells. Additionally, we also evaluated the multiplicatively interactive effects between genotypes and AFB1-exposure years according to the following formula: OReg > OReg’ × ORe’g[25]. Some evidence of multiplicatively interaction was observed (2.13 > 1.30 × 1.37).
Table 5.
Joint effects of XPC polymorphism and AFB1-exposure years on AFB1-DNA adducts
| AFB1-exposure years | XPC genotype | Low-level adducts | High-level adducts | OR (95% CI)a | P | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| n | % | n | % | ||||
| Short | LL | 387 | 25.2 | 180 | 17.6 | Reference | |
| LG | 388 | 25.2 | 228 | 22.3 | 1.26 (0.99-1.61) | 0.06 | |
| GG | 96 | 6.2 | 80 | 7.8 | 1.81 (1.28-2.56) | 7.89 × 10-4 | |
| LG/GG | 484 | 31.5 | 308 | 30.2 | 1.37 (1.09-1.72) | 6.59 × 10-3 | |
| Long | LL | 313 | 20.4 | 189 | 18.5 | 1.30 (1.01-1.68) | 0.04 |
| LG | 281 | 18.3 | 250 | 24.5 | 1.94 (1.52-2.49) | 1.29 × 10-7 | |
| GG | 72 | 4.7 | 94 | 9.2 | 2.87 (2.01-4.09) | 6.64 × 10-9 | |
| LG/GG | 353 | 23.0 | 344 | 33.7 | 2.13 (1.69-2.69) | 1.69 × 10-10 | |
Adjusted for age, sex, HBsAg, anti-HCV, and race.
Abbreviations: LL, XPC genotype with codon 939 Lys alleles; LG, XPC genotype with codon 939 Lys and Gln alleles; GG, XPC genotype with codon 939 Gln alleles; LG/GG, the combination of LG and GG genotypes.
Discussion
To the best our knowledge, no studies have investigated the role of DNA-repair gene XPC codon 939 polymorphisms in the risk of the levels of AFB1-DNA adducts, especially from AFB1-exposure areas. In this study, we analyzed the association between this polymorphism and the levels of AFB1-DNA adducts among Guangxi population, from an high AFB1-exposure area. The results showed that XPC genotypes with codon 939 Gln alleles were related with higher levels of AFB1-DNA adducts (OR = 1.49, 95% CI = 1.27-1.76). These results may suggest that XPC codon 939 polymorphism may have functional significance in the AFB1-induced DNA damage.
AFB1, an important chemical carcinogen, is mainly metabolized by cytochrome P450 into the genotoxic metabolic AFBO which can bind to DNA and cause the formation of AFB1-guanine adducts. This kind of AFB1-DNA adducts, including 8,9-dihydro-8-(N7-guanyl)-9-hydroxy AFB1-DNA (AFB1-N7-Gua-DNA) adduct, and formamidopyridine-AFB1-DNA (AFB1-FAPy-DNA) adduct [2], if not repaired, might induce DNA damage such as base damage and oxidative DNA damage [2,26,27].
While XPC gene spans 33kb on chromosome 3p25, and consists of 16 exons and 15 introns. This gene encodes a 940-amino acid protein, an important DNA damage recognition molecule which plays an important role in NER pathway. XPC protein binds tightly with another important NER protein HR23B to form a stable XPC-HR23B complex, the first protein component that recognizes and binds to the DNA damage sites. XPC-HR23B complex can recognize a variety of DNA adducts formed by exogenous carcinogens such as AFB1 and binds to the DNA damage sites. Therefore, it may play a role in the formation process of AFB1-DNA adducts [3-5].
More than one hundred polymorphisms in the XPC gene have been identified and more and more evidence has expressed that the polymorphisms of this gene are associated with the function of DNA repair capacity [6,7,11]. In this study, we only analyzed XPC codon 939 polymorphism because this polymorphism changes the amino acids coded, which may be associated with decreased DNA repair capacity [7,8,13-18,20,21,28], increased frequency of p53 mutations [29,30], and increased tumor risk [6,11,15]. Recent some studies have shown that low DNA repair capacity resulting from the genetic mutation of XPC codon 939 polymorphism can progress AFB1-induced HCC [31-33], suggesting that XPC codon 939 polymorphism may be important in the repair of AFB1-DNA adducts. Our present data not only supported this hypothesis, but also we found that this polymorphism would be able to interact with AFB1-exposure years, especially long-year AFB1 exposure, in the formation of AFB1-DNA adducts. Possibly, differences in the AFB1-exposure years reflect differences in cumulative exposure information. In tissues and cells with longer-years AFB1 exposure, AFB1-DNA adducts are cumulated because of the deficiency of DNA repair ability.
Although some clues of the interactive effects between either HBV or HCV infection and XPC codon 939 genotypes on the levels of AFB1-DNA adducts were found in this study, the effect seems to be greatest at the genotype of XPC-GG under the conditions of positive infection history. This may be because of low detoxification capacity resulting from chronic liver diseases history, and low DNA repair ability, which results in the formation of AFB1-DNA adducts.
Conclusion
To the best of our knowledge, this is the first report to investigation associations between the polymorphism at the codon 939 of XPC and the levels of AFB1-DNA adducts. We found evidence to suggest that the XPC codon 939 Gln alleles are associated with increased levels of DNA damage that may be due to reduced detoxification and DNA repair function. However, Selection bias might have occurred through the selection of hospital-based subjects. Furthermore, liver disease (resulting from virus infection) itself may affect the metabolism of aflatoxin and modify the levels of aflatoxin-DNA adducts. Additionally, other polymorphisms (such as the polymorphisms of XRCC4) might be able to further modify the effect of XPC polymorphism on AFB1-DNA adducts. Therefore, future studies need to characterize the role of the XPC codon 939 Gln alleles in functional detoxification and DNA repair assays and to test to see whether they affect the levels of other biomarkers of DNA damage. Given that high AFB1-DNA adducts positively associates with liver cancer risk, the finding of a genetic susceptibility (if confirmed) may have implications for cancer screening and prevention.
Acknowledgements
We thank Zhou Lin Deng, Yi Ping Wei, Ai Ming Ma and Yin Qin for DNA sample extraction and management; Feng Liang, and Qun Qing Yu for molecular biochemical technique. We also thank all members of Department of Medical Test, Medicine, and the Infective Control, Affiliated Hospital of Youjiang Medical College for Nationalities for their help. This study was supported in part by the National Natural Science Foundation of China (No. 81372639, 81160255, 81472243, and 81460423), the Innovation Program of Guangxi Municipal Education Department (No. 201204LX674), Innovation Program of Shanghai Municipal Education Commission (No.13YZ035), the Natural Science Foundation of Guangxi (No. 2013GXNSFAA019251, 2014GXNSFDA118021, and 2014GXNSFAA118144), Key Discipline and Specialty Foundation of Shanghai Municipal Commission of Health and Family Planning, and “Shu Guang” project (supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation, NO. 13SG19).
Disclosure of conflict of interest
None.
References
- 1.Guengerich FP, Johnson WW, Shimada T, Ueng YF, Yamazaki H, Langouet S. Activation and detoxication of aflatoxin B1. Mutat Res. 1998;402:121–128. doi: 10.1016/s0027-5107(97)00289-3. [DOI] [PubMed] [Google Scholar]
- 2.Wang JS, Groopman JD. DNA damage by mycotoxins. Mutat Res. 1999;424:167–181. doi: 10.1016/s0027-5107(99)00017-2. [DOI] [PubMed] [Google Scholar]
- 3.Araujo SJ, Wood RD. Protein complexes in nucleotide excision repair. Mutat Res. 1999;435:23–33. doi: 10.1016/s0921-8777(99)00042-7. [DOI] [PubMed] [Google Scholar]
- 4.Batty DP, Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 5.Sugasawa K. XPC: its product and biological roles. Adv Exp Med Biol. 2008;637:47–56. doi: 10.1007/978-0-387-09599-8_6. [DOI] [PubMed] [Google Scholar]
- 6.Qiu L, Wang Z, Shi X, Wang Z. Associations between XPC polymorphisms and risk of cancers: A meta-analysis. Eur J Cancer. 2008;44:2241–2253. doi: 10.1016/j.ejca.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 7.Zhang D, Chen C, Fu X, Gu S, Mao Y, Xie Y, Huang Y, Li Y. A meta-analysis of DNA repair gene XPC polymorphisms and cancer risk. J Hum Genet. 2008;53:18–33. doi: 10.1007/s10038-007-0215-5. [DOI] [PubMed] [Google Scholar]
- 8.Liang J, Gu A, Xia Y, Wu B, Lu N, Wang W, Lu C, Zheng Q, Wang S, Wang X. XPC gene polymorphisms and risk of idiopathic azoospermia or oligozoospermia in a Chinese population. Int J Androl. 2009;32:235–241. doi: 10.1111/j.1365-2605.2007.00842.x. [DOI] [PubMed] [Google Scholar]
- 9.Xia Q, Huang XY, Xue F, Zhang JJ, Zhai B, Kong DC, Wang C, Huang ZQ, Long XD. Genetic polymorphisms of DNA repair genes and DNA repair capacity related to aflatoxin b1 (AFB1)-induced DNA damages. In: Chen C, editor. New Research Directions in DNA Repair. 1. Rijeka: InTech; 2013. pp. 377–412. [Google Scholar]
- 10.Long XD, Yao JG, Zeng Z, Huang CH, Huang ZS, Huang YZ, Ban FZ, Huang XY, Yao LM, Fan LD, Fu GH. DNA repair capacity-related to genetic polymorphisms of DNA repair genes and aflatoxin B1-related hepatocellular carcinoma among Chinese population. In: Kruman I, editor. DNA Repair. Rijeka: InTech; 2011. pp. 505–524. [Google Scholar]
- 11.Long XD, Ma Y, Zhou YF, Ma AM, Fu GH. Polymorphism in xeroderma pigmentosum complementation group C codon 939 and aflatoxin B1-related hepatocellular carcinoma in the Guangxi population. Hepatology. 2010;52:1301–1309. doi: 10.1002/hep.23807. [DOI] [PubMed] [Google Scholar]
- 12.Caronia D, Patino-Garcia A, Milne RL, Zalacain-Diez M, Pita G, Alonso MR, Moreno LT, Sierrasesumaga-Ariznabarreta L, Benitez J, Gonzalez-Neira A. Common variations in ERCC2 are associated with response to cisplatin chemotherapy and clinical outcome in osteosarcoma patients. Pharmacogenomics J. 2009;9:347–353. doi: 10.1038/tpj.2009.19. [DOI] [PubMed] [Google Scholar]
- 13.De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, Velghe A, Van Meerbeeck J, Thierens H. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631:101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Dong Z, Guo W, Zhou R, Wan L, Li Y, Wang N, Kuang G, Wang S. Polymorphisms of the DNA repair gene XPA and XPC and its correlation with gastric cardiac adenocarcinoma in a high incidence population in North China. J Clin Gastroenterol. 2008;42:910–915. doi: 10.1097/MCG.0b013e3180f6262c. [DOI] [PubMed] [Google Scholar]
- 15.Francisco G, Menezes PR, Eluf-Neto J, Chammas R. XPC polymorphisms play a role in tissue-specific carcinogenesis: a meta-analysis. Eur J Hum Genet. 2008;16:724–734. doi: 10.1038/ejhg.2008.6. [DOI] [PubMed] [Google Scholar]
- 16.Hansen RD, Sorensen M, Tjonneland A, Overvad K, Wallin H, Raaschou-Nielsen O, Vogel U. XPA A23G, XPC Lys939Gln, XPD Lys751Gln and XPD Asp312Asn polymorphisms, interactions with smoking, alcohol and dietary factors, and risk of colorectal cancer. Mutat Res. 2007;619:68–80. doi: 10.1016/j.mrfmmm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Mechanic LE, Millikan RC, Player J, de Cotret AR, Winkel S, Worley K, Heard K, Tse CK, Keku T. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis. 2006;27:1377–1385. doi: 10.1093/carcin/bgi330. [DOI] [PubMed] [Google Scholar]
- 18.Shore RE, Zeleniuch-Jacquotte A, Currie D, Mohrenweiser H, Afanasyeva Y, Koenig KL, Arslan AA, Toniolo P, Wirgin I. Polymorphisms in XPC and ERCC2 genes, smoking and breast cancer risk. Int J Cancer. 2008;122:2101–2105. doi: 10.1002/ijc.23361. [DOI] [PubMed] [Google Scholar]
- 19.Takebayashi Y, Nakayama K, Kanzaki A, Miyashita H, Ogura O, Mori S, Mutoh M, Miyazaki K, Fukumoto M, Pommier Y. Loss of heterozygosity of nucleotide excision repair factors in sporadic ovarian, colon and lung carcinomas: implication for their roles of carcinogenesis in human solid tumors. Cancer Lett. 2001;174:115–125. doi: 10.1016/s0304-3835(01)00690-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Zhang Z, Yan W. Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta. 2005;359:150–155. doi: 10.1016/j.cccn.2005.03.047. [DOI] [PubMed] [Google Scholar]
- 21.Zhou RM, Li Y, Wang N, Zhang XJ, Dong XJ, Guo W. [Correlation of XPC Ala499Val and Lys939Gln polymorphisms to risks of esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma] . Ai Zheng. 2006;25:1113–1119. [PubMed] [Google Scholar]
- 22.Long XD, Ma Y, Wei YP, Deng ZL. The polymorphisms of GSTM1, GSTT1, HYL1*2, and XRCC1, and aflatoxin B1-related hepatocellular carcinoma in Guangxi population, China. Hepatol Res. 2006;36:48–55. doi: 10.1016/j.hepres.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh LL, Hsu SW, Chen DS, Santella RM. Immunological detection of aflatoxin B1-DNA adducts formed in vivo. Cancer Res. 1988;48:6328–6331. [PubMed] [Google Scholar]
- 24.Long XD, Ma Y, Qu de Y, Liu YG, Huang ZQ, Huang YZ, Lin ZH, Wei NB, Zhou SC. The polymorphism of XRCC3 codon 241 and AFB1-related hepatocellular carcinoma in Guangxi population, China. Ann Epidemiol. 2008;18:572–578. doi: 10.1016/j.annepidem.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Brennan P. Gene-environment interaction and aetiology of cancer: what does it mean and how can we measure it? Carcinogenesis. 2002;23:381–387. doi: 10.1093/carcin/23.3.381. [DOI] [PubMed] [Google Scholar]
- 26.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 27.Wilson DM rd, Thompson LH. Life without DNA repair. Proc Natl Acad Sci U S A. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laczmanska I, Gil J, Karpinski P, Stembalska A, Trusewicz A, Pesz K, Ramsey D, Schlade-Bartusiak K, Blin N, Sasiadek MM. Polymorphism in nucleotide excision repair gene XPC correlates with bleomycin-induced chromosomal aberrations. Environ Mol Mutagen. 2007;48:666–671. doi: 10.1002/em.20333. [DOI] [PubMed] [Google Scholar]
- 29.Ryk C, Kumar R, Sanyal S, de Verdier PJ, Hemminki K, Larsson P, Steineck G, Hou SM. Influence of polymorphism in DNA repair and defence genes on p53 mutations in bladder tumours. Cancer Lett. 2006;241:142–149. doi: 10.1016/j.canlet.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 30.Sakano S, Matsumoto H, Yamamoto Y, Kawai Y, Eguchi S, Ohmi C, Matsuyama H, Naito K. Association between DNA repair gene polymorphisms and p53 alterations in Japanese patients with muscle-invasive bladder cancer. Pathobiology. 2006;73:295–303. doi: 10.1159/000099124. [DOI] [PubMed] [Google Scholar]
- 31.Strom SS, Estey E, Outschoorn UM, Garcia-Manero G. Acute myeloid leukemia outcome: role of nucleotide excision repair polymorphisms in intermediate risk patients. Leuk Lymphoma. 2010;51:598–605. doi: 10.3109/10428190903582804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan SG, Yamanegi K, Zheng ZM, Boyle J, Imoto K, Oh KS, Baker CC, Gozukara E, Metin A, Kraemer KH. XPC branch-point sequence mutations disrupt U2 snRNP binding, resulting in abnormal pre-mRNA splicing in xeroderma pigmentosum patients. Hum Mutat. 2010;31:167–175. doi: 10.1002/humu.21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gangwar R, Mandhani A, Mittal RD. XPC gene variants: a risk factor for recurrence of urothelial bladder carcinoma in patients on BCG immunotherapy. J Cancer Res Clin Oncol. 2010;136:779–786. doi: 10.1007/s00432-009-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


